Abstract
Cryptosporidium spp. cause acute gastrointestinal disease that can be fatal for immunocompromised individuals. These protozoan parasites are resistant to conventional antiparasitic chemotherapies and the currently available drugs to treat these infections are largely ineffective. Genomic studies suggest that, unlike other protozoan parasites, Cryptosporidium is incapable of de novo pyrimidine biosynthesis. Curiously, these parasites possess redundant pathways to produce dTMP, one involving thymidine kinase (TK) and the second via thymidylate synthase-dihydrofolate reductase. Here we report the expression and characterization of TK from C. parvum. Unlike other TKs, CpTK is a stable trimer in the presence and absence of substrates and the activator dCTP. Whereas the values of kcat = 0.28 s−1 and Km,ATP = 140 μm are similar to those of human TK1, the value of Km(thymidine) = 48 μm is 100-fold greater, reflecting the abundance of thymidine in the gastrointestinal tract. Surprisingly, the antiparasitic nucleosides AraT, AraC, and IDC are not substrates for CpTK, indicating that Cryptosporidium possesses another deoxynucleoside kinase. Trifluoromethyl thymidine and 5-fluorodeoxyuridine are good substrates for CpTK, and both compounds inhibit parasite growth in an in vitro model of C. parvum infection. Trifluorothymidine is also effective in a mouse model of acute disease. These observations suggest that CpTK-activated pro-drugs may be an effective strategy for treating cryptosporidiosis.
Keywords: Antiviral Agents, Nucleoside Nucleotide Analogs, Nucleoside Nucleotide Metabolism, Nucleotide, Parasite, Pyrimidine, Cryptosporidium, Nucleoside Prodrugs, Thymidine Kinase, Trifluoromethylthymidine
Introduction
Cryptosporidium spp. are intracellular parasitic protozoa that cause cryptosporidiosis, an acute gastrointestinal disease characterized by severe diarrhea. Cryptosporidium is a major cause of malnutrition in the developing world (1). The disease is typically short term and self-limiting in healthy individuals; however, the infection can be chronic and life-threatening in children and immunocompromised patients. Cryptosporidium infection is transmitted by oocysts found in contaminated drinking and recreational water (2). These oocysts are resistant to standard water treatment. Cryptosporidium hominis, which is specific for humans, and Cryptosporidium parvum, which infects a wide range of hosts, are responsible for most human disease. The prevalence of C. parvum infections in calves raises serious concerns that C. parvum oocysts could be easily obtained and used as a bioterrorism agent (3).
No effective treatment exists for cryptosporidiosis (4). Cryptosporidium is resistant to conventional antiparasitic chemotherapies such as antifolates; several drug targets are either absent or highly divergent in the parasite (5). In addition, Cryptosporidium contains many putative drug efflux transporters, which may also protect the parasite (6). Drug development is further impeded by the inability to continuously culture Cryptosporidium in vitro.
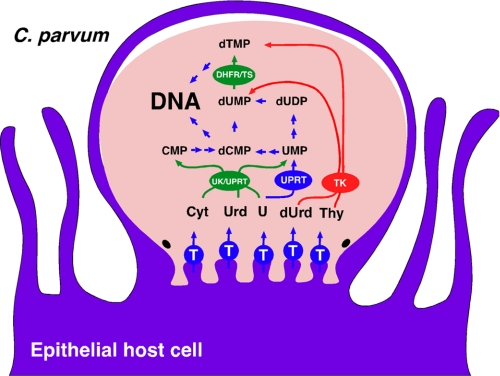
Nucleotide metabolism in general, and dTMP biosynthesis in particular, provides a rich source of drug targets for many diseases. Genomic analysis suggests that, unlike most protozoan parasites, Cryptosporidium lacks de novo pyrimidine nucleotide biosynthetic pathways and instead salvages pyrimidines from the host, producing dTMP via thymidine kinase (TK)3 (E.C. number 2.7.1.21) and thymidylate synthase-dihydrofolate reductase (TS-DHFR) (Fig. 1). Although several laboratories are developing inhibitors of Cryptosporidium TS-DHFR (7–9), the presence of these redundant pathways calls to question whether blocking dTMP biosynthesis is a viable strategy for anti-cryptosporidial chemotherapy. Indeed, the intestinal parasites Giardia lamblia (10) and Entamoeba histolytica (11) lack TS and appear to obtain dTMP entirely via the phosphorylation of thymidine. Intriguingly, Cryptosporidium is the only Apicomplexan that possesses a TK (12), and it appears to have obtained its TK gene from bacteria via horizontal gene transfer. Together, these observations suggest that Cryptosporidium may have adapted its metabolism to exploit a unique abundance of thymidine in the gastrointestinal environment (13).
FIGURE 1.
C. parvum pyrimidine nucleotide salvage pathways. Enzymes that appear to have been obtained via horizontal gene transfer are shown in red (bacteria) and green (plant). T, nucleoside transporter.
TK plays a critical role in the activation of deoxynucleoside prodrugs used in antiviral and anticancer chemotherapy (14). These compounds are converted to the mono-, di-, and triphosphates and then incorporated into DNA and RNA. TFT, AraT, IUdR, IDC, and AraC, but not AZT and acyclovir, have been reported to have anticryptosporidial activity in a cell culture model of infection (15). All of these nucleosides are potential substrates for CpTK, suggesting that CpTK may have very broad specificity like a viral TK. Importantly, the antiparasitic effects are observed at concentrations that do not affect the host cells, which is surprising because the host cells also contain these same deoxynucleoside salvage enzymes. Perhaps this selective antiparasitic activity derives from the catalytic properties of CpTK.
We have expressed and characterized CpTK as part of a program to identify potential drug targets in the nucleotide salvage pathways of C. parvum. FUdR, TFT, and AZT are good substrates for CpTK. In contrast, AraT, AraC, and IDC are not substrates for CpTK, suggesting that Cryptosporidium possess another deoxynucleoside kinase. Importantly TFT inhibits C. parvum growth in a mouse model of C. parvum infection, suggesting that the pyridimidine deoxynucleoside salvage pathways can be subverted for chemotherapy.
EXPERIMENTAL PROCEDURES
Materials
3′-Amino-3′-deoxythymidine was purchased from Toronto Research Chemicals (Ontario, Canada). Activated CH-Sepharose 4B beads were purchased from Amersham Biosciences. [methyl-3H]Thymidine (2 Ci/mmol) was purchased from PerkinElmer Life Sciences. DEAE paper discs were purchased from Whatman (Piscataway, NJ). β-l-Thymidine was purchased from Carbosynth (Berkshire, UK). All other reagents were purchased from Sigma.
Synthesis of Nucleoside Analogs
The syntheses of KP296-8 were reviewed previously (16). KP1308 and KP1309 were synthesized as previously described (17). The syntheses of KP330, KP332-4, and KP1283 are described in Ref. 17.
Cloning
The coding sequence of CpTK was PCR amplified from plasmid MTT1-CpTK-GFP using the following primers: ATTCGGATCCATGGCAAAATTATACTTTTACTATTC (forward) and TCAGGTCGACTTAGAAATTGTATTCTTCACAATTAATT (reverse) and subsequently subcloned into pMAL-c2X (New England Biolabs). The resulting construct will be referred to as pMAL-CpTK.
Additionally, the coding sequence of CpTK was PCR amplified from pMAL-CpTK using the following primers: TGGTGCCTCGTGGTAGCCATGCAAAATTATACTTTTACTATTCAGCAGCAAAATTATACTTTTACTATTCAGCA (forward) and CTCAGCTTCCTTTCGGGCTTTGTTATTAGAAATTGTATTCTTCACAATTAATTATATGA (reverse) and subsequently cloned into pET28a. The resulting construct will be referred to as pET-CpTK.
Cell-free Translation
Untagged CpTK was expressed using a wheat germ cell-free transcription-translation system as previously described (18). The coding sequence of CpTK was amplified from pMAL-CpTK vector by PCR using the following primers: CAGGACTCGAGATGGCAAAATTATACTTTTACTATTCAGCA (forward) and CATGGTCCCGGGTTAGAAATTGTATTCTTCACAATTAATTAT (reverse) and subsequently cloned into a cell-free vector that carries SP6 RNA polymerase promoter and Ω sequence for the wheat germ ribosome binding site. Transcription and translation were carried out as described (18). Wheat germ lysate with CpTK was kept at −80 °C until purification.
Bacterial Expression
Alternatively, His6-tagged CpTK was expressed in bacteria. Briefly, pET-CpTK was transformed into chemically competent BL21(DE3) cells and grown in LB medium at 30 °C until A600 ∼1.0. Cultures were induced with isopropyl 1-thio-β-d-galactopyranoside to a final concentration of 0.5 mm and allowed to grow overnight at 16 °C. Cells were harvested and cell pellets were stored at −80 °C until purification.
Purification of CpTK
Untagged CpTK was purified by affinity chromatography on 3′-aminothymidine-Sepharose resin. 3′-Aminothymidine-Sepharose resin was synthesized as previously described (19, 20). Wheat germ lysate was loaded onto the thymidine column and purification was carried out as previously described (19, 20). Elution fractions were assayed for enzyme activity and fractions with activity were pooled and concentrated and stored at 4 °C. Protein concentration was determined using the Bio-Rad protein assay dye reagent according to the manufacturer's instructions. Prior to assaying, CpTK was passed through a Centri-Sep desalting column (Princeton Separations) to remove thymidine from the enzyme sample.
Alternatively, His6-tagged CpTK was purified by affinity chromatography using a HisTrap column (Amersham Biosciences) on an ÄKTA Purifier (GE Healthcare). Briefly, cells were resuspended in Binding Buffer (20 mm Na2HPO4, pH 7.8, 500 mm NaCl, 20 mm imidazole) and lysed by sonication. Cell lysate was loaded onto the HisTrap column and His6-tagged CpTK was eluted using a gradient to 500 mm imidazole. Purified enzyme was used immediately.
Enzyme Assays
CpTK activity was measured using a radiometric assay as previously described (21, 22). Reactions were carried out in a final volume of 100 μl at 25 °C. Conversion of thymidine to dTMP was measured by the DEAE disc method as described (21, 22). Additionally, a coupled spectrophotometric assay was used where the production of ADP was coupled to the production of NADH via pyruvate kinase/lactate dehydrogenase reactions and monitored at 340 nm (23). Reactions were carried out in a final volume of 500 μl and the decrease in absorbance at 340 nm was measured on a Hitachi U-2000 spectrophotometer. Kinetic parameters were determined using the SigmaPlot Enzyme Kinetics Module by fitting to the Michaelis-Menten equation.
Determination of Oligomeric State
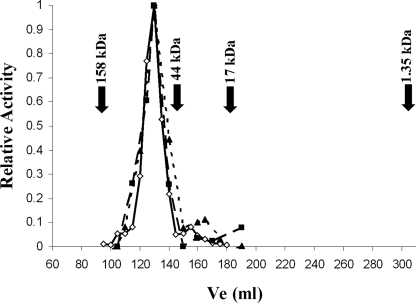
The oligomeric state of CpTK was determined by size-exclusion chromatography using a Sephacryl S-200 HR column (Amersham Biosciences). CpTK oligomerization was determined in the absence and presence of substrates (thymidine and ATP) and an allosteric effector (dCTP). The CpTK elution profile was created by assaying fractions for enzyme activity. The molecular mass of the CpTK oligomer was estimated from a calibration curve obtained from standard proteins in the Bio-Rad Gel Filtration Standard kit: bovine γ-globulin (158 kDa), chicken ovalbumin (44 kDa), equine myoglobin (17 kDa), and vitamin B12 (1.35 kDa). Blue dextran was used to determine the void volume of the column.
Cell Culture Model of C. parvum Infection
Human ileocecal adenocarcinoma epithelial cell line HCT-8 was infected with C. parvum oocysts (24). Parasite growth was measured using a real-time PCR assay as previously described (25) or a high content imaging assay.4 The values of EC50 were calculated using the Hill-Slope model (Equation 1) using Prism version 5 (GraphPad Software Inc., La Jolla, CA),
where n is the Hill coefficient. The host cell cytotoxicity was determined using the LIVE/DEAD assay (Molecular Probes, Carlsbad, CA).
Mouse Model of C. parvum Infection
The anticryptosporidial activity of TFT was assessed in the interleukin (IL)-12 knock-out mouse model that resembles the acute human disease (26, 27). Mice were inoculated with 1,000 oocysts and treated with phosphate-buffered saline, 200 mg/kg of TFT, or 2000 mg/kg of paromomycin starting 4 h postinfection. Compounds were given for 7 days and mice were sacrificed on day 8 (peak infection). Mice (10 per group) were treated by gavage given in a split dose. Parasite load was quantified by fluorescence-activated cell sorter assays for the presence of the oocysts in feces at days 0, 4, and 7. Fecal pellets from the mice were routinely collected daily and homogenized in adjusted volumes of 2.5% potassium dichromate. Aliquots (200 μl) of vortexed samples were processed over microscale sucrose gradients essentially as previously described (28). The oocyst-containing fraction was collected and washed. Purified oocysts were incubated with a fluorescein-labeled oocyst-specific monoclonal antibody (OW5O-fluorescein isothiocyanate) for 20 min. Samples were adjusted to 600 μl and assayed with a 102-s sampling interval (100 μl) using logical gating of forward/side scatter and a OW5O-fluorescein isothiocyanate fluorescence signal on a BD Biosciences FACScan flow cytometer (29).
Statistical Analysis
Flow cytometry data were evaluated by analysis of variance (Microsoft Excel; Microsoft Corp., Redmond, WA).
RESULTS
Expression and Purification of Recombinant CpTK
CpTK was initially expressed using a cell-free wheat germ transcription/translation system. The enzyme was purified using thymidine affinity chromatography (supplemental Fig. S1). One milliliter of wheat germ lysate yielded 520 μg of ∼90% pure CpTK. Alternatively, His6-tagged CpTK was expressed in bacteria using the pET system. One liter of culture yielded ∼10 mg of ∼90% pure enzyme (supplemental Fig. S1).
Kinetic Characterization
The CpTK reaction exhibited classic Michaelis-Menten kinetics (supplemental Fig. S2). Wheat germ lysate CpTK has a Km of 48 μm for dT and kcat = 0.28 s−1 when the production of dTMP was monitored at saturating concentrations of ATP. Similar values are obtained for His-tagged CpTK, indicating that the His tag has no effect on enzyme function (Table 1). In addition, similar values are obtained when the production of ADP was monitored (Table 1), indicating that the enzyme preparation does not contain a contaminating ATPase. A Km value of 140 ± 30 μm was observed for ATP at saturating concentrations of dT (Table 1).
TABLE 1.
Kinetic parameters for CpTK
All reactions were 100 μm dCTP unless otherwise noted. Reactions characterizing phosphate acceptors were carried out with 5 mm ATP and reactions characterizing phosphate donors were carried out with 250 μm dT.
| Substrate | Km | kcat | kcat/Km |
|---|---|---|---|
| μm | s−1 | m−1s−1 | |
| Thymidinea | 48 ± 3 | 0.28 ± 0.05 | (4.8 ± 0.5) × 103 |
| Thymidineb | 89 ± 6 | 0.28 ± 0.007 | (3.2 ± 0.1) × 103 |
| Thymidinec | 40 ± 2 | 0.3 ± 0.1 | (6.7 ± 2) × 103 |
| Thymidine (−dCTP)d | 45 ± 15 | 0.04 ± 0.004 | (8.8 ± 0.8) × 102 |
| ATPd | 140 ± 30 | 0.28 ± 0.05 | (2.3 ± 0.1) × 103 |
| ATP (−dCTPd) | 110 ± 50 | 0.04 ± 0.004 | (3.5 ± 0.3) × 102 |
| dURc | 118 ± 1 | 0.53 ± 0.06 | (4.5 ± 0.5) × 103 |
| TFTc | 22 ± 3 | 0.36 ± 0.01 | (6.4 ± 0.2) × 103 |
| FUdRc | 40 ± 2 | 0.24 ± 0.02 | (5.8 ± 0.8) × 103 |
| AZTc | 32 ± 4 | 0.17 ± 0.02 | (5.0 ± 0.5) × 103 |
| β-l-Thymidinec | 290 ± 30 | 0.007 ± 0.0002 | (2.4 ± 0.6) × 10 |
| D4Tc | 50 ± 10 | 0.03 ± 0.002 | (6.4 ± 0.2) × 102 |
a Radioactive assay with untagged CpTK.
b Radioactive assay with His-tagged CpTK.
c Spectrophotometric assay with His-tagged CpTK.
d Spectrophotometric assay with untagged CpTK. Values are the average and standard deviations of two to three independent experiments.
Allosteric Regulation
CpTK was activated by dCTP and dCDP by 7- and 2.5-fold, respectively. dCTP is not a phosphate donor, but instead activates CpTK by increasing kcat; it does not change the values of Km of either dT or ATP (Table 1). Unlike Escherichia coli TK, CpTK was not activated by dATP or dGTP. Like other TKs, CpTK is inhibited by dTTP (data not shown).
Oligomeric State
The oligomeric state of CpTK was determined using size-exclusion chromatography. CpTK eluted at a size of 70 ± 10 kDa (Fig. 2), which is most consistent with a trimer (monomeric molecular mass of 23.3 kDa). Importantly, the presence of the activator dCTP or substrates dT and ATP did not change the oligomeric state of CpTK.
FIGURE 2.
Determination of the CpTK oligomeric state using gel filtration chromatography. Untagged CpTK elutes at a molecular mass of ∼70 kDa, consistent with being a trimer under the three conditions tested: enzyme alone (diamonds), CpTK + 100 μm dCTP (triangles), and CpTK + 100 μm Thd + 1 mm ATP (squares). The elution volumes of the standards are as follow: blue dextran, 96 ml; bovine γ-globulin, 158 kDa, 107 ml; chicken ovalbumin, 44 kDa, 142 ml; equine myoglobin, 17 kDa, 180 ml; and vitamin B12, 1.35 kDa, 303 ml.
Activation of Naturally Occurring Nucleosides
CpTK efficiently phosphorylates both dT and dU with similar values of kcat/Km (Table 1). dG and dC are poor substrates, and no phosphorylation was observed for the other naturally occurring nucleosides or for dTMP (Table 2). CpTK utilizes a wide range of NTPs as the phosphate donor, with the exception of CTP. The preference for phosphate donors is: ATP = dGTP > GTP > UTP (Table 2). PPi cannot serve as the phosphate donor.
TABLE 2.
Substrate specificity of CpTK
Untagged CpTK was used for all assays.
| Substrate (100 μm) | % Activity |
|---|---|
| Phosphate acceptorsa | |
| dT | 100 |
| dU | 69 |
| dA | <1 |
| dG | <1 |
| dC | 7 |
| G | 1 |
| U | <1 |
| A | <1 |
| C | <1 |
| dTMP | <1 |
| Phosphate donorsb | |
| ATP | 100 |
| dGTP | 99 |
| GTP | 42 |
| UTP | 17 |
| CTP | <1 |
| PPi | <1 |
a For phosphate acceptors: spectrophotometric assay; all reactions contained 5 mm ATP; activity is relative to 100 μm dT.
b For phosphate donors: radioactive assay; all reactions contained 250 μm dT; activity is relative to 5 mm ATP.
Activation of Prodrugs and Other Nucleoside Analogs
The specificity of CpTK was further evaluated by testing the ability of antiviral and anticancer drugs to serve as substrates (Table 3). TFT, FUdR, AZT, IUdR, ClUdR, and bromodeoxyuridine, all close analogs of dT, are efficiently phosphorylated by CpTK (Table 3). The Michaelis-Menten parameters for TFT, FUdR, and AZT are equivalent to those of dT (Table 1). D4T is a moderately good substrate; the value of Km for D4T is equivalent to that of dT, but the value of kcat is lower by a factor of 10. β-l-Thymidine is a poor substrate; the value of kcat/Km for β-l-thymidine is less than that of dT by a factor of 200, which derives primarily from a decrease in the value of kcat.
TABLE 3.
CpTK activation of antimetabolites
Conditions used were 100 μm phosphoacceptor, 5 mm ATP. Activity is relative to dT.
| Compound | % Activity |
|---|---|
| TFT | 130 |
| 5-I-dU | 89 |
| 5-Cl-dU | 110 |
| 5-Bromodeoxyuridine | 84 |
| AZT | 60 |
| FudR | 120 |
| D4T | 12 |
| β-l-Thymidine | <5 |
| Acyclovir | <1 |
| AraC | <1 |
| AraT | <1 |
| IDC | <1 |
Although AraC, AraT, and IDC are reported to display anti-cryptosporidial activity (15), none of these compounds are substrates for CpTK, nor do they inhibit the phosphorylation of dT. Acyclovir also was not phosphorylated by CpTK, which can explain the resistance of C. parvum to this compound.
We screened a collection of pyrimidine nucleoside analogs to more thoroughly probe the specificity requirements of CpTK (Table 4). In general, only modest substitutions of similar or smaller size are tolerated in both the sugar and base components. A 2′-deoxy sugar is strongly preferred: the 2′ position cannot tolerate OH or F in ara configuration (AraT, KP296-8) and the 3′ position can be N3 but not acetyl (KP1280). Few substitutions are allowed on the base. The 5 positions can tolerate hydrogen, fluorine, bromine, chlorine, iodine, and CF3, all of which are of similar or smaller size compared with the CH3 of dT. At the 4 position replacement of the carbonyl group by N4-OH (KP1308, KP1309) removes activity. Surprisingly, and in contrast to dU, 2′-deoxypseudouridine (KP320) is not a substrate, but addition of a 1-methyl to give KP332 (an analog of dT) produces a good substrate, with kcat comparable with that for dT and a 3-fold higher Km. Addition of CH3 to the 3 position of 2-deoxypseudouridine (KP333) abrogates activity. These requirements suggest that the CpTK active site is very restrictive and discriminating of phosphate acceptors.
TABLE 4.
CpTK activation of synthetic thymidine and cytidine analogs
Conditions used were 100 μm phosphoacceptor, 5 mm ATP. Activity is relative to dT. na, not applicable.
Effect of Prodrugs on C. parvum Growth in the Cell Culture Model of Infection
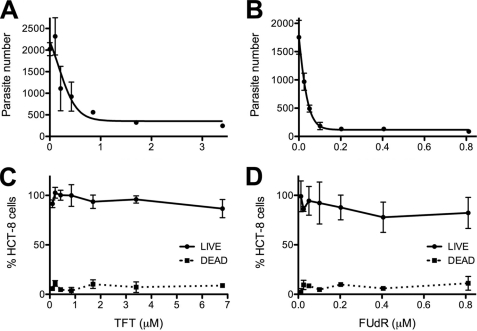
The antiparasitic activities of AZT, TFT, and FUdR were tested by infecting monolayers of HCT-8 cells with C. parvum oocysts. AZT is a poor inhibitor of C. parvum growth with EC50 > 370 μm, consistent with a previous report (15) (supplemental Fig. S1). Both TFT and FUdR showed potent antiparasitic activity (Fig. 3, A and B). TFT has an EC50 = 410 nm, somewhat better than a previous report (EC50 = 900 nm (15)). No cytotoxicity was observed at 6.8 μm TFT (Fig. 3C), similar to other studies (15, 30). FUdR has an EC50 = 28 nm, and no cytotoxicity was observed at 0.8 μm (Fig. 3D). In previous reports, FUdR inhibited the proliferation of colon cancer cell lines with values of EC50 ranging from 10 to 209 μm (30). These observations suggest that sufficient therapeutic windows may exist for both TFT and FUdR.
FIGURE 3.
Effect of TFT and FUdR on parasite and host cell growth in a cell culture model of C. parvum infection. A and B, C. parvum growth was assayed by high-content imaging. A, TFT; B, FUdR. C and D, host cell cytotoxicity was assessed using the LIVE/DEAD® assay (Invitrogen). Data are representative of two independent experiments. C, TFT; D, FUdR.
Effect of TFT on C. parvum Growth in a Mouse Model
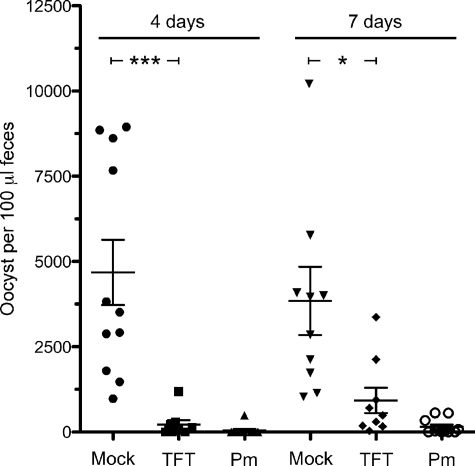
To determine whether thymidine analogs can compete effectively with thymidine for uptake into C. parvum in the gastrointestinal tract, TFT was tested in the IL-2 knock-out mouse model of acute infection (26, 27). Paromomycin treatment (2000 mg/kg) causes a >99% reduction in the oocysts recovered from feces in this model system (p < 0.0001, Fig. 4). Similarly, TFT treatment (200 mg/kg) caused a 95% reduction after 4 days of treatment (p < 0.0006, Fig. 4). A significant decrease of parasite load (76% inhibition, p < 0.02) was still observed with continued treatment through day 7 (Fig. 4).
FIGURE 4.
Treatment with TFT in a IL-12 mouse model of C. parvum infection. Number of parasite oocysts in mouse feces 4 and 7 days postinfection with treatment with phosphate-buffered saline (Mock), 200 mg/kg of TFT, and 2000 mg/kg of paromomycin (Pm) is shown. ***, p < 0.0006; *, p < 0.02. The bars denote the averages and S.E.
DISCUSSION
The anti-cryptosporidial activity of TFT, AraT, IUdR, IDC, and AraC suggested that that CpTK might have the broad specificity of a viral TK (15). However, the above experiments demonstrate that CpTK has narrow substrate specificity for dT and dU and does not phosphorylate AraT, AraC, and IDC. These observations suggest that Cryptosporidium must possess another deoxynucleoside kinase. Two other nucleoside kinases have been identified in the genome: adenosine kinase and uridine kinase/uracil phosphoribosyl transferase (UK/UPRT). Although Cryptosporidium-adenosine kinase phosphorylates AraA (13), it appears to have strict specificity for purine nucleosides (31). Curiously, CpUK/UPRT appears to have been obtained from a plant or algae (13). Little is known about the substrate preferences of plant UK/UPRTs (32); perhaps CpUK/UPRT is responsible for the activation of these prodrugs.
Surprisingly, AZT does not exhibit antiparasitic activity despite being an excellent substrate for CpTK. The phosphorylation of AZT-MP is rate-limiting in human cells (33); this step may also be difficult in C. parvum. Failure to be converted to the triphosphate could account for the poor parasitic activity of AZT. Intriguingly, D4T-MP is a good substrate for dTMP kinase, suggesting that antiviral D4T (stavudine) may be an effective anti-parasitic agent even though it is a relatively poor substrate for CpTK.
The presence of CpTK, together with the observation that other gastrointestinal parasites do not require TS, raises important questions about the effectiveness of targeting CpTS-DHFR for anticryptosporidial chemotherapy. Indeed, the abundance of thymidine in the gastrointestinal tract is reflected in the high value of Km for dT for both the C. parvum and E. coli enzymes (Km = 48 and 17 μm for CpTK and EcTK, respectively (21)), suggesting that dT is abundant in the gastrointestinal tract. In contrast, the value of Km = 0.5 μm for human TK1 (34, 35) reflects the low concentrations of dT in plasma (36).
Deoxynucleoside kinases form two superfamilies: the thymidine kinase 1 (TK1)-like family and the thymidine kinase 2 (TK2)-like family, so named for their homology to human cytosolic TK1 (hTK1) and mitochondrial TK2 (hTK2), respectively (37). CpTK is a member of the TK1 family and is 62% identical to E. coli TK (EcTK) and 23% identical to hTK1. Narrow specificity for phosphate acceptors, broad specificity for phosphate donors, and allosteric regulation by dCTP and dTTP observed for CpTK are typical of TK1s in general and most similar to EcTK, reflecting the bacterial origins of the parasite enzyme. CpTK is notably different from hTK1 in two respects. First, CpTK is allosterically regulated by dCTP and dCDP in addition to dTTP. Similar allosteric regulation is observed in EcTK, which is regulated by a number of nucleoside di- and triphosphates (21, 38). Second, CpTK is a trimer regardless of the presence of substrates or allosteric activator, whereas both hTK1 and EcTK are regulated via changes in oligomerization (39, 40). In this respect, CpTK is similar to Caenorhabditis elegans TK, another member of the TK1 family (41). Importantly, CpTK displays very different functional properties than hTK2, which has broad specificity for phosphate acceptor and narrow specificity for phosphate donors characteristic of the TK2 family (37, 42). These observations suggest that it might be possible to develop selective inhibitors of CpTK, although redundancy of the pyrimidine salvage pathways argues against this strategy.
Although CpTK is not likely to be a drug target per se, our results suggest that TK-activated prodrugs may provide an effective strategy to treat cryptosporidiosis. Both TFT and FUdR display selective antiparasitic effects in vitro; C. parvum is at least 20-fold more sensitive to TFT and 400-fold more sensitive to FUdR than host cells, which is comparable or better than the therapeutic index of ribavirin (TI = 2–30 (43)), so a sufficient therapeutic window may exist for these compounds despite their well known cytotoxicity. Moreover, TFT causes a dramatic reduction in oocyst load in the IL-2 knock-out mouse model of infection, demonstrating that the prodrug can compete effectively in a thymidine-rich environment. TFT is rapidly metabolized in both mice and humans by the action of uridine and thymidine phosphorylases (44), so it is possible that more effective anticryptosporidial activity could be obtained with different dosing. Alternatively, a thymidine phosphorylase inhibitor could be used to increase drug stability (44–46). Importantly, TAS-102, a combination of TFT and thymidine phosphorylase inhibitor, is currently in clinical trials for colon cancer (46).
The mechanistic basis for the parasite selectivity of TFT and FUdR is unclear. Viral TKs, such as herpes simplex virus type 1 TK, often have broader specificity than human TK1 and TK2 (14), which explains the antiviral properties of compounds such as acyclovir. However, the specificity of CpTK is very similar to human TK1, and both TFT and FUdR are also good substrates for the human enzymes. Perhaps the salvage pathways are simply more efficient in the parasite than the host. The cytotoxicity of both TFT and FUdR is attributed to the incorporation of the corresponding triphosphates into DNA (47, 48). Therefore rapidly dividing cells are more sensitive to these compounds, so antiparasitic activity could also reflect faster proliferation of the parasite.
TFT-MP and FUdR-MP have an additional mode of action: both inhibit TS (49, 50), which could further decrease the dTTP pool, facilitating the incorporation of TFT-TP and FUdR-TP into DNA. Intriguingly, Cryptosporidium TS-DHFR is more active than human TS (51), suggesting that CpTS-DHFR could also be more sensitive to TFT-MP and FUdR-MP.
Cryptosporidium contains a plethora of transporters, many of which are associated with drug resistance (6, 52). Although the role of these transporters in drug action has yet to be elucidated, their mere presence suggests that drug accumulation may be a significant obstacle in the development of chemotherapy for cryptosporidiosis. Nucleoside prodrugs offer an appealing strategy to circumvent this problem. Because the parasite relies on salvage pathways, Cryptosporidium must have efficient uptake systems for thymidine that will also uptake prodrugs such as TFT. The efficacy of TFT in the IL-2 knock-out mouse model demonstrates that the pyrimidine salvage pathways can be subverted for treatment. Therefore nucleoside prodrugs offer a promising route to new anticryptosporidial drugs.
Supplementary Material
Acknowledgment
We acknowledge Nina McNair for technical assistance.
This work was supported, in whole or in part, by National Institutes of Health Grants RO1 AI055268 (to B. S.), U01AI75466 (to L. H.), and AI082617 (to P. K. R.), and the Veterans Affairs and the Atlanta Research and Education Foundation (to J. M.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S3.
L. Sharling, X. Liu, D. R. Gollapalli, S. K. Maurya, L. Hedstrom, and B. Striepen, manuscript in preparation.
- TK
- thymidine kinase
- CpTK
- Cryptosporidium parvum thymidine kinase
- EcTK
- E. coli thymidine kinase
- TS
- thymidylate synthase
- DHFR
- dihydrofolate reductase
- TFT
- trifluoromethylthymidine
- AraT
- thymine 1-β-d-arabinofuranoside
- IUdR
- 5-iodo-2′-deoxyuridine
- AraC
- 1-β-d-arabinofuranosylcytosine (cytarabine)
- AZT
- 3′-azido-2′,3′-dideoxythymidine
- dT
- thymidine
- dU
- deoxyuridine
- dG
- deoxyguanosine
- dC
- deoxycytidine
- UK/UPRT
- uridine kinase/uridine phosphoribosyl transferase
- FUdR
- 5-fluoro-2′-deoxyuridine
- ClUdR
- 5-chloro-2′-deoxyuridine
- D4T
- 2′,3′-didehydro-2′,3′-dideoxythymidine
- IDC
- 5-iodo-2′-deoxycytidine
- IL
- interleukin
- TK1
- thymidine kinase 1
- MP
- monophosphate.
REFERENCES
- 1.Huang D. B., White A. C. (2006) Gastroenterol. Clin. North Am. 35, 291–314, viii [DOI] [PubMed] [Google Scholar]
- 2.Carey C. M., Lee H., Trevors J. T. (2004) Water Res. 38, 818–862 [DOI] [PubMed] [Google Scholar]
- 3.DuPont H. L., Chappell C. L., Sterling C. R., Okhuysen P. C., Rose J. B., Jakubowski W. (1995) N. Engl. J. Med. 332, 855–859 [DOI] [PubMed] [Google Scholar]
- 4.Mead J. R. (2002) Drug Resist. Updat. 5, 47–57 [DOI] [PubMed] [Google Scholar]
- 5.Abrahamsen M. S., Templeton T. J., Enomoto S., Abrahante J. E., Zhu G., Lancto C. A., Deng M., Liu C., Widmer G., Tzipori S., Buck G. A., Xu P., Bankier A. T., Dear P. H., Konfortov B. A., Spriggs H. F., Iyer L., Anantharaman V., Aravind L., Kapur V. (2004) Science 304, 441–445 [DOI] [PubMed] [Google Scholar]
- 6.Sauvage V., Aubert D., Escotte-Binet S., Villena I. (2009) Mol. Biochem. Parasitol. 167, 81–94 [DOI] [PubMed] [Google Scholar]
- 7.Martucci W. E., Udier-Blagovic M., Atreya C., Babatunde O., Vargo M. A., Jorgensen W. L., Anderson K. S. (2009) Bioorg. Med. Chem. Lett. 19, 418–423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pelphrey P. M., Popov V. M., Joska T. M., Beierlein J. M., Bolstad E. S., Fillingham Y. A., Wright D. L., Anderson A. C. (2007) J. Med. Chem. 50, 940–950 [DOI] [PubMed] [Google Scholar]
- 9.Vásquez J. R., Goozé L., Kim K., Gut J., Petersen C., Nelson R. G. (1996) Mol. Biochem. Parasitol. 79, 153–165 [DOI] [PubMed] [Google Scholar]
- 10.Morrison H. G., McArthur A. G., Gillin F. D., Aley S. B., Adam R. D., Olsen G. J., Best A. A., Cande W. Z., Chen F., Cipriano M. J., Davids B. J., Dawson S. C., Elmendorf H. G., Hehl A. B., Holder M. E., Huse S. M., Kim U. U., Lasek-Nesselquist E., Manning G., Nigam A., Nixon J. E., Palm D., Passamaneck N. E., Prabhu A., Reich C. I., Reiner D. S., Samuelson J., Svard S. G., Sogin M. L. (2007) Science 317, 1921–1926 [DOI] [PubMed] [Google Scholar]
- 11.Loftus B., Anderson I., Davies R., Alsmark U. C., Samuelson J., Amedeo P., Roncaglia P., Berriman M., Hirt R. P., Mann B. J., Nozaki T., Suh B., Pop M., Duchene M., Ackers J., Tannich E., Leippe M., Hofer M., Bruchhaus I., Willhoeft U., Bhattacharya A., Chillingworth T., Churcher C., Hance Z., Harris B., Harris D., Jagels K., Moule S., Mungall K., Ormond D., Squares R., Whitehead S., Quail M. A., Rabbinowitsch E., Norbertczak H., Price C., Wang Z., Guillén N., Gilchrist C., Stroup S. E., Bhattacharya S., Lohia A., Foster P. G., Sicheritz-Ponten T., Weber C., Singh U., Mukherjee C., El-Sayed N. M., Petri W. A., Jr., Clark C. G., Embley T. M., Barrell B., Fraser C. M., Hall N. (2005) Nature 433, 865–868 [DOI] [PubMed] [Google Scholar]
- 12.Zhu G. (2008) in Cryptosporidium and Cryptosporidiosis (Fayer R., Xiao L. eds) pp. 57–77, Second Ed., CRC Press, Boca Raton, FL [Google Scholar]
- 13.Striepen B., Pruijssers A. J., Huang J., Li C., Gubbels M. J., Umejiego N. N., Hedstrom L., Kissinger J. C. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 3154–3159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Al-Madhoun A. S., Tjarks W., Eriksson S. (2004) Mini. Rev. Med. Chem. 4, 341–350 [DOI] [PubMed] [Google Scholar]
- 15.Woods K. M., Upton S. J. (1998) FEMS Microbiol. Lett. 168, 59–63 [DOI] [PubMed] [Google Scholar]
- 16.Pankiewicz K. W. (2000) Carbohydr. Res. 327, 87–105 [DOI] [PubMed] [Google Scholar]
- 17.Felczak K., Miazga A., Poznański J., Bretner M., Kulikowski T., Dzik J. M., Gołos B., Zieliński Z., Cieśla J., Rode W. (2000) J. Med. Chem. 43, 4647–4656 [DOI] [PubMed] [Google Scholar]
- 18.Mudeppa D. G., Pang C. K., Tsuboi T., Endo Y., Buckner F. S., Varani G., Rathod P. K. (2007) Mol. Biochem. Parasitol. 151, 216–219 [DOI] [PubMed] [Google Scholar]
- 19.Kokoris M. S., Black M. E. (2002) Protein Sci. 11, 2267–2272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tung P. P., Respass J., Summers W. C. (1996) Yale J. Biol. Med. 69, 495–503 [PMC free article] [PubMed] [Google Scholar]
- 21.Chen M. S., Prusoff W. H. (1978) Methods Enzymol. 51, 354–360 [DOI] [PubMed] [Google Scholar]
- 22.Furlong N. B. (1963) Anal. Biochem. 5, 515–522 [DOI] [PubMed] [Google Scholar]
- 23.Schelling P., Folkers G., Scapozza L. (2001) Anal. Biochem. 295, 82–87 [DOI] [PubMed] [Google Scholar]
- 24.Umejiego N. N., Gollapalli D., Sharling L., Volftsun A., Lu J., Benjamin N. N., Stroupe A. H., Riera T. V., Striepen B., Hedstrom L. (2008) Chem. Biol. 15, 70–77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cai X., Woods K. M., Upton S. J., Zhu G. (2005) Antimicrob. Agents Chemother. 49, 4437–4442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Campbell L. D., Stewart J. N., Mead J. R. (2002) J. Parasitol. 88, 1014–1016 [DOI] [PubMed] [Google Scholar]
- 27.Ehigiator H. N., Romagnoli P., Borgelt K., Fernandez M., McNair N., Secor W. E., Mead J. R. (2005) Parasite Immunol. 27, 17–28 [DOI] [PubMed] [Google Scholar]
- 28.Arrowood M. J., Donaldson K. (1996) J. Eukaryot. Microbiol. 43, 89S. [DOI] [PubMed] [Google Scholar]
- 29.Arrowood M. J., Hurd M. R., Mead J. R. (1995) J. Parasitol. 81, 404–409 [PubMed] [Google Scholar]
- 30.de Bruin M., van Capel T., Van der Born K., Kruyt F. A., Fukushima M., Hoekman K., Pinedo H. M., Peters G. J. (2003) Br. J. Cancer 88, 957–964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Galazka J., Striepen B., Ullman B. (2006) Mol. Biochem. Parasitol. 149, 223–230 [DOI] [PubMed] [Google Scholar]
- 32.Islam M. R., Kim H., Kang S. W., Kim J. S., Jeong Y. M., Hwang H. J., Lee S. Y., Woo J. C., Kim S. G. (2007) Plant Mol. Biol. 63, 465–477 [DOI] [PubMed] [Google Scholar]
- 33.Lavie A., Konrad M. (2004) Mini. Rev. Med. Chem. 4, 351–359 [DOI] [PubMed] [Google Scholar]
- 34.Munch-Petersen B., Cloos L., Jensen H. K., Tyrsted G. (1995) Adv. Enzyme Regul. 35, 69–89 [DOI] [PubMed] [Google Scholar]
- 35.Birringer M. S., Perozzo R., Kut E., Stillhart C., Surber W., Scapozza L., Folkers G. (2006) Protein Expr. Purif. 47, 506–515 [DOI] [PubMed] [Google Scholar]
- 36.Shields A. F., Coonrod D. V., Quackenbush R. C., Crowley J. J. (1987) J. Nucl. Med. 28, 1435–1440 [PubMed] [Google Scholar]
- 37.Arnér E. S., Eriksson S. (1995) Pharmacol. Ther. 67, 155–186 [DOI] [PubMed] [Google Scholar]
- 38.Okazaki R., Kornberg A. (1964) J. Biol. Chem. 239, 275–284 [PubMed] [Google Scholar]
- 39.Munch-Petersen B., Tyrsted G., Cloos L. (1993) J. Biol. Chem. 268, 15621–15625 [PubMed] [Google Scholar]
- 40.Iwatsuki N., Okazaki R. (1967) J. Mol. Biol. 29, 139–154 [DOI] [PubMed] [Google Scholar]
- 41.Skovgaard T., Munch-Petersen B. (2006) Nucleosides Nucleotides Nucleic Acids 25, 1165–1169 [DOI] [PubMed] [Google Scholar]
- 42.Pérez-Pérez M. J., Priego E. M., Hernández A. I., Familiar O., Camarasa M. J., Negri A., Gago F., Balzarini J. (2008) Med. Res. Rev. 28, 797–820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Markland W., McQuaid T. J., Jain J., Kwong A. D. (2000) Antimicrob. Agents Chemother. 44, 859–866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fukushima M., Suzuki N., Emura T., Yano S., Kazuno H., Tada Y., Yamada Y., Asao T. (2000) Biochem. Pharmacol. 59, 1227–1236 [DOI] [PubMed] [Google Scholar]
- 45.Emura T., Suzuki N., Fujioka A., Ohshimo H., Fukushima M. (2005) Int. J. Oncol. 27, 449–455 [PubMed] [Google Scholar]
- 46.Temmink O. H., Emura T., de Bruin M., Fukushima M., Peters G. J. (2007) Cancer Sci. 98, 779–789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Emura T., Nakagawa F., Fujioka A., Ohshimo H., Yokogawa T., Okabe H., Kitazato K. (2004) Int. J. Mol. Med. 13, 249–255 [PubMed] [Google Scholar]
- 48.Yin M. B., Rustum Y. M. (1991) Cancer Commun. 3, 45–51 [PubMed] [Google Scholar]
- 49.Danenberg P. V., Lockshin A. (1981) Pharmacol. Ther. 13, 69–90 [DOI] [PubMed] [Google Scholar]
- 50.Eckstein J. W., Foster P. G., Finer-Moore J., Wataya Y., Santi D. V. (1994) Biochemistry 33, 15086–15094 [DOI] [PubMed] [Google Scholar]
- 51.Atreya C. E., Anderson K. S. (2004) J. Biol. Chem. 279, 18314–18322 [DOI] [PubMed] [Google Scholar]
- 52.Benitez A. J., McNair N., Mead J. (2007) Parasitol. Res. 101, 1611–1616 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.