Abstract
Human chromosome end-capping and telomerase regulation require POT1 (Protection of Telomeres 1) and TPP1 proteins, which bind to the 3′ ssDNA extension of human telomeres. POT1–TPP1 binding to telomeric DNA activates telomerase repeat addition processivity. We now provide evidence that this POT1–TPP1 activation requires specific interactions with telomerase, rather than it being a DNA substrate-specific effect. First, telomerase from the fish medaka, which extends the same telomeric DNA primer as human telomerase, was not activated by human POT1–TPP1. Second, mutation of a conserved glycine, Gly100 in the TEN (telomerase essential N-terminal) domain of TERT, abolished the enhancement of telomerase processivity by POT1–TPP1, in contrast to other single amino acid mutations. Chimeric human–fish telomerases that contained the human TEN domain were active but not stimulated by POT1–TPP1, showing that additional determinants of processivity lie outside the TEN domain. Finally, primers bound to mouse POT1A and human TPP1 were activated for extension by human telomerase, whereas mPOT1A–mTPP1 was most active with mouse telomerase, indicating that these mammalian telomerases have specificity for their respective TPP1 proteins. We suggest that a sequence-specific interaction between TPP1 in the TPP1–POT1–telomeric DNA complex and the G100 region of the TEN domain of TERT is necessary for high-processivity telomerase action.
Keywords: Chromosome end-capping, POT1–TPP1, processivity, telomerase, telomeres
Telomeres, the very ends of linear chromosomes, are potential sites of genome instability. Chromosome ends are therefore capped by proteins that bind to either the double-stranded telomeric DNA repeats or the single-stranded 3′ extension at the very end of the chromosome. The importance of 3′ end-capping is illustrated by the dramatic phenotype of deletion of pot1 (Protection of Telomeres 1) in Schizosaccharomyces pombe: Telomeric DNA is eroded, chromosome ends undergo random fusion, most cells die, and the rare survivors have all three of their chromosomes circularized, which allows vegetative life without telomeres (Baumann and Cech 2001; Wang and Baumann 2008).
The situation is more subtle in mammals. RNAi knockdown of human POT1 in transformed human cells causes an increase in telomere associations that are thought to indicate chromosome end fusions (Veldman et al. 2004), although the frequency of such events is much less than that obtained by inhibition of the double-stranded telomeric DNA-binding protein TRF2 (Hockemeyer et al. 2005). In addition, upon reduction of hPOT1, all telomeres show a transient DNA damage response in the G1 phase of the cell cycle (Hockemeyer et al. 2005), and similar results have been obtained in chicken cells (Churikov et al. 2006). In the mouse, where a complete knockout can be studied, there are two POT1 genes (Hockemeyer et al. 2006). POT1A (but not POT1B) is required to repress a DNA damage signal at telomeres (Hockemeyer et al. 2006; Wu et al. 2006). Mouse POT1A represses the DNA damage response that occurs through ATR (ataxia telangiectasia and Rad3-related) kinase signaling, whereas TRF2 represses DNA damage response through ATM (ataxia telangiectasia mutated) kinase signaling (Denchi and de Lange 2007).
Another telomeric protein, TPP1 (Houghtaling et al. 2004; Liu et al. 2004; Ye et al. 2004), is the heterodimeric partner of POT1 (Wang et al. 2007; Xin et al. 2007). The POT1–TPP1 complex is analogous to the TEBP α–β heterodimer in the ciliated protozoan Oxytricha nova (Gray et al. 1991; Horvath et al. 1998). A substantial portion of POT1–TPP1 is associated with the dsDNA-binding proteins TRF1 and TRF2 through a bridge formed by TIN2 (Kim et al. 1999; O'Connor et al. 2006; Sarthy et al. 2009; Takai et al. 2010). A sixth protein, RAP1, is critical for inhibiting nonhomologous end-joining of mammalian telomeres (Sarthy et al. 2009). Unlike budding yeast Rap1 (Konig et al. 1996), the mammalian RAP1 does not bind DNA on its own, but is brought to the telomere by binding to TRF2. The six-protein mammalian telomere complex has been termed “shelterin” (de Lange 2005).
In addition to their role in chromosome end protection, telomeric proteins provide a major level of telomerase regulation. In budding yeast, where this regulation has been studied in great detail, it is mediated by both double-stranded and single-stranded telomere-binding proteins (Moretti et al. 1994; Evans and Lundblad 1999; Teixeira et al. 2004). In human cells, hPOT1 has been reported to be a telomerase-dependent positive regulator or negative regulator of telomere elongation (Colgin et al. 2003; Loayza and De Lange 2003), and TPP1 also contributes to telomere length control, at least in part by recruitment of POT1 (Ye et al. 2004).
A detailed mechanistic understanding of the role of POT1–TPP1 has benefited from studies with purified proteins. The crystal structure of the POT1–telomeric DNA complex shows the DNA buried in a groove formed by two OB (oligonucleotide-binding) folds (Lei et al. 2004), and, as expected, this complex is refractory to extension by telomerase (Kelleher et al. 2005; Lei et al. 2005). When TPP1 is added, the apparent Kd is decreased by 10-fold, while the protein-binding site on the DNA is unchanged; yet, when POT1–TPP1 is bound upstream of the DNA 3′ end, leaving a single-stranded 3′ tail, telomerase extends it with enhanced repeat addition processivity (hereafter termed “processivity”) (Wang et al. 2007). That is, multiple telomeric repeats are synthesized on a single DNA molecule prior to dissociation. Although it is not straightforward to compare in vitro and in vivo results, this enhanced processivity means that a single round of telomerase action is within a factor of 2 of accounting for the amount of extension of a telomere seen in a single cell cycle (Zhao et al. 2009).
We provided evidence that POT1–TPP1 bound to a DNA primer stimulates telomerase processivity by two mechanisms: inhibiting primer dissociation from telomerase, and facilitating the translocation of the primer/template (Latrick and Cech 2010). These activities of POT1–TPP1 could be explained if it bound to telomerase and, in fact, an interaction with TERT has been reported (Xin et al. 2007). Alternatively, or in addition, stimulation of processivity might be a DNA substrate-specific effect. For example, the continued interaction of a G-rich portion of the primer with the anchor site of telomerase might be helpful for initiation but inhibitory to processivity, and POT1–TPP1 binding to the nascent DNA could prevent its interaction with the anchor site. In this study, we develop a new approach involving mixing and matching telomerase and telomere components from different organisms to show that POT1–TPP1 stimulation of processivity does not act at the level of the DNA substrate, but requires an interaction with telomerase. Furthermore, using site-specific mutagenesis, we identify a site in the TEN (telomerase essential N-terminal) domain of TERT, in a subregion known as the DAT (dissociates activities of telomerase) domain (Armbruster et al. 2001), that is required for this stimulation.
Results
TPP1 activation is species-specific
Telomerase of the fish medaka synthesizes the same TTAGGG telomeric repeats as human telomerase (Xie et al. 2008). Its telomerase RNA (TR) has a size and secondary structure similar to that of human telomerase, and the two TERT proteins are 36.6% identical and 50.6% similar (Yap et al. 2005; Xie et al. 2008). When the medaka TERT and TR were expressed in a rabbit reticulocyte lysate (RRL) and presented with a telomeric oligodeoxynucleotide and dNTPs, the primer was extended to give a ladder of telomeric repeats (Fig. 1), as expected from previous work (Xie et al. 2008). The extension pattern was similar to that obtained with reconstituted human TERT and TR (Fig. 1, cf. lanes 3 and 7). Because these reactions were performed in a vast excess of primer over enzyme, the appearance of an extended ladder of products indicates processivity, as confirmed rigorously elsewhere (Bryan et al. 2000; Latrick and Cech 2010).
Figure 1.
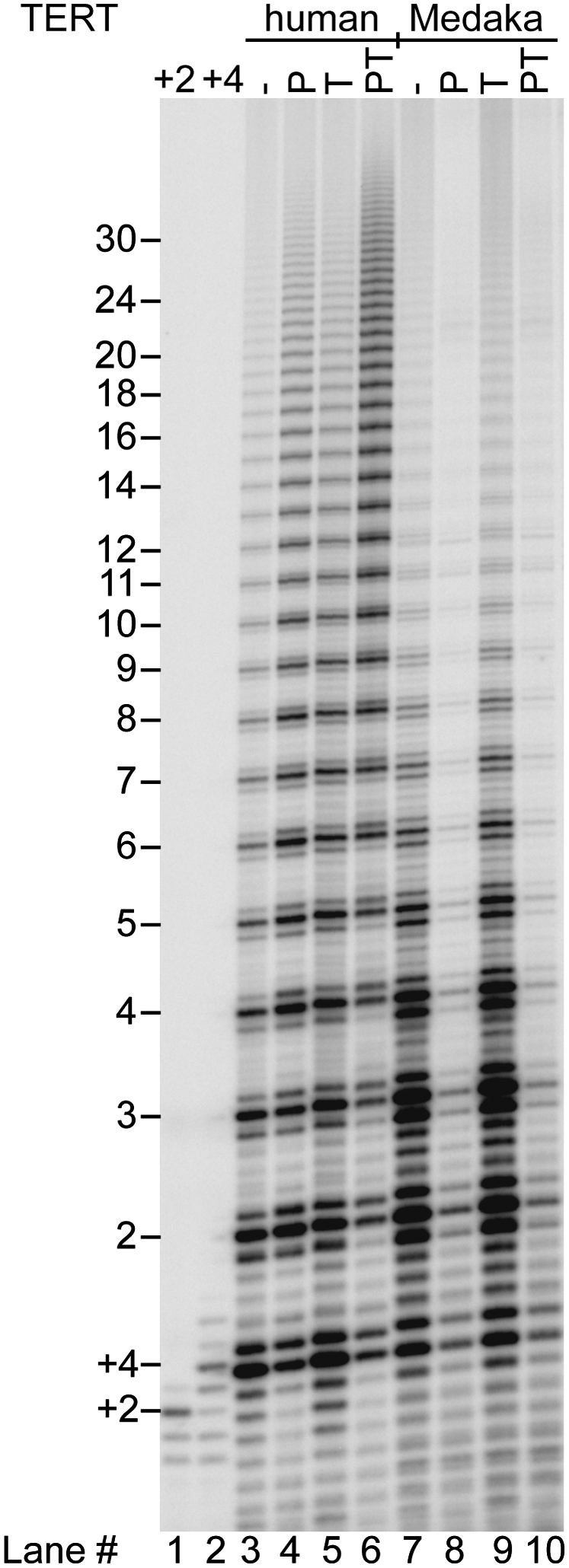
Human POT1–TPP1 bound to a telomeric DNA primer enhances the processivity of human but not medaka telomerase. TERT and TR were expressed in RRLs and telomerase was immunopurified. Primer a5 (TTAGGGTTAGCGTTAGGG, 50 nM) has a single base mutation that positions POT1 on the 10 nt at the primer 5′ end (Lei et al. 2005). Direct telomerase assay (1 h) in the presence of 32P-dGTP with primer alone (−), or in the presence of 500 nM POT1 (P), 500 nM TPP1 (T), or 500 nM of each protein (PT). Lanes +2 and +4 are markers synthesized by telomerase extension of primer (GGTTAG)3 in the presence of dGTP only (+2) or dGTP and dTTP but no dATP (+4). They consistently run slightly higher than the corresponding products in the experimental lanes because of the slight difference in base composition of the primers. The numbers 2–30 at left indicate the number of telomeric repeats synthesized.
When the two telomerases were presented with a human POT1–primer complex with a protruding 7-nucleotide (nt) 3′ tail, they responded differently. This DNA tail length was sufficient for extension by human telomerase (Fig. 1, lane 4), which extended the primer with slightly increased processivity as documented previously (Lei et al. 2005). Medaka telomerase, on the other hand, was substantially but not completely inhibited (90% inhibition) (Fig. 1, lane 8). This inhibition could be overcome by adding excess primer (Supplemental Fig. 1) or by changing the sequence of the TR template and primer to a non-POT1-binding sequence (data not shown), which argue against the POT1 directly inactivating medaka telomerase; presumably, the human POT1–primer complex is a poor substrate for medaka telomerase. Human TPP1 alone had little effect on either medaka or human telomerase (Fig. 1, cf. lanes 3 and 5, and lanes 7 and 9), which is expected because TPP1 by itself does not bind DNA. In the key experiment, TPP1 was unable to stimulate the reaction of medaka telomerase on POT1–DNA (in Fig. 1, lane 10 is unchanged from lane 8). This was in contrast to the positive control with human telomerase, where POT1–TPP1 consistently gave more than twofold enhanced processivity (Fig. 1, cf. lanes 6 and 4). (The TPP1 proteins used throughout this study are the fully active N-terminal portions, TPP1-N [see the Materials and Methods].)
The medaka versus human telomerase experiments allow a strong conclusion, because most of the components of the two reactions were identical. The same POT1–TPP1–DNA complex was tested in both cases, and the components of the RRL were the same. The only differences were the TERT and TR. If POT1–TPP1 were acting through binding to the DNA substrate without interaction with telomerase, it should have affected both reactions identically. If some component of the RRL were displacing POT1–TPP1 from the DNA, both reactions should have been activated. Instead, the different effect of TPP1 on the two reactions suggests that elements of TERT or TR that differ between medaka and human interact with TPP1.
Mutation in the TEN domain prevents TPP1 activation
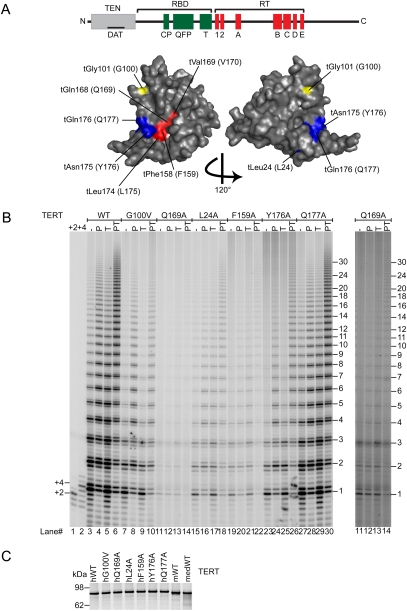
The TEN domain of TERT has been implicated in processivity in human, yeast, and Tetrahymena telomerases (Moriarty et al. 2004; Autexier and Lue 2006; Zaug et al. 2008). In an attempt to identify a specific region of telomerase involved in a functional interaction with TPP1, we made a series of single amino acid mutations in the TEN domain of hTERT and tested them for telomerase activity with and without POT1–TPP1. We made use of the crystal structure of the TEN domain of Tetrahymena TERT (Jacobs et al. 2006); we assumed that conserved amino acids (see alignment in Supplemental Fig. 2) would occupy similar positions in the Tetrahymena and human structures. We then concentrated our mutagenesis on surface residues (Fig. 2A), where substitutions would be less likely to disrupt the structure of the TEN domain but might interfere with putative intermolecular interactions with TPP1.
Figure 2.
Mutations in the TEN domain of hTERT uncouple POT1–TPP1 activation from basal activity. (A) Domain structure of TERT and crystal structure of the TEN domain of Tetrahymena TERT, shown as a surface representation. Based on the assumption that conserved amino acids occupy similar positions in the human and Tetrahymena proteins, the amino acids that define the anchor site are shown in red, and other amino acids mutated in this study are also colored. The numbering system is given as Tetrahymena TERT (human TERT). (B) Telomerase activity assays for hTERT mutants complexed with hTR. The conditions are the same as in Figure 1. (C) SDS-PAGE showing the integrity and equal concentration of the 35S-labeled hTERT proteins used in B, as well as the medaka (medWT) and mouse (mWT) TERTs.
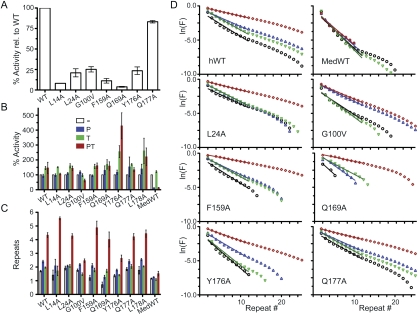
Mutations in the TEN domain decreased telomerase activity to different extents (Figs. 2B, 3A). (Throughout this study, we use “activity” to indicate the total incorporation of nucleotides per unit time for a constant amount of TERT, whereas “processivity” indicates the average number of repeats added following a single binding event.) Furthermore, like the wild-type enzyme, most of the mutants were stimulated slightly when POT1 was bound to the 5′ end of the DNA primer, leaving a 7-nt 3′ tail. There was also slight stimulation by TPP1 alone, a phenomenon that has not been investigated further. In the presence of both POT1 and TPP1, most mutant telomerases extended the DNA with the enhanced processivity characteristic of wild-type telomerase. Especially dramatic was Q169A, a mutant of a highly conserved amino acid in the DNA-binding groove of TEN. Q169A telomerase had only 4.3% ± 0.6% (mean ± SEM, n = 5) of wild-type activity on a naked DNA primer, but, when quantified, it was clear that both its activity and processivity were enhanced by POT1–TPP1 (Figs. 2B, 3).
Figure 3.
Quantitation of activity and processivity of hTERT mutants confirms the exceptional nature of G100V. (A) Activity of mutant telomerases relative to wild type. Error bars represent standard error, typically five independent measurements. (B) Change in activity when primer is bound to hPOT1 (P), hTPP1 (T), or both (PT), with each activity measurement normalized to the activity of the same telomerase in the absence of hPOT1 and hTPP1. The wild-type (WT) medaka telomerase shown as the last set. (C) Processivity of mutant telomerases, with standard error as above. (D) Analysis of processivity from the gel of Figure 2B, one of the data sets used for C. Medaka data from the gel of Figure 4. (F) Fraction of telomerase products extended beyond the indicated repeat number. See the Materials and Methods for details.
Unique among all the mutants tested was G100V, because it was unaffected by TPP1. Telomerase activity was decreased somewhat by the G100V mutation, measured as 25% ± 3% of wild-type (Fig. 3A). G100V telomerase showed some enhancement of activity and processivity by POT1 alone, but not by TPP1 alone; importantly, there was no increase of activity or processivity by POT1–TPP1 relative to POT1 alone (Figs. 2B, 3B–D). When G100V TERT and hTR were overexpressed in human 293T cells (Cristofari and Lingner 2006), the telomerase assembled in cells acted just like that assembled in RRLs: It had reduced activity relative to wild-type telomerase, and there was no increase of activity or processivity by POT1–TPP1 relative to POT1 alone (Supplemental Fig. 3).
TEN domain is not sufficient
Given the deleterious effect of the G100V mutation on telomerase processivity, it seemed possible that the TEN domain of TERT might be not only necessary but also sufficient for activation by POT1–TPP1. We therefore constructed a chimeric TERT that had the human TEN domain and the remainder of the protein sequence from medaka. As shown in Figure 4 (lane 5), this chimeric TERT was active with medaka TR, as expected, because its RNA-binding domain was derived from medaka TERT. It was not active with the human TR, suggesting that some of the TERT–TR interactions involve sequences that differ between these two homologous systems.
Figure 4.
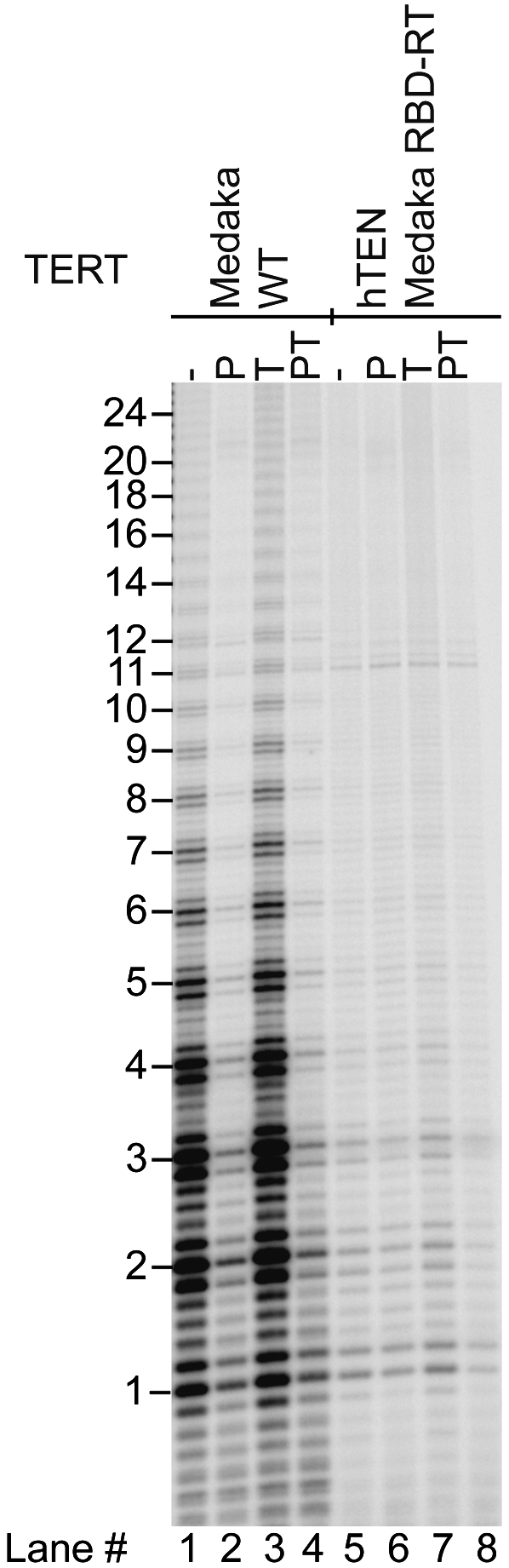
Chimeric human–medaka TERT is active but is not stimulated by hPOT1–TPP1. The chimeric TERT has the TEN domain of hTERT (amino acids 1–187) fused to the remainder of medaka TERT (starting with Phe185), reconstituted with medaka TR. Lanes 1–4 show activity of wild-type (WT) medaka TERT–medaka TR for comparison. Primer a5 is at 50 nM; each protein is at 500 nM as in previous figures.
Unlike medaka telomerase, the chimeric human–medaka telomerase was not inhibited by POT1 bound to the primer (Fig. 4, cf. lanes 6 and 2). Thus, the proposed steric hindrance between medaka telomerase and human POT1 appears to be localized to the TEN domain. Importantly, when the chimeric human–medaka telomerase was tested with a POT1–TPP1-bound primer, it gave the normal ladder of extension products, but did not show the enhanced processivity seen with human telomerase (Fig. 4, lane 8). Thus, while the TEN domain is necessary for POT1–TPP1 activation, it is not sufficient. We hypothesize that enhanced processivity by POT1–TPP1 requires the TEN domain to interact productively with some other region of TERT or TR, and that these interaction partners differ between human and medaka.
Human and mouse TPP1 activate their respective telomerases
Having identified a site within human telomerase required for its functional interaction with human POT1–TPP1, we wished to explore the specificity with respect to the telomeric proteins. Is the interaction through POT1 or TPP1, and how species-specific is it? However, the medaka telomeric proteins have not been characterized. We therefore explored the mouse system, hoping that it would have diverged enough from the human system to give some information about specificity of interactions. Indeed, the mouse has two POT1 proteins, which provide nonoverlapping functions in telomere maintenance (Hockemeyer et al. 2006). Furthermore, coimmunoprecipitation experiments indicate that mouse POT1A can form a complex with human TPP1, and human POT1 with mouse TPP1, although not as efficiently as the homologous combinations (Hockemeyer et al. 2007).
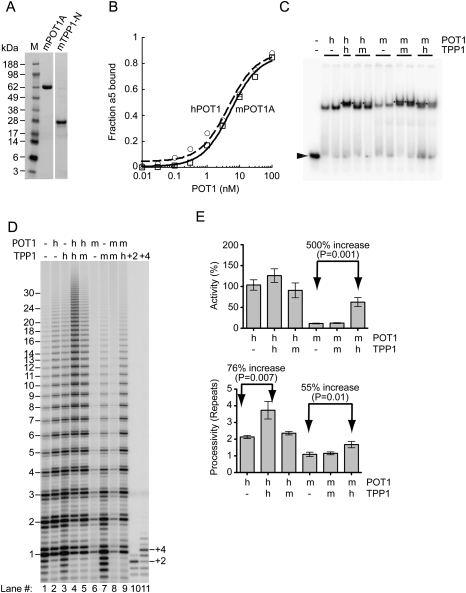
Mouse POT1A and mouse TPP1 were overexpressed in Escherichia coli and purified essentially to homogeneity (Fig. 5A). The mouse POT1A bound to the telomeric primer a5 used for telomerase assays with a similar equilibrium dissociation constant as human POT1: Kd = 5 nM (Fig. 5B). Human TPP1 formed the expected ternary complex with hPOT1 on a telomeric oligonucleotide, while the hPOT1–mTPP1 complex was formed inefficiently (Fig. 5C). When mTPP1 was substituted for hTPP1 in telomerase assays containing hPOT1 bound to primer a5, it did not give stimulation of processivity (Fig. 5D, lane 5 does not show the high processivity of lane 4, and is instead the same as hPOT1 alone in lane 2). The lack of function may simply be due to inefficient complex formation by the heterologous hPOT1–mTPP1 combination.
Figure 5.
Human but not mouse TPP1 can activate human telomerase even in a heterologous POT1–TPP1 complex. (A) Typical purity of the mouse POT1A and TPP1 determined by SDS-PAGE. (B) Binding of mouse POT1A (squares, solid line) and human POT1 (circles, dashed line) to a trace amount of radiolabeled a5 primer determined by filter binding. (C) Formation of POT1–DNA binary complexes and POT1–TPP1–DNA ternary complexes determined by an electrophoretic mobility shift assay. Radiolabeled primer T8GGTTAGGGTTAG (position indicated by arrow), where the eight T nucleotides are necessary to reveal the subtle gel shift obtained when TPP1 (an acidic protein) joins the complex. Human (h) and mouse (m) proteins, each at 50 nM. All combinations of POT1 and TPP1 gave a stable ternary complex (indicated by an asterisk to the left of the band), with complex formation being complete in the case of human/human (h/h), half-complete in the cases of mouse/mouse (m/m) and mouse/human (m/h), and only a low extent in the case of human/mouse (h/m). (D) Telomerase activity assays with 50 nM primer a5 in the absence (−) or presence of telomere proteins (500 nM each) as indicated. (E) Quantitation of total human telomerase activity (relative to activity of primer a5 in the absence of telomere proteins) and of processivity (in units of number of telomeric repeats) for the data shown in D and independent experiments not shown. Error bars represent standard deviation; n = 3.
Human and mouse POT1 bound to similar positions on the telomeric primer a5, in all cases binding mostly to the 5′ end of the DNA, leaving a 7-nt 3′ tail (Supplemental Fig. 4). This tail length is sufficient for human telomerase to extend a5 DNA bound to hPOT1 (Lei et al. 2005), yet binding of mouse POT1A to telomeric primer a5 substantially inhibited telomerase action (Fig. 5D, cf. lane 6 and control lane 1). We do not know if the inhibitory effect of mouse POT1A may involve a non-DNA-binding portion of POT1A or greater steric hindrance of mouse POT1A. Mouse TPP1 alone had no effect on telomerase action (Fig. 5D, lane 7 is identical to lane 1), consistent with the fact that it does not bind DNA by itself. The addition of mTPP1 did not rescue the inhibition of human telomerase by mPOT1A (Fig. 5D, lane 8 is identical to lane 6), in stark contrast to many studies in which hTPP1 promotes the synthesis of long repeats on hPOT1-inhibited primers (Wang et al. 2007; Latrick and Cech 2010).
The combination of mPOT1A and hTPP1 gave a striking result: Telomerase activity was increased by fivefold relative to mPOT1A alone (Fig. 5D [cf. lanes 9 and 6], E [top]). Furthermore, processivity was increased significantly upon addition of hTPP1 (Fig. 5E, bottom), the 55% increase being almost as large as the 76% increase seen upon addition of hTPP1 to the hPOT1 reaction. Thus, the mPOT1A–mTPP1 combination is functionally incompatible with human telomerase, but the mPOT1A–hTPP1 combination is functional, despite the less efficient complex formation upon substitution of the heterologous mouse protein (Fig. 5C). The simplest explanation is that hTPP1 provides a sequence-specific interaction with human telomerase. We cannot discount more complex models, such as hTPP1 inducing a conformational change in POT1 (both hPOT1 and mPOT1A) and the interaction with telomerase then being mediated through the POT1 subunit.
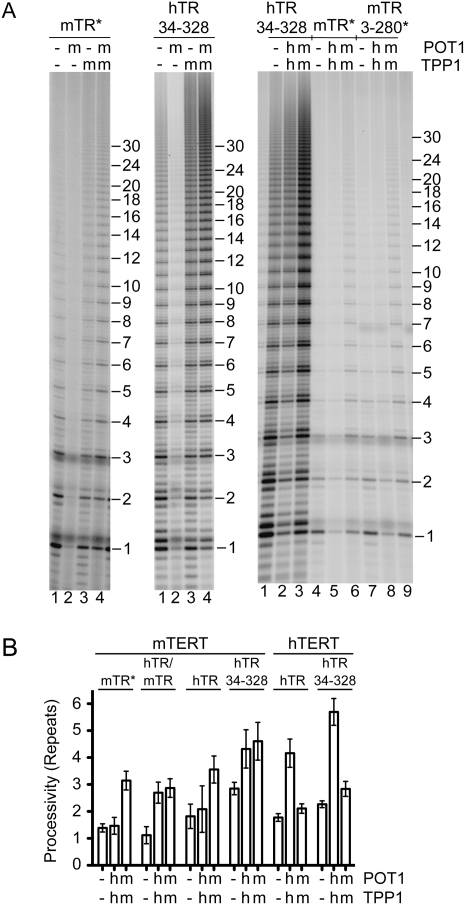
On the basis of our model that TPP1 coevolved with TERT to stimulate its processivity, we hypothesized that the mouse POT1–TPP1 complex might stimulate processivity of mouse telomerase. The mouse telomerase core enzyme reconstituted from TERT and TR gives only a single round of extension in vitro because the RNA has too short a primer alignment sequence (Supplemental Fig. 5A; Chen and Greider 2003). Binding mouse POT1–TPP1 to the primer does not rescue this defect (Supplemental Fig. 5B). However, mouse telomerase can be converted into a processive enzyme by substituting human TR for the mouse TR, or substituting just the template-containing pseudoknot domain of hTR, or by two single nucleotide mutations in the mouse RNA (mTR*) that increase base-pairing between the primer and the alignment region of mTR (Chen and Greider 2003). We assembled each of these hybrid and mutant mouse telomerases and confirmed that they were active and now gave processive extension (Fig. 6). In all cases, activity was inhibited 75% by binding of mPOT1A to the primer, analogous to the effect of mPOT1A on the activity of the human enzyme described above. However, further binding of mTPP1 not only reversed this inhibition, but also gave a substantial increase in processivity (2.2-fold increase in the case of the mTERT–mTR* telomerase) (for other versions of mouse telomerase, see Fig. 6B). In contrast, the human POT1–TPP1 did not affect the processivity of the mTERT–mTR* telomerase (although it did stimulate two of the telomerases that contained mTERT and portions of hTR) (Fig. 6B). Thus, the activation of telomerase processivity by cognate TPP1 telomere proteins may be a general feature of telomerase regulation in mammals.
Figure 6.
Telomerases that contain mouse TERT have their processivity activated by mouse POT1A–TPP1. (A) Representative activity data, with 50 nM primer a5 and 500 nM each indicated protein. All telomerases were comprised of mTERT plus the RNA indicated; mTR* is full-length mouse TR, with the asterisk indicating two point mutations in the primer alignment region that improve primer binding (see Supplemental Fig. 5B). For other RNAs, numbers represent the nucleotides present in the transcript. Activity of telomerases in the two left panels was consistently low, so these data are shown with longer exposure than in the right panel. (B) Quantitation of telomerase processivity (in units of number of telomeric repeats) for the data shown in A and independent experiments not shown. hTR/mTR indicates a two-piece RNA system consisting of nucleotides 34–192 of hTR and domains CR4 and CR5 of mTR. Error bars represent standard deviation; n ranged from 3 to 6.
Discussion
In addition to its role in chromosome end-capping, the POT1–TPP1 telomeric protein complex increases the processivity of primer elongation by telomerase (Wang et al. 2007). This activation does not require POT1–TPP1 binding continuously to the nascent ssDNA, but does require at least one binding site upstream of the part of the primer that is bound to telomerase's RNA template (Latrick and Cech 2010). This activity of POT1–TPP1 suggests a physical interaction with telomerase, but the interaction may be transient, as our attempts to show stable binding by pull-down experiments or by photocross-linking were unsuccessful. Here we took advantage of the recent discoveries of telomerases and telomeric proteins from other vertebrate species and used mix-and-match experiments to provide evidence that there is indeed a specific interaction between human TPP1 and human telomerase, as well as between mouse TPP1 and mouse telomerase.
We also tested site-specific mutants of TERT using telomerase enzymatic assays, an approach that has the power of identifying functionally important sites even if they interact only in a short-lived intermediate or in the transition state. This approach identified the G100V mutant as having the special property of retaining activity but being inert to TPP1 activation. G100 is in a subregion of the TEN domain of TERT called DAT (Fig. 2A); DAT mutants dissociate two activities of telomerase: catalytic activity, and the ability to function in vivo (Armbruster et al. 2001).
There could be direct contact between TPP1 and the G100 region of TERT that either prevents the primer from dissociating during a vulnerable moment in the reaction cycle or causes an allosteric change in the enzyme conformation. However, the sort of genetic evidence we present here cannot distinguish a direct interaction from an indirect interaction. Thus, the interaction partner of TPP1 could be elsewhere, in TERT or TR or both, and that interaction could induce a conformational change that involves G100. Furthermore, even if the interaction is directly with G100, it cannot be sufficient to activate telomerase; medaka TERT has a G99 that aligns with human G100 (Supplemental Fig. S2), yet it is unable to respond to POT1–TPP1. In substantiation of this point, we constructed a chimeric TERT containing the human TEN domain fused to medaka RNA-binding and RT domains and found it to be active, but not further activated by POT1–TPP1. Furthermore, the activation of telomerase processivity appears to involve the RNA subunit as well: Note that mouse telomerase (mTERT–mTR*) was activated only by mouse POT1–TPP1, but two of the mTERT telomerases that contained portions of hTR were activated by both mouse and human POT1–TPP1 (Fig. 6B).
The biological activity of G100V telomerase has been tested in primary human HA5 cells by David Sealey, Michael Taboski, and Lea Harrington (pers comm.). The transfected mutant and wild-type TERT genes were expressed at approximately the same level as judged by mRNA analysis, but it was not possible to measure the TERT protein levels. No G100V telomerase activity could be detected by the TRAP assay in cell extracts or after immunopurification, indicating that the mutant had decreased activity; however, the sensitivity of the experiment was such that 25% residual activity could have gone undetected. Transfection of G100V TERT failed to immortalize HA5 cells or to maintain telomere length, in contrast to wild-type TERT. These data clearly indicated that the G100V mutation was deleterious to telomerase function in vivo. The data could not distinguish if the effect was simply at the level of decreased protein stability, or if substantial amounts of telomerase were assembled but were functionally defective.
One alternative model that we can now discount is that POT1–TPP1 enhances telomerase processivity entirely through its interaction with the DNA. A priori, such an explanation would seem quite reasonable. For example, human telomeric DNA repeats are prone to form G-quadruplex structures, and these can be deleterious to telomerase extension (Zahler et al. 1991; Zaug et al. 2005). Binding of POT1 is known to disrupt G quadruplexes in favor of formation of the protein–DNA complex (Zaug et al. 2005), and POT1–TPP1 would be expected to have the same ability. Or there could be an interaction between the upstream G-rich sequence and the telomerase anchor site that might need to be disrupted to give maximum processivity, and coating of the DNA with POT1–TPP1 could prevent such an interaction. However, any such DNA substrate-level effects should have enhanced the activity of both human and medaka telomerases. Our finding that medaka telomerase is not activated argues against this model.
Recruitment of telomerase by telomere-binding proteins has been reported in budding yeast, where the telomerase-associated Est1 protein interacts with the Cdc13 telomeric DNA-binding protein (Qi and Zakian 2000). Est1–Cdc13 fusion proteins direct yeast telomerase to telomeres, consistent with a natural function of these proteins in mediating telomerase access (Evans and Lundblad 1999). A subregion of the human TEN domain called the DAT domain, when fused to the telomere DNA-binding protein TRF2, redirects DAT mutants of TERT to telomeres, suggesting a telomere recruitment function for the DAT domain (Armbruster et al. 2003). DAT mutants can also be rescued by fusing the mutant hTERT to hPOT1 (Armbruster et al. 2004). Our new data provide strong support for the Counter laboratory model (Armbruster et al. 2004) that the DAT domain of TERT mediates telomere–telomerase association, and they suggest a more detailed interpretation as follows: Wild-type telomerase may associate with telomeres through interaction of the DAT domain with TPP1, which in turn binds to POT1, and the rescue of DAT mutants by fusing the mutant hTERT to hPOT1 is successful because the fusion supplants the need for having the TPP1 “bridge” in this interaction. Thus, the interaction between the G100 region of TERT and TPP1 described here may be involved not only in telomerase processivity, but also in telomerase recruitment to telomeres.
Materials and methods
Mutagenesis
Mutations in the human TEN domain of TERT were made using the QuickChange II XL site-directed mutagenesis kit (Stratagene) with primers designed as described in Zheng et al. (2004).
In vitro reconstitution of telomerase
C-terminal HA-tagged human TERT was expressed from phTERT-HA2, and hTR was expressed from phTR, using the TnT quick-coupled transcription/translation system (Promega). Each 500-μL reaction contained 400 μL of TnT quick mix, 8 μL of 35S-methionine (Perkin-Elmer), 10 μL of PCR enhancer (Promega), 10 μL of 1 mM methionine, 10 μg of supercoiled phTERT-HA2, and 10 μg of hTR, which was transcribed from Fok I-cut pHTR plasmid DNA, and water to a total of 500 μL. After incubation for 2 h at 30°C, the reconstituted telomerase complex was affinity-purified on anti-HA F7 agarose beads (Santa Cruz Biotechnologies). Anti-HA F7 agarose beads, washed with 1× human telomerase assay buffer (defined below) without KCl, were added for immunopurification for 2 h at 4°C. The beads were washed with 1× human telomerase buffer with 30% glycerol four times and then resuspended in 1× human telomerase assay buffer with 30% glycerol. The quantity of 35S-hTERT was determined. Unused beads were quick-frozen in liquid nitrogen and stored at −80°C. T7-tagged medaka TERT was expressed from pCite-MedTERT, medaka TR was expressed from Fok I-cut pMedTR, and C-terminal HA-tagged mouse TERT was expressed from pCite-mTERT (J. Chen, Arizona State University). mTR and mTR* were expressed from PCR-generated templates using the TnT quick-coupled transcription/translation system, with other experimental details as described above for human TERT and TR.
Telomerase activity assays
Activity of the immunopurified human telomerase complex reconstituted in vitro was determined by a direct assay modified from a published protocol (Wang et al. 2007). The reaction mixture (20 μL) contained 1× human telomerase assay buffer (50 mM Tris-HCl at pH 8.0, 50 mM KCl, 1 mM MgCl2, 5 mM 2-mercaptoethanol, 1 mM spermidine), 0.05 μM telomeric DNA primer, 0.5 mM dATP, 0.5 mM dTTP, 2.92 μM dGTP, and 0.33 μM 32P-dGTP (3000 Ci/mmol, 1 Ci = 37GBq), with 6 μL of immunpurified telomerase complex. For medaka TERT telomerase assays, the concentration of cold dGTP was increased to 50 μM. Reactions were incubated for 1 h at 30°C, which was determined to be a time when activity was still increasing with time, but when processivity had plateaued. Reactions were stopped with the addition of 100 μL of 3.6 M NH4OAc containing 20 μg of glycogen. Ethanol (500 μL) was added for precipitation. After incubating for 1 h at −80°C, samples were centrifuged for 15 min at 4°C. Pellets were washed with 70% ethanol and resuspended in 10 μL of H2O followed by 10 μL of 2× loading buffer (94% formamide, 0.1× TBE, 0.1% bromophenol blue, 0.1% xylene cyanol). The heat-denatured samples were loaded onto a 10% polyacrylamide/7 M urea/1× TBE denaturing gel for electrophoresis. After electrophoresis, the gel was dried and quantified by using a PhosphorImager (GE Healthcare).
Protein expression and purification
Proteins hPOT1 and hTPP1-N (amino acids 89–334) were expressed and purified as reported previously (Lei et al. 2004; Wang et al. 2007). The ORFs encoding mPOT1A and mTPP1-N (amino acids 1–246) (designed based on sequence alignment with hTPP1-N) were PCR-amplified from cDNA and cloned into a pET-His-Smt3 expression vector (Mossessova and Lima 2000). The proteins were overexpressed in Bl21 (DE3) cells. His-Smt3-mPOT1A and His-Smt3-mTPP1-N were purified from the soluble cell lysates using Ni-affinity chromatography (Qiagen), and the His-Smt3 tag was removed. Untagged mPOT1A and mTPP1-N were further purified by gel exclusion (Superdex200 [GE] and Superdex75 [GE], respectively) and anion exchange (MonoQ, GE) chromatography, and the fractions containing pure proteins were flash-frozen in liquid nitrogen and stored at −80°C. Concentrations were determined by the Bio-Rad Protein Assay, based on the Bradford method; hPOT1 was typically ∼40%–50% active and mPOT1A was typically 25% active, as determined by quantitative DNA-binding titrations.
Processivity quantitation
The number of counts for each repeat in a lane of a telomerase activity gel was determined using PhosphorImager scanning and ImageQuant software. The counts for a band were corrected by dividing by the total number of radiolabeled guanosines in that extension product. Then, for each band n, the fraction F of the chains that continued past n was calculated (Latrick and Cech 2010). The graph of ln F versus repeat number (see examples in Fig. 3D) was fit with a linear regression, and processivity = −0.693/slope. The first repeat was not included in the fit because telomerase synthesizes one or two repeats before synthesis is linear.
Acknowledgments
We are grateful to David C.F. Sealey, Michael A.S. Taboski, and Lea A. Harrington (University of Toronto and University of Edinburgh) for sharing unpublished information, and to Julian Chen (Arizona State University) and Chris Counter (Duke University) for helpful discussions. We also thank Julian Chen for plasmids encoding the medaka and mouse TERT and TR, and Sandy Chang (University of Texas, M.D. Anderson Cancer Center) for a plasmid encoding mouse POT1A. J.N. is a Howard Hughes Medical Institute fellow of the Helen Hay Whitney Foundation.
Footnotes
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.1881810.
Supplemental material is available at http://www.genesdev.org.
References
- Armbruster BN, Banik SS, Guo C, Smith AC, Counter CM. N-terminal domains of the human telomerase catalytic subunit required for enzyme activity in vivo. Mol Cell Biol. 2001;21:7775–7786. doi: 10.1128/MCB.21.22.7775-7786.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armbruster BN, Etheridge KT, Broccoli D, Counter CM. Putative telomere-recruiting domain in the catalytic subunit of human telomerase. Mol Cell Biol. 2003;23:3237–3246. doi: 10.1128/MCB.23.9.3237-3246.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armbruster BN, Linardic CM, Veldman T, Bansal NP, Downie DL, Counter CM. Rescue of an hTERT mutant defective in telomere elongation by fusion with hPot1. Mol Cell Biol. 2004;24:3552–3561. doi: 10.1128/MCB.24.8.3552-3561.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Autexier C, Lue NF. The structure and function of telomerase reverse transcriptase. Annu Rev Biochem. 2006;75:493–517. doi: 10.1146/annurev.biochem.75.103004.142412. [DOI] [PubMed] [Google Scholar]
- Baumann P, Cech TR. Pot1, the putative telomere end-binding protein in fission yeast and humans. Science. 2001;292:1171–1175. doi: 10.1126/science.1060036. [DOI] [PubMed] [Google Scholar]
- Bryan TM, Goodrich KJ, Cech TR. A mutant of Tetrahymena telomerase reverse transcriptase with increased processivity. J Biol Chem. 2000;275:24199–24207. doi: 10.1074/jbc.M003246200. [DOI] [PubMed] [Google Scholar]
- Chen JL, Greider CW. Determinants in mammalian telomerase RNA that mediate enzyme processivity and cross-species incompatibility. EMBO J. 2003;22:304–314. doi: 10.1093/emboj/cdg024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Churikov D, Wei C, Price CM. Vertebrate POT1 restricts G-overhang length and prevents activation of a telomeric DNA damage checkpoint but is dispensable for overhang protection. Mol Cell Biol. 2006;26:6971–6982. doi: 10.1128/MCB.01011-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colgin LM, Baran K, Baumann P, Cech TR, Reddel RR. Human POT1 facilitates telomere elongation by telomerase. Curr Biol. 2003;13:942–946. doi: 10.1016/s0960-9822(03)00339-7. [DOI] [PubMed] [Google Scholar]
- Cristofari G, Lingner J. Telomere length homeostasis requires that telomerase levels are limiting. EMBO J. 2006;25:565–574. doi: 10.1038/sj.emboj.7600952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Lange T. Shelterin: The protein complex that shapes and safeguards human telomeres. Genes & Dev. 2005;19:2100–2110. doi: 10.1101/gad.1346005. [DOI] [PubMed] [Google Scholar]
- Denchi EL, de Lange T. Protection of telomeres through independent control of ATM and ATR by TRF2 and POT1. Nature. 2007;448:1068–1071. doi: 10.1038/nature06065. [DOI] [PubMed] [Google Scholar]
- Evans SK, Lundblad V. Est1 and Cdc13 as comediators of telomerase access. Science. 1999;286:117–120. doi: 10.1126/science.286.5437.117. [DOI] [PubMed] [Google Scholar]
- Gray JT, Celander DW, Price CM, Cech TR. Cloning and expression of genes for the Oxytricha telomere-binding protein: Specific subunit interactions in the telomeric complex. Cell. 1991;67:807–814. doi: 10.1016/0092-8674(91)90075-a. [DOI] [PubMed] [Google Scholar]
- Hockemeyer D, Sfeir AJ, Shay JW, Wright WE, de Lange T. POT1 protects telomeres from a transient DNA damage response and determines how human chromosomes end. EMBO J. 2005;24:2667–2678. doi: 10.1038/sj.emboj.7600733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hockemeyer D, Daniels JP, Takai H, de Lange T. Recent expansion of the telomeric complex in rodents: Two distinct POT1 proteins protect mouse telomeres. Cell. 2006;126:63–77. doi: 10.1016/j.cell.2006.04.044. [DOI] [PubMed] [Google Scholar]
- Hockemeyer D, Palm W, Else T, Daniels JP, Takai KK, Ye JZ, Keegan CE, de Lange T, Hammer GD. Telomere protection by mammalian Pot1 requires interaction with Tpp1. Nat Struct Mol Biol. 2007;14:754–761. doi: 10.1038/nsmb1270. [DOI] [PubMed] [Google Scholar]
- Horvath MP, Schweiker VL, Bevilacqua JM, Ruggles JA, Schultz SC. Crystal structure of the Oxytricha nova telomere end binding protein complexed with single strand DNA. Cell. 1998;95:963–974. doi: 10.1016/s0092-8674(00)81720-1. [DOI] [PubMed] [Google Scholar]
- Houghtaling BR, Cuttonaro L, Chang W, Smith S. A dynamic molecular link between the telomere length regulator TRF1 and the chromosome end protector TRF2. Curr Biol. 2004;14:1621–1631. doi: 10.1016/j.cub.2004.08.052. [DOI] [PubMed] [Google Scholar]
- Jacobs SA, Podell ER, Cech TR. Crystal structure of the essential N-terminal domain of telomerase reverse transcriptase. Nat Struct Mol Biol. 2006;13:218–225. doi: 10.1038/nsmb1054. [DOI] [PubMed] [Google Scholar]
- Kelleher C, Kurth I, Lingner J. Human protection of telomeres 1 (POT1) is a negative regulator of telomerase activity in vitro. Mol Cell Biol. 2005;25:808–818. doi: 10.1128/MCB.25.2.808-818.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SH, Kaminker P, Campisi J. TIN2, a new regulator of telomere length in human cells. Nat Genet. 1999;23:405–412. doi: 10.1038/70508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konig P, Giraldo R, Chapman L, Rhodes D. The crystal structure of the DNA-binding domain of yeast RAP1 in complex with telomeric DNA. Cell. 1996;85:125–136. doi: 10.1016/s0092-8674(00)81088-0. [DOI] [PubMed] [Google Scholar]
- Latrick C, Cech T. POT1–TPP1 enhances telomerase processivity by slowing primer dissociation and aiding translocation. EMBO J. 2010 doi: 10.1038/emboj.2009.409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei M, Podell ER, Cech TR. Structure of human POT1 bound to telomeric single-stranded DNA provides a model for chromosome end-protection. Nat Struct Mol Biol. 2004;11:1223–1229. doi: 10.1038/nsmb867. [DOI] [PubMed] [Google Scholar]
- Lei M, Zaug AJ, Podell ER, Cech TR. Switching human telomerase on and off with hPOT1 protein in vitro. J Biol Chem. 2005;280:20449–20456. doi: 10.1074/jbc.M502212200. [DOI] [PubMed] [Google Scholar]
- Liu D, Safari A, O'Connor MS, Chan DW, Laegeler A, Qin J, Songyang Z. PTOP interacts with POT1 and regulates its localization to telomeres. Nat Cell Biol. 2004;6:673–680. doi: 10.1038/ncb1142. [DOI] [PubMed] [Google Scholar]
- Loayza D, De Lange T. POT1 as a terminal transducer of TRF1 telomere length control. Nature. 2003;423:1013–1018. doi: 10.1038/nature01688. [DOI] [PubMed] [Google Scholar]
- Moretti P, Freeman K, Coodly L, Shore D. Evidence that a complex of SIR proteins interacts with the silencer and telomere-binding protein RAP1. Genes & Dev. 1994;8:2257–2269. doi: 10.1101/gad.8.19.2257. [DOI] [PubMed] [Google Scholar]
- Moriarty TJ, Marie-Egyptienne DT, Autexier C. Functional organization of repeat addition processivity and DNA synthesis determinants in the human telomerase multimer. Mol Cell Biol. 2004;24:3720–3733. doi: 10.1128/MCB.24.9.3720-3733.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mossessova E, Lima CD. Ulp1–SUMO crystal structure and genetic analysis reveal conserved interactions and a regulatory element essential for cell growth in yeast. Mol Cell. 2000;5:865–876. doi: 10.1016/s1097-2765(00)80326-3. [DOI] [PubMed] [Google Scholar]
- O'Connor MS, Safari A, Xin H, Liu D, Songyang Z. A critical role for TPP1 and TIN2 interaction in high-order telomeric complex assembly. Proc Natl Acad Sci. 2006;103:11874–11879. doi: 10.1073/pnas.0605303103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi H, Zakian VA. The Saccharomyces telomere-binding protein Cdc13p interacts with both the catalytic subunit of DNA polymerase α and the telomerase-associated est1 protein. Genes & Dev. 2000;14:1777–1788. [PMC free article] [PubMed] [Google Scholar]
- Sarthy J, Bae NS, Scrafford J, Baumann P. Human RAP1 inhibits non-homologous end joining at telomeres. EMBO J. 2009;28:3390–3399. doi: 10.1038/emboj.2009.275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takai KK, Hooper S, Blackwood S, Gandhi R, de Lange T. In vivo stoichiometry of shelterin components. J Biol Chem. 2010;285:1457–1467. doi: 10.1074/jbc.M109.038026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teixeira MT, Arneric M, Sperisen P, Lingner J. Telomere length homeostasis is achieved via a switch between telomerase- extendible and -nonextendible states. Cell. 2004;117:323–335. doi: 10.1016/s0092-8674(04)00334-4. [DOI] [PubMed] [Google Scholar]
- Veldman T, Etheridge KT, Counter CM. Loss of hPot1 function leads to telomere instability and a cut-like phenotype. Curr Biol. 2004;14:2264–2270. doi: 10.1016/j.cub.2004.12.031. [DOI] [PubMed] [Google Scholar]
- Wang X, Baumann P. Chromosome fusions following telomere loss are mediated by single-strand annealing. Mol Cell. 2008;31:463–473. doi: 10.1016/j.molcel.2008.05.028. [DOI] [PubMed] [Google Scholar]
- Wang F, Podell ER, Zaug AJ, Yang Y, Baciu P, Cech TR, Lei M. The POT1–TPP1 telomere complex is a telomerase processivity factor. Nature. 2007;445:506–510. doi: 10.1038/nature05454. [DOI] [PubMed] [Google Scholar]
- Wu L, Multani AS, He H, Cosme-Blanco W, Deng Y, Deng JM, Bachilo O, Pathak S, Tahara H, Bailey SM, et al. Pot1 deficiency initiates DNA damage checkpoint activation and aberrant homologous recombination at telomeres. Cell. 2006;126:49–62. doi: 10.1016/j.cell.2006.05.037. [DOI] [PubMed] [Google Scholar]
- Xie M, Mosig A, Qi X, Li Y, Stadler PF, Chen JJ. Structure and function of the smallest vertebrate telomerase RNA from teleost fish. J Biol Chem. 2008;283:2049–2059. doi: 10.1074/jbc.M708032200. [DOI] [PubMed] [Google Scholar]
- Xin H, Liu D, Wan M, Safari A, Kim H, Sun W, O'Connor MS, Songyang Z. TPP1 is a homologue of ciliate TEBP-β and interacts with POT1 to recruit telomerase. Nature. 2007;445:559–562. doi: 10.1038/nature05469. [DOI] [PubMed] [Google Scholar]
- Yap WH, Yeoh E, Brenner S, Venkatesh B. Cloning and expression of the reverse transcriptase component of pufferfish (Fugu rubripes) telomerase. Gene. 2005;353:207–217. doi: 10.1016/j.gene.2005.04.038. [DOI] [PubMed] [Google Scholar]
- Ye JZ, Hockemeyer D, Krutchinsky AN, Loayza D, Hooper SM, Chait BT, de Lange T. POT1-interacting protein PIP1: A telomere length regulator that recruits POT1 to the TIN2/TRF1 complex. Genes & Dev. 2004;18:1649–1654. doi: 10.1101/gad.1215404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zahler AM, Williamson JR, Cech TR, Prescott DM. Inhibition of telomerase by G-quartet DNA structures. Nature. 1991;350:718–720. doi: 10.1038/350718a0. [DOI] [PubMed] [Google Scholar]
- Zaug AJ, Podell ER, Cech TR. Human POT1 disrupts telomeric G-quadruplexes allowing telomerase extension in vitro. Proc Natl Acad Sci. 2005;102:10864–10869. doi: 10.1073/pnas.0504744102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaug AJ, Podell ER, Cech TR. Mutation in TERT separates processivity from anchor-site function. Nat Struct Mol Biol. 2008;15:870–872. doi: 10.1038/nsmb.1462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, Sfeir AJ, Zou Y, Buseman CM, Chow TT, Shay JW, Wright WE. Telomere extension occurs at most chromosome ends and is uncoupled from fill-in in human cancer cells. Cell. 2009;138:463–475. doi: 10.1016/j.cell.2009.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng L, Baumann U, Reymond JL. An efficient one-step site-directed and site-saturation mutagenesis protocol. Nucleic Acids Res. 2004;32:e115. doi: 10.1093/nar/gnh110. [DOI] [PMC free article] [PubMed] [Google Scholar]