Abstract
The development of anther and pollen is important for male reproduction, and this process is coordinately regulated by many external and internal cues. In this study, we systematically examined the male reproductive phenotypes of a series of brassinosteroid biosynthetic and signaling mutants and found that, besides the expected cell-expansion defects, these mutants also showed reduced pollen number, viability, and release efficiency. These defects were related with abnormal tapetum and microspore development. Using both real-time quantitative RT-PCR and microarray experiments, we found that the expression of many key genes required for anther and pollen development was suppressed in these mutants. ChIP analysis demonstrated that BES1, an important transcription factor for brassinosteroid signaling, could directly bind to the promoter regions of genes encoding transcription factors essential for anther and pollen development, SPL/NZZ, TDF1, AMS, MS1, and MS2. Taken together, these data lead us to propose that brassinosteroids control male fertility at least in part via directly regulating key genes for anther and pollen development in Arabidopsis. Our work provides a unique mechanism to explain how a phytohormone regulates an essential genetic program for plant development.
Keywords: male fertility, regulatory network, signaling, chromatin immunoprecipitation
Most angiosperm plants have hermaphroditic flowers that often undergo cross-fertilization by pollinators. The amount of pollens produced and the rate of pollen dispersal largely determine male reproductive success (1). Therefore, the development of anther and pollen is critical for successful reproduction in plant's life cycle (2). Previous molecular genetic studies have established a major genetic program controlling anther and pollen development in Arabidopsis (3, 4). At an early anther developmental stage, SPL/NZZ is required for sporocyte formation and cell proliferation (5, 6). EMS1/EXS is essential for tapetum formation (7, 8). DYT1 is required for early tapetal development (9), and TDF1, which is highly expressed in tapetum, meiocytes, and microspores, is likely to regulate callose dissolution around microspores and exine formation of pollen wall (10). AMS, encoding a putative MYC transcription factor, is specifically expressed in tapetum and microspores and required for microspore mitosis (11). AtMYB103 is a member of the R2R3 MYB gene family, only expressed in tapetum and required for tapetal development and microsporogenesis (12). MS1 acts downstream of AtMYB103 and plays a critical role in regulating exine formation and pollen coat development (12–14). MS2 is involved in sporopollenin synthesis and normal exine patterning (15). Further analyses have revealed genetic interactions among these genes. DYT1 acts downstream of SPL/NZZ and EMS1/EXS, and is required for normal expression of AMS, MS1, and other tapetum-preferential genes (9). TDF1 acts downstream of DYT1 and upstream of AMS and AtMYB103 in the transcriptional regulatory networks of tapetal development (10).
Besides the above essential components in the anther and pollen developmental network, many phytohormones, including auxin, gibberellins, ethylene, cytokinins, and jasmonic acids, can affect male fertility (16–20), and many biosynthetic or signaling mutants of phytohormones often have reduced male fertility. For example, auxin biosynthetic double mutants yuc2 yuc6 (21) and auxin signaling quadruple mutant tir1 afb1 afb2 afb3 (22) have dramatically decreased male fertility. Mutations in other hormone pathways, such as gibberellins (23), ethylene (18), and jasmonic acid (19), also lead to reduced male fertility. It was recently reported that auxin synthesized in anthers is essential for coordinating pollen maturation and anther dehiscence, whereas auxin transport contributes to late stamen development and preanthesis filament elongation (17). DELLA proteins, RGA, RGL1, and RGL2 repress stamen growth and anther development in the ga1-3 mutant, which exhibits male sterility (23). Gibberellins also impact pollen viability and pollen tube elongation (20, 24). Furthermore, constitutive activation of ethylene signaling in the ctr1 mutant leads to defects in anther elongation (18); cytokinins are also involved in anther development (16).
Brassinosteroids (BRs) are also essential for male fertility in plants. First, pollen is a rich source of endogenous BRs (25). Second, during lily pollen development, the conjugated teasterone is found at the microspore stage, and its decrease is correlated with the increased level of free BRs (26). Third, experiments with Prunus avium indicated that BRs induce pollen tube elongation (27). More importantly, genetic studies in Arabidopsis indicated that the BR mutants, such as cpd (28), bin2 (29), and bri1-201 (30), have little or no male fertility. In the BR-deficient cpd mutant, the failure of pollen tube elongation is a likely cause of male sterility (28). In another BR-deficient mutant, dwf4, male sterility may be caused by a significantly reduced filament elongation, leading to a failure of pollen delivery to the stigma (31). We also found that the reduced lengths of male and female organs might be a partial explanation for male sterility in the BR-related mutants (Fig. S1 A and B). However, the underlying molecular mechanism of BRs in regulating anther/pollen development is completely unknown.
Many key components in the BR signaling pathway have been identified and provide essential tools to investigate the mechanisms of BR regulating male fertility. BRs are perceived by a cell-surface receptor, named BRI1 (32). BAK1 might function as a positive coreceptor of BRI1 (33), although BKI1 acts negatively in the BR signaling by inhibiting BRI1’s activity (33, 34). BIN2 negatively affects BR signaling by phosphorylating BES1/BZR1, which are positively controlled via dephosphorylation by the phosphatase BSU1 (33, 35). The dephosphorylated BES1/BZR1 then regulates gene expression by binding to their cis- elements, such as E-box and BRRE (36, 37).
In this study, we examined representative BR mutants for their male fertility phenotypes and observed the morphology of developing anthers via light microscopy and transmission electron microscopy (TEM), and pollen shape via SEM. We found that these mutants not only exhibited significantly reduced filament length, but also produced dramatically fewer pollen grains, and displayed abnormal tapetal development and pollen exine patterns. Using microarray analysis and real-time quantitative RT-PCR (qRT-PCR), we found that the expression of many key regulators in pollen and anther development, including AMS, MYB103, MS2, MS1, and some downstream genes of MS1, was dramatically reduced in the BR mutants. Furthermore, ChIP experiments demonstrated that BES1 could directly bind to the promoter regions of SPL/NZZ, TDF1, MS2, MYB103, MS1, and At3g23770. Taking these data together, we conclude that BRs can regulate male fertility by controlling the expression of a set of key genes involved in multiple steps of anther and pollen development.
Results
Pollen Viability Is Slightly Reduced in the BR-Related Mutants.
Although some male-sterile mutants can produce pollen, they show reduced pollen viability (13, 38, 39). To test pollen viability in the BR biosynthetic and signaling mutants, we stained pollen grains with Alexander's solution (40), and found that nearly 99% of pollen grains from wild-type and a weak BR-biosynthetic mutant, det2-1 were viable, but the viability of pollen grains from a severe BR-biosynthetic mutant, cpd, a severe BR-perception mutant, bri1-116, a severe BR-signaling mutant, bin2-1, and myriBKI1-YFP-OX (34), was decreased to 95, 90, 94, and 95%, respectively (Fig. S2A). However, unlike severe male-sterile mutants, such as ams (11) and dyt1 (9), which produce no viable pollens, the pollen grains from the BR mutants still were mostly viable, indicating that pollen viability might not be a major cause for the dramatically decreased fertility in these BR mutants.
Number of Pollen Grains in BR-Related Mutants Is Greatly Reduced.
In the above experiments, we also noticed that anthers of the BR mutants usually contained fewer pollen grains than that of the wild type. Because it is difficult to count the exact number of pollen grains in each stamen, we estimated the relative pollen number through multiplying anther length by the pollen number per locule in each transverse section of anthers at stages 11 and 12. We found that bri1-116 and cpd only produced about 20% of pollens per anther of the wild type (Fig. 1J). This dramatically reduced number of pollen grains might account for much of the reduced male fertility in these mutants.
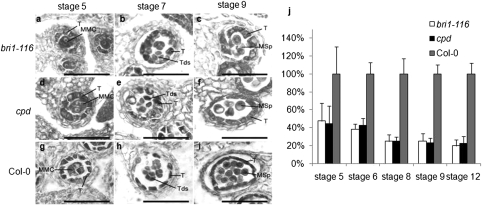
Fig. 1.
Comparison of developing anthers and microspores among bri1-116, cpd, and the wild type. MMC, microspore mother cell; Msp, microspore; T, tapetum; Tds, tetrads. Micrographs of 6-μm thick anther transverse sections at different anther stages in bri1-116, cpd, and the wild type (Col-0). (A–C) bri1-116; (D–F) cpd; (G–I) Col-0. (A, D, and G) stage 5; (B, E, and H) stage 7; (C, F, and I) stage 9. (Scale bars, 50 μm.) (J) Microspore number per locule at different stages of anther development in bri1-116, cpd, and the wild type. Relative number per locule was calculated through multiplying anther diameter by pollen number per locule counted with longitudinal sections of anther stages 5, 6, 8, and 9; at stage 12 the pollen number was calculated with transverse sections (Experimental Procedures). MMC was counted at stage 5, meiocytes were counted at stage 6, microspores were counted at stages 8 and 9, and pollen grains were counted at stage 12.
Defects in Pollen Tube Elongation Are Not a Major Cause of Male Sterility in BR-Related Mutants.
Failure of pollen tube elongation of cpd causing male sterility has been reported (28), so we examined whether pollen tube growth is inhibited in both bri1-116 and cpd. A callose-specific dye, Aniline blue, was used to stain growing pollen tubes in pistils. At 3 h after hand-pollination, the wild-type pollen tubes grew 52.3 μm, on average, and most of them reached three-quarters of the pistil length from the wild-type stigma (Fig. S2B). Surprisingly, there was no significant difference in pollen tube growth between cpd and the wild type during self-pollination, or between cpd and wild-type pollen tubes growing on either cpd or wild-type pistils (Fig. S2B), indicating that pollen tube elongation is unlikely the cause of male sterility in cpd. Although the pollen tube of bri1-116 was slightly shorter than that of the wild type when growing them in bri1-116 pistils (Fig. S2B), the pollen tubes could still reach the end of pistils after 12 h growth, suggesting that pollen tube growth is unlikely a reason for the reduced male fertility in the BR mutants.
BR-Related Mutants Are Defective in Pollen Release and Exine Pattern Formation.
When we conducted hand pollination, we experienced difficulties in pollen release from bri1-116 and cpd anthers. We then ascertained whether pollen grains still adhered to the anther walls after pollination using SEM. The dehiscent anthers of bri1-116, cpd, and the wild type all contained pollen grains before pollination (Fig. 2 A–C). After hand pollination, pollen grains were rarely seen on the inner wall of the wild-type anther, but a considerable number of pollen grains could be seen in bri1-116 and cpd anthers (Fig. 2 D–F), indicating that these mutants were deficient in pollen release after anther dehiscence.
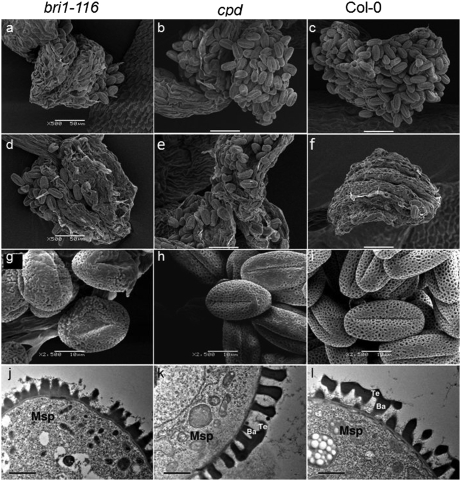
Fig. 2.
bri1-116 and cpd are defective in exine pattern formation. (A–C) Scanning electron micrographs of dehiscent anthers of bri1-116 (A), cpd (B) and Col-0 (C). (Scale bars, 50 μm.) (D–F) After pollination, more pollen grains are retained on the inner wall of the anthers of bri1-116 (D) and cpd (E) than Col-0 (F). (Scale bars, 50 μm.) (G–I) Scanning electron micrographs of mature pollen grains: wild type (I), bri1-116 (G), and cpd (H). (Scale bars, 10 μm.) (J–L) Transmission electron micrographs of transverse section of microspores at stage 8: bri1-116 (J), cpd (K), and wild type (L). Ba, bacula; Msp, microspore; Te, tectum. (Scale bars, 1 μm.)
Because the inner surface of the wild-type, bri1-116, and cpd anther walls appeared similar (Fig. 2 D–F), it is likely that the defects of pollen dispersal in the mutants resulted from a defect of pollen grains. Therefore, we examined the exine structure using TEM. The wild-type exine has a network-like structure with a large number of lacunae and three narrow apertures (Fig. 2I). In contrast, the lacunae of bri1-116 exine was incomplete, as indicated by the loss of network-like structure, and the pollen apertures were also bigger than those of the wild type (Fig. 2G). In addition, cpd pollen grains exhibited similar but less severe phenotypes than bri1-116 (Fig. 2H). We further examined exine development using TEM analysis. Compared with the wild type (Fig. 2I), although exine structure of the cpd mutant appeared normal (Fig. 2K), bri1-116 and cpd lacked an obvious bacula/tectum structure (Fig. 2 J and K). This abnormal structure of exine in the BR mutants might have caused more pollen grains adhering to the inner surface of anther wall after pollination.
Tapetal Development Is Abnormal in Both cpd and bri1-116.
To investigate the cellular and molecular mechanisms that lead to pollen-grain defects in these mutants, we further examined the morphology of bri1-116 and cpd anthers using paraffin sections. We observed a difference of developing anthers between the mutants and the wild type. At stage 5, the mutant tapetal cells were greatly larger and more vacuolated than the wild-type cells (Fig. 1 A, D, and G). At stage 7, mutant tetrads were apparently normal in shape and size, although tapetal cells of the mutants were extremely vacuolated and larger than that of the wild type (Fig. 1 B, E, and H). At stage 9, a similar tapetum phenotype persisted in the mutants, and surprisingly, microspores in the mutant anthers were significantly fewer, larger, and more vacuolated than those in wild type (Fig. 1 C, F, and I). This finding explains why the mutants produced fewer pollen grains than the wild type. At stage 10, tapetal cells in both the mutants and the wild type began to degenerate, showing no significant difference. From stage 11 to stage 14, compared with the wild type, much fewer pollen grains were observed in the mutants.
We then counted the number of microspore mother cells and developing pollen grains at stages 5, 6, 8, 9, and 12 (Fig. 1J). Compared with the wild type, bri1-116 and cpd only had, respectively, about 47.34 and 44.64% of microspore mother cells at stage 5, and 38.36 and 42.81% at stage 6; and only produced about 24.77 and 25.15% of pollen grains at stage 8, 25.15 and 22.83% at stage 9, and 19.85 and 22.71% at stage 12 (Fig. 1J).
BRs Regulate Expression of Key Genes in Tapetum and Microspore Development.
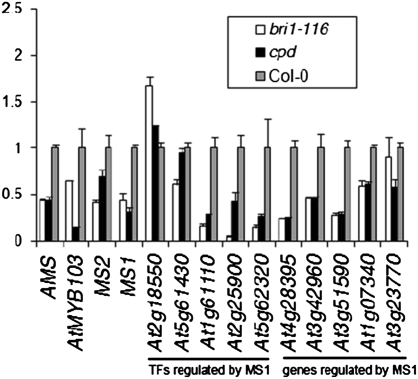
The defects of anther development and exine pattern in these BR mutants were similar to the phenotypes of the ProMS1:MS1-SRDX transgenic lines, which had partially inhibited MS1 function (14). Therefore, we hypothesized that BRs may regulate anther development by controlling the expression of some genes in this network. Among the known genes in this network, AMS, AtMYB103, and MS1 (11, 12, 41) are expressed in the tapetum of anthers at stage 9, which is the period of the smallest buds we could identify in bri1-116 and cpd. SPL and DYT1 are expressed primarily at stage 5 or earlier in anther cells (6, 9), and TDF1 is mainly expressed from stage 5 to early stage 9 (10). In addition, abnormal tapetal cells and microspores were observed in the mutants at stage 9. Therefore, we first checked the expression of SPL/NZZ, DYT1, TDF1, AMS, AtMYB103, MS1, and MS2 in the buds approximately containing stages 7 to 9 anther, and found that the expression levels of most genes in bri1-116 and cpd were significantly lower than that in the wild type (Fig. 3). Previous studies have identified a set of genes regulated by MS1 (14, 41), so we further investigated the expression levels of 10 of the highly induced genes by MS1, and found that most of these genes were down-regulated in bri1-116 and cpd (Fig. 3). We also created a BES1-GFP overexpression line (BES1-OX) driven by 35S promoter, and found that the expression of AMS, MYB103, MS1, and At4G28395 was significantly higher in BES1-OX than in the wild-type Col-0 (Fig. S3). Our results strongly suggest that the down-regulation of many genes in anther and pollen development might account for the defects of pollen development in the BR mutants.
Fig. 3.
The expression of MS1 and its target genes, and several essential genes in anther and pollen development, is regulated by BRs. Real-time qRT-PCR analysis of AMS, AtMYB103, MS2, MS1, and target genes of MS1 in bri1-116, cpd, and Col-0. The floral buds approximately containing anthers at stages 8 to 10 were collected and used for this analysis. The expression level of each gene in Col-0 was defined as “1.” Bars indicate the SE.
To more broadly investigate the differential gene expression among bri1-116, cpd, and the wild type during floral development, we used Agilent Microarray 4 × 44K slide format containing 43,803 Arabidopsis thaliana probes and RNA samples from the floral buds containing approximately stages 7 to 9 anther, to perform microarray experiments. We found that most of these known genes in anther and pollen development were down-regulated more than 2-fold in the BR mutants (Fig. 4), consistent with the results of real-time qRT-PCR analysis.
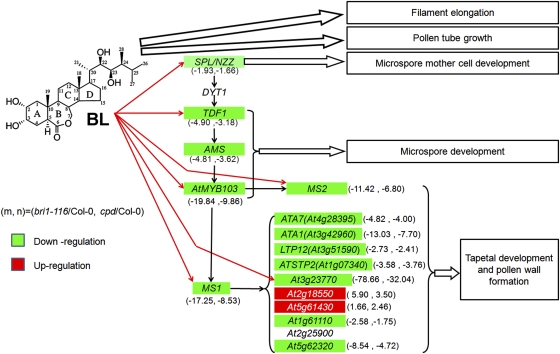
Fig. 4.
A model to illustrate the roles of BRs in regulating male fertility in Arabidopsis. Besides the roles in regulating filament elongation and pollen tube growth via promoting cell elongation, the BR signaling also controls the tapetum and microspore development in Arabidopsis by directly regulating the expression of SPL/NZZ, TDF1, MYB103, MS2, MS1, and MS1-target genes. Solid arrows indicate a direct regulation on gene expression. Open arrows indicate the positive effect on the final developmental process. Genes highlighted with green indicate a down-regulation in BR biosynthetic or signaling mutants, and genes highlighted with red indicate an up-regulation. The first and second numbers in the parenthesis indicate fold-changes of gene expression in bri1-116 and cpd, respectively, compared to wild type.
BES1 Directly Binds to the Promoter Regions of Several Key Anther and Pollen Developmental Genes.
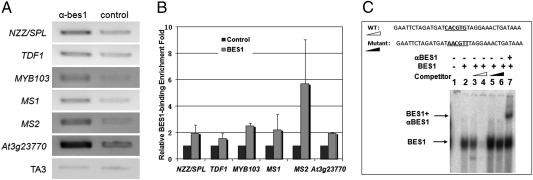
BR signaling is known to be mainly mediated by transcription factor BES1 and its homologs. To test whether the expression of anther and pollen developmental genes was primarily or secondarily regulated by BR signaling, we conducted the ChIP experiments using wild-type plants. We observed that BES1 can directly bind to the promoter regions of SPL/NZZ, TDF1, MS1, MS2, AtMYB103, and a highly differentially expressed MS1 target gene, At3g23770 (42) with a 1.5- to more than 5-fold enrichment compared with a negative control (Fig. 5 A and B). We further conducted an in vitro binding experiment with BES1 to the MS2 promoter using gel mobility shift assay, and found that BES1 can specifically bind to this promoter (Fig. 5C). These results suggest that multiple anther genes are direct targets of BES1 or its related transcription factors.
Fig. 5.
BES1 directly binds to the promoter regions of key regulatory genes in anther and pollen development. (A) Anti-BES1 antibodies or an unrelated antibody as negative control were used to precipitate chromatin prepared from 5-week-old Col-0 adult plants. TA3 was used as an internal control. (B) Real-time qPCR analysis of ChIP products. The fold-changes were calculated based on the relative change in anti-BES1 compared with an unrelated antibody, which was used as a control and defined as “1.” (C) BES1 binds to MS2 promoter in vitro. Without probe (Lane 1), with labeled probe (Lane 2), labeled probe with 50× or 250× unlabeled wild-type (Lanes 3 and 4) or the mutant sequences (Lanes 5 and 6). Anti-BES1 antibody was added (Lane 7).
Discussion
After systematic analyses of the male-fertility phenotypes of the BR-deficient and signaling mutants, and a transgenic line, we obtained strong evidence that supports the regulation of BR signaling on pollen and anther development. First, the pollen number is greatly reduced. Compared to the wild type, the BR mutants, bri1-116 and cpd, only produced fewer than half of the normal number of microspore mother cells, and produce about 25% of pollen grains at stages 8 and 9, and about 20% at stage 12 (Fig. 1J). Therefore, the reduced pollen number in the BR mutants is likely caused by the decreased microspore mother cells and early abortion of some microspores. Second, the pollen grains produced in the BR mutants had an abnormal exine pattern, as indicated by lacking an obvious bacula/tectum structure (Fig. 2 J–L), resulting in rare release from anther locules. Third, tapetal development of the BR mutants was abnormal, indicated by the extremely vacuolated and enlarged tapetal cells (Fig. 1), providing a reasonable explanation for the abnormal deposition of pollen wall components and abnormal pollen exine patterning.
Our results strongly suggest that BRs affect anther and pollen development at several stages by controlling the expression of SPL/NZZ, TDF1, AMS, MYB103, MS2, MS1, and many downstream genes of MS1. First, in the BR mutants, the expression of SPL/NZZ is significantly inhibited. This decrease might account for the reduced number of microspore mother cells in the mutants. Interestingly, it was observed that the expression of BRI1 in the spl/nzz mutant was 3-fold higher than that in the wild type (4), suggesting that a feedback regulatory mechanism may also be involved in the BR signaling and the default regulator network in pollen and anther development. Furthermore, the expression of multiple genes, including TDF1, AMS, MYB103, and MS1, involved in pollen development is significantly inhibited in the mutants, which might cause the early abortion of some microspores and finally lead to the greatly reduced number of pollen grains. Finally, the expression of MS2 and many MS1-regulated genes, which directly regulate the structure and composition of pollen coat, is also suppressed in the BR mutants. Therefore, BRs function from the early microspore mother formation to late pollen maturation to regulate anther development by controlling the expression of SPL/NZZ, TDF1, MYB103, MS1, and MS2.
Although it is well known that many genes involved in wall loosing and cell elongation are activated by the BR-regulated transcription factors, such as BES1/BZR1 (37, 43), it is poorly understood whether the BR-activated transcription factors can directly regulate genes involved in other developmental processes. For instance, BES1 and BIM1 can directly bind to the promoter regions of SAUR-AC1 and regulate its expression (37). A recent study found that AtMYB30, encoding a MYB family transcription factor, is a direct target gene of BES1, which may play an important role in the early stage of plant development (44). It was reported that BRs may play an important role in directing root epidermal cells to differentiate into hair cells or mature hairless cells in Arabidopsis (45). In this work, we found that the promoter regions of many genes involved in pollen and anther development contain multiple binding sites of BES1/BZR1 (Fig. S4), and our ChIP experiments demonstrated that at least one site in the promoter regions of NZZ/SPL, TDF1, MS1, MS2, MYB103, and AT3g23770 can directly bind to BES1. Therefore, this study provides a unique example to demonstrate that BRs regulate cell division and differentiation, leading to a specific developmental process, which is largely beyond the known BR-regulated cell expansion.
Based on the current and previous findings, we summarize a model to illustrate the cellular and molecular basis of BRs in regulating male fertility in Arabidopsis. As shown in Fig. 4, BR signaling can positively regulate filament and pollen tube elongation, which are related to cell elongation. More importantly, BRs control multiple steps of anther development, including microspore mother cell formation, microspore development, tapetal development, and pollen coat formation. The regulation is largely through the direct binding of BES1 to the promoter regions of NZZ/SPL, TDF1, AMS, MYB103, MS2, MS1, and MS1-target genes. Many external and internal stimuli can also regulate male fertility. For example, a high temperature treatment on 3-week-old Arabidopsis seedlings with four to five mature flowers led to the failure of separation of pollen mother cells, inhibition of microspore differentiation and male meiotic processes, and finally, failure to produce any pollen grains (46). However, the underlying molecular mechanisms for such regulation are still largely unknown. Moreover, although the hand pollination with bri1-116 pollen can lead to successful fertilization under optimal conditions, we did observe the slower growth of bri1-116 pollen tubes than the wild-type pollen tubes. Therefore, the large amount of BRs in mature pollens may play an important role in regulating efficiency of fertilization under adverse conditions. Our study provides important insights into the mechanisms by which an internal signal interacts with an essential genetic program to regulate the corresponding developmental process. Furthermore, various stimuli, including multiple phytohormones, may coordinately regulate several steps of anther and pollen development; thus, further studies on the interaction of different signals are needed to understand this regulatory network.
Experimental Procedures
Plant Material.
Arabidopsis thaliana ecotype Columbia (Col-0) was used as the wild-type control. The BR-related mutants, bri1-116, cpd, bin2, det2, a transgenic myriBKI1-YFP line, and BES1-GFP overexpression line (BES1-OX) are all in the Col-0 background. Seeds were imbibed for 4 days at 4 °C and sown on soil; plants were grown at 22 °C under a 16-h light/8-h dark cycle.
Light and Electron Microscopy Observation.
To measure pollen viability, freshly dehiscent anthers were collected, stained with Alexander's staining solution (40), and pressed to release pollen grains. Pollen grains were then viewed with an Olympus BH-2 light microscope (Olympus Co., Ltd). SEM analysis was performed as described previously (47), and the samples were observed by a JSM-6360LV SEM (JEOL Ltd). For TEM analysis, the buds approximately containing stage-8 anthers were fixed and dehydrated as described by Zhang et al. (48), imbedded in a mixture of Spurr's Resin to acetone with a ratio of 1:1 for 2 h and Spurr's Resin alone for 2 h, and the material was polymerized in molds at 35 °C for 16 h and then 60 °C for 48 h. Ultrathin sections (90–100 nm thick) were observed with a JEM-2100 TEM (JEOL Ltd).
Paraffin Sections.
Inflorescences were incubated for at least 12 h in FAA fixing solution (47.5% ethanol, 5% acetic acid, and 10% formaldehyde in water), and sequentially dehydrated at room temperature in 20% l-butanol with 50% ethanol (2–4 h), 35% l-butanol with 50% ethanol (2–4 h), 55% l-butanol with 40% ethanol (2–4 h), 75% l-butanol and 25% ethanol (4–6 h), and100% l-butanol in water(4–6 h), and finally twice in 100% paraffin at 62 °C with 45 min each. Samples were then embedded and sectioned (6 μm) with a Leica RM2135 slicing machine (Leica Geosystems). Sections were roasted at 65 °C for 2 h, de-waxed by reversing the steps of dehydration procedure described above, stained with 15% toluidine blue for 40 s, and sealed with neutral balsam (Ri-Chu BioScience Co., Ltd).
Estimation of Relative Pollen Number per Locule.
Relative pollen number per locule at stages 11 and 12 is calculated by the formula N = (L × n)/(Lw × nw), in which N represents relative pollen number per locule, L represents anther length, n represents pollen number per locule counted with transverse sections of anthers, and “w” indicates the data from the wild type. The relative pollen number per locule at stages 5 to 9 is calculated through multiplying anther diameter by pollen number per locule with longitudinal sections of anthers, and a similar formula was used for the transverse sections.
In Vivo Pollen Tube Growth Assays and Aniline Blue Staining.
Dehiscent anthers were removed and used to brush pollens onto the stigmas. The pistils at 3 h after pollination were dissected and fixed. Aniline blue (Ri-Chu BioScience Co., Ltd) staining was performed as described by Mori et al. (49) to visualize pollen tube growth. Pollen tubes were observed using an Olympus IX71 fluorescence microscope (Olympus Co., Ltd).
Real-Time qRT-PCR Analysis.
Total RNA was extracted from floral buds containing approximately stages 7 to 9 anthers or from inflorescences containing stages 1 to 8 anthers using Tiangen RNAprep plant kit (Tiangen Biotech Co., Ltd). The first-trand cDNA was then synthesized using Takara PrimeScript First-Strand cDNA Synthesis kit (Takara Bio Inc.), and then used for real-time qRT-PCR. Primer sequences for each gene are listed in Table S1. Real-time qRT-PCR was performed in triplicate using Bio-Rad iCycler (Bio-Rad Labratories) and data were collected and analyzed with Bio-Rad MyiQ Single-Color Real-Time PCR Detection System. In addition, a U-BOX gene (At5g15400) was used as a control to normalize the level of total RNA.
Microarray Experiments.
Total RNA was extracted from flower buds approximately at anther stages 7 to 9 of bri1-116, cpd, and Col-0, and used for microarray analysis. See the SI Experimental Procedures for details.
ChIP Experiments.
ChIP was performed primarily based on the method described by Pikaard's laboratory (www.biology.wustl.edu/pikaard). Open flowers and buds from 5-week-old Col-0 adult plants were cross-linked and used for ChIP assays. About 3 μg of purified anti-BES1 antibody (or an unrelated antibody as negative control) was used to immunoprecipitate chromatin sample extracted from 500-mg tissues. The ChIP products were purified and used for PCR with primers from these candidate target genes (Table S2). Two independent biological replica were performed.
Gel Mobility Shift Assay.
The BES1 protein purification and GMSA procedure were performed essentially as described by Li et al., (44). About 0.5-ng labeled probe and 100 ng of purified BES1 protein were used in each binding reaction. A DNA fragment (−219 to −185 from the MS2 promoter) was used as a probe.
Supplementary Material
Acknowledgments
We thank W. Zhang for advice on making paraffin sections, H. Saiyin and D. Chen for assistance in preparing paraffin sections, and Y. Zhao for critically reading the manuscript. This work was supported by a start-up fund of Fudan University to X.W., Grants 30871330 and 90817004 of the National Natural Science Foundation of China (to X.W.), Grant 08018 of the Chun-Tsung Chinese Undergraduate Research Endowment (to Q.Y.), and an Advanced Visiting Scholar by State Key Laboratory of Genetic Engineering to Y. Zhao.
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/cgi/content/full/0912333107/DCSupplemental.
References
- 1.Allison AS, Paul OL. Reproductive traits and male fertility in plants: empirical approaches. Annu Rev Ecol Syst. 1993;24:331–351. [Google Scholar]
- 2.Goldberg RB, Beals TP, Sanders PM. Anther development: basic principles and practical applications. Plant Cell. 1993;5:1217–1229. doi: 10.1105/tpc.5.10.1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ma H. Molecular genetic analyses of microsporogenesis and microgametogenesis in flowering plants. Annu Rev Plant Biol. 2005;56:393–434. doi: 10.1146/annurev.arplant.55.031903.141717. [DOI] [PubMed] [Google Scholar]
- 4.Wijeratne AJ, et al. Differential gene expression in Arabidopsis wild-type and mutant anther: insights into anther cell differentiation and regulatory networks. Plant J. 2007;52:14–29. doi: 10.1111/j.1365-313X.2007.03217.x. [DOI] [PubMed] [Google Scholar]
- 5.Balasubramanian S, Schneitz K. NOZZLE regulates proximal-distal pattern formation, cell proliferation and early sporogenesis during ovule development in Arabidopsis thaliana. Development. 2000;127:4227–4238. doi: 10.1242/dev.127.19.4227. [DOI] [PubMed] [Google Scholar]
- 6.Yang WC, Ye D, Xu J, Sundaresan V. The SPOROCYTELESS gene of Arabidopsis is required for initiation of sporogenesis and encodes a novel nuclear protein. Genes Dev. 1999;13:2108–2117. doi: 10.1101/gad.13.16.2108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Canales C, Bhatt AM, Scott R, Dickinson H. EXS, a putative LRR receptor kinase, regulates male germline cell number and tapetal identity and promotes seed development in Arabidopsis. Curr Biol. 2002;12:1718–1727. doi: 10.1016/s0960-9822(02)01151-x. [DOI] [PubMed] [Google Scholar]
- 8.Zhao DZ, et al. The EXCESS MICROSPOROCYTES1 gene encodes a putative leucine-rich repeat receptor protein kinase that controls somatic and reproductive cell fates in the Arabidopsis anther. Genes Dev. 2002;16:2021–2031. doi: 10.1101/gad.997902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang W, et al. Regulation of Arabidopsis tapetum development and function by DYSFUNCTIONAL TAPETUM1 (DYT1) encoding a putative bHLH transcription factor. Development. 2006;133:3085–3095. doi: 10.1242/dev.02463. [DOI] [PubMed] [Google Scholar]
- 10.Zhu J, et al. Defective in Tapetal Development and Function 1 is essential for anther development and tapetal function for microspore maturation in Arabidopsis. Plant J. 2008;55:266–277. doi: 10.1111/j.1365-313X.2008.03500.x. [DOI] [PubMed] [Google Scholar]
- 11.Sorensen A, et al. The Arabidopsis ABORTED MICROSPORES (AMS) gene encodes a MYC class transcription factor. Plant J. 2003;33:413–423. doi: 10.1046/j.1365-313x.2003.01644.x. [DOI] [PubMed] [Google Scholar]
- 12.Higginson T, Li SF, Parish RW. AtMYB103 regulates tapetum and trichome development in Arabidopsis thaliana. Plant J. 2003;35:177–192. doi: 10.1046/j.1365-313x.2003.01791.x. [DOI] [PubMed] [Google Scholar]
- 13.Wilson Z, Morroll S, Dawson J, Swarup R, Tighe P. The Arabidopsis MALE STERILITY1 (MS1) gene is a transcriptional regulator of male gametogenesis, with homology to the PHD-finger family of transcription factors. Plant J. 2001;28:27–39. doi: 10.1046/j.1365-313x.2001.01125.x. [DOI] [PubMed] [Google Scholar]
- 14.Ito T, et al. Arabidopsis MALE STERILITY1 encodes a PHD-Type transcription factor and regulates pollen and tapetum development. Plant Cell. 2007;19:3549–3562. doi: 10.1105/tpc.107.054536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Aarts MG, et al. The Arabidopsis MALE STERILITY 2 protein shares similarity with reductases in elongation/condensation complexes. Plant J. 1997;12:615–623. doi: 10.1046/j.1365-313x.1997.00615.x. [DOI] [PubMed] [Google Scholar]
- 16.Huang S, et al. Transgenic studies on the involvement of cytokinin and gibberellin in male development. Plant Physiol. 2003;131:1270–1282. doi: 10.1104/pp.102.018598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cecchetti V, Altamura M, Falasca G, Costantino P, Cardarelli M. Auxin regulates Arabidopsis anther dehiscence, pollen maturation, and filament elongation. Plant Cell. 2008;20:1760–1774. doi: 10.1105/tpc.107.057570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kieber J, Rothenberg M, Roman G, Feldmann K, Ecker J. CTR1, a negative regulator of the ethylene response pathway in Arabidopsis, encodes a member of the raf family of protein kinases. Cell. 1993;72:427–441. doi: 10.1016/0092-8674(93)90119-b. [DOI] [PubMed] [Google Scholar]
- 19.Park J, et al. A knock-out mutation in allene oxide synthase results in male sterility and defective wound signal transduction in Arabidopsis due to a block in jasmonic acid biosynthesis. Plant J. 2002;31:1–12. doi: 10.1046/j.1365-313x.2002.01328.x. [DOI] [PubMed] [Google Scholar]
- 20.Singh D, Jermakow A, Swain S. Gibberellins are required for seed development and pollen tube growth in Arabidopsis. Plant Cell. 2002;14:3133–3147. doi: 10.1105/tpc.003046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cheng Y, Dai X, Zhao Y. Auxin biosynthesis by the YUCCA flavin monooxygenases controls the formation of floral organs and vascular tissues in Arabidopsis. Genes Dev. 2006;20:1790–1799. doi: 10.1101/gad.1415106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dharmasiri N, et al. Plant development is regulated by a family of auxin receptor F box proteins. Dev Cell. 2005;9:109–119. doi: 10.1016/j.devcel.2005.05.014. [DOI] [PubMed] [Google Scholar]
- 23.Cheng H, et al. Gibberellin regulates Arabidopsis floral development via suppression of DELLA protein function. Development. 2004;131:1055–1064. doi: 10.1242/dev.00992. [DOI] [PubMed] [Google Scholar]
- 24.Chhun T, et al. Gibberellin regulates pollen viability and pollen tube growth in rice. Plant Cell. 2007;19:3876–3888. doi: 10.1105/tpc.107.054759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Grove M, et al. Brassinolide, a plant growth-promoting steroid isolated from Brassica napus pollen. Nature. 1979;281:216–217. [Google Scholar]
- 26.Asakawa S, Abe H, Nishikawa N, Natsume M, Koshioka M. Purification and identification of new acyl-conjugated teasterones in lily pollen. Biosci Biotechnol Biochem. 1996;60:1416–1420. [Google Scholar]
- 27.Hewitt F, et al. Effect of brassinolide and other growth regulators on the germination and growth of pollen tubes of Prunus avium using a multiple hanging-drop assay. Aust J Plant Physiol. 1985;1:201–211. [Google Scholar]
- 28.Szekeres M, et al. Brassinosteroids rescue the deficiency of CYP90, a cytochrome P450, controlling cell elongation and de-etiolation in Arabidopsis. Cell. 1996;85:171–182. doi: 10.1016/s0092-8674(00)81094-6. [DOI] [PubMed] [Google Scholar]
- 29.Li J, Nam K, Vafeados D, Chory J. BIN2, a new brassinosteroid-insensitive locus in Arabidopsis. Plant Physiol. 2001;127:14–22. doi: 10.1104/pp.127.1.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bouquin T, Meier C, Foster R, Nielsen M, Mundy J. Control of specific gene expression by gibberellin and brassinosteroid. Plant Physiol. 2001;127:450–458. [PMC free article] [PubMed] [Google Scholar]
- 31.Kim T, et al. Arabidopsis CYP85A2, a cytochrome P450, mediates the Baeyer-Villiger oxidation of castasterone to brassinolide in brassinosteroid biosynthesis. Plant Cell. 2005;17:2397–2412. doi: 10.1105/tpc.105.033738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li J, Chory J. A putative leucine-rich repeat receptor kinase involved in brassinosteroid signal transduction. Cell. 1997;90:929–938. doi: 10.1016/s0092-8674(00)80357-8. [DOI] [PubMed] [Google Scholar]
- 33.Li J, Jin H. Regulation of brassinosteroid signaling. Trends Plant Sci. 2007;12:37–41. doi: 10.1016/j.tplants.2006.11.002. [DOI] [PubMed] [Google Scholar]
- 34.Wang X, Chory J. Brassinosteroids regulate dissociation of BKI1, a negative regulator of BRI1 signaling, from the plasma membrane. Science. 2006;313:1118–1122. doi: 10.1126/science.1127593. [DOI] [PubMed] [Google Scholar]
- 35.Yin Y, et al. BES1 accumulates in the nucleus in response to brassinosteroids to regulate gene expression and promote stem elongation. Cell. 2002;109:181–191. doi: 10.1016/s0092-8674(02)00721-3. [DOI] [PubMed] [Google Scholar]
- 36.He J, et al. BZR1 is a transcriptional repressor with dual roles in brassinosteroid homeostasis and growth responses. Science. 2005;307:1634–1638. doi: 10.1126/science.1107580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yin Y, et al. A new class of transcription factors mediates brassinosteroid-regulated gene expression in Arabidopsis. Cell. 2005;120:249–259. doi: 10.1016/j.cell.2004.11.044. [DOI] [PubMed] [Google Scholar]
- 38.Procissi A, et al. Five gametophytic mutations affecting pollen development and pollen tube growth in Arabidopsis thaliana. Genetics. 2001;158:1773–1783. doi: 10.1093/genetics/158.4.1773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Teng C, et al. Serine palmitoyltransferase, a key enzyme for de novo synthesis of sphingolipids, is essential for male gametophyte development in Arabidopsis. Plant Physiol. 2008;146:1322–1332. doi: 10.1104/pp.107.113506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Alexander MP. Differential staining of aborted and nonaborted pollen. Biotech Histochem. 1969;44:117–122. doi: 10.3109/10520296909063335. [DOI] [PubMed] [Google Scholar]
- 41.Yang C, Vizcay-Barrena G, Conner K, Wilson Z. MALE STERILITY1 is required for tapetal development and pollen wall biosynthesis. Plant Cell. 2007;19:3530–3548. doi: 10.1105/tpc.107.054981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kang J, Zhang G, Bonnema G, Fang Z, Wang X. Global analysis of gene expression in flower buds of Ms-cd1 Brassica oleracea conferring male sterility by using an Arabidopsis microarray. Plant Mol Biol. 2008;66:177–192. doi: 10.1007/s11103-007-9261-9. [DOI] [PubMed] [Google Scholar]
- 43.Li L, Deng XW. It runs in the family: regulation of brassinosteroid signaling by the BZR1–BES1 class of transcription factors. Trends Plant Sci. 2005;10:266–268. doi: 10.1016/j.tplants.2005.04.002. [DOI] [PubMed] [Google Scholar]
- 44.Li L, et al. Arabidopsis MYB30 is a direct target of BES1 and cooperates with BES1 to regulate brassinosteroid-induced gene expression. Plant J. 2009;58:275–286. doi: 10.1111/j.1365-313X.2008.03778.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kuppusamy KT, Chen AY, Nemhauser JL. Steroids are required for epidermal cell fate establishment in Arabidopsis roots. Proc Natl Acad Sci USA. 2009;106:8073–8076. doi: 10.1073/pnas.0811633106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kim SY, Hong CB, Lee I. Heat shock stress causes stage-specific male sterility in Arabidopsis thaliana. J Plant Res. 2001;114:301–307. [Google Scholar]
- 47.Guan Y, et al. RUPTURED POLLEN GRAIN1, a member of the MtN3/saliva gene family, is crucial for exine pattern formation and cell integrity of microspores in Arabidopsis. Plant Physiol. 2008;147:852–863. doi: 10.1104/pp.108.118026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhang Z, et al. Transcription factor AtMYB103 is required for anther development by regulating tapetum development, callose dissolution and exine formation in Arabidopsis. Plant J. 2007;52:528–538. doi: 10.1111/j.1365-313X.2007.03254.x. [DOI] [PubMed] [Google Scholar]
- 49.Mori T, Kuroiwa H, Higashiyama T, Kuroiwa T. GENERATIVE CELL SPECIFIC 1 is essential for angiosperm fertilization. Nat Cell Biol. 2005;8:64–71. doi: 10.1038/ncb1345. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.