Summary
Gene duplication provides an important source of genetic raw material for phenotypic diversification [1, 2], but few studies have detailed the mechanisms through which duplications produce evolutionary novelty within species [3–6]. Here, we investigate how a set of recently duplicated homologs of the floral inducer FLOWERING LOCUS T (FT) has contributed to sunflower domestication. We find that changes in expression of these duplicates are associated with differences in flowering behavior between wild and domesticated sunflower. In addition, we present genetic and functional evidence demonstrating that a frameshift mutation in one paralog, Helianthus annuus FT 1 (HaFT1), underlies a major QTL for flowering time and experienced a selective sweep during early domestication. Notably, this dominant-negative allele delays flowering through interference with action of another paralog, HaFT4. Together, these data reveal that changes affecting the expression, sequence, and gene interactions of HaFT paralogs have played key roles during sunflower domestication. Our findings also illustrate the important role that evolving interactions between new gene family members may play in fostering phenotypic change.
Highlights
After recent expansion, the sunflower FT gene family members have met diverse fates.
HaFT expression changes are associated with a shift to early, long-day flowering.
A frameshift in domesticated HaFT1 impacts flowering by interfering with HaFT4.
HaFT1 experienced a selective sweep during early sunflower domestication.
Results and Discussion
In plants, lineage-specific duplications in the FLOWERING LOCUS T-like (FT-like) gene family are often found and may be important substrates for evolutionary innovation [7–13]. FT plays a crucial and widely conserved role in regulation of flowering time by environmental cues [9, 10, 12, 14–17]. Briefly, genes in the photoperiod pathway integrate cues from the circadian clock and light signaling such that FT is highly expressed in the leaf only under inductive photoperiods [16, 18–20]. FT protein travels from the leaf through the phloem to the shoot apical meristem [12, 21, 22], where, through interactions with additional proteins, it initiates a gene regulatory cascade that promotes reproductive meristem identity [23–25]. Divergence in spatial, developmental or photoperiod-specific gene expression patterns has been observed among lineage-specific FT-like paralogs [8, 11, 12, 26, 27], and two rice FT paralogs differ in how they promote flowering in different photoperiods [13]. Natural variants in one or more FT-like genes have also been linked to variation in flowering in A. thaliana and domesticated cereals [11, 17, 28–31]. Here, we have investigated the evolution of FT-like genes in the common sunflower, Helianthus annuus, and the roles these genes played during domestication.
Recent Expansion and Diverse Fates
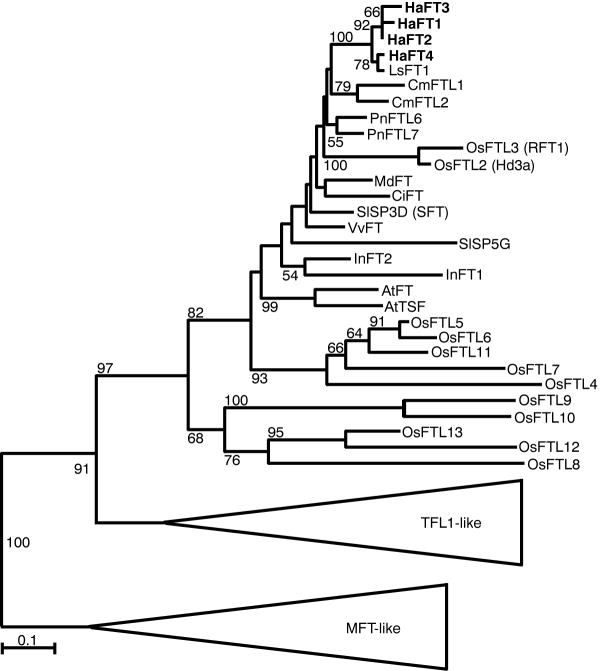
Four sunflower FT-like paralogs—HaFT1, HaFT2, HaFT3, and HaFT4—were isolated from H. annuus by PCR and hybridization-based methods. A phylogeny constructed with FT homologs from a diverse set of plants revealed these sunflower paralogs are all more closely related to each other than to homologs from other species (Figure 1), indicating they arose by a series of relatively recent duplications. All paralogs have highly similar sequences and exon-intron structure (Figure 2A, Figure S1, Table S1), though HaFT4 is more divergent and one amino acid shorter than the remaining paralogs. All four duplicates have the conserved FT amino acid at two residues that functionally distinguish FT from TERMINAL FLOWER 1 (TFL1) [32, 33].
Figure 1. Recent Duplications of Sunflower FT-like Genes.
Maximum likelihood phylogeny based on amino acid sequences of plant FT proteins. Bootstrap percentages > 50% shown above branches. Species abbreviations: Antirrhinum majus, Am; Arabidopsis thaliana, At; Citrus unshiu, Ci; Cucurbita maxima, Cm; Helianthus annuus, Ha; Ipomea nil, In; Lactuca sativa, Ls; Malus x domestica, Md; Oryza sativa, Os; Populus nigra, Pn; Solanum lycopersicum, Sl; and Vitis vinifera, Vv. See also Table S1 and Table S6.
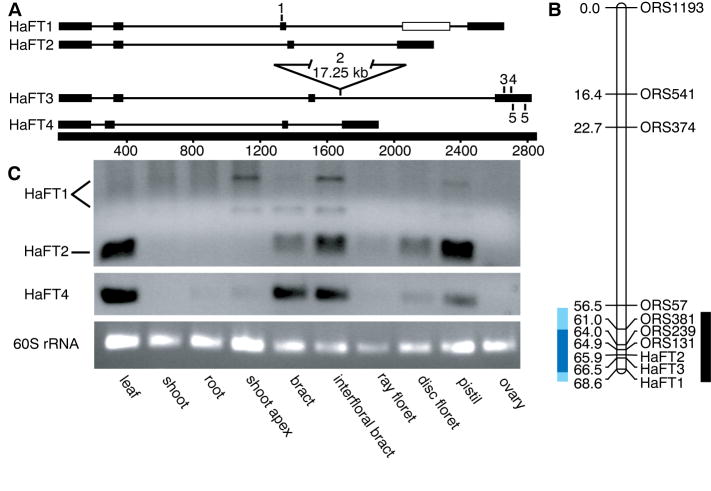
Figure 2. Gene Structure, Map Position, and Spatial Expression of HaFT paralogs.
(A) Exon-intron structure of HaFT coding sequences shown to scale. The open rectangle in HaFT1 denotes an alternatively spliced exon. Numbered sites mark the locations of the frameshift in HaFT1 (1) and putative loss-of-function mutations in HaFT3 segregating in natural populations (2–5). HaFT3 mutations include a 17.25 kb insert in the third intron (2), a 7 bp deletion (3), a 1 bp deletion (4), and two cosegregating premature stop mutations (5).
(B) Genetic map of LG6 indicating the map positions of HaFT1, HaFT2, and HaFT3 relative to previously mapped QTL region (black) [35], and relative to the wild introgression into a domesticated background in NILs (blue, introgression end points occur within light blue regions).
(C) Spatial expression of HaFT1, HaFT2, and HaFT4 in wild sunflower assayed by RT-PCR. See also Figure S1 and Table S3.
The synonymous substitution rate between two sequences, Ks, provides a measure of time since divergence as synonymous sites are expected to evolve neutrally. Ks is ~0.45 for all comparisons between HaFT4 and the other paralogs (Table S1), a magnitude consistent with duplication during a polyploidy event at the base of the Heliantheae tribe [34]. Ks comparisons of HaFT1, HaFT2, and HaFT3 with each other are much lower (~0.04–0.08), indicating these duplications likely occurred within the genus Helianthus. HaFT4 maps to linkage group (LG) 14 of the sunflower genetic map, whereas the other three paralogs all map to the same end of LG6 (Figure 2B), suggesting the recent duplications occurred in tandem. Since gene conversion may homogenize tandem duplicates, further sequencing from additional Heliantheae species will be required to precisely time and order the more recent events.
After duplication, gene copies may have several evolutionary fates including non-functionalization, retention for additional dosage, neofunctionalization, subfunctionalization, and differential improvement of ancestral functions [2]. We surveyed spatial expression patterns of the HaFT genes in wild H. annuus to examine what processes have acted on these paralogs (Figure 2C). Because HaFT1, HaFT2, and HaFT3 have extremely similar coding sequences, primers that uniquely amplify each copy could not be designed. Instead, HaFT2 expression was distinguished by restriction digest. All PCR reactions exhibiting expression were also cloned, and 24 clones per reaction were sequenced to verify the contributing paralog(s). HaFT4 and HaFT2 exhibited similar expression patterns; however, HaFT1 expression diverged in two notable ways. HaFT1 was not expressed in leaves but, unlike HaFT2 and HaFT4, was expressed in the shoot apex. Sequencing of cloned PCR products confirmed these results. HaFT1 also exhibited an alternative splice form retaining part of the third intron, which, though in frame, contains premature stop codons. Depending on whether the single copy ancestor of these genes was expressed in the shoot apex, either subfunctionalization or neofunctionalization may have preserved HaFT1, though gain of a new expression domain is the more parsimonious explanation.
HaFT3 expression was not detected by sequencing RT-PCR products from any tissue (0/192 clones sequenced total across 8 tissues), and four mutations likely to disrupt function (Figure 2A) were found in 54 of 60 wild and domesticated accessions of H. annuus surveyed and in sister species H. argophyllus (Table S2 and S3). These findings provide strong evidence consistent with non-functionalization of HaFT3.
Robust, efficient, and universal sunflower transformation protocols are as yet undeveloped; however, since FT activity is widely conserved, HaFT function is testable by heterologous complementation. To determine whether the three expressed HaFT genes were functionally equivalent to A. thaliana FT, we overexpressed the coding region of each paralog from the CaMV35S promoter in Columbia-0 plants and ft-1 mutants raised in long days. Wild alleles of all three sunflower paralogs accelerated flowering in both wild-type and mutant backgrounds (Figure 3, Table S4, Figure S2), indicating these sunflower paralogs all encode functional copies of FT.
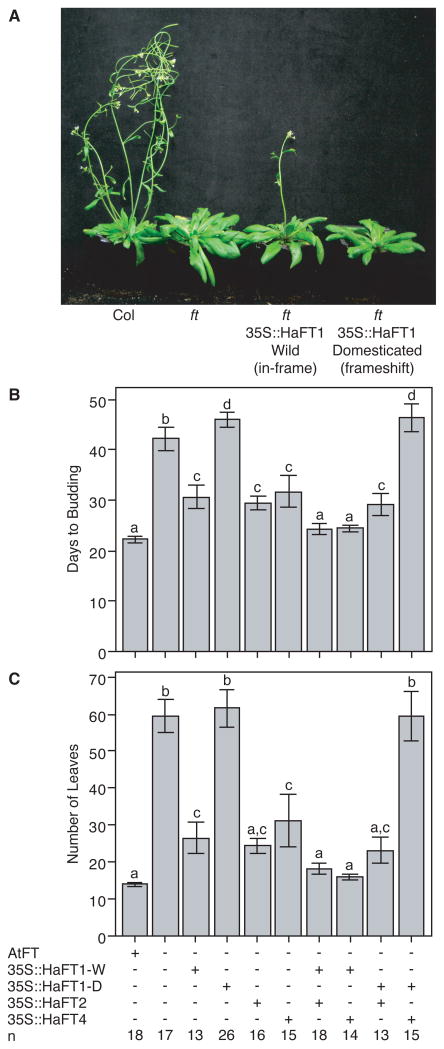
Figure 3. Frameshift-Carrying Domesticated HaFT1 Allele Has Dominant-Negative Effect.
(A) Overexpression of in-frame wild allele of HaFT1 complements the A. thaliana ft mutant whereas overexpression of the frameshift-carrying domesticated allele does not. Plants photographed 36 days after germination.
(B and C) Days to budding (B) and rosette leaf number (C) of plants overexpressing 0, 1, or 2 HaFT paralogs in an ft background. 35S::HaFT1-W plants overexpressed the wild, in-frame HaFT1 allele; 35S::HaFT1-D plants overexpressed the domesticated, frameshift-carrying HaFT1 allele. Means for each genotype were compared with a general linear model, and pairwise comparisons were performed with Tukey's multiple comparison test. Different letters above the 95% confidence intervals denote significantly different phenotype distributions. See also Figure S2 and Table S4.
QTL Characterization
The three HaFT paralogs that map to LG6 co-localize with a QTL explaining 7–36% of flowering time variation, depending on cross and environment, in crosses between wild and domesticated sunflower (Figure 2B) [35–37]. While HaFT3 appears non-functional, HaFT1 and HaFT2 are strong candidates for the gene underlying the QTL. Because FT is involved in photoperiodic floral induction, we characterized the photoperiod response of parents of a wild x elite-crop QTL population. Plants from the wild parent’s population, Ann1238, exhibited a short-day response, flowering ~10 days earlier in short and intermediate days than under long days (Figure 4A). The domesticated parent line, CMSHA89, had the opposite behavior—long-day response—flowering ~10 days earlier in long days. Comparing flowering time in each group’s inductive photoperiod, the domesticated line flowered ~10 days earlier than the wild population.
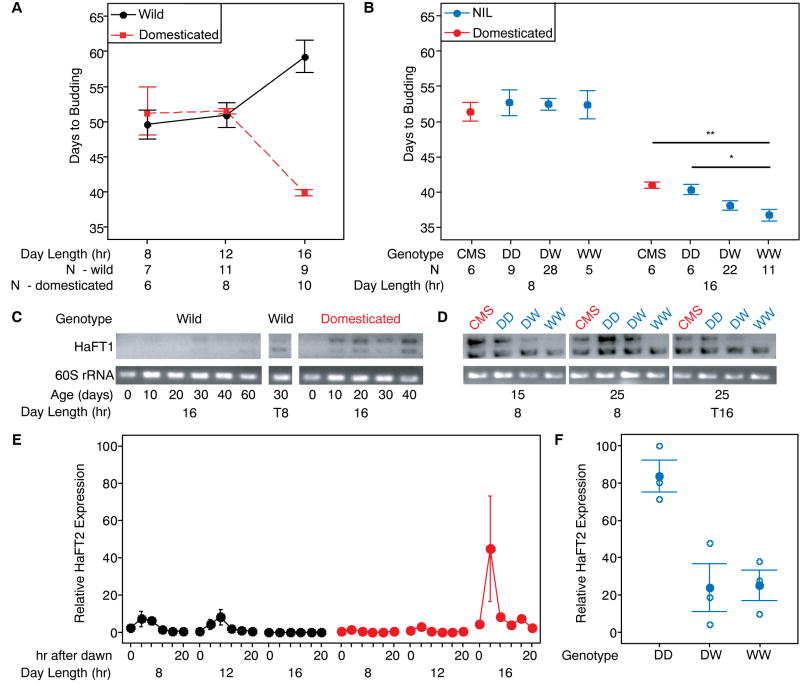
Figure 4. Flowering Time, HaFT1 Expression, and HaFT2 Expression in Parental and NIL Plants.
(A) Days to budding of wild (black) and domesticated (orange) parents in three photoperiods. Mean ± SE shown. (B) Days to budding of domesticated (CMS, orange) and NIL plants (blue) homozygous for the domesticated LG6 QTL region (DD), heterozygous (DW), and homozygous wild (WW) in short and long days. 95% confidence intervals shown. Differences among genotypes tested by general linear model corrected with Tukey’s multiple comparison test, **, p < 0.02; *, p = 0.058. (C) HaFT1 shoot apex expression in developing wild and domesticated parents in long days. Age measured as days after sowing. T8 plants experienced 20 long days followed by 10 short days. (D) HaFT1 shoot apex expression in CMS, DD, DW, and WW plants 15 and 25 days after sowing in short days. T16 plants experienced 15 short days followed by 10 long days. (E) HaFT2 leaf expression in wild (black) and domesticated (orange) parents every 4 hr on the 30th day after sowing. Mean ± SE for three biological replicates shown. (F) HaFT2 leaf expression in long-day grown DD, DW, and WW plants 4 hr after dawn, 30 days after sowing. Mean of three technical replicates per biological replicate (open circles) and mean ± SE for three biological replicates (filled circles) shown. Relative expression expressed as delta-delta-Ct normalized to 60S rRNA and scaled to the highest individual measurement. See also Figure S3 and Table S5.
To determine what aspect of this divergence in flowering behavior the LG6 QTL controls, we developed a near isogenic line (NIL) for this region by genotypic selection during backcrossing for five generations of a descendant of the original recombinant panel to CMSHA89 followed by selfing (Figure 2B). Photoperiod response was then characterized for individuals homozygous for the domesticated allele in the LG6 QTL region (DD), heterozygous (DW), and homozygous for the wild allele (WW).
Like the domesticated parent, all genotypes showed a long-day response (Figure 4B). Thus, this QTL does not mediate the photoperiod response difference between the parents. A photoperiod-specific difference in flowering time was observed between genotypes however. WW plants budded ~4 days earlier than DD plants or the domesticated parent only in the inductive long-day photoperiod (Figure 4B). These results were similar in direction and magnitude to the QTL effect observed previously [35]. Similar results were obtained on replication (Table S5), and a larger sample size provided sufficient statistical power to show the DW phenotype was intermediate. Additional phenotyping revealed photoperiod-specific differences for 9 of 13 traits including a 7-day difference in anthesis date and 1 cm difference in disc diameter between DD and WW plants in long days. As it is unlikely that many genes in a small genomic region would independently affect diverse traits in a photoperiod-specific manner, we expect these additional differences could be direct pleiotropic effects or indirect effects of the change in developmental timing caused by allelic variation in a single gene.
Evaluation of Candidate Paralogs
The NIL phenotypes suggest a model for the action of the gene underlying the QTL. First, the photoperiod-specific effect of the QTL on flowering suggests a gene with photoperiod-specific expression or action. Second, since DD plants flower later than WW plants, the domesticated allele likely contains a loss of function or dominant negative mutation.
To evaluate whether HaFT1 or HaFT2 fit these criteria, we compared their expression and cDNA sequence in the parental genotypes and NIL plants. In the wild parent, expression of both candidates was upregulated by the inductive photoperiod, short days (Figure 4C and 4E). In the domesticated line, the inductive photoperiod (long days) also upregulated HaFT2 but HaFT1 was expressed in both photoperiods, and similar patterns were observed for DD, DW, and WW plants (Figures 4C and 4F, Figure S3C). Thus, regulatory changes responsible for expression differences of these genes between the parents mostly act in trans-. There was one notable difference between the genotypes: HaFT2 peak abundance was 3–4 fold higher in DD plants than in DW or WW plants, consistent with a 4–5 fold increase in the domesticated parent relative wild parent (Figures 4E and 4F) and indicative of a cis-regulatory difference affecting HaFT2 peak abundance. This change is unlikely to underlie the QTL effect, however. Increased HaFT2 expression in DD plants should accelerate flowering, but these plants flowered later than WW plants. DD, DW, and WW plants differed in relative abundance of the longer HaFT1 splice form; however, these differences were not photoperiod-specific and thus also unlikely to cause the observed phenotypic differences.
We did find a coding sequence difference that could explain the LG6 QTL effect. While the wild and domesticated HaFT2 amino acid sequences were identical, the domesticated allele of HaFT1 differed from the wild allele by a frameshift mutation (TG -> C) in the third exon (Figure 2A). The frameshift does not create a premature stop codon, but rather leads to a protein 17 amino acids longer than wild-type that is half novel sequence (Figure S1). No premature stop mutations are present in the long splice form of this allele, meaning differential nonsense mediated decay could explain expression variation of this form in the NILs.
The frameshift mutation in the domesticated allele is consistent with the observation that DD plants flowered later than WW plants, but it is more difficult to explain the QTL’s photoperiod-specific effect on phenotype since HaFT1 was expressed in both photoperiods in all NIL genotypes. If HaFT1 affects flowering whenever expressed and the frameshift is a simple loss of function, the effect of the QTL on flowering would be expected in both photoperiods. Alternatively, if the frameshift mutation has a dominant negative effect, then it could have photoperiod-specific action through interference with the function of other FT paralogs. Since HaFT2 and HaFT4 were expressed only in long days in the CMSHA89 background (Figure S3), dominant negative action of HaFT1 would be photoperiod-specific.
To determine whether the domesticated and wild HaFT1 alleles are functionally distinct, we took a heterologous complementation approach. Full-length cDNAs of the in-frame wild allele and frameshift-carrying domesticated allele were overexpressed in Columbia-0 and ft mutant A. thaliana backgrounds. The two constructs differed only by the frameshift mutation.
While overexpression of the in-frame wild allele accelerated flowering in both backgrounds, overexpression of the frameshift-carrying domesticated allele did not (Figure 3A, Table S4). Indeed, ft transformants carrying the frameshift allele were slightly delayed in flowering relative to untransformed ft mutants (Figure 3B), though a similar delay was not observed in the wild-type background nor evident from leaf counts (Table S4). These results indicate that the frameshift mutation functionally alters the HaFT1 protein and provided us a preliminary indication that it may not be a simple loss of function.
To test whether the HaFT1 frameshift could cause a dominant negative effect with photoperiod-specific action by interfering with other HaFT paralogs, we crossed ft plants overexpressing the HaFT1 frameshift allele to ft plants overexpressing either HaFT2 or HaFT4. We predicted that if the frameshift allele interferes with HaFT2 or HaFT4 function, then it should suppress the complementation of late flowering in ft mutants by these paralogs. Transgenic ft mutants overexpressing the in-frame allele of HaFT1 were also crossed to plants overexpressing HaFT2 or HaFT4 for comparison. The genotypes of cross progeny and transgene overexpression were confirmed by RT-PCR (Figure S2D).
When overexpressed in either the ft HaFT2 or ft HaFT4 background, the wild allele further accelerated flowering and reduced leaf number relative to plants without this transgene (Figure 3B and 3C, Table S4). This is consistent with its expected effects and demonstrates that transgenics carrying multiple HaFT transgenes do not exhibit any generalized cosuppression. In the ft HaFT2 background, overexpressing the HaFT1 frameshift allele did not alter flowering time. In contrast, overexpressing this allele in the ft HaFT4 background significantly delayed flowering, fully suppressing complementation of ft by HaFT4 (Figure 3B and 3C, Table S4).
Thus, the photoperiod-specific effect of the LG6 QTL region on NIL phenotypes may be mechanistically explained by dominant negative action of the HaFT1 frameshift mutation. Though HaFT1 is expression is not photoperiod-specific, HaFT4 is only expressed under long-day conditions (Figure S3). Therefore, dominant negative interference of the domesticated allele of HaFT1 with HaFT4 and a consequent delay in flowering only occurs in the inductive photoperiod. Whether the long form of the domesticated allele shares this functionality is an uninvestigated though intriguing possibility. Several aspects of HaFT paralog expression, transport, and redundancy in sunflower were not recapitulated in our experiments, and the eventual development of sunflower transformants will be required confirm our findings. One potential mechanism for the dominant negative interaction between the domesticated allele of HaFT1 and HaFT4 could be interference with binding of HaFT4 to shoot apex proteins required for floral induction. Similar interference mechanisms have been proposed to explain the effects of chimeric FT/TFL1 constructs or TFL1 on flowering also based on transgenic studies [33, 38].
Selection During Early Domestication
We examined the historical importance of the HaFT1 frameshift mutation during domestication by sequencing a 711bp region containing the third exon and surrounding intron sequence from a diversity panel of elite-bred, Native American landrace, and wild H. annuus (Figure 5, Table S2). Nearly all elite (36/36) and landrace (36/38) alleles contained the frameshift mutation while nearly all wild alleles (44/46) were in-frame. This dramatic allele frequency change between wild populations (q = 0.05) and early domesticates (q = 0.95) strongly suggests selection acting during early domestication. Of the four exceptions, the two landraces heterozygous for the in-frame allele may result from recent admixture between domesticated and wild H. annuus or an incomplete selective sweep. Both wild lines segregating for the frameshift allele were from Oklahoma. These may indicate recent admixture, but could also represent the allele’s area of origin.
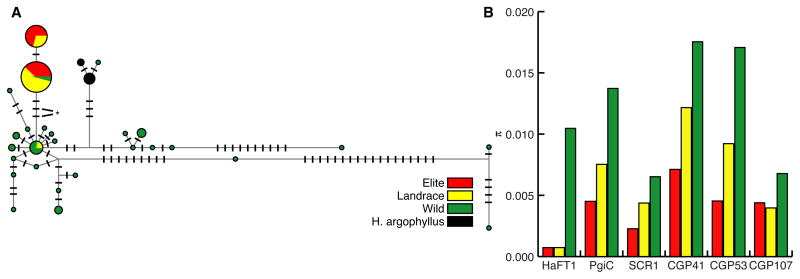
Figure 5.
HaFT1 Frameshift Distinguishes Wild From Domesticated Accessions and Experienced a Selective Sweep During Domestication. (A) Median joining haplotype network constructed from 711bp region of HaFT1 sequenced from elite-bred (red), landrace (yellow), and wild (green) H. annuus and wild H. argophyllus (black). TG→C frameshift mutation (*) and a noncoding SNP define a branch separating nearly all domesticated lines from nearly all wild accessions. The number of hatchmarks on a branch indicates the number of substitutions. (B) Average pairwise nucleotide diversity (π) for HaFT1 and five putative neutral loci in samples of elite-bred, Native American landrace, and wild H. annuus. See also Table S2.
Other aspects of HaFT1 sequence diversity support these conclusions (Figure 5A). A substitution in the second intron was in complete linkage disequilibrium with the frameshift. A single nucleotide polymorphism unique to domesticated lines was also found, suggesting the sweep occurred sufficiently long ago and the allele remained sufficiently isolated from wild germplasm that new variation has accumulated without passing into wild plants by gene flow.
We tested whether HaFT1 shows a signature of a selective sweep using a maximum-likelihood adaptation of the Hudson-Kreitman-Aguadé (MLHKA) test [39]. The test compares the likelihood of a strictly neutral model to a model where a candidate gene is under selection. We sequenced portions of five putatively neutral control loci on the same diversity panel. A separate multi-locus HKA test verified that these genes did not depart from neutrality. MLHKA tests were then conducted for elite, landrace, and wild datasets separately to determine the timing of selection. These tests found that HaFT1 was evolving neutrally in wild H. annuus populations (p = 0.281) but under selection in landrace (p = 0.048) and elite lines (p = 0.015). Reduction of HaFT1 average pairwise nucleotide diversity (π) with domestication was much greater than reductions in neutral gene nucleotide diversity with domestication, consistent with this result (Figure 5B).
Linkage disequilibrium decays rapidly in domesticated sunflower (r2=0.32 at 5kb, r2=0.1 at ~100kb [40]). As neither HaFT2 nor HaFT3 is present in BAC sequence 60 kb upstream or 51 kb downstream of HaFT1, it is unlikely this sweep affected all three paralogs. Indeed, MLHKA analysis of HaFT3 sequences we obtained revealed no signature of selection during early domestication (p = 0.095).
Conclusions
Our results demonstrate that multiple recently evolved FT-like duplicates have met a variety of fates and have played diverse roles in flowering time divergence between wild and domesticated sunflower. Expression divergence in one paralog, HaFT2, caused by both cis- and trans-regulatory effects, is associated with a shift to earlier, long-day responsive flowering. A frameshift mutation in a second paralog, HaFT1, maps to a major flowering QTL, and affects developmental timing through interference with the function of a third paralog, HaFT4. Furthermore, comparison of sequence diversity to neutrally evolving loci revealed a signature of selection on HaFT1 during early domestication.
Together, to our knowledge, our findings provide the first functional and population genetic evidence identifying an early domestication gene in sunflower. Like most previously identified early domestication genes, HaFT1 is involved in transcriptional regulation and the domesticated allele is not a simple null [41]. Retention of the frameshift allele through modern breeding despite selection for earlier, more synchronous flowering likely occurred due to absence of genetic variation in modern crop progenitors and possibly also favorable pleiotropic effects. Selection on flowering time during modern breeding must have acted at other loci, perhaps including HaFT2 where we located cis-regulatory differences predicted to promote early flowering.
Our findings also illustrate how gene duplication may foster evolutionary change by creating an opportunity for new gene-gene interactions within gene families to evolve and produce natural variation. As many transcriptional regulators and signaling molecules participate in gene complexes, often as homo- or heterodimers, we speculate that origin and modulation of within-gene family interactions made possible by gene family expansion is an important contributor to biological diversity [42].
Supplementary Material
Acknowledgments
We thank Z. Lai, A. Posto, K. Turner, D. Rasmussen, L. Washington, T. Tsui, S. Tang, Indiana University Greenhouse staff, and several undergraduate volunteers for technical assistance; D. Wills, J. Burke, A. Heesacker, and S. Knapp for providing mapping panel DNAs; and L. Moyle, M. Hahn, and the Rieseberg, Michaels, and Moyle labs for scientific input. This work was supported by NIH (GM059065) and NSF (DBI0421630 and DBI0820451) grants to LHR, a NIH Ruth L. Kirschstein Postdoctoral Fellowship (5F32GM072409-02) to JLS, and a NSF Doctoral Dissertation Improvement Grant to BKB (DEB0608118). GenBank accession numbers of deposited sequences: GQ884199–GQ885119 (Table S6).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Ohno S. Evolution by Gene Duplication. Berlin: Springer-Verlag; 1970. [Google Scholar]
- 2.Conant GC, Wolfe KH. Turning a hobby into a job: How duplicated genes find new functions. Nat Rev Genet. 2008;9:938–950. doi: 10.1038/nrg2482. [DOI] [PubMed] [Google Scholar]
- 3.Kroymann J, Donnerhacke S, Schnabelrauch D, Mitchell-Olds T. Evolutionary dynamics of an Arabidopsis insect resistance quantitative trait locus. Proceedings of the National Academy of Sciences of the United States of America. 2003;100:14587–14592. doi: 10.1073/pnas.1734046100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Xiao H, Jiang N, Schaffner E, Stockinger EJ, van der Knaap E. A Retrotransposon-Mediated Gene Duplication Underlies Morphological Variation of Tomato Fruit. Science. 2008;319:1527–1530. doi: 10.1126/science.1153040. [DOI] [PubMed] [Google Scholar]
- 5.Parker HG, VonHoldt BM, Quignon P, Margulies EH, Shao S, Mosher DS, Spady TC, Elkahloun A, Cargill M, Jones PG, et al. An Expressed Fgf4 Retrogene Is Associated with Breed-Defining Chondrodysplasia in Domestic Dogs. Science. 2009;325:995–998. doi: 10.1126/science.1173275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kim M, Cui ML, Cubas P, Gillies A, Lee K, Chapman MA, Abbott R, Coen E. Regulatory genes control a key morphological and ecological trait transferred between species. Science. 2008;322:1116–1119. doi: 10.1126/science.1164371. [DOI] [PubMed] [Google Scholar]
- 7.Hecht V, Foucher F, Ferrándiz C, Macknight R, Navarro C, Morin J, Vardy ME, Ellis N, Beltrán JP, Rameau C, et al. Conservation of Arabidopsis flowering genes in model legumes. Plant Physiol. 2005;137:1420–1434. doi: 10.1104/pp.104.057018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Danilevskaya ON, Meng X, Hou Z, Ananiev EV, Simmons CR. A genomic and expression compendium of the expanded PEBP gene family from maize. Plant Physiol. 2008;146:250–264. doi: 10.1104/pp.107.109538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hayama R, Agashe B, Luley E, King R, Coupland G. A circadian rhythm set by dusk determines the expression of FT homologs and the short-day photoperiodic flowering response in Pharbitis. Plant Cell. 2007;19:2988–3000. doi: 10.1105/tpc.107.052480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bohlenius H, Huang T, Charbonnel-Campaa L, Brunner AM, Jansson S, Strauss SH, Nilsson O. CO/FT regulatory module controls timing of flowering and seasonal growth cessation in trees. Science. 2006;312:1040–1043. doi: 10.1126/science.1126038. [DOI] [PubMed] [Google Scholar]
- 11.Kikuchi R, Kawahigashi H, Ando T, Tonooka T, Handa H. Molecular and Functional Characterization of PEBP Genes in Barley Reveal the Diversification of Their Roles in Flowering. Plant Physiol. 2009;149:1341–1353. doi: 10.1104/pp.108.132134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lin MK, Belanger H, Lee YJ, Varkonyi-Gasic E, Taoka K, Miura E, Xoconostle-Cázares B, Gendler K, Jorgensen RA, Phinney B, et al. FLOWERING LOCUS T protein may act as the long-distance florigenic signal in the cucurbits. Plant Cell. 2007;19:1488–1506. doi: 10.1105/tpc.107.051920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Komiya R, Yokoi S, Shimamoto K. A gene network for long-day flowering activates RFT1 encoding a mobile flowering signal in rice. Development. 2009:dev.040170. doi: 10.1242/dev.040170. [DOI] [PubMed] [Google Scholar]
- 14.Kardailsky I, Shukla VK, Ahn JH, Dagenais N, Christensen SK, Nguyen JT, Chory J, Harrison MJ, Weigel D. Activation tagging of the floral inducer FT. Science. 1999;286:1962–1965. doi: 10.1126/science.286.5446.1962. [DOI] [PubMed] [Google Scholar]
- 15.Kobayashi Y, Kaya H, Goto K, Iwabuchi M, Araki T. A Pair of Related Genes with Antagonistic Roles in Mediating Flowering Signals. Science. 1999;286:1960–1962. doi: 10.1126/science.286.5446.1960. [DOI] [PubMed] [Google Scholar]
- 16.Hayama R, Yokoi S, Tamaki S, Yano M, Shimamoto K. Adaptation of photoperiodic control pathways produces short-day flowering in rice. Nature. 2003;422:719–722. doi: 10.1038/nature01549. [DOI] [PubMed] [Google Scholar]
- 17.Yan L, Fu D, Li C, Blechl A, Tranquilli G, Bonafede M, Sanchez A, Valarik M, Yasuda S, Dubcovsky J. The wheat and barley vernalization gene VRN3 is an orthologue of FT. Proc Natl Acad Sci USA. 2006;103:19581–19586. doi: 10.1073/pnas.0607142103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Suárez-López P, Wheatley K, Robson F, Onouchi H, Valverde F, Coupland G. CONSTANS mediates between the circadian clock and the control of flowering in Arabidopsis. Nature. 2001;410:1116–1120. doi: 10.1038/35074138. [DOI] [PubMed] [Google Scholar]
- 19.Valverde F, Mouradov A, Soppe W, Ravenscroft D, Samach A, Coupland G. Photoreceptor regulation of CONSTANS protein in photoperiodic flowering. Science. 2004;303:1003–1006. doi: 10.1126/science.1091761. [DOI] [PubMed] [Google Scholar]
- 20.Sawa M, Nusinow DA, Kay S, Imaizumi T. FKF1 and GIGANTEA complex formation is required for day-length measurement in Arabidopsis. Science. 2007;318:261–265. doi: 10.1126/science.1146994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Corbesier L, Vincent C, Jang S, Fornara F, Fan Q, Searle I, Giakountis A, Farrona S, Gissot L, Turnbull C, et al. FT protein movement contributes to long-distance signaling in floral induction of Arabidopsis. Science. 2007;316:1030–1033. doi: 10.1126/science.1141752. [DOI] [PubMed] [Google Scholar]
- 22.Tamaki S, Matsuo S, Wong HL, Yokoi S, Shimamoto K. Hd3a protein is a mobile flowering signal in rice. Science. 2007;316:1033–1036. doi: 10.1126/science.1141753. [DOI] [PubMed] [Google Scholar]
- 23.Pnueli L, Gutfinger T, Hareven D, Ben-Naim O, Ron N, Adir N, Lifschitz E. Tomato SP-interacting proteins define a conserved signaling system that regulates shoot architecture and flowering. Plant Cell. 2001;13:2687–2702. doi: 10.1105/tpc.010293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Abe M, Kobayashi Y, Yamamoto S, Daimon Y, Yamaguchi A, Ikeda Y, Ichinoki H, Notaguchi M, Goto K, Araki T. FD, a bZIP protein mediating signals from the floral pathway integrator FT at the shoot apex. Science. 2005;309:1052–1056. doi: 10.1126/science.1115983. [DOI] [PubMed] [Google Scholar]
- 25.Wigge PA, Kim MC, Jaeger KE, Busch W, Schmid M, Lohmann JU, Weigel D. Integration of spatial and temporal information during floral induction in Arabidopsis. Science. 2005;309:1056–1059. doi: 10.1126/science.1114358. [DOI] [PubMed] [Google Scholar]
- 26.Yamaguchi A, Kobayashi Y, Goto K, Abe M, Araki T. TWIN SISTER OF FT (TSF) Acts as a Floral Pathway Integrator Redundantly with FT. Plant Cell Physiol. 2005;46:1175–1189. doi: 10.1093/pcp/pci151. [DOI] [PubMed] [Google Scholar]
- 27.Igasaki T, Watanabe Y, Nishiguchi M, Kotoda N. The FLOWERING LOCUS T/TERMINAL FLOWER 1 family in Lombardy poplar. Plant Cell Physiol. 2008;49:291–300. doi: 10.1093/pcp/pcn010. [DOI] [PubMed] [Google Scholar]
- 28.Takahashi Y, Teshima KM, Yokoi S, Innan H, Shimamoto K. Variations in Hd1 proteins, Hd3a promoters, and Ehd1 expression levels contribute to diversity of flowering time in cultivated rice. Proc Natl Acad Sci USA. 2009 doi: 10.1073/pnas.0812092106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hagiwara WE, Uwatoko N, Sasaki A, Matsubara K, Nagano H, Onishi K, Sano Y. Diversification in flowering time due to tandem FT-like gene duplication, generating novel Mendelian factors in wild and cultivated rice. Molecular Ecology. 2009;18:1537–1549. doi: 10.1111/j.1365-294X.2009.04119.x. [DOI] [PubMed] [Google Scholar]
- 30.Bonnin I, Rousset M, Madur D, Sourdille P, Dupuits C, Brunel D, Goldringer I. FT genome A and D polymorphisms are associated with the variation of earliness components in hexaploid wheat. Theor Appl Genet. 2008;116:383–394. doi: 10.1007/s00122-007-0676-0. [DOI] [PubMed] [Google Scholar]
- 31.Schwartz C, Balasubramanian S, Warthmann N, Michael T, Lempe J, Sureshkumar S, Maloof J, Borevitz J, Chory J, Weigel D. Cis-regulatory changes at FLOWERING LOCUS T mediate natural variation in flowering responses of Arabidopsis thaliana. 2009 doi: 10.1534/genetics.109.104984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hanzawa Y, Money T, Bradley D. A single amino acid converts a repressor to an activator of flowering. Proc Natl Acad Sci USA. 2005;102:7748–7753. doi: 10.1073/pnas.0500932102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ahn J, Miller D, Winter VJ, Banfield MJ, Lee JH, Yoo SY, Henz S, Brady RL, Weigel D. A divergent external loop confers antagonistic activity on floral regulators FT and TFL1. EMBO J. 2006;25:605–614. doi: 10.1038/sj.emboj.7600950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Barker MS, Kane NC, Matvienko M, Kozik A, Michelmore RW, Knapp SJ, Rieseberg LH. Multiple paleopolyploidizations during the evolution of the Compositae reveal parallel patterns of duplicate gene retention after millions of years. Mol Biol Evol. 2008;25:2445–2455. doi: 10.1093/molbev/msn187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Burke JM, Tang S, Knapp SJ, Rieseberg LH. Genetic analysis of sunflower domestication. Genetics. 2002;161:1257–1267. doi: 10.1093/genetics/161.3.1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Baack EJ, Sapir Y, Chapman MA, Burke JM, Rieseberg LH. Selection on domestication traits and quantitative trait loci in crop-wild sunflower hybrids. Mol Ecol. 2008;17:666–677. doi: 10.1111/j.1365-294X.2007.03596.x. [DOI] [PubMed] [Google Scholar]
- 37.Wills DM, Burke JM. Quantitative trait locus analysis of the early domestication of sunflower. Genetics. 2007;176:2589–2599. doi: 10.1534/genetics.107.075333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shalit A, Rozman A, Goldshmidt A, Alvarez JP, Bowman JL, Eshed Y, Lifschitz E. The flowering hormone florigen functions as a general systemic regulator of growth and termination. Proceedings of the National Academy of Sciences. 2009;106:8392–8397. doi: 10.1073/pnas.0810810106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wright SI, Charlesworth B. The HKA test revisited: a maximum-likelihood-ratio test of the standard neutral model. Genetics. 2004;168:1071–1076. doi: 10.1534/genetics.104.026500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fusari C, Lia V, Hopp HE, Heinz R, Paniego N. Identification of Single Nucleotide Polymorphisms and analysis of Linkage Disequilibrium in sunflower elite inbred lines using the candidate gene approach. BMC Plant Biology. 2008;8:7. doi: 10.1186/1471-2229-8-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Doebley J, Gaut BS, Smith BD. The molecular genetics of crop domestication. Cell. 2006;127:1309–1321. doi: 10.1016/j.cell.2006.12.006. [DOI] [PubMed] [Google Scholar]
- 42.Pereira-Leal J, Levy E, Kamp C, Teichmann S. Evolution of protein complexes by duplication of homomeric interactions. Genome Biology. 2007;8:R51. doi: 10.1186/gb-2007-8-4-r51. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.