Abstract
The human pathogen Staphylococcus aureus secretes several complement evasion molecules to combat the human immune response. Extracellular complement-binding protein (Ecb) binds to the C3d domain of C3 and thereby blocks C3 convertases of the alternative pathway and C5 convertases via all complement pathways. Inhibition of C5 convertases results in complete inhibition of C5a generation and subsequent neutrophil migration. Here, we show that binding of Ecb to the C3d domain of C3b is crucial for inhibition of C5 convertases. Ecb does not interfere with substrate binding to convertases but prevents formation of an active convertase enzyme.
Keywords: Bacteria, Complement, Convertases, Inflammation, Innate Immunity, Evasion
Introduction
Staphylococcus aureus is a Gram-positive bacterium and an important human pathogen that causes community- and hospital-derived infections. Infections can range from mild wound infections and light food poisoning to severe diseases such as bacteremia and endocarditis (1). S. aureus permanently colonizes 20% of the population, and another 60% are intermediate carriers (2). Bacteria need to be able to survive in the human host to become a pathogen. Therefore, they express adhesion factors, adapt to intracellular milieus, or make a thick capsule (3). Next to these mechanisms, S. aureus secretes several small molecules that effectively inhibit the innate immune system, and a vast majority appear to have evolved to specifically avoid complement attack (3–5). Inhibition of the complement system is very important because its activation leads to coverage of bacteria with C3b, resulting in phagocytosis, and formation of C5a, which is important for chemotaxis of neutrophils toward the site of infection. The complement system comprises three activation routes: the classical pathway (CP),2 the lectin pathway (LP), and the alternative pathway (AP). All activation routes converge in the formation of C3 convertases that cleave C3 into C3a and C3b. Next to its importance in phagocytosis, C3b deposition is essential to the downstream formation of C5 convertases that cleave C5 into the strong chemoattractant C5a and C5b, which is essential to the formation of the membrane attack complex (6–10).
Recently, we described the complement inhibitory properties of extracellular fibrinogen-binding protein (Efb) and extracellular complement-binding protein (Ecb) (11) (also known as Ehp) (12). Although Efb originally was described as a fibrinogen-binding protein (13, 14), further study has revealed a modular nature to this molecule: the N terminus of Efb is responsible for fibrinogen binding, but the C terminus contributes to complement inhibition by binding the C3d domain of C3, C3b, iC3b, and C3dg (11, 15, 16). Ecb is a homologue of the C-terminal domain of Efb; although it lacks the ability to bind fibrinogen, it binds with greater affinity to the C3d domain of C3 (11, 12). Efb and Ecb specifically bind to C3b-containing convertases, which are the C3 convertase of the AP and the C5 convertases of all complement pathways. They both inhibit C3b deposition via the AP, but most importantly they strongly inhibit C5a generation via all pathways, which was demonstrated in vivo as well (11). Although Efb and Ecb completely inhibit C5a generation, the molecular mechanism behind C5 convertase inhibition is not well understood. Because Ecb is the most effective inhibitor of C5a generation and because it exclusively inhibits the complement system as it cannot bind to fibrinogen (11, 12), we used this protein and an inactive, site-directed mutant in Ecb to study the mechanism of action on C5 convertases in greater detail.
EXPERIMENTAL PROCEDURES
Protein Expression and Purification
Preparation of Ecb (9 kDa) and Ecb N63E/R75E/N82E was described previously. Ecb was expressed with an N-terminal cleavable His tag to be able to use Ecb with or without the His tag. The His tag was removed by Enterokinase cleavage. Ecb N63E/R75E/N82E was prepared only in the form where its His tag had been proteolytically removed (11, 12). C3 and fB were purified from freshly isolated human plasma and C3b was generated as described (17). Human IgG was isolated from pooled serum of healthy volunteers via protein G purification according to the manufacturer's protocol (GE Healthcare). C1q, C2, C4, C5, properdin, and fD were obtained commercially (Calbiochem). Recombinant CRIg-S, the short form of CRIg which binds C3b and iC3b, was obtained from Genentech, Inc.
C5a Generation via CP/LP and AP
For C5a analyses, heat-killed S. aureus (to prevent formylmethionylleucylphenylalanine production) were incubated with 10% human serum in HBS2+ (Hepes-buffered saline: 20 mm Hepes, 140 mm NaCl, 5 mm CaCl2, and 2.5 mm MgCl2) for 30 min at 37 °C in the presence of Ecb or Ecb N63E/R75E/N82E. Subsequently, collected supernatants were tested for the presence of C5a. Freshly isolated neutrophils were incubated with the supernatant, and calcium mobilization was measured as described (18). Exclusive activation of the CP/LP was performed in fD-deficient serum (19), whereas activation of the AP was analyzed in human serum in HBS-MgEGTA (Hepes-buffered saline, 20 mm Hepes, 140 mm NaCl, 5 mm MgCl2, and 10 mm EGTA).
CP Activation on S. aureus with Purified Proteins
S. aureus strain Wood 46 was cultured overnight on blood agar plates (BD Pharmingen) and resuspended in HBS2+ with 0.1% BSA. To obtain C4b-coated bacteria, 50 μl of 1 × 108/ml bacteria were incubated with IgG (50 μl at 100 μg/ml, 30 min at 37 °C), human C1 (50 μl at 30 μg/ml, 40 min at 4 °C), and human C4 (100 μl at 30 μg/ml, 40 min at 37 °C). Bacteria were washed between every step with HBS2+ with 0.1% BSA. Subsequently, the bacteria were incubated with 50 μl of C2 (5 μg/ml), several concentrations of C3 in the presence or absence of His-Ecb or His-chemotaxis inhibitory protein of S. aureus (CHIPS) (both at 10 μg/ml) for 30 min at 37 °C. C3b deposition was detected using fluorescein isothiocyanate-conjugated (Fab′)2 anti-human C3 antibodies (Protos Immunoresearch, San Francisco) and flow cytometry. Binding of His-Ecb and His-CHIPS was detected using mouse anti-His tag antibodies (Qiagen) followed by phycoerythrin-labeled goat anti-mouse IgG (DakoCytomation) and flow cytometry. To determine C5a formation by the CP convertase, C4b-coated bacteria were incubated with C2, C3, and C5 in the presence or absence of Ecb (10 μg/ml) for 30 min at 37 °C, and supernatants were collected by centrifugation. Collected supernatants were tested for calcium mobilization with U937-C5aR cells (as above).
AP Activation on S. aureus with Purified Components
Fifty μl of S. aureus strain Wood 46 (1 × 108/ml) was opsonized with 1% human serum in HBS-MgEGTA with 0.1% BSA for 30 min at 37 °C. To decay surface-bound convertases, bacteria were washed, taken up in 500 μl of PBS and incubated for 1 h at 37 °C. Preopsonized bacteria were then incubated in 50 μl of HBS-MgEGTA containing fB (10 μg/ml), fD (1 μg/ml), and several concentrations of C3 in the presence or absence of His-Ecb or His-CHIPS (both at 10 μg/ml) for 30 min at 37 °C. C3b deposition was detected by fluorescein isothiocyanate-conjugated F(ab′)2 anti-human C3 antibodies. To investigate C5a generation, preopsonized bacteria were incubated with 50 μl pf HBS-MgEGTA containing fB (10 μg/ml), fD (1 μg/ml), C3 (25 μg/ml), and several concentrations of C5 in the presence or absence of Ecb (10 μg/ml) for 30 min at 37 °C. Then, supernatants were collected by centrifugation and were tested for C5a calcium mobilization with U937-C5aR cells.
C5 Binding Enzyme-linked Immunosorbent Assays
Binding of (C3b)2 to C5 was studied as described previously (20) with minor modifications. Microtiter plates were coated with 3 μg/ml anti-C5a antibody clone 2077 (Hycult Biotechnology) and blocked with PBS with 4% BSA and 0.1% Tween. Between every step, plates were washed with PBS containing 0.05% Tween. Subsequently, C5 (30, 10, and 3 μg/ml) was added after washing. Incubation with (C3b)2 followed in the presence of absence of Ecb or CRIg (both at 10 μg/ml). Binding of (C3b)2 to C5 was detected using peroxidase-labeled anti-C3 antibodies (Protos Immunoresearch). To determine C5 binding to convertases formed on microtiter plates, complement was activated as described with minor modifications (21, 22). Microtiter plates were coated with IgM (CP) or lipopolysaccharide (AP) and blocked with PBS containing 4% BSA and 0.1% Tween 20. Formation of convertases was achieved by addition of human serum for 1 h at 37 °C followed by another round of serum incubation for 1 h at 37 °C. After washing, convertases were decayed for 1 h at 37 °C. Purified C5 was added for 1 h at 37 °C in the presence or absence of Ecb or CRIg. C5 binding was detected using an anti-C5a antibody clone 2077 followed by peroxidase-conjugated goat anti-mouse IgG (Southern Biotechnology Associates, Birmingham, AL).
Conversion of fB
Conversion of fB into Bb was analyzed by incubation of 50 μl of zymosan (1 mg/ml; Sigma) with 3% human serum in HBS2+ and 0.1% BSA. To decay surface-bound convertases, zymosan was washed and incubated for 1 h at 37 °C in PBS. Washed zymosan was resuspended in 50 μl of fB (20 μg/ml) and fD (2 μg/ml) with or without Ecb in HBS2+ for 30 min at 37 °C. Washed pellets were boiled in SDS sample buffer, and surface-bound Bb was analyzed by immunoblotting using anti-fB polyclonal goat antibody (Quidel) and peroxidase-conjugated donkey anti-goat IgG (Santa Cruz Biotechnology). To study fB stabilization on the surface, opsonized zymosan was incubated with Ecb (10 μg/ml) during the decay period of 1 h, and samples were analyzed by Western blotting right after decay. To analyze fluid phase conversion of fB into Bb, C3b (10 μg/ml) was incubated with fB (5 μg/ml) and fD (0.5 μg/ml) in the presence or absence of Ecb or Ecb N63E/R75E/N82E (both at 10 μg/ml) in HBS2+ for 5 min at 37 °C. Conversion of fB into Bb was analyzed by immunoblotting as described above.
Stabilization of fB via Biacore
Interactions between fB and C3b were analyzed by surface plasmon resonance as described previously (23). C3b (±9000 RU) was covalently attached to the surface of a CM5 biosensor chip (GE Healthcare) in 10 mm NaAC, pH 4.5 (using standard amine coupling). These experiments were performed in HBS-P (0.01 m Hepes, 0.15 m NaCl, 0.005% surfactant P20, pH 7.4; GE Healthcare) at a flow rate of 30 μl/ml. First Ecb (10 μg/ml, ± 5 s) was injected to bind (±500 RU), then fB (50 μg/ml) was injected across the surface for 2 min followed by dissociation of 2 min. To study the impact of Ecb on formation of active convertases, we covalently linked C3b (±10,000 RU) on a CM5 chip as previously described (24). In short, 1000 RU of C3b was coupled to a CM5 chip via amine coupling and subsequently, 50 μl of fB (50 μg/ml) and fD (5 μg/ml) was injected followed by 100 μl of C3 (100 μg/ml) in Hepes-Ni2+ (Hepes-buffered saline, 20 mm Hepes, 140 nm NaCl, 1 mm NiCl) at a flow rate of 20 μl/min. These cycles were repeated until the appropriate amount (10,000 RU) of C3b was covalently attached to the surface. Ecb was bound to one flow path as described above. Then, 100 μl of fB (50 μg/ml) and fD 5 (μg/ml) was injected. After 600 s, C3 (50 μl, 100 μg/ml) was injected across the surface followed by injection of 50 μl of 0.1 m citrate, 1 m NaCl, 1 mm EDTA, pH 5, to remove noncovalently bound C3/C3b.
RESULTS
Ecb Binding to C3d Domain of C3b Is Essential for C5 Convertase Inhibition
C5 convertases are generated by the binding of C3b to preexisting C3 convertases resulting in the CP/LP C5 convertase (C4b2aC3b) and the AP C5 convertase C3b2Bb (7, 8). We previously observed that Ecb potently blocks C5 convertases. Because Ecb is also known to bind C3b, we wondered whether C3b binding is crucial to C5 convertase inhibition. The binding of Ecb to C3b is well defined: amino acids Asn63, Arg75, and Asn82 of Ecb bind to the C3d domain of C3b (12). Subsequently, a C3d-binding mutant of Ecb lacking these residues (Ecb N63E/R75E/N82E) does not bind C3b. We used this mutant to study the importance of this interaction in C5 convertase inhibition. We incubated bacteria with human serum in the presence of Ecb or Ecb N63E/R75E/N82E and analyzed C5a generation via calcium mobilization of U937-C5aR. Fig. 1 shows that Ecb N63E/R75E/N82E is unable to block C5a generation via the CP/LP (A) and the AP (B). Thus, binding of Ecb to the C3d domain of C3b is essential for inhibition of C5 convertases.
FIGURE 1.
Ecb binding to the C3d domain of C3b is essential for C5 convertase inhibition. C5a generation in the presence of Ecb or its C3d-binding mutant is shown. Bacteria were incubated with human serum, and C5a activity in supernatants was analyzed by calcium mobilization of neutrophils. A, C5a generation via the CP/LP in ΔfD serum. B, C5a generation via the AP in human serum in the presence of MgEGTA. Both panels represent the mean ± S.E. (error bars) of three separate experiments.
Ecb Binds to C3b on Bacterial Surface and Specifically Inhibits C3b-containing Convertases
The data above suggest that Ecb inhibits C5 convertases through its interaction with C3b. Recently, we showed that Ecb inhibits C3 convertases of the AP and the C5 convertases of all pathways. Because these experiments were performed in a serum environment we could only speculate how Ecb may block these convertases. To pinpoint the exact mechanism of action, we now studied the inhibitory properties of Ecb in a purified system. Formation of C3 and C5 convertases is a multistep process, and to determine exactly at which stage Ecb binds and blocks convertases we generated surface-bound convertases with purified components and added Ecb at every single stage to look for inhibition of C3b deposition or C5a generation. To generate CP convertases, bacteria were first opsonized with purified human IgG and subsequently incubated with C1. These particles were then incubated with C4 to enable C1-mediated activation of C4 into C4b, which covalently binds to the bacterial surface. The C4b-coated bacteria were incubated with C2 and C3 resulting in formation of C3 convertases (C4b2a) that cleave C3 into C3a and C3b. When C4b-coated particles were incubated with C2 and C3 in the presence of His-Ecb and His-CHIPS (as a negative control) (18) we observed that both proteins failed to inhibit C3b deposition on the bacterial surface (Fig. 2A). However, we observed a C3-dependent binding of His-Ecb to the surface, indicating that Ecb binds to bacterium-bound C3b molecules during opsonization (Fig. 2B). Addition of His-Ecb during incubation of bacteria with IgG, C1, C4, or C2 alone did not result in inhibition of C3b deposition or detectable Ecb binding to the bacterial surface (data not shown). Finally, we studied the effect of Ecb on the CP C5 convertase by incubating C4b-coated particles with C2, C3, and C5 in the presence of Ecb and measuring C5a generation in supernatants by calcium mobilization. Fig. 2C shows that Ecb blocks C5a generation by purified CP C5 convertases.
FIGURE 2.
Ecb binds to C3b on the bacterial surface and inhibits C3b-containing convertases. A, C4b-coated bacteria were opsonized with C2 and C3 in the presence or absence of His-Ecb or His-CHIPS (both at 10 μg/ml). B, protein binding to the bacterial surface and C3b deposition on the bacterial surface was measured. C3-dependent binding of His-Ecb (10 μg/ml) is shown. C, C5a generation via the CP is shown. C4b-coated bacteria were incubated with C2, C3, and C5 in the presence or absence of Ecb. C5a generation was analyzed by calcium mobilization using U937-C5aR cells. D, C3b deposition via the AP is shown. Preopsonized bacteria were incubated with fB, fD, and C3 in the presence or absence of Ecb (10 μg/ml). E, C5a generation via the AP is shown. Opsonized bacteria were incubated with fB, fD, C3, and C5 in the presence of Ecb (10 μg/ml). C5a generation in the supernatant was tested by calcium mobilization. A, B, D, and E represent the mean ± S.E. (error bars) of three separate experiments. C is a representative of three separate experiments.
To generate AP convertases we first opsonized bacteria with human serum. After decay of 1 h, preopsonized bacteria were incubated with fB, fD, and C3, which resulted in formation of AP convertases on the bacterial surface (C3bBb and C3b2Bb). When C3b-coated bacteria were incubated with fB, fD, and C3 in the presence of Ecb or CHIPS we observed that Ecb, but not CHIPS, blocked C3b deposition via the AP (Fig. 2D). This is consistent with recent published data in whole serum where Ecb was shown to inhibit C3b deposition via the AP as well (11). To study the effect of Ecb on AP C5 convertases, C3b-coated particles were incubated with fB, fD, C3, and C5 in the presence or absence of Ecb, and C5a generation was measured. Similarly to the CP, we observed that Ecb inhibits C5a generation by purified AP C5 convertases (Fig. 2E). Similar inhibition was observed when AP convertases were generated in the presence of human properdin, a positive convertase regulator (supplemental Fig. 1). Together, these studies show that Ecb binds to C3b molecules deposited on the bacterial surface, which results in inhibition of C5 convertases of the CP and inhibition of C3 and C5 convertases of the AP.
Ecb Does Not Inhibit Binding of C5 to Convertases
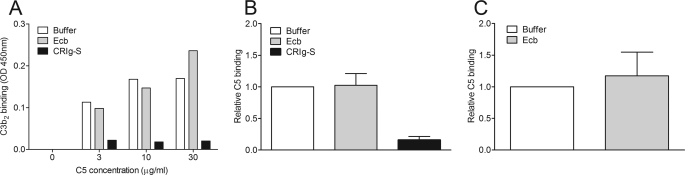
Because Ecb inhibits conversion of C5 by C5 convertases, we analyzed whether Ecb can inhibit C5 binding to convertases. The cleavage of C5 by C5 convertases likely occurs in a stepwise process: (a) C5 binds to the C4b-C3b (CP/LP) or C3b-C3b (AP) dimeric component of the convertase (25); (b) the protease component, C2a (CP/LP) or Bb (AP) cleaves C5. Therefore, C5 binding to convertases can be studied in an enzyme-linked immunosorbent assay format where C5 binds to C4b-C3b generated by decay of convertases on a surface or to preformed (C3b)2 (7). As a control, we used CRIg, a complement receptor found on liver Kupffer cells that recognizes opsonized particles via C3b and iC3b (26). A soluble form of CRIg functions as a complement inhibitor because it binds to C3b and inhibits substrate binding (C3 or C5) to AP convertases (20). Here, we used CRIg-S, a shorter form of CRIg with similar functions. Fig. 3A shows that (C3b)2 binding to C5, captured on microtiter plates, is not inhibited by Ecb, whereas CRIg-S does inhibit binding. To investigate C5 binding to naturally formed convertases rather than in a mimicked form, we allowed C5 binding to decayed convertases originated by complement activation on microtiter plates. Again, we observed that in contrast to CRIg-S, Ecb does not reduce C5 binding to AP convertases (Fig. 3B). Similar results were obtained for the CP as Ecb cannot inhibit C5 binding to decayed CP convertases (Fig. 3C). All of these data provide evidence that Ecb does not interfere with substrate binding to convertases.
FIGURE 3.
Ecb does not inhibit C5 binding to convertases. A, C3b2 binding to C5. C5 was captured on anti-C5a-coated microtiter plates. Binding of C3b2 to C5 in the presence or absence of Ecb or CRIg-S (both at 10 μg/ml) was analyzed. B, C5 binding to decayed AP convertases in the presence of Ecb or CRIg-S (both at 10 μg/ml). C, C5 binding to decayed CP convertases in the presence or absence of Ecb (10 μg/ml). A is a representative of three separate experiments; B and C represent the mean ± S.E. (error bars) of three experiments.
Ecb Stabilizes fB Binding to C3b and Prevents C3bBb Formation
Because Ecb inhibits C3b-containing convertases without affecting substrate binding, we explored whether Ecb affects the formation of convertases. C3b was covalently coupled to the surface of a CM5 biosensor chip, and Ecb was injected to bind ±500 RU. Recent studies show that the binding between C3b and Ecb is highly stable and can be compared with the affinity for Efb-C with C3b (8.8 nm) (15). Subsequent injection of fB revealed a 4-fold increase of fB binding to C3b chips treated with Ecb (Fig. 4A). Next to that, the interaction between fB and C3b appeared more stable in the presence of Ecb. This stabilizing effect was not due to a direct interaction between Ecb and fB because we determined by enzyme-linked immunosorbent assay that Ecb does not bind fB (supplemental Fig. 2). To investigate whether binding of Ecb to C3b prevented formation of C3bBb on the Biacore chip, we first injected fB and fD on a C3b chip treated with or without Ecb. Subsequently, we injected C3 allowing covalent deposition of C3b on the control chip. However, no C3b was deposited on the C3b chip pretreated with Ecb (Fig. 4b). This indicates that Ecb stabilizes fB on C3b and prevents formation of C3bBb.
FIGURE 4.
Ecb stabilizes fB binding to C3b and prevents C3bBb formation. A, fB binding to C3b via Biacore. C3b was covalently coupled to the surface of a CM5 biosensor chip where after Ecb or buffer was injected, and fB binding was analyzed. B, C3b deposition on the Biacore surface in the presence or absence of Ecb. The panels are representatives of three separate flow paths.
Ecb Prevents fB Conversion on Zymosan Surface
To investigate whether Ecb can stabilize fB on a natural surface as well, we incubated zymosan with 1% human serum to allow formation of AP convertases and proconvertases (C3bB). We then studied the decay of surface-bound convertases in the presence or absence of Ecb. We allowed decay of convertases and proconvertases for 1 h at 37 °C. Although no fB or Bb was detected on the zymosan surface, we found more fB on the surface in the presence of Ecb. This indicates that Ecb stabilizes C3bB on the zymosan surface as well (Fig. 5A).
FIGURE 5.
Ecb prevents fB conversion on the zymosan surface. A, fB stabilization on zymosan in the presence or absence of Ecb (10 μg/ml). Zymosan was opsonized with human serum whereupon Ecb was added during decay. fB binding was detected by immunoblotting. B, fluid phase fB conversion. C3b, fB, and fD were incubated with or without Ecb or Ecb N63E/R75E/N82E (10 μg/ml), and Bb formation was analyzed by immunoblotting. C, fB conversion on zymosan. Preopsonized zymosan was incubated with fB and fD in the presence or absence of Ecb or Ecb N63E/R75E/N82E (10 μg/ml). Bb formation was analyzed by immunoblotting. All figures are representatives of three separate experiments.
To study how Ecb-mediated stabilization of C3bB affects convertase formation and to see whether the C3d-binding part of Ecb is essential, we generated fluid phase AP C3 convertases by incubation of C3b, fB, and fD in the presence or absence of Ecb or Ecb N63E/R75E/N82E, and we analyzed Bb formation. Ecb strongly inhibits the formation of Bb in fluid phase conditions (Fig. 5B and supplemental Fig. 3), whereas the C3d-binding mutant does not. To analyze the influence of Ecb or the C3d-binding mutant on conversion of fB into Bb on the surface, preopsonized zymosan was incubated with fB and fD in the presence or absence of Ecb or Ecb N63E/R75E/N82E, and Bb formation was analyzed. Fig. 5C shows that Ecb, but not the C3d-binding mutant, inhibits the formation of Bb on zymosan. These data suggest that Ecb binding to C3b blocks formation of an active convertase enzyme.
DISCUSSION
The covalent deposition of C3b molecules on bacterial surfaces is a key event in the innate immune response (9, 10). Cleavage of C3 into C3b leads to remarkable conformational changes in the C3b molecule, which relocate the 35-kDa C3d domain and allow covalent binding of its thioester group to the target surface (27, 28). Deposited C3b triggers phagocytosis but also forms an essential part of the convertase enzymes that cleave C3 and C5 (7, 29, 30). S. aureus has developed several ways to survive in the human host. One of these mechanisms is secretion of small molecules that effectively inhibit the complement system (4, 5, 31).
Among the complement evasion strategies of S. aureus, we find a large number of proteins that specifically interact with C3b or C3. The C3 convertase of the AP is a dimeric complex of surface-bound C3b and Bb (Fig. 6A). During activation of C3, C3b forms a heterodimer with C3 and thus positions the attached Bb molecule to cleave C3 (17). The AP C3 convertase can be inhibited by several S. aureus C3b-binding molecules (11, 17, 32, 33). The staphylococcal complement inhibitor (SCIN) binds C3b in the α′N-terminal tail and domains MG6 and MG7 which are close to the Bb binding site (17). Because SCIN also binds to Bb, it forms a bridge between C3b and Bb and prevents movement of Bb toward C3 (Fig. 6B) (17, 34). Staphylococcal immunoglobulin-binding protein binds to C3 (32, 35, 36) and factor H, and seems to block convertase activity by formation of a tripartite complex of Sbi·FH·C3. The mechanism by which Ecb blocks AP C3 convertases is distinct from SCIN and Sbi. Ecb binds to bacterium-bound C3b during the opsonization process and subsequently stabilizes the C3bB proconvertase. This stabilized complex cannot be activated into C3bBb. Because the C3d domain of C3b is located far from the fB binding site (28, 37) (Fig. 6B), a direct interaction between Ecb and fB seems unlikely. Furthermore, Ecb does not seem to bind to other parts of the C3b molecule except to C3d because the C3d-binding mutant is not capable of inhibiting Bb formation. Binding of Ecb to C3d could alter the orientation of the C3b molecule on the bacterial surface. However, conversion of fB into Bb is blocked in fluid phase as well. Most likely, Ecb binding to C3d results in conformational change in the C3b molecule (as proposed for Efb) (15), causing enhanced binding of fB to C3b. This enhanced binding of fB to C3b could result in inhibition of Bb formation if fD is unable to bind and/or cleave fB in this conformation. Because Efb binds to C3d as well (15) and has similar complement inhibitory functions (11, 38), we expect that Efb functions in a similar way.
FIGURE 6.
Convertase inhibition by S. aureus. A, schematic representation of the covalent attached C3b on the target surface (left), the active C3 convertase (C3bBb, center), and the binding of C3 to the convertase (right). B, schematic representation of SCIN bound to the AP C3 convertase (left) and Ecb bound to the convertase (right).
Next to its effect on the AP C3 convertase, Ecb also blocks the C5 convertases of all pathways. In the CP/LP C5 convertase, C3b incorporates into the preexisting C3 convertase (C4b2a) to generate the C5 convertase (C3bC4b2a) (8, 39–41). Although C4b is inefficient in capturing C5, the deposited C3b increases the affinity of the enzyme for C5 and thereby alters substrate specificity of the enzyme from C3 into C5 (30). Because Ecb blocks C3bC4b2a but not C4b2a, we previously suggested that Ecb might block C5 binding to C3b (11). Here, although we demonstrate in more detail that Ecb blocks C5 convertase activity by binding to C3b via its C3d domain, our data show that C5 can still bind to the C5 convertase in the presence of Ecb. Because the structural organization of C5 convertases is presently unknown, it is difficult to speculate how Ecb blocks C5 convertase enzymes of the CP/LP (8, 42–44). Further research is needed to understand how binding to C3b can inhibit the CP/LP C5 convertase. Our data provide compelling evidence that the interaction with C3d is critical for C5 convertase inhibition by Ecb. This indicates that the conformation or spatial orientation of C3b on the surface is important for C5 convertase activity.
This work was supported by NIAID/National Institutes of Health Grant AI071028 (to B. V. G.). This work was also supported by the Council for Medical Sciences of the Netherlands Organization for Scientific Research (S. H. M. R. and J. A. G. v. S.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1–3.
- CP
- classical pathway
- LP
- lectin pathway
- AP
- alternative pathway
- Efb
- extracellular fibrinogen-binding protein
- Ecb
- extracellular complement-binding protein
- fB
- factor B
- fD
- factor D
- CRIg
- complement receptor of the immunoglobulin superfamily
- BSA
- bovine serum albumin
- CHIPS
- chemotaxis inhibitory protein of S. aureus
- PBS
- phosphate-buffered saline
- RU
- relative units
- SCIN
- staphylococcal complement inhibitor
- Bb
- activated fragment of fB.
REFERENCES
- 1.Lowy F. D. (1998) N. Engl. J. Med. 339, 520–532 [DOI] [PubMed] [Google Scholar]
- 2.Peacock S. J., de Silva I., Lowy F. D. (2001) Trends Microbiol. 9, 605–610 [DOI] [PubMed] [Google Scholar]
- 3.Rooijakkers S. H., van Kessel K. P., van Strijp J. A. (2005) Trends Microbiol. 13, 596–601 [DOI] [PubMed] [Google Scholar]
- 4.Foster T. J. (2005) Nat. Rev. Microbiol. 3, 948–958 [DOI] [PubMed] [Google Scholar]
- 5.Lambris J. D., Ricklin D., Geisbrecht B. V. (2008) Nat. Rev. Microbiol. 6, 132–142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gasque P. (2004) Mol. Immunol. 41, 1089–1098 [DOI] [PubMed] [Google Scholar]
- 7.Pangburn M. K., Rawal N. (2002) Biochem. Soc. Trans. 30, 1006–1010 [DOI] [PubMed] [Google Scholar]
- 8.Rawal N., Pangburn M. K. (2001) Int. Immunopharmacol. 1, 415–422 [DOI] [PubMed] [Google Scholar]
- 9.Walport M. J. (2001) N. Engl. J. Med. 344, 1140–1144 [DOI] [PubMed] [Google Scholar]
- 10.Walport M. J. (2001) N. Engl. J. Med. 344, 1058–1066 [DOI] [PubMed] [Google Scholar]
- 11.Jongerius I., Köhl J., Pandey M. K., Ruyken M., van Kessel K. P., van Strijp J. A., Rooijakkers S. H. (2007) J. Exp. Med. 204, 2461–2471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hammel M., Sfyroera G., Pyrpassopoulos S., Ricklin D., Ramyar K. X., Pop M., Jin Z., Lambris J. D., Geisbrecht B. V. (2007) J. Biol. Chem. 282, 30051–30061 [DOI] [PubMed] [Google Scholar]
- 13.Bodén M. K., Flock J. I. (1989) Infect. Immun. 57, 2358–2363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bodén M. K., Flock J. I. (1992) Microb. Pathog. 12, 289–298 [DOI] [PubMed] [Google Scholar]
- 15.Hammel M., Sfyroera G., Ricklin D., Magotti P., Lambris J. D., Geisbrecht B. V. (2007) Nat. Immunol. 8, 430–437 [DOI] [PubMed] [Google Scholar]
- 16.Lee L. Y., Liang X., Höök M., Brown E. L. (2004) J. Biol. Chem. 279, 50710–50716 [DOI] [PubMed] [Google Scholar]
- 17.Rooijakkers S. H., Wu J., Ruyken M., van Domselaar R., Planken K. L., Tzekou A., Ricklin D., Lambris J. D., Janssen B. J., van Strijp J. A., Gros P. (2009) Nat. Immunol. 10, 721–727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rooijakkers S. H., Ruyken M., van Roon J., van Kessel K. P., van Strijp J. A., van Wamel W. J. (2006) Cell Microbiol. 8, 1282–1293 [DOI] [PubMed] [Google Scholar]
- 19.Rooijakkers S. H., Ruyken M., Roos A., Daha M. R., Presanis J. S., Sim R. B., van Wamel W. J., van Kessel K. P., van Strijp J. A. (2005) Nat. Immunol. 6, 920–927 [DOI] [PubMed] [Google Scholar]
- 20.Wiesmann C., Katschke K. J., Yin J., Helmy K. Y., Steffek M., Fairbrother W. J., McCallum S. A., Embuscado L., DeForge L., Hass P. E., van Lookeren Campagne M. (2006) Nature 444, 217–220 [DOI] [PubMed] [Google Scholar]
- 21.Roos A., Bouwman L. H., Munoz J., Zuiverloon T., Faber-Krol M. C., Fallaux-van den Houten F. C., Klar-Mohamad N., Hack C. E., Tilanus M. G., Daha M. R. (2003) Mol. Immunol. 39, 655–668 [DOI] [PubMed] [Google Scholar]
- 22.Seelen M. A., Roos A., Wieslander J., Mollnes T. E., Sjöholm A. G., Wurzner R., Loos M., Tedesco F., Sim R. B., Garred P., Alexopoulos E., Turner M. W., Daha M. R. (2005) J. Immunol. Methods 296, 187–198 [DOI] [PubMed] [Google Scholar]
- 23.Hourcade D. E. (2006) J. Biol. Chem. 281, 2128–2132 [DOI] [PubMed] [Google Scholar]
- 24.Jokiranta T. S., Westin J., Nilsson U. R., Nilsson B., Hellwage J., Löfås S., Gordon D. L., Ekdahl K. N., Meri S. (2001) Int. Immunopharmacol. 1, 495–506 [DOI] [PubMed] [Google Scholar]
- 25.Rawal N., Pangburn M. K. (2000) J. Immunol. 164, 1379–1385 [DOI] [PubMed] [Google Scholar]
- 26.Helmy K. Y., Katschke K. J., Jr., Gorgani N. N., Kljavin N. M., Elliott J. M., Diehl L., Scales S. J., Ghilardi N., van Lookeren Campagne M. (2006) Cell 124, 915–927 [DOI] [PubMed] [Google Scholar]
- 27.Janssen B. J., Huizinga E. G., Raaijmakers H. C., Roos A., Daha M. R., Nilsson-Ekdahl K., Nilsson B., Gros P. (2005) Nature 437, 505–511 [DOI] [PubMed] [Google Scholar]
- 28.Janssen B. J., Christodoulidou A., McCarthy A., Lambris J. D., Gros P. (2006) Nature 444, 213–216 [DOI] [PubMed] [Google Scholar]
- 29.Rawal N., Pangburn M. (2001) J. Immunol. 166, 2635–2642 [DOI] [PubMed] [Google Scholar]
- 30.Rawal N., Pangburn M. K. (2003) J. Biol. Chem. 278, 38476–38483 [DOI] [PubMed] [Google Scholar]
- 31.Rooijakkers S. H., van Strijp J. A. (2007) Mol. Immunol. 44, 23–32 [DOI] [PubMed] [Google Scholar]
- 32.Burman J. D., Leung E., Atkins K. L., O'Seaghdha M. N., Lango L., Bernadó P., Bagby S., Svergun D. I., Foster T. J., Isenman D. E., van den Elsen J. M. (2008) J. Biol. Chem. 283, 17579–17593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ricklin D., Tzekou A., Garcia B. L., Hammel M., McWhorter W. J., Sfyroera G., Wu Y. Q., Holers V. M., Herbert A. P., Barlow P. N., Geisbrecht B. V., Lambris J. D. (2009) J. Immunol. 183, 2565–2574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Garcia B. L., Tzekou A., Ramyar K. X., McWhorter W. J., Ricklin D., Lambris J. D., Geisbrecht B. V. (2009) Acta Crystallogr. Sect. F. Struct. Biol. Cryst. Commun. 65, 482–485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Upadhyay A., Burman J. D., Clark E. A., Leung E., Isenman D. E., van den Elsen J. M., Bagby S. (2008) J. Biol. Chem. 283, 22113–22120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Haupt K., Reuter M., van den Elsen J., Burman J., Hälbich S., Richter J., Skerka C., Zipfel P. F. (2008) PLoS Pathog. 4, e1000250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Janssen B. J., Read R. J., Brünger A. T., Gros P. (2007) Nature 448, E1–E2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ricklin D., Ricklin-Lichtsteiner S. K., Markiewski M. M., Geisbrecht B. V., Lambris J. D. (2008) J. Immunol. 181, 7463–7467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Daha M. R., Fearon D. T., Austen K. F. (1976) J. Immunol. 117, 630–634 [PubMed] [Google Scholar]
- 40.Medicus R. G., Schreiber R. D., Götze O., Müller-Eberhard H. J. (1976) Proc. Natl. Acad. Sci. U.S.A. 73, 612–616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Medicus R. G., Götze O., Müller-Eberhard H. J. (1976) Scand. J. Immunol. 5, 1049–1055 [DOI] [PubMed] [Google Scholar]
- 42.Kinoshita T., Takata Y., Kozono H., Takeda J., Hong K. S., Inoue K. (1988) J. Immunol. 141, 3895–3901 [PubMed] [Google Scholar]
- 43.Kozono H., Kinoshita T., Kim Y. U., Takata-Kozono Y., Tsunasawa S., Sakiyama F., Takeda J., Hong K., Inoue K. (1990) J. Biol. Chem. 265, 14444–14449 [PubMed] [Google Scholar]
- 44.Sandoval A., Ai R., Ostresh J. M., Ogata R. T. (2000) J. Immunol. 165, 1066–1073 [DOI] [PubMed] [Google Scholar]