Abstract
The biomedical literature is an important source of information about the biological activity and effects of chemicals. We present an application that extracts terms indicating biological activity of chemicals from Medline records, associates them with chemical name and stores the terms in a repository called ChemoText. We describe the construction of ChemoText and then demonstrate its utility in drug research by employing Swanson’s ABC discovery paradigm. We reproduce Swanson’s discovery of a connection between magnesium and migraine in a novel approach that uses only proteins as the intermediate B terms. We validate our methods by using a cutoff date and evaluate them by calculating precision and recall. In addition to magnesium, we have identified valproic acid and nitric oxide as chemicals which developed links to migraine. We hypothesize, based on protein annotations, that zinc and retinoic acid may play a role in migraine. The ChemoText repository has promise as a data source for drug discovery.
Keywords: Literature-based discovery, Drug discovery, Text mining
1. Introduction
A central endeavor in drug research is determining the biological effects and activities of a chemical. Effects are observed and measured in a variety of venues from the test tube to the human body, from high throughput studies to those involving a single individual. The data from these experiments is increasingly being deposited in publicly available repositories (e.g. PubChem [1]), but even so, a large part of information about biological effects of chemicals is recorded only in the biomedical literature. We have developed a methodology to extract terms which indicate biological effect from Medline [2] and house them in a repository where they can be analyzed and mined. We call this repository ChemoText and have described the early development of the methodology in previous work[3].
1.1 Previous Work
Mining the literature for new drug therapies is a growing field. The earliest and best known research into using literature to find new treatments for disease is the work of Don Swanson. A researcher in information science, Swanson developed a methodology for literature-based discovery based on his observations of scientific literature[4]. He noted that the increasing specialization of scientists was paralleled by an increasing specialization in scientific journals. He described a situation where scientific domains no longer interacted through the reading and publishing of their literatures: researchers reading and publishing in one set of journals were not aware of articles in other journals. The literatures become islands and, in Swanson’s terms, noninteractive. This situation according to Swanson creates the potential for knowledge to go unconnected, relationships not recognized, and inferences not made, a situation he termed undiscovered public knowledge. Swanson demonstrated that these connections could be established through literature mining. Using his literature mining technique, often termed the ABC method, Swanson made several discoveries, among them a connection between Raynaud’s disease and fish oil [5] and the potential of magnesium to treat migraines[6]. Swanson emphasized that literature mining methods only assisted with hypothesis generation or hypothesis support, and that any hypothesis derived from the literature, must, like any other, be substantiated by experimental science.
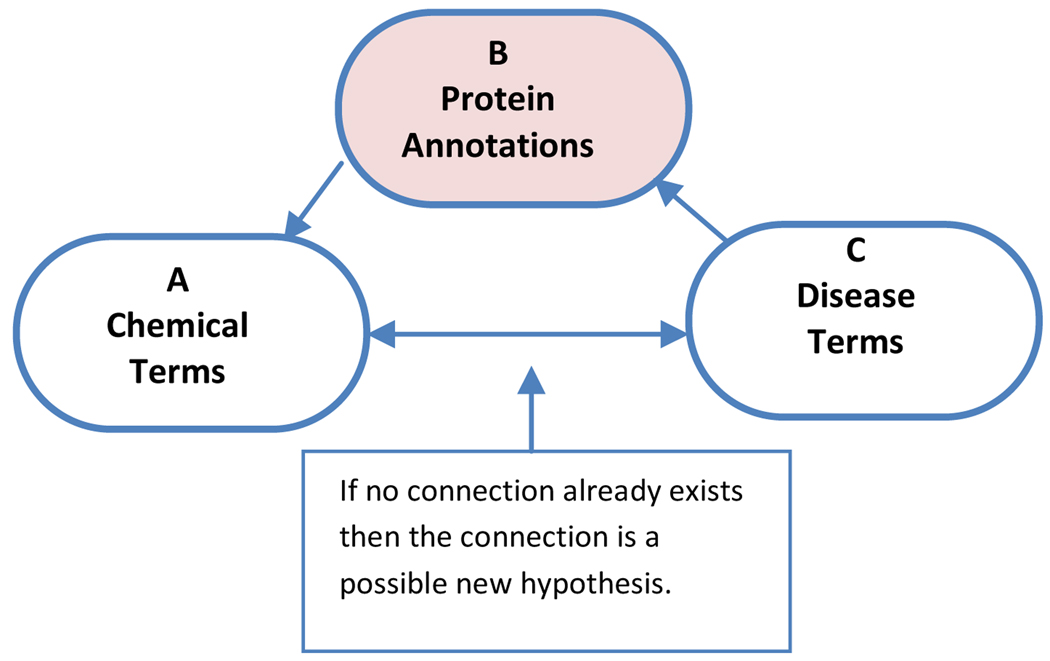
Swanson’s ABC methodology starts with identifying a disease or condition of interest. As an example we will consider migraine. (See Figure 1.) The term migraine becomes the C term. In the next step the literature is searched for terms which co-occur with migraine. These are the intermediary B terms and include in the case of migraine terms such as spreading cortical depression, vasoconstriction, and vasodilation. The B terms can be seen as terms for physiological conditions or states or processes which underlie the disease state. In the next step potential treatments – the A terms – are identified which are associated with the B terms. Next the C – A connection is tested and the only potential treatments retained for further examination are those which have not yet been explicitly linked to migraine.
Figure 1.
Swanson’s ABC Paradigm
Many researchers have followed in Swanson’s footsteps and constructed applications for discovery based on the ABC paradigm, but differing in other particulars. Swanson extended his original manual methods in collaboration with Smalheiser and created an automated version of their work called Arrowsmith[7]. Lindsay and Gordon broadened the corpus from titles to include abstracts and employed lexical methods and statistical measures to evaluate and limit the terms[8]. Weeber et al. [9] developed an application that used lexical methods and made use of the Unified Medical Language System (UMLS) [10], a suite of tools and knowledge sources available from the NLM for identifying, mapping, and understanding medical language. Srinivasan [11] also employed the UMLS but chose MeSH [12] as her corpus and developed ranking and weighting metrics to help narrow down the lengthy B term lists. Wren et al. [13] used a network paradigm and co-occurrence metrics, ranking on terms extracted from titles and abstracts. The ABC paradigm was described in graph language by Narayanasamy et al. [14] who used the concept of transitivity to describe the A–C connection. They applied the methods to find relationships between breast cancer genes. Yetisgen-Yildiz and Pratt [15] created an application called LitLinker based on MeSH terms and also using the UMLS for term selection and reduction steps. Seki and Mostafa [16] employed an inference network model and applied it to find implicit connections between genes and diseases. Petrič et al. emphasized rare terms in their application in order to find novel and innovative connections [17].
1.2 Evaluation of literature-based discovery systems
Evaluating results achieved through literature-based discovery methods is a challenge. Reproducing Swanson magnesium or fish oil discoveries has been a validation approach taken by several groups[8,9,18]. These discoveries are considered the gold standard because they have been confirmed by clinical studies. Comparing data from two time periods is also considered an important validation method [19]. Yetisgen-Yildiz and Pratt [15] and Hristovski et al. [20] used recall and precision metrics to score overall the predictions made in the earlier time baseline period with results from a later time period. Seki and Mostafa in [16] used an external data source to validate their predicted connections between genes and disease. In an experimental approach to validation, Wren et al. [13] performed in vitro cell assays to substantiate their literature-based claim that chlorpromazine can treat cardiac hypertrophy. Medical experts evaluated the results in [21,22].
Because disparate methods have been used by authors to evaluate their LBD systems there has been to date no way to compare the efficacy of applications. In a very recent paper (too recent to influence the design of this study) Yetisgen-Yildiz and Pratt [23]describe promising methodologies to remedy this situation. These include principles to consider when designing LBD research such as conducting multiple experiments and keeping the methods independent of prior knowledge. The authors also introduce metrics that will enable the evaluation of the ranking of the hypothesis set, not just the precision and recall of the entire set.
In this work we briefly review the construction of the ChemoText repository, and then we demonstrate its utility in drug research by reproducing Swanson’s discovery connecting magnesium to the treatment of migraine. The significant component of our implementation of the ABC method is that we have limited the B terms to protein annotations (see Figure 2). We apply this limitation not only to reduce the volume of data, but also because proteins are the agents behind most physiological processes and are therefore studied both by scientists investigating disease and by scientists looking for drugs. Because these very different groups of scientists may not be aware of each other’s work, there must be a strong potential for finding undiscovered implicit relationships between drugs (A terms) and diseases (C terms) via proteins (B terms).
Figure 2.
Swanson’s ABC using ChemoText
Other researchers in literature-based discovery have made use of the vital connections between drugs, proteins, and disease. Ahlers et al. [22] for instance extract text from Medline records and process it semantically to extract very specific information about the relationship between proteins, drugs, and disease. They use this information to postulate the mechanism of action of antipsychotic agents. The mechanism of action is carried out by the proteins that are found to be intermediary terms between disease and drug. In our work we use this relationship to hypothesize new therapies for disease.
2. Methods
2.1 Extraction of MeSH terms
The goal in developing ChemoText was to build a repository of chemicals associated with terms extracted from the literature that represented the chemicals’ biological activity or effect. The strategy was to extract these activity terms from Medline annotations. (See Figure 3.) Three categories of annotations were identified that indicated activity: MeSH drug effects annotations, MeSH disease annotations, and the proteins listed in the RN and MeSH section of the Medline record. MeSH or medical subject headings [12] are annotations assigned by indexers at the National Library of Medicine (NLM). Drug effects were extracted by finding all the drug effects subheadings and extracting the corresponding MeSH heading. The proteins and diseases were identified by looking up the terms in the MeSH Tree file. Tree categories C and F in this file were used to identify diseases, and the category D12 identified proteins. (The category D12 contains amino acids and peptides in addition to proteins; for brevity we will refer to this group as proteins.)
Figure 3. Medline processing into data tables.
The top part of the figure shows selected MeSH annotations in the Medline record for PubMed ID 16640785. The bottom of the figure shows the database entries in ChemoText that result from the processing of this Medline record.
2.2 Identification of subject chemicals
The Medline record can list more than one chemical. One or more of them may be the subject of the research, while other chemicals are peripheral, perhaps discussed or used in the experimental procedure, but not the central object of study. In order to reduce the volume of data we chose to extract the chemicals that were the subjects of study and then associate the activity terms only with those chemical(s). We developed a heuristic algorithm that evaluates the MeSH subheadings or qualifiers occurring with the chemical annotations and identifies the chemicals most likely to be the subjects. The heuristic follows a rule-based stepwise procedure, a procedure developed based on the detailed analysis of 125 Medline records. In this process, the annotations from each Medline record were examined to see if more than one chemical was annotated and identified as a major topic. If only one chemical was found and major, it was tagged as the subject chemical. If more than one chemical was identified as major, then the subheadings or qualifiers of each were examined. If the subheadings were the same for each of the chemicals, then they were all tagged as subjects. Preliminary analysis of the small test set had shown that certain subheadings were more commonly associated with subjects then other headings. (See Table 1.) Pharmacology, therapeutic use, and administration & dosage, for instance, are subheadings commonly annotated to the subject chemical, while the subheadings metabolism and biosynthesis are less common annotations for subject chemicals. We assembled a hierarchy of subheadings, starting with those most commonly associated with subjects to those rarely seen associated with subjects. We used this hierarchy to compare the chemicals in the remainder of the records and tag those most likely to be subjects. Medline records with more than one subject are common. Forty percent have more than one subject chemical, and the average number of subject chemicals per Medline record is 1.65. In the next step of the processing each of the subject chemicals was associated with the previously extracted activity and effects terms.
Table 1. Hierarchy of MeSH subheadings used when establishing subject chemicals.
Only chemicals flagged as major in at least one of their subheadings are used as input to the algorithm. If a subheading from level one is found, the associated chemical(s) are designated subjects. Only if no chemical has a subheading from the first group does the algorithm look at subheadings from the second group. If no chemicals have been identified annotated with subheadings from the first two groups, then chemicals tagged with a subheading from level 3 are tagged as subjects.
| Level | MeSH subheadings |
|---|---|
| 1 |
Pharmacology OR Adverse Effects OR Therapeutic Use OR Administration & Dosage OR Toxicity OR Pharmacokinetics |
| 2 | Any subheadings except Biosynthesis, Metabolism, Chemistry |
| 3 | Biosynthesis OR Metabolism OR Chemistry |
2.3 Complete repository
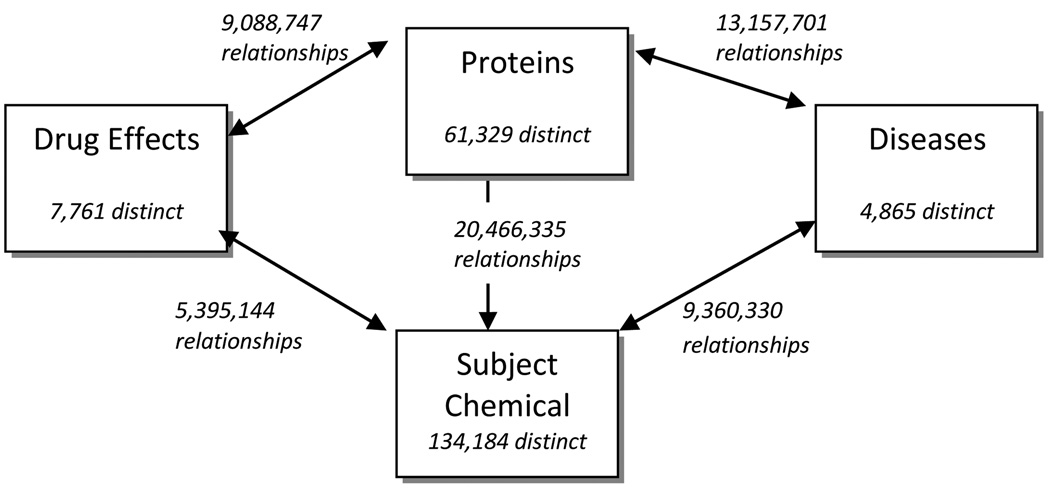
The 2008 Medline baseline file was downloaded from the NLM and used as the corpus for extraction routines. The extract routines were written in Perl. The data was loaded into a MySQL database and subsequent processing was performed in SQL or Microsoft Access. The completed data base depicted as a network is shown in Figure 4. The diagram shows the number of unique entities in each category as well as the number of relationships between entities stored in ChemoText. The baseline file contained 16,880,015 records; 6,635,344 records had identified subject chemicals and were included in ChemoText.
Figure 4.
Schematic view of ChemoText
There are other repositories that contain combinations of drug, disease, and protein information. STITCH (Search Tool for Interactions of Chemicals) contains small molecule chemicals and proteins[24]. The curated relationships in this resource come from both publicly available assay databases and from literature extraction. The cBioC resource relies on text-mining and community curation to establish and vet its protein-protein and protein-disease connections[25][26]. KEGG[27] and DrugBank [28] are two other sources of drug and protein information. The focus of KEGG is protein pathways while the focus of DrugBank is drugs and their protein targets. Both are highly curated.
In contrast to these resources, the data in ChemoText is extracted automatically and undergoes no manual curation. While the quality of the data in ChemoText may not rival a curated source, its breadth of coverage is more extensive, mirroring the broad reach of PubMed.
2.4 Literature-based discovery methods
We next explored the potential of using ChemoText for drug discovery. Our goal was to generate a list of chemicals linked implicitly but not explicitly to a particular disease through the literature. Such a list or hypothesis set may contain chemicals important to drug research either as new treatments or as key chemicals in the physiology of the disease. To generate the hypotheses, the ABC methodology of Swanson [6] was adopted.
The ChemoText database was queried for all articles published before 1985 in which migraine disorders, migraine with aura, or migraine without aura were included in the MeSH annotations. (The first article which first directly connected magnesium to migraines was published in 1985. We limited ourselves to evidence before that year for the baseline data.) These were the C terms. In the next step each protein annotation included in any of these articles was extracted. This was the pool of proteins associated with migraine. (B terms) This pool contained 131 proteins and included names for specific proteins as well as protein families (e.g. Receptors, Adrenergic).
In the next step the link between chemical and protein was examined. All chemicals were identified which, in the baseline period before 1985, appeared as a subject chemical in an article with the annotation of any of the migraine pool proteins. Chemical family names such as Amines or Lactones were eliminated to reduce the data volume. The resulting set of terms were the A terms. The number of migraine pool proteins associated with each chemical was counted. In the next step the link between the chemical and disease in the baseline period was examined. All chemicals were identified that appeared as a subject chemical in an Medline record before 1985 with the annotation of migraine. These records represented already known connections between the chemical and disease and were eliminated. The entire ChemoText database was examined to determine which chemicals predicted to have a link to migraine based on the evidence of the baseline period did indeed have literature evidence of a connection in the test period. The most common MeSH subheadings appearing with these chemicals when they were annotated with migraine were also extracted to help elucidate what kind of link emerged.
3. Results
3.1 Hypothesis set and validation
Our experiment produced a list of 4,725 chemicals potentially connected with migraine. (See Table 2 Part A.) We term this list our hypothesis set. When the set was ranked by protein count (Prot Ct), magnesium appeared near the top of the list at position 3. This closely reproduces Swanson’s discovery.
Table 2. Comparing baseline and test period results.
Ranked by protein count the top 12 chemicals out of 4,725 that are predicted to have a connection to migraine based on their associations with migraine proteins before 1985. Part A contains information available in Medline during the baseline period before 1985. Part B contains data extracted from Medline records in the test period from 1985 through 2007.
| A. Baseline Data: 1984 and before | B. Test Data: After 1984 | |||||
|---|---|---|---|---|---|---|
| Rank | Chemical Name | Prot Ct | First Yr | Article Ct | Disease Qualifier | Chemical Qualifier |
| 1 | Sodium | 104 | 2006 | 1 | blood | cerebrospinal fluid |
| 2 | Zinc | 93 | 0 | 0 | ||
| 3 | Magnesium | 91 | 1985 | 39 | blood | blood |
| 4 | Copper | 88 | 1986 | 1 | etiology | adverse effects |
| 5 | Corticosterone | 86 | 0 | 0 | ||
| 6 | Prednisolone | 84 | 2007 | 1 | complications | therapeutic use |
| 7 | Cysteine | 81 | 1994 | 3 | radionuclide imaging | analogs & derivatives |
| 8 | Edetic Acid | 80 | 1989 | 1 | physiopathology | admin & dosage |
| 9 | Lead | 79 | 0 | 0 | ||
| 10 | Colchicine | 77 | 0 | 0 | ||
| 11 | Cyclic GMP | 76 | 1995 | 4 | physiopathology | physiology |
| 12 | Nicotine | 75 | 1999 | 3 | drug therapy | adverse effects |
Many researchers have reproduced Swanson’s magnesium – migraine discovery; thus our observation is not novel, but can be viewed as a method validation. However, the design of ChemoText enabled us to extend this analysis in a novel direction. For each chemical in the hypothesis set the ChemoText database was searched for any link between the chemical and migraine after 1984. These results were summarized and combined with the results from the baseline period. Table 2 Part B contains these new columns: First Year (abbreviated First Yr, the first year an article appeared directly associating the chemical to migraine), Article Count (abbreviated Article Ct, the count of articles with this direct association) and the most common qualifiers or subheadings appearing in the annotations of the disease and the chemical with migraine (Disease Qualifier and Chemical Qualifier). Magnesium was first connected-to migraine in 1985 and has had 39 articles since connecting it to migraine. Both the most common disease qualifier and the most common chemical qualifier occurring in records in which migraine and magnesium occur together were blood, indicating the blood levels of magnesium are important in migraine.
The set was visually examined to see what general observations could be made. The set contains many types of chemicals. Sodium, zinc, copper and magnesium are elements. Cysteine is an amino acid and cyclic GMP is a nucleotide. Pharmaceuticals become more common as one scans down the list. The disease and chemical qualifiers indicate that the connections between the chemicals and migraine were varied. A number of chemicals were annotated indicating they treat migraine. Some chemicals like copper apparently cause migraine, and some appear to be involved in the physiological mechanisms of migraine (e.g. cyclic GMP).
The total set contained 154 chemicals which had no connection to migraine in the baseline period but developed a connection by 2007. Among the top 12 chemicals eight (66%) have developed links to migraine since 1984. The Article Count element was adopted as a rough indicator of the significance of a chemical’s connection to migraine. Magnesium has had 39 articles linking it to migraine since 1985 while copper has only one since its first connection in 1986. Sodium has only one article linking it directly to migraine, but the article is recent therefore the connection is newly established and its significance as of today is understandably low.
Based on the article count metric, two chemicals, valproic acid and nitric oxide, warrant further discussion. (See Table 3.) Valproic acid, found in position 105, has only 43 migraine-related proteins. The first article discussing its therapeutic use in migraine appeared in 1988 and by 2007, 83 articles linked valproic acid to migraine, twice as many as magnesium. Valproic acid is an example of drug re-profiling. It was used for many years as an anti-epileptic drug before being tried in migraine prophylaxis[29]. Valproic acid developed the strongest link to migraine based on the article count metric yet it did not appear as high as magnesium in the hypothesis set based on baseline protein count.
Table 3.
Baseline and test period results for valproic acid and nitric oxide
| A. Baseline data: 1984 and before | B. Test Data: After 1984 | |||||
|---|---|---|---|---|---|---|
| Rank | Chemical Name | Prot Ct | First Yr | Article Ct | Disease Qualifier | Chemical Qualifier |
| 103 | Mannitol | 44 | 0 | 0 | ||
| 104 | Penicillin G | 43 | 0 | 0 | ||
| 105 | Valproic Acid | 43 | 1988 | 83 | drug therapy | therapeutic use |
| 106 | Deuterium | 43 | 0 | 0 | ||
| 107 | Aluminum | 42 | 0 | 0 | ||
| 108 | Orotic Acid | 42 | 0 | 0 | ||
| … | 0 | 0 | ||||
| 598 | Quartz | 11 | 0 | 0 | ||
| 599 | Nitric Oxide | 11 | 1991 | 40 | physiopathology | physiology |
| 600 | Orciprenaline | 11 | 0 | 0 | ||
| 601 | Methaqualone | 11 | 0 | 0 | ||
Nitric oxide appears relatively low in the list as well at position 599, linked to only 11 proteins in common with the pool of migraine-linked proteins, but by 2007 it had 40 articles linking it to migraine, one more than magnesium. The most common qualifiers indicate that nitric oxide is important in the physiology of migraine.
3.2 Evaluation
Precision and recall were calculated using the following formulas.
| (1) |
HS is the number of entries in the hypothesis set and FL stands for the number of chemicals which will develop a future link to migraine. Future linked chemicals are those that existed in the baseline period, and had no direct link to migraine during that period, but by the end of the 1985–2007 test period had developed a direct link to migraine. We chose to use the terms FL and HS instead of adopting the True Positive (TP), True Negative (TN), etc. terminology because the latter scheme implies a certainty of outcome that our experiment could not support. The term True Positive, for example, sounds definitive, but all the links between drugs and a disease are not definitely established at a particular point in time. The links evolve over time as the result of ongoing research and publication.
The search of the entire ChemoText determined that there were 177 future linked chemicals; our routines found 154 of them. The 23 chemicals were missed because they did not have proteins linked to them from the migraine protein pool. In other words, the B – C connection did not pick up these chemicals. The intersection of the hypothesis set and the future linked (FL) chemicals gives the number of future linked chemicals found by our experiments.
The results for recall and precision are as follows.
The recall results are high. Selecting migraine drugs based on proteins identified 87% of the future chemicals connected to migraine. Our precision results, however, are weak. Only 3.3% of the chemicals in the hypothesis set developed a connection to migraine after 1984.
One likely reason for the low precision is that the 131 proteins connected to migraine include many protein families. These annotations can be very general and therefore have the likelihood of being annotated with many chemicals. For instance, Adenosine Triphosphatases and Peptide Hydrolases are two protein annotations from the migraine protein pool. While these families certainly have a connection to migraine, they are so broad that they will have connections to many other diseases and chemicals. As a result they will likely increase our hypothesis set significantly with chemicals of little potential connection to migraine. Not all protein families can be discounted, however. Receptors, Serotonin is also a protein family, but it has a well-known importance to the physiology of migraine and should not be undervalued. In future work we hope to develop other metrics which attribute a weight to the protein annotations that will reflect their importance to the disease being investigated.
We hypothesized that those chemicals with a weak connection to migraine will have fewer protein annotations from the migraine protein pool. We investigated the use of protein count thresholds to improve our results.
3.3 Increasing Precision
We investigated the relationship between protein count and the strength of the connection of a chemical to migraine. To reflect the importance of the connection between a chemical and migraine we continued the use of the article count metric. This metric acts as a weighted count, giving chemicals a weight equal to the number of publications connecting them with migraine. Counting co-occurrences to estimate relationship strength is a common technique in text mining (e.g. [30]). Using article count, however, does have limitations. It is a direct measure of publication activity, and publications may not always accurately reflect significance of a chemical. (It is even difficult to define the significance of a chemical.) Publication rates may increase, for instance, if a certain drug is suspected of having dangerous side effects. Additionally, a chemical which has ten articles connecting it to migraine cannot be said to be ten times more important than a chemical with only one article. Despite these limitations we will use the article count metric as a rough indicator for the importance of a connection between a chemical and migraine.
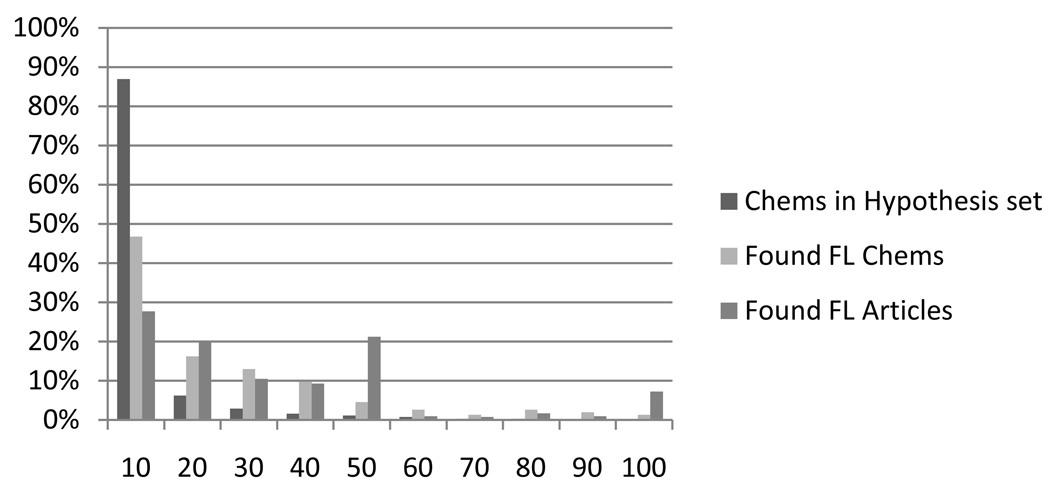
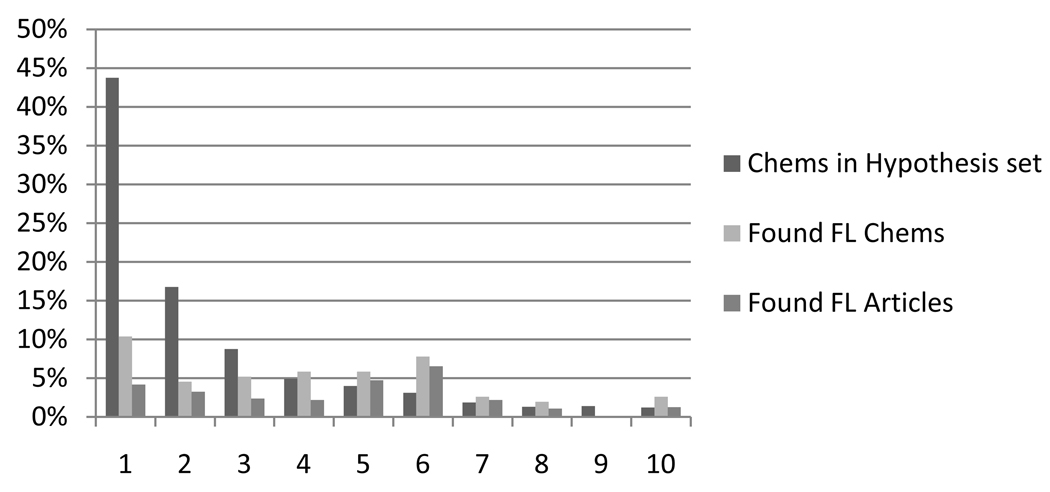
For a graphic understanding of these relationships between protein count, future linked (FL) count, and article count, we created a bar chart which grouped the hypothesis set by protein count ranges. (See Figure 5.) For each protein count range, the following percentages were depicted as bars: the percentage of the hypothesis set, percentage of future linked (FL) chemicals, and percentage of future linked articles. The bars in the first group, 10 proteins and under, show that over 80% of the hypothesis set chemicals have fewer than 10 proteins linking them to migraine. This large group has around 40% of the future linked chemicals. This group however has only around 25% of the articles linking chemicals to migraine. Because so many chemicals in the hypothesis set had fewer than 10 proteins, a separate bar chart (Figure 6) was created to look at the 0–10 range in detail. This graph shows that over 40% of the chemicals in the hypothesis set had only one protein from the migraine protein pool. This large group contained only 10% of the true migraine chemicals and less than 5% of the migraine articles. Eliminating this group of chemicals could improve precision without significantly degrading recall. To test this idea, precision and recall were recalculated as the chemicals with the lowest protein counts were consecutively eliminated. The results are contained in Table 4.
Figure 5.
Bar chart showing percentages by protein count
Figure 6.
Bar chart showing percentages by protein count for chemicals with 10 or fewer associated proteins
Table 4. Precision and recall results as thresholds are applied.
Hypothesis Set Count – number of chemicals in hypothesis set, Found FL Chemicals – number of future linked chemicals found by our process, Found FL Articles – number of articles associated with the found future linked chemicals. Precision, Recall, and Article Recall are calculated from the hypothesis set when the protein count (protct) threshold is applied.
| Threshold Applied |
Hypothesis Set Count |
Found FL Chemicals |
Found FL Articles |
Precision | Recall | Article Recall |
|---|---|---|---|---|---|---|
| none | 4725 | 154 | 552 | 0.03 | 0.870 | 0.909 |
| protct > 1 | 2658 | 138 | 529 | 0.05 | 0.780 | 0.871 |
| protct > 2 | 1867 | 131 | 511 | 0.07 | 0.740 | 0.842 |
| protct > 3 | 1454 | 123 | 498 | 0.08 | 0.695 | 0.820 |
| protct > 4 | 1223 | 114 | 486 | 0.09 | 0.644 | 0.801 |
| protct > 5 | 1034 | 105 | 460 | 0.10 | 0.593 | 0.758 |
| protct > 6 | 888 | 93 | 424 | 0.10 | 0.525 | 0.699 |
| protct > 7 | 801 | 89 | 412 | 0.11 | 0.503 | 0.679 |
| protct > 8 | 739 | 86 | 406 | 0.12 | 0.486 | 0.669 |
| protct > 9 | 674 | 86 | 406 | 0.13 | 0.486 | 0.669 |
| protct > 10 | 617 | 82 | 399 | 0.13 | 0.463 | 0.657 |
This table includes a new element: Article Recall. To calculate this we used the following formula.
| (2) |
We will illustrate this formula using the results from the entire hypothesis set.
The numerator in this equation is the number of articles associated with the 154 chemicals from our hypothesis set that did indeed develop a future link (FL) to migraine. The denominator is the number of articles for the chemicals in our hypothesis set that developed a future link to migraine in addition to the 55 articles associated with the 23 chemicals that our routines did not find. Article recall overall was 90.9%. Article recall is higher than chemical recall because the chemicals we did find had on average more articles associated with them then the chemicals we did not find.
Table 4 records the change in precision and recall as protein count thresholds were applied to the hypothesis set. The elimination of each group of chemicals caused an increase in precision and a decrease in recall. By eliminating all chemicals with 10 or fewer proteins, the hypothesis set contains 617 chemicals. Of these 82 or 13% are future linked. While the chemical recall was decreased to 46.3%, the article recall decreased only to 65.7%, showing that the chemicals remaining had a more significant connection to migraine as measured by article count. The three chemicals which eventually developed the strongest link to migraine (magnesium, nitric oxide, and valproic acid) are all included in the set of 617, although nitric oxide, with only 11 chemicals from the protein pool, was close to the cutoff. Our results on the whole compare favorably to other similar studies [15,20].
4. Discussion
In this proof of concept study, our strategy of using proteins as the intermediary terms in the ABC paradigm was very effective in finding chemicals in the literature prior to 1985 that later developed a link to migraine. The reason for this likely lies in the central role proteins play in both disease and drug research. The study of disease increasingly focuses on the physiology of the disease state at the molecular level, a level in which observations of proteins and their interactions with other molecules are central. Drug research focuses on proteins as well, searching for drugs that will modulate the behavior of proteins involved in the disease pathway.
Restricting the B terms to proteins has also allowed us to reduce the size and complexity of the datasets we work with. A count of protein annotations in our database showed that they comprise roughly 12% of the MeSH annotations in the subset of Medline records stored in ChemoText (those with annotated chemicals). This represents a significant reduction in data volume, and likely a reduction in noise, while the signal in the data remains strong enough for the purposes of our study.
While drawing connections based on common proteins is effective in recall, the utility of the protein count variable is not so clear. Chemicals with the lowest protein counts can be eliminated without significant deterioration in recall, and chemicals with the highest protein counts are more likely to be connected to migraine than the chemicals overall. Eight out of the top 12 chemicals from the hypothesis set developed a link to migraine, a much higher proportion than the 3.3% overall. In between the high and low extremes, however, the correlation between protein count and strength of the connection to migraine becomes less apparent. Table 5 calculates protein and article counts based on data retrieved from the entire ChemoText database. Part A on the left ranks the chemicals connected to migraine by article count. Sumatriptan has overwhelmingly the highest article count, but a protein count of only 69. The related triptan drugs which are also highly written about have even lower protein counts. The right hand side of the table ranks the chemicals by protein count. The article counts do not approach the 675 articles of sumatriptan; with 230 articles serotonin comes the closest.
Table 5. View of ChemoText data through 2007.
Part A is ranked by article count; Part B is ranked by protein count.
| Part A. Ranked by Article Count (Art Ct) | Part B. Ranked by Protein Count (Prot Ct) | ||||||
|---|---|---|---|---|---|---|---|
| ChemName | Prot Ct |
First Yr |
Art Ct | Chem Name | Prot Ct |
First Yr |
Art Ct |
| Sumatriptan | 69 | 1988 | 675 | Calcium | 478 | 1950 | 11 |
| Ergotamine | 72 | 1962 | 314 | Ethanol | 433 | 1969 | 6 |
| Serotonin | 404 | 1959 | 230 | Nitric Oxide | 423 | 1991 | 40 |
| Propranolol | 256 | 1968 | 165 | Estradiol | 416 | 1971 | 23 |
| Methysergide | 81 | 1963 | 151 | Cyclic AMP | 408 | 1976 | 6 |
| Flunarizine | 66 | 1980 | 136 | Serotonin | 404 | 1959 | 230 |
| rizatriptan | 14 | 1996 | 126 | Dexamethasone | 395 | 1967 | 11 |
| Dihydroergotamine | 47 | 1974 | 112 | Norepinephrine | 394 | 1954 | 25 |
| Aspirin | 328 | 1953 | 104 | Dopamine | 394 | 1970 | 24 |
| Caffeine | 246 | 1950 | 99 | Cysteine | 382 | 1994 | 3 |
| Valproic Acid | 230 | 1988 | 83 | Adenosine Triphosphate | 377 | 1979 | 7 |
| zolmitriptan | 16 | 1996 | 79 | Oxygen | 375 | 1980 | 6 |
| Metoclopramide | 105 | 1974 | 63 | Progesterone | 361 | 1951 | 33 |
| eletriptan | 17 | 1998 | 62 | Testosterone | 358 | 1955 | 6 |
| Acetaminophen | 203 | 1972 | 61 | Sodium | 355 | 2006 | 1 |
| naratriptan | 9 | 1997 | 61 | Potassium | 354 | 1981 | 3 |
| Histamine | 348 | 1950 | 54 | Hydrocortisone | 353 | 1979 | 10 |
| Clonidine | 211 | 1970 | 54 | Nicotine | 353 | 1999 | 3 |
| Pizotyline | 17 | 1974 | 51 | Histamine | 348 | 1950 | 54 |
| Indomethacin | 284 | 1964 | 46 | Cholesterol | 348 | 1973 | 13 |
| Nitric Oxide | 423 | 1991 | 40 | Acetylcholine | 338 | 1959 | 5 |
| Magnesium | 316 | 1985 | 39 | Morphine | 333 | 1960 | 9 |
| Cinnarizine | 45 | 1977 | 39 | Adenosine | 332 | 1953 | 7 |
| Tyramine | 146 | 1967 | 37 | Aspirin | 328 | 1953 | 104 |
| Nitroglycerin | 150 | 1968 | 35 | Epinephrine | 325 | 1950 | 15 |
| Amitriptyline | 147 | 1965 | 34 | Cyclosporine | 324 | 1994 | 4 |
| Metoprolol | 115 | 1980 | 34 | Sodium Chloride | 322 | 1951 | 4 |
| Progesterone | 361 | 1951 | 33 | Magnesium | 316 | 1985 | 39 |
We have observed that protein count seems more indicative of a connection to migraine for endogenous chemicals than for exogenous ones. Endogenous molecules are those that occur naturally in the body. Exogenous molecules are foreign to the body, and therefore drugs belong to this category. (Many drugs are forms or derivatives of endogenous chemicals so this is not a strict definition.) We can speculate that endogenous chemicals are likely to be involved in multiple pathways in the body and will therefore be over time studied for their relationship to many diseases and will accumulate protein annotations. The goal in creating a drug, on the other hand, is to make its action as targeted as possible in order to reduce unwanted effects. Often a drug targets a single protein like a receptor. The literature annotations will likely include other proteins as well as the upstream, downstream, and off-target effects are elucidated. In future work we plan to divide the chemicals if possible into endogenous and exogenous groups to test the significance of the protein count variable in each group.
We have shown that applying protein count cutoffs can work as dial to select different levels of recall and precision. In practice the decision as to what levels of precision and recall are acceptable depends on the purpose and resources of the researcher. Achieving the best possible recall may be most important to drug researchers who have other information resources on hand to limit the hypothesis set. These researchers can limit the set to exogenous molecules and then examine external data such as toxicity and patent information to cull unlikely candidates. These researchers may even augment the hypothesis set with structurally similar molecules and then screen the whole set in silico or in vitro. Relatively higher precision, on the other hand, may be more important to other researchers who do not have screening resources.
One of the main challenges in developing ChemoText and in implementing Swanson’s ABC discovery paradigm lies in the designation of chemicals in MeSH. The first challenge is that the name of a chemical may change over time. While NLM maintains helpful records mapping names to earlier designations, we have not written or implemented all the routines necessary to trace the history of a chemical and relate all the names to a unique identifier. The second hurdle is that chemicals may be categorized in several ways. Again the NLM provides the very helpful Tree database [31], but the complexity of chemicals makes them difficult to categorize. For instance, many endogenous molecules (including proteins) are synthesized and used as drug therapies. It is not possible from to know from the annotations whether a reference is to the endogenous or the exogenous form of the molecule.
Our definition of a direct connection between a chemical and a disease consists of a cooccurrence of a subject chemical and the annotated disease. This definition is restrictive and leaves out co-mention of a chemical with a disease in an abstract or title. It also omits possibly informative MeSH co-occurrences. Our ChemoText database is limited by time as well. We currently update it on a yearly basis when the new baseline data is available from the National Library of Medicine. The MeSH vocabulary is also updated on a yearly basis, and therefore can lag behind the results being reported.
We have found that the key relationships and entities important to computational drug discovery show strong presence in the MeSH annotations that we do include. This key information includes chemicals, diseases, and proteins. The limitations in the scope of the data also reduce its size. The insights we have gained from data streamlined enough to move back and forth in time to understand the evolution of a drug or disease treatments are valuable enough to risk missing connections. Because our methods involve inference – taking a set of data and predicting new things based on it – we do not need the newest information to construct a hypothesis set. We would however need the newest and most complete information available on PubMed to validate any predictions we would make based the hypothesis set.
Magnesium provides a good example of the restrictiveness of our procedures and what they would and would not consider a relationship. Entering the query “magnesium and migraine” in PubMed Entrez gives 128 articles (as of 08/20/2008). In three of the four articles before 1985 though magnesium occurs in the Medline record, magnesium is not the main topic. The Altura 1984 [32] article does meet our criteria for magnesium to be the subject drug, but as the article is about strokes, migraine is only mentioned in the abstract and not annotated. The 1973 German article linking migraine therapy to magnesium glutamate specified glutamates as the main topic[33]; no abstract is provided so it is difficult to assess the accuracy of that annotation. The 1985 Altura article [34] about the calcium antagonist properties of magnesium is the first article we include in ChemoText with a direct link between magnesium and migraine.
Predictions
The analysis that produced Table 5 Part B was rerun to include all chemicals, those with and those without a direct link to migraine in ChemoText. When the list was sorted by protein count, only three chemicals among the top-ranked 35 showed no link to migraine: zinc, tetradecanoylphorbol acetate, and retinoic acid (MeSH term Tretinoin). Tetradecanoylphorbol acetate is a plant derivative and, because we have noted a stronger link between protein count and endogenous molecules and tetradecanoylphorbol is exogenous (as well as a known carcinogen), we will not predict that it has a connection to migraine. We predict that zinc and retinoic acid have a connection to migraine. We will briefly discuss some of the literature evidence here.
Zinc is an important nutrient in the human diet. In the body it plays many roles both in structure as a component of many proteins, but also in cell signaling. In [35] Frederickson et al. review the role of zinc in neurobiology. Several of the roles they outline for zinc in the nervous system have possible links to migraine. Zn2+, the ionic form of zinc, is a neurotransmitter and is stored in and released from a neuron in the brain that also releases glutamate, a neurotransmitter known to be involved in the physiology of migraine. Zinc has been shown to be active with regard to at least two key receptors in migraine physiology: the NMDA receptor and GABA receptor. The level of free zinc in cells, particularly in pathological conditions, is modulated by nitric oxide, a molecule with direct links to the etiology of migraine.
Retinoic acid is a form of Vitamin A and an important molecule in regulating gene transcription. In the nervous system it has been studied extensively for its role in development of the embryo and its link to maintaining and remodeling the nervous system is also under investigation[36]. Excessive Vitamin A can cause a number of conditions including idiopathic intracranial hypertension, a condition with symptoms very similar to migraine including severe head pain and visual disturbances[37]. Neither Vitamin A nor retinoic acid has a direct link to migraine in ChemoText, however isotretinoin, a isomer of retinoic acid, has one link[38]. In this case study a woman with unilateral Darier’s disease was prescribed isotretinoin to treat her skin eruptions. She also complained of migraines. During the treatment with isotretinoin the headaches ceased, but once the treatment concluded and she stopped taking isotretinoin, the migraines returned. Retinoic acid also has a link to nitric oxide: in keratinocytes retinoic acid has been shown to reduce inflammation through inhibiting the synthesis of nitric oxide[39].
5. Conclusion
In this research we have developed a methodology for inferring drug-disease associations based on a novel implementation of Swanson’s ABC text mining paradigm. The novelty of our approach is that we use only MeSH protein annotations as the intermediate B terms. This approach gives our work the following advantages over other implementations of Swanson’s model. First, limiting the B terms to proteins lowers the volume and dimensionality of our data and makes it more tractable. This allows us to combine data from two time periods not only to validate our findings but also to understand what kinds of connections have emerged between the chemical and the disease. Using proteins additionally obviates the need to have a scientist review the intermediary results and make decisions about how to proceed, a requisite step in some other literature-based discovery applications. In our application, human effort is saved for evaluation of resulting hypotheses. Additionally, using proteins as the intermediary terms also has sound biological footing: proteins are frequently the intermediary between disease and drugs. This consideration justifies their use as functional B terms in the ABC approach. In this proof of concept and methods development study, we have demonstrated the utility of our approach by reproducing Swanson’s well known connection between magnesium and migraine, as well as by predicting several other known links between drugs and disease.
Our ChemoText data repository is well-suited to finding implicit relationships. One of its strengths comes from identifying the subject chemical of a Medline record. This is a novel technique that not only reduces the volume of data, but reduces the noise associated with term co-occurrence.
Article count was introduced as a rough metric for the importance or significance of a connection between a chemical and a disease. Although we are hoping to use a more sophisticated measure of significance in our future work, the article count metric has allowed us to identify two chemicals with comparable significance to magnesium: valproic acid and nitric oxide. Despite the many literature mining projects endeavoring to reproduce Swanson’s migraine-magnesium connection, no one, as far as we know, has identified the strong link between these chemicals and migraine. (Swanson himself however in [6] noted a connection between epilepsy and migraine.) Valproic acid and nitric oxide should be included with magnesium as a gold standard for future literature-based discovery research.
Based on the importance of protein count for endogenous molecules, we have predicted that zinc and retinoic acid have a connection to migraine.
Our approach to literature-based discovery has several limitations. Connections between biological entities which occur in the title, abstract, or full text of the article will not be picked up. Additionally, the identification of the subject chemical is performed by a heuristic algorithm and therefore not always accurate. The principle of assuming that two biological entities are related because terms referring to them co-occur in the same Medline record has its limitations and can produce false connections.
By its distillation of a large body of chemical and disease research, ChemoText offers many rich avenues for exploration. (See Figure 4.) We hope to extend our techniques to a wider scope of drug-disease associations. We also aim to improve on our understanding of the patterns residing in the data so that we can develop procedures and metrics that will lead to higher precision and models with improved predictive abilities. In order to improve evaluation, we hope in the future to adopt the guidelines described in[23]. As the biomedical literature grows in volume and continues to segment into specialties, the need for tools to combine literatures in rational, useful ways will become increasingly critical to scientists in drug discovery. We have shown that ChemoText represents a promising addition to the field of literature-based drug discovery.
Acknowledgements
This work has been partially funded by NIH grant P20-HG003898. NCB would like to thank Alex Tropsha for his support and help. The authors would also like to thank Jane Rosov and the National Library of Medicine for their work providing Medline.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.NLM. PubChem. 2006 Available at: http://pubchem.ncbi.nlm.nih.gov/, 2008.
- 2.National Library of Medicine. MEDLINE. doi: 10.1080/15360280801989377. Available at: http://www.nlm.nih.gov/pubs/factsheets/medline.html. [DOI] [PubMed]
- 3.Extracting Drug Activity Terms from Medline Annotations. Proceedings: Summit on Translational Bioinformatics. American Medical Informatics Association; 2008. Mar, [Google Scholar]
- 4.Swanson DR. Medical literature as a potential source of new knowledge. Bull.Med.Libr.Assoc. 1990 Jan;78(1):29–37. [PMC free article] [PubMed] [Google Scholar]
- 5.Swanson DR. Fish oil, Raynaud's syndrome, and undiscovered public knowledge. Perspect.Biol.Med. 1986 Autumn;30(1):7–18. doi: 10.1353/pbm.1986.0087. [DOI] [PubMed] [Google Scholar]
- 6.Swanson DR. Migraine and magnesium: eleven neglected connections. Perspect.Biol.Med. 1988 Summer;31(4):526–557. doi: 10.1353/pbm.1988.0009. [DOI] [PubMed] [Google Scholar]
- 7.Smalheiser NR, Swanson DR. Using ARROWSMITH: a computer-assisted approach to formulating and assessing scientific hypotheses. Comput.Methods Programs Biomed. 1998 Nov;57(3):149–153. doi: 10.1016/s0169-2607(98)00033-9. [DOI] [PubMed] [Google Scholar]
- 8.Lindsay RK, Gordon MD. Literature-based discovery by lexical statistics. J Am Soc Inf Sci. 1999;50(7):574–587. [Google Scholar]
- 9.Weeber M, Klein H, de Jong-van den Berg LTW, Vos R. Using Concepts in Literature-Based Discovery: Simulating Swanson's Raynaud-Fish Oil and Migraine-Magnesium Discoveries. J Am Soc Inf Sci Tech. 2001;52(7):548–557. [Google Scholar]
- 10.National Library of Medicine. Unified Medical Language System Fact Sheet. Available at: http://www.nlm.nih.gov/pubs/factsheets/umls.html.
- 11.Srinivasan P. MeSHmap: a text mining tool for MEDLINE. Proc.AMIA.Symp. 2001:642–646. [PMC free article] [PubMed] [Google Scholar]
- 12.Medical Subject Headings. Available at: http://www.nlm.nih.gov/mesh/meshhome.html.
- 13.Wren JD, Bekeredjian R, Stewart JA, Shohet RV, Garner HR. Knowledge discovery by automated identification and ranking of implicit relationships. Bioinformatics. 2004 Feb 12;20(3):389–398. doi: 10.1093/bioinformatics/btg421. [DOI] [PubMed] [Google Scholar]
- 14.Narayanasamy V, Mukhopadhyay S, Palakal M, Potter DA. TransMiner: mining transitive associations among biological objects from text. J.Biomed.Sci. 2004 Nov-Dec;11(6):864–873. doi: 10.1007/BF02254372. [DOI] [PubMed] [Google Scholar]
- 15.Yetisgen-Yildiz M, Pratt W. Using statistical and knowledge-based approaches for literature-based discovery. J.Biomed.Inform. 2006 Dec;39(6):600–611. doi: 10.1016/j.jbi.2005.11.010. [DOI] [PubMed] [Google Scholar]
- 16.Seki K, Mostafa J. Discovering implicit associations between genes and hereditary diseases. Pac.Symp.Biocomput. 2007:316–327. [PubMed] [Google Scholar]
- 17.Petrič I, Urbančič T, Cestnik B, Macedoni-Lukšič M. Literature mining method RaJoLink for uncovering relations between biomedical concepts. J.Biomed.Inform. 2008 doi: 10.1016/j.jbi.2008.08.004. [DOI] [PubMed] [Google Scholar]
- 18.Srinivasan P, Libbus B. Mining MEDLINE for implicit links between dietary substances and diseases. Bioinformatics. 2004 Aug 4;20 Suppl 1:i290–i296. doi: 10.1093/bioinformatics/bth914. [DOI] [PubMed] [Google Scholar]
- 19.Bekhuis T. Conceptual biology, hypothesis discovery, and text mining: Swanson's legacy. Biomed.Digit.Libr. 2006 Apr 3;3:2. doi: 10.1186/1742-5581-3-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hristovski D, Stare J, Peterlin B, Dzeroski S. Supporting discovery in medicine by association rule mining in Medline and UMLS. Medinfo. 2001;10(Pt 2):1344–1348. [PubMed] [Google Scholar]
- 21.Weeber M, Vos R, Klein H, De Jong-Van Den Berg LT, Aronson AR, Molema G. Generating hypotheses by discovering implicit associations in the literature: a case report of a search for new potential therapeutic uses for thalidomide. J.Am.Med.Inform.Assoc. 2003 May-Jun;10(3):252–259. doi: 10.1197/jamia.M1158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ahlers CB, Hristovski D, Kilicoglu H, Rindflesch TC. Using the literature-based discovery paradigm to investigate drug mechanisms. AMIA.Annu.Symp.Proc. 2007:6–10. [PMC free article] [PubMed] [Google Scholar]
- 23.Yetisgen-Yildiz M, Pratt W. A new evaluation methodology for literature-based discovery systems. J.Biomed.Inform. 2009 Aug;42(4):633–643. doi: 10.1016/j.jbi.2008.12.001. [DOI] [PubMed] [Google Scholar]
- 24.Kuhn M, von Mering C, Campillos M, Jensen LJ, Bork P. STITCH: interaction networks of chemicals and proteins. Nucleic Acids Res. 2008 Jan;36(Database issue):D684–D688. doi: 10.1093/nar/gkm795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Baral C, Gonzalez G, Gitter A, Teegarden C, Zeigler A, Joshi-Tope G. CBioC: beyond a prototype for collaborative annotation of molecular interactions from the literature. Comput.Syst.Bioinformatics Conf. 2007;6:381–384. doi: 10.1142/9781860948732_0038. [DOI] [PubMed] [Google Scholar]
- 26.Gonzalez G, Uribe JC, Tari L, Brophy C, Baral C. Mining gene-disease relationships from biomedical literature: weighting protein-protein interactions and connectivity measures. Pac.Symp.Biocomput. 2007:28–39. [PubMed] [Google Scholar]
- 27.Kanehisa M, Goto S. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000 Jan 1;28(1):27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wishart DS, Knox C, Guo AC, Cheng D, Shrivastava S, Tzur D, et al. DrugBank: a knowledgebase for drugs, drug actions and drug targets. Nucleic Acids Res. 2008 Jan;36(Database issue):D901–D906. doi: 10.1093/nar/gkm958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sorensen KV. Valproate: a new drug in migraine prophylaxis. Acta Neurol.Scand. 1988 Oct;78(4):346–348. doi: 10.1111/j.1600-0404.1988.tb03667.x. [DOI] [PubMed] [Google Scholar]
- 30.Stapley BJ, Benoit G. Biobibliometrics: information retrieval and visualization from cooccurrences of gene names in Medline abstracts. Pac.Symp.Biocomput. 2000:529–540. doi: 10.1142/9789814447331_0050. [DOI] [PubMed] [Google Scholar]
- 31.MeSH Trees File. Available at: http://www.nlm.nih.gov/mesh/2009/download/mtr_abt.html.
- 32.Altura BT, Altura BM. Interactions of Mg and K on cerebral vessels--aspects in view of stroke. Review of present status and new findings. Magnesium. 1984;3(4–6):195–211. [PubMed] [Google Scholar]
- 33.Vosgerau H. Migraine therapy with magnesium glutamate. Ther.Ggw. 1973 Apr;112(4):640. passim. [PubMed] [Google Scholar]
- 34.Altura BM. Calcium antagonist properties of magnesium: implications for antimigraine actions. Magnesium. 1985;4(4):169–175. [PubMed] [Google Scholar]
- 35.Frederickson CJ, Koh JY, Bush AI. The neurobiology of zinc in health and disease. Nat.Rev.Neurosci. 2005 Jun;6(6):449–462. doi: 10.1038/nrn1671. [DOI] [PubMed] [Google Scholar]
- 36.Maden M. Retinoic acid in the development, regeneration and maintenance of the nervous system. Nat.Rev.Neurosci. 2007 Oct;8(10):755–765. doi: 10.1038/nrn2212. [DOI] [PubMed] [Google Scholar]
- 37.Volcy-Gomez M, Uribe CS. Headaches in idiopathic intracranial hypertension. A review of ten years in a Columbian hospital. Rev.Neurol. 2004 Sep 1–15;39(5):419–423. [PubMed] [Google Scholar]
- 38.Rotunda AM, Cotliar J, Haley JC, Craft N. Unilateral Darier's disease associated with migraine headache responsive to isotretinoin. J.Am.Acad.Dermatol. 2005 Jan;52(1):175–176. doi: 10.1016/j.jaad.2004.08.011. [DOI] [PubMed] [Google Scholar]
- 39.Becherel PA, Le Goff L, Ktorza S, Chosidow O, Frances C, Issaly F, et al. CD23-mediated nitric oxide synthase pathway induction in human keratinocytes is inhibited by retinoic acid derivatives. J.Invest.Dermatol. 1996 Jun;106(6):1182–1186. doi: 10.1111/1523-1747.ep12347939. [DOI] [PubMed] [Google Scholar]