This study describes the creation of a binary protein–protein interaction map of core cell cycle proteins of Arabidopsis thaliana using two complementary interaction assays, yeast two-hybrid and bimolecular fluorescence complementation. It integrates this map with expression data and describes 357 protein–protein interactions, of which 293 are previously unreported.
Abstract
As in other eukaryotes, cell division in plants is highly conserved and regulated by cyclin-dependent kinases (CDKs) that are themselves predominantly regulated at the posttranscriptional level by their association with proteins such as cyclins. Although over the last years the knowledge of the plant cell cycle has considerably increased, little is known on the assembly and regulation of the different CDK complexes. To map protein–protein interactions between core cell cycle proteins of Arabidopsis thaliana, a binary protein–protein interactome network was generated using two complementary high-throughput interaction assays, yeast two-hybrid and bimolecular fluorescence complementation. Pairwise interactions among 58 core cell cycle proteins were tested, resulting in 357 interactions, of which 293 have not been reported before. Integration of the binary interaction results with cell cycle phase-dependent expression information and localization data allowed the construction of a dynamic interaction network. The obtained interaction map constitutes a framework for further in-depth analysis of the cell cycle machinery.
INTRODUCTION
Cell division is an essential biological process that is regulated by an evolutionarily conserved from fungi to mammals molecular machinery (Nurse, 1990). Transition through the different phases of the cell cycle relies on the activity of a group of Ser/Thr kinases, designated cyclin-dependent kinases (CDKs). These CDKs regulate cell cycle progression through the phosphorylation of key substrates, such as histones, the retinoblastoma oncoprotein, and the transcription factors E2F-DP (reviewed in Humphrey and Pearce, 2005; Inzé and De Veylder, 2006). To ensure that the cell division program is executed correctly, CDKs associate with regulatory subunits called cyclins. Because cyclins accumulate and degrade cyclically during the cell cycle, they are the major factors that determine the timing of the CDK activity (Hunt, 1991; Morgan, 1997). In addition, cyclins regulate the substrate specificity of the CDK/cyclin (CYC) complex by directing the CDK subunits to subcellular spaces (Ubersax et al., 2003; Wittenberg, 2005). Depending on their structural similarity and periodic accumulation patterns, cyclins are grouped into G1-specific (or D-type in higher eukaryotes) and mitotic (A- and B-types) cyclins (Pines, 1993; Fung and Poon, 2005; Inzé and De Veylder, 2006).
Besides cyclins, other regulatory proteins, including the CDK subunit (CKS) and CDK inhibitory (CKI) proteins, steer CDK activity by direct association with the CDK/CYC complexes. CKS proteins function as docking factors for substrates of the CDK/CYC complex (Patra and Dunphy, 1998; Patra et al., 1999), whereas the CKI proteins bind and inhibit these complexes in response to antimitogenic and developmental signals (LaBaer et al., 1997; Cheng et al., 1999). Furthermore, phosphorylation by the WEE1 and CDK-activating kinases and dephosphorylation by the CDC25 phosphatase control the CDK activity (McGowan and Russell, 1995; Kiyokawa and Ray, 2008). In addition, the CDK activating kinases are regulated by their association with H-type cyclins (Fisher and Morgan, 1994; Kaldis, 1999; Yamaguchi et al., 2000; Shimotohno et al., 2004).
In the attempt to determine all core cell cycle proteins encoded by the Arabidopsis thaliana genome, a high-quality-based annotation protocol identified 61 proteins: 12 CDKs (classified into six classes from A to F), two CKS proteins, 30 cyclins (10 A-type, nine B-type, 10 D-type, and one H-type), one retinoblastoma-related (RBR) protein, one WEE kinase, seven Kip-related proteins (KRPs) (distantly related to mammalian KIP/CIP and CKI proteins), and eight E2F-DP transcription factors (Vandepoele et al., 2002). Among the latter, three unknown cell cycle regulators were found, designated DP-E2F-like (DEL) proteins that probably act as competitors of the E2F-DP proteins (Kosugi and Ohashi, 2002; Mariconti et al., 2002; Vandepoele et al., 2002).
Although the molecular components of the cell cycle control machinery appear to be highly conserved across eukaryotes, not all data obtained for mammalian systems can be directly extrapolated to plants. Most notably, plants have extended families of cell cycle proteins; for example, Arabidopsis has more cyclins than mammals (Vandepoele et al., 2002). It still remains to be analyzed whether the large number of cyclins reflects functional redundancy or specialized functions for some cyclin subclasses. Moreover, in contrast with mammals, plants lack the canonical D-type cyclin binding CDK4/CDK6 (Bryja et al., 2008) but possess a unique class of CDKs, designated as B-type, that contains a plant-specific cyclin binding motif (Joubès et al., 2000). In addition, the KRPs also correspond to a plant-specific class of cell cycle regulators (Ormenese et al., 2004; Wang et al., 2008). Cell cycle regulatory mechanisms unique to plants might be attributed to a number of features related to plant growth, such as the indeterminate mode of development, the absence of cell migration, and the responsiveness of growth rates and development toward changes in environmental conditions (Burssens et al., 2000; de Jager et al., 2005).
Cell cycle progression is controlled by the constant assembly and disassembly of protein complexes that are regulated by gene transcription, posttranslational modifications, and cellular localization (de Lichtenberg et al., 2005a). Despite the increasing knowledge on plant cell cycle control, little is known about the composition of the different CDK/CYC complexes and their spatio-temporal occurrence. Although protein–protein interaction (PPI) data are available, derived from coimmunoprecipitation, in vitro pull-down, yeast two-hybrid (Y2H), and tandem affinity purification assays, a binary interaction network is missing. Here, we analyzed systematically core cell cycle complexes in Arabidopsis by generating a binary PPI map with two complementary interaction assays, bimolecular fluorescence complementation (BiFC) and Y2H. The resulting interactome network was correlated with available cell cycle phase-dependent gene expression data and subcellular localization information, revealing distinct cell cycle networks operating at different cell division stages. In addition, we created an online database, gathering confocal images of the binary interactions acquired from the BiFC assay and the localization data for each tested protein pair.
RESULTS
Building the Binary Arabidopsis Core Cell Cycle Interactome
To generate a binary Arabidopsis core cell cycle PPI map, 58 cell cycle regulatory proteins (see Supplemental Data Set 1A online) were tested for a selected number of interactions by applying independently the BiFC and the Y2H assays. These proteins included 55 core cell cycle proteins previously annotated by Vandepoele et al. (2002), two anaphase-promoting complex (APC) activators (CCS52A2 and CCS52B) (Fülöp et al., 2005; Li and Zhang, 2009), and one identified member of the Arabidopsis CDK family, CDKG;2, (Menges et al., 2005). Four core cell cycle proteins (CYCA1;2, CYCA3;3, CYCD7;1, and CYCB2;4) were excluded from the Vandepoele et al. (2002) set because no open reading frames (ORFs) were available, and two (CDKC;1 and CDKC;2) were excluded because they had been shown to function in the spliceosomal machinery rather than in the cell cycle control (Kitsios et al., 2008).
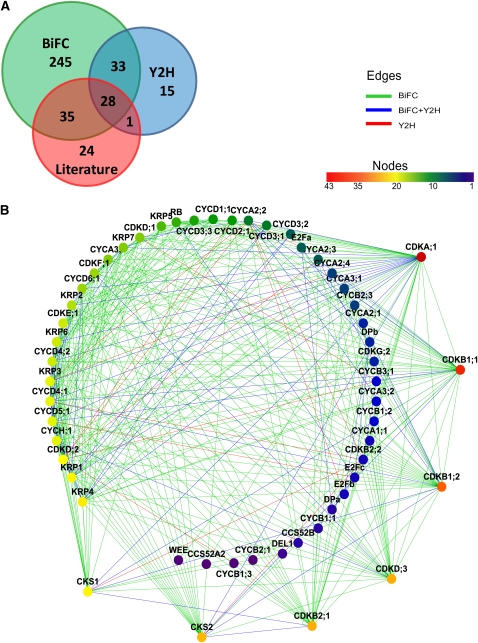
The BiFC vectors were created with the MultiSite Gateway technology that combined the cell cycle ORFs and the green fluorescent protein (GFP) fragments downstream of the constitutive cauliflower mosaic virus 35S promoter (Karimi et al., 2007). Each core cell cycle protein was C-terminally tagged with either the N- (nGFP; 1 to 155 amino acids) or the C-terminal (cGFP; 156 to 720 amino acids) part of the GFP. The constructs were transiently coexpressed in leaf epidermal cells of tobacco (Nicotiana benthamiana) by Agrobacterium tumefaciens–mediated leaf infiltration (Wydro et al., 2006). Binary interactions were screened when one of the core cell cycle proteins was tagged with nGFP and the other protein with cGFP. Only when the first orientation of the tags gave a negative result, was the reciprocal interaction tested (see Supplemental Data Set 1B online). Of the total 917 pairwise interactions tested, 341 (37%) interacting pairs were identified (Figure 1A; see Supplemental Data Set 1C online). The overall quality of the BiFC data was validated by testing for interaction between 40 pairs of unlikely interacting proteins, such as S and M phase cyclins (see Supplemental Table 1 online). None of these cyclin-cyclin pairs yielded a fluorescent signal, indicating that the probability of false positives in our experimental setup is low.
Figure 1.
The Core Cell Cycle Binary PPI Screen.
(A) Comparison of the BiFC and the Y2H results and overlap between interactions detected in the two binary screens and the literature-based data.
(B) Graphical representation of the network. Node color represents the connectivity of each protein. A rainbow scale is used, with red as the most connected through yellow and green to blue as the least connected. Edge colors show the type of assay that detected the interaction for each couple: BiFC (green), Y2H (red), and both methods (blue). The corresponding Cytoscape file is available as Supplemental Data Set 2 online.
The Y2H assay was performed by applying the mating method in a microtiter plate format (Soellick and Uhrig, 2001) with the same set of cell cycle proteins as that used in the BiFC assay excluding CDKG;2, CYCA3;4, CYCD3;3, and CYCB1;2 (see Supplemental Data Set 1A online). For A- and B-type CDKs, a kinase death allele was used as well, whereas for most of the mitotic cyclins an N-terminally truncated (deleting the destruction box) version was included (see Supplemental Data Set 1A online), as these mutations had been found to stabilize the interaction with their binding partners (Boudolf et al., 2009). The cell cycle ORFs were cloned in frame at the C terminus of the binding and activating domains of GAL4 to create translational fusion constructs. Twelve activation domain-containing constructs resulted in self-activation and therefore were only used as preys (see Supplemental Data Set 1A online). The interaction between two proteins was considered positive when both reporter genes were activated (the biosynthetic histidine gene HIS3, resulting in histidine prototrophy, and the β-galactosidase-encoding gene LacZ, giving rise to blue colonies in the presence of 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside) (see Supplemental Data Set 1D online). In total, 733 interactions tested by the BiFC were covered by the Y2H analysis, resulting in 77 (11% of the total number tested) interactions (Figure 1A; see Supplemental Data Set 1C online). As shown in Figure 1A, the overlap between the two binary PPI assays was rather small, illustrating that the techniques are highly complementary in detecting pairwise protein–protein interactions. BiFC detected 264 interactions more than did Y2H. From a total of 357 interactions, 61 (17%) were detected by both PPI methods. The number of interacting pairs detected exclusively in the BiFC and the Y2H assays was 280 (78%) and 16 (4.5%), respectively. For four proteins, namely, CYCB1;4, CYCB2;2, DEL2, and DEL3, no interacting partners were detected in either of the assays.
To estimate the quality of the binary PPI network, we compared it with 88 known pairwise Arabidopsis core cell cycle interactions (a positive reference set) from publicly available databases (see Methods; see Supplemental Data Set 1C online). Sixty-four (73%) of these interactions were confirmed in our data set, of which 35 were detected by BiFC only, 28 by both BiFC and Y2H, and one by Y2H only (Figure 1A). In conclusion, the PPI screens confirmed the majority of data available in the literature and yielded 293 previously undetected binary interactions, the latter corresponding to 82% of all detected interactions.
Visualization of the Cell Cycle Binary PPI Network
The results from the two binary PPI screens were integrated into an interactome network map (Figure 1B), consisting of 54 nodes (representing the individual proteins) that were interconnected by 348 edges (each representing one pairwise interaction). To determine the statistical significance of the network, we performed a randomization experiment (see Methods), which illustrated a significant dissimilarity between the experimental and random PPI networks.
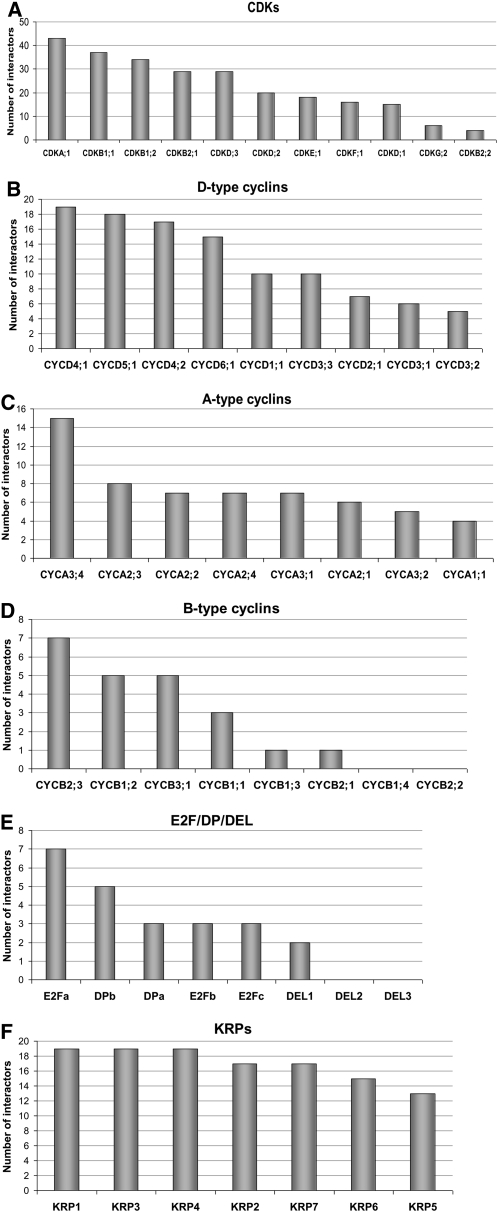
When all analyzed core cell cycle proteins were sorted according to the number of their binding partners (Figure 1B; see Supplemental Data Set 1A online), several highly connected proteins, qualifying as hubs, were identified (e.g., CDKA;1, CDKB1, CDKD;3, CKS, and CYCD4). Next, the number of interactions among different protein families was analyzed. As shown in Figure 2A, the most interconnected nodes were CDKA;1, CDKB1;1, and the two other B-type CDKs (CDKB1;2 and CDKB2;1). By contrast, CDKB2;2 interacted with only nine cell cycle proteins. CKS2 and CKS1 proteins were similar to each other in terms of number of interacting proteins (see Supplemental Data Set 1A online). In general, D-type cyclins and CYCH;1 had more interacting partners than other cyclins (Figures 2B to 2D). On average, the number of interactors was the lowest for B-type cyclins (Figure 2D). E2Fa was the most interconnected transcription factor (Figure 2E). Within the KRP family, the proteins had similar numbers of interactions (Figure 2F).
Figure 2.
Classification of Cell Cycle Proteins According to Their Connectivity in the Network per Protein Family.
(A) CDKs.
(B) D-type cyclins.
(C) A-type cyclins.
(D) B-type cyclins.
(E) E2F/DP/DEL transcription factors.
(F) KRPs.
The number of interactions per protein is given in the ordinate.
Coexpression Analysis of the Cell Cycle Binary PPI Network
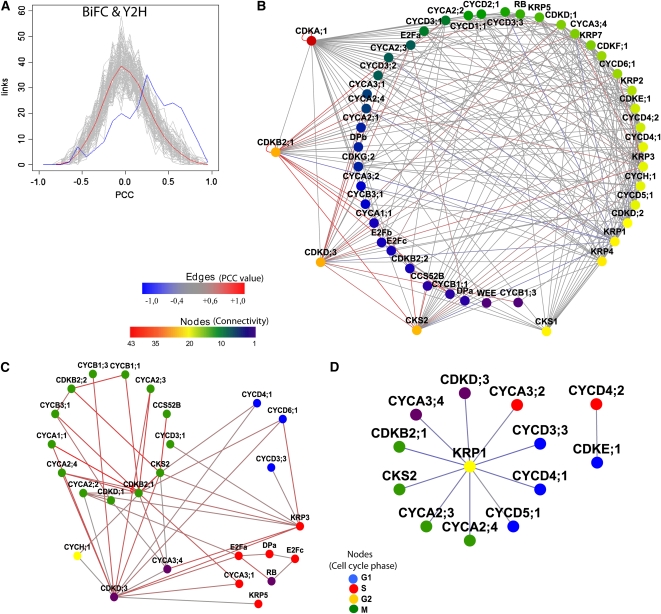
Studies in yeast and human cells indicated a correlation between transcript and protein levels, especially for the periodically transcribed cell cycle genes (Futcher, 1999; Boutros and Byrne, 2005; de Lichtenberg et al., 2005b; Shakir et al., 2008). Therefore, networks of coexpressed genes can provide a framework to predict gene functions because genes with similar expression patterns are most probably involved in a common biological process (Wolfe et al., 2005). To investigate to what extent the PPI data correlated with transcription patterns, the Pearson correlation coefficient (PCC) values were calculated for 264 (74%) detected interactions based on the Arabidopsis ATH1 microarray compendium of 518 experiments focused on cell cycle or plant growth and development (see Supplemental Data Set 1E online). PCC values between 0.6 and 1.0 indicated a highly positive correlation (Aoki et al., 2007), a criterion met in 19% (50) of all detected interactions with assigned PCC values; values between −0.4 and 0.6, no significant correlation for 76% (202) of all detected interactions, and values below −0.4, highly anticorrelated pairs for 5% (12). For 26% (93) of the detected interactions, no PCC values were available. The BiFC and Y2H assays generated similar ratios between positively correlated PCC and anticorrelated values (see Supplemental Figures 1A and 1B online), while literature-curated PPIs have only positive or noncorrelated PCC values (see Supplemental Figure 1C online). When the distribution of PCC values for our PPI network was compared with that of a set of 1000 random networks composed of the same set of 58 cell cycle proteins (see Methods), a significant enrichment (P value < 1.6E-9, one-sided Wilcoxon signed rank test) of interactions with high positive correlation and anticorrelation was observed for the PPI network (Figure 3A). When juxtaposed with random Gaussian distribution, the experimental PPI network was shifted toward the highly positively correlated protein pairs, demonstrating the specificity of the binary interactions. The total PCC curve was similar to the BiFC one, due to the higher number of PPIs detected with this method. Interestingly, after calculating the transcript PCC values for both the BiFC and Y2H PPI networks, we observed that the anticorrelation peak resulted from the BiFC data set (see Supplemental Figures 2A and 2B online). The average transcript PCC value for both BiFC and Y2H data sets equaled 0.25, which is considerably higher than that of a randomized data set (0.015) (see Supplemental Figure 2C online). Such a result increased the confidence in the identified protein pairs.
Figure 3.
Transcript Coexpression Information for the PPI Network.
(A) Comparison of the binary PPI network (blue curve) with a Gaussian distribution of the randomized networks (gray curves and red line for the average of the random data sets).
(B) Binary PPI network merged with pairwise coexpression data. The edge color represents the correlation in transcript expression for each pair. Red depicts highly positively correlated pairs; blue, highly anticorrelated pairs; and gray, protein pairs without significant transcript correlation (interacting pairs for which no transcriptional PCC value was available are not displayed).
(C) Interaction network of highly positive transcript PCC values.
(D) Interaction network with negative transcript PCC values (indicating anticorrelated expression data). Nodes in (C) and (D) are colored based on phase of the cell cycle in which the gene is expressed.
The Cytoscape file corresponding to (B) is available as Supplemental Data Set 3 online and that corresponding to (C) and (D) is available as Supplemental Data Set 4 online.
Subsequently, the PPI network was merged with the PCC data. Colors were attributed to the edges of the binary PPI network according to the PCC value of the protein pair (Figure 3B). Subnetworks of positively and negatively correlated PPIs were observed. The highest PCC values were assigned to the M phase–specific protein pairs, CDKB2;1 with A1-, A2-, B1-, and B3-type cyclins; and CKS2 with CCS52B and CDKB2;1 (Figure 3C). A highly positive transcript correlation occurred also for the S phase–specific RBR1-E2F-DP complex, but a negative correlation was seen for the CDKE;1-CYCD4;2 pair (Figure 3D). The highest anticorrelation was attributed to KRP1 in pairs with CDKB2;1, CDKD;3, CKS2, and D3-, D4-, A2-, and A3-type cyclins (Figure 3D), suggesting a role for KRP1 as a potent CDK/CYC inhibitor. Interestingly, other representatives of the CKI family, KRP3 and KRP5, were highly positively correlated with proteins from both S (e.g., CYCD3;3 and CYCD6;1) and G2/M phases (e.g., CKS2, B-type CDKs, and CYCD3;1) (Figure 3C). Because all anticorrelated pair interactions were detected by the BiFC assay only, we sought to confirm them by another in vivo imaging technique, such as fluorescence lifetime imaging microscopy (FLIM) to determine Förster resonance energy transfer (FRET) (Russinova et al., 2004). Therefore, the interactions between KRP1 and its anticorrelated mitotic partners (CKS2 and CYCA2;4) and partners from other cell cycle phases (such as CYCA3;4) were analyzed. The interaction between KRP1 and CDKA;1 was used as a positive control. The fluorescence lifetime was measured for the donor cyan fluorescent protein (CFP) molecules separately and then compared with the fluorescence lifetime of the donor CFP molecules in the presence of the acceptor yellow fluorescent protein (YFP) molecules. We observed a reproducible reduction in the fluorescence lifetime for KRP1-CFP when coexpressed with either CDKA;1-Venus or CKS2-Venus and a reduction in the fluorescence lifetime for CYCA2;4-CFP when coexpressed with KRP1-Venus (see Supplemental Table 2 online; Figures 4A to 4F), suggesting interactions between these proteins. Interestingly, the strongest reduction in the fluorescence lifetime of CYCA2;4-CFP in the presence of KRP1-Venus was observed in the subnuclear structures, a pattern reminiscent of that of the BiFC KRP1/CYCA2;4 fluorescent complex (Figures 4G to 4I). No reduction in the fluorescence lifetime of CYCA3;4-CFP in the presence of KRP1-CFP was detected (see Supplemental Table 2 online).
Figure 4.
FLIM Assay for the Selected Core Cell Cycle Protein Pairs in Vivo in Tobacco Epidermis.
(A) to (F) Fluorescence intensity images acquired by FLIM are shown as gray-scale pictures (left). Lifetime images (right) are represented as pseudocolor according to the color code ranging from 1 ns (blue) to 3 ns (red). The respective lifetime values measured for KRP1-CFP expressed alone ([A] and [B] as a control in two different experiments) or with CDKA;1-Venus (D) or CKS2-Venus (E), and for CYCA2:4-CFP expressed alone (C) or with KRP1-Venus (F) are indicated on the color scales. Coexpression of two fusion proteins strongly reduces the fluorescence lifetime of the donor protein.
(G) to (I) Subcellular localization of the BiFC complex between KRP1-nGFP and CYCA2;4-cGFP in tobacco epidermal cells.
(G) Images show the complex localizing in bright subnuclear speckles.
(H) and (I) Subcellular localization of KRP1-GFP and CYCA2;4-GFP expressed separately in tobacco epidermis.
Connectivity Patterns and Modular Composition of the Cell Cycle Binary PPI Network
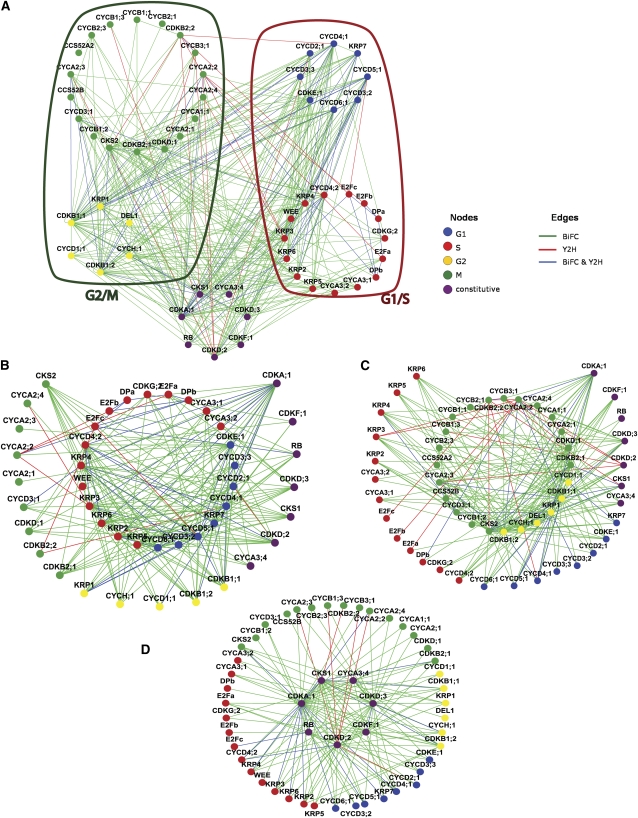
To uncover protein complexes that operate during different cell cycle transitions, the PPI network was overlaid with cell cycle phase-dependent expression data (Menges et al., 2003, 2005). When a transcript was found to oscillate, a protein was assigned to one of the four cell cycle phases, dependent on its main peak of expression. Noncycling genes were considered as constitutively expressed, resulting in a five-cluster network (Figure 5A). Because the PPI network constitutes a small subnet of the global Arabidopsis interactome and thus is not scale free (Stumpf et al., 2005), it might appear that nodes are excessively and randomly connected. To evaluate the connectivity between different clusters, several network parameters were calculated, taking only the edges between proteins from different cell cycle clusters into account (see Supplemental Data Set 1F online). The number of connected components, being a subcluster in which all nodes are connected to each other, characterizes the network topology. Dispersed and less connected networks will have many connected components (Dong and Horvath, 2007). G1/S, S/G2, and G2/M clusters had a low number of connected components, suggesting a crosstalk between these cell cycle phases (see Supplemental Data Set 1F online). Surprisingly, this number was low for the G1/G2 network as well. Similar patterns were detected when the network density was calculated (see Supplemental Data Set 1F online). The density values for the S/M and M/G1 networks were very low (0.047 and 0.055, respectively), whereas for G1/S, S/G2, and G2/M, the values were above 0.11. Again, the value for the G1/G2 network was strikingly high (0.25), pointing to a link between these phases. The network heterogeneity calculations confirmed a higher tendency to contain hubs for the G1/S and M/G2 subnetworks in comparison with the other cell cycle cluster combinations.
Figure 5.
Coexpression Analysis of the PPI Network.
(A) Global network of interaction among all detected interacting protein pairs. The red and green loops include the G1/S and G2/M subnetworks, respectively. The node and edge colors depict the cell cycle phase when the expression of a gene peaks and the assay (green for BiFC; red for Y2H; and blue for both methods) that detected particular interaction pairs, respectively.
(B) Set of G1/S proteins interacting among each other (blue-red circle) and with their first neighbors (directly interacting proteins) from other cell cycle phases.
(C) G2/M proteins interacting among each other (yellow-green circle) and with their first neighbors from other cell cycle phases.
(D) Constitutively expressed proteins at the nexus of cell cycle phase-specific PPIs. The Cytoscape file corresponding to this figure is available as Supplemental Data Set 5 online.
When the first interacting neighbors of the G1/S network were plotted (Figure 5B), a highly interconnected E2F/DP cluster and a cluster including multiple D-type cyclins were revealed. KRPs bound to multiple D-type cyclins, confirming their regulatory role in the cell cycle entry (Nakai et al., 2006). A3-type cyclins were connected to the E2F-RBR hub, in line with their function as S phase cyclins. In addition, the G1/S network interacted with noncycling proteins, such as CDKA;1, D-type CDKs, CDKF;1, CYCA3;4, and CKS1. RBR1 interacted with the E2F subunits but not with the DP proteins. Also, the G2/M regulator CDKB1;1 was linked with the G1/S subnetwork, corroborating the idea that this particular CDK plays a role earlier in the cell cycle than anticipated based on its expression peak during G2 (Boudolf et al., 2004).
To dissect the PPI network among proteins that are most abundant at the G2-to-M transition, the G2/M subnetwork, including its first interacting neighbors, was plotted (Figure 5C). The network displayed connections between the B-type CDKs and the A1-, A2-, and B1-type cyclins. CYCA1;1 interacted not only with the mitotic CDKB2;1, but also with CDKA;1 and CDKB1;1, which both had a broader expression spectrum. All A2-type cyclins bound to three of the B-type CDKs (CDKB1;1, CDKB1;2, and CDKB2;1). CYCB1;1 and CYCB1;2 exhibited a similar interaction pattern. Interestingly, A-type cyclins were found to interact with several KRPs. The G2/M network also included the APC activator proteins CCS52A2 and CCS52B, in accordance with their function in the destruction of mitotic cyclins.
Proteins corresponding to constitutively expressed genes (CDKA;1, CKS1, CDKD, CDKF;1, CYCA3;4, and RBR1) formed a network with each other, but interacted as well with proteins from all cell cycle phases (Figure 5D). For example, CDKA;1 bound the G1-specific D-type cyclins, the S phase–specific A3-type cyclins, and the M phase–specific A2-type cyclins, suggesting a sequential interaction with these cyclins during the cell cycle. CDKA;1 also formed complexes with the CKIs that probably control the entry to the cell cycle. Interestingly, the constitutive CYCA3;4 interacted with the KRPs produced during the S and G2 phases. CKS1 had a wide spectrum of interactors, including CDKs, KRPs, and different cyclins.
In Vivo Localization of the Binary Cell Cycle Complexes
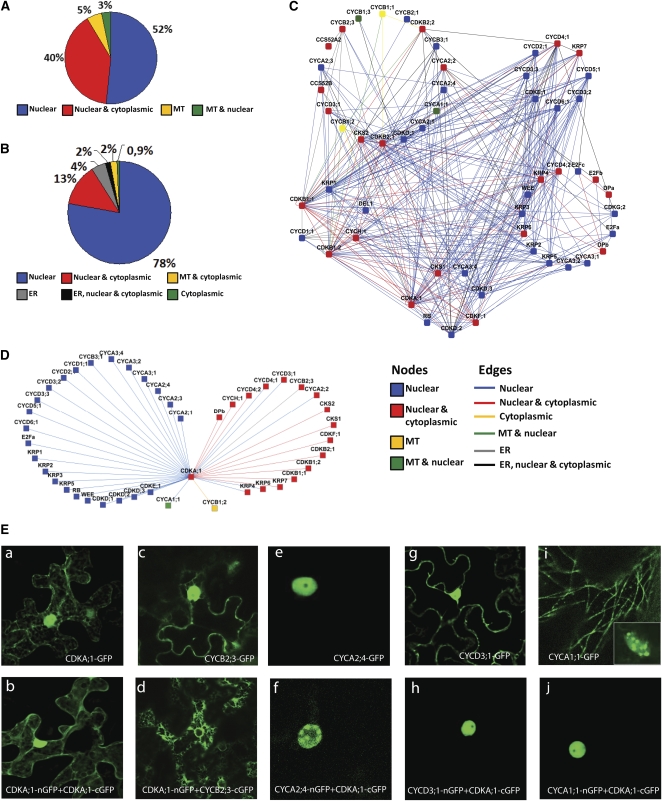
The BiFC assay enables the in vivo spatial detection of the formed binary protein complexes (Kerppola, 2008). We took advantage of this feature and analyzed the localization of the individual proteins and the respective complexes in leaf epidermal cells of tobacco. When transiently expressed, the majority of the 58 core cell cycle proteins localized either exclusively in the nucleus (52%) or in both nucleus and cytoplasm (40%) (Figure 6A; see Supplemental Data Set 1A online). A few proteins (5%) associated with either cortical microtubules (MTs) or were localized in both the nucleus and MTs (3%). The 341 detected complexes were found in the nucleus, cytoplasm, and other compartments, including the endoplasmic reticulum and MTs (Figure 6B; see Supplemental Data Set 1C online). The localization information (http://www.psb.ugent.be/split-gfp/interactome.html) was plotted against the PPI network (Figure 5A) and grouped into the cell cycle phase subnetworks (Figure 6C). The G1 and S networks were clearly enriched for proteins exclusively present and interacting in the nucleus (see Supplemental Figure 3A online), whereas all MT-localized and most of the cytoplasmic complexes were found in the M phase cluster (see Supplemental Figure 3B online).
Figure 6.
Localization of the Binary PPI Network.
(A) Localization distribution of all 58 cell cycle proteins in tobacco leaves. MT, microtubules; ER, endoplasmic reticulum.
(B) Localization distribution of all 341 cell cycle binary protein complexes in tobacco leaves.
(C) Localization map of the proteins and complexes of the binary PPI network. Node colors represent the localization of single GFP-tagged proteins, and edge colors mark the localization of particular complexes.
(D) Connectivity and localization network of CDKA;1 complexes. The Cytoscape file corresponding to (C) and (D) is available as Supplemental Data Set 6 online.
(E) Subcellular localization of CDKA;1/cyclin complexes. Confocal images of tobacco leaves transiently expressing CDKA;1-GFP (a), CDKA;1 homodimer (b), and each of the cyclins separately ([c], [e], [g], and [i]) and the split GFP complexes formed between CDKA;1 and an appropriate cyclin ([d], [f], [h], and [j]): CYCB2;3-GFP ([c] and [d]), CYCA2;4-GFP ([e] and [f]), CYCD3;1-GFP ([g] and [h]), and CYCA1;1-GFP ([i] and [j]).
As CDKA;1 was the most interconnected node in the PPI network, we analyzed the spatial distribution of its complexes in more detail (Figure 6D). CDKA;1 localized as a homodimer in both nucleus and cytoplasm (Figures 6Ea and 6Eb). Interestingly, CDKA;1 formed complexes in distinct compartments when interacting with specific cyclins. For example, the CDKA;1-CYCB2;3 complex localized to the endoplasmic reticulum (Figure 6Ed), whereas CYCB2;3 alone was both nuclear and cytoplasmic (Figure 6Ec). Alternatively, CDKA;1-CYCA2;4 complex formation occurred in the nucleus with more pronounced subnuclear dots, probably corresponding to the chromocenters (Figure 6Ef), although alone, CYCA2;4 localized homogenously to the nucleus (Figure 6Ee). While CDKA;1 and CYCD3;1 both displayed a nuclear and cytoplasmic localization (Figures 6Ea and 6Eg), the CDKA;1-CYCD3;1 complex was detected only in the nucleus (Figure 6Eh). When CYCA1;1 was expressed alone, it localized in the nucleus and on MTs (Figure 6Ei), but the CDKA;1-CYCA1;1 complex was formed exclusively in the nucleus (Figure 6Ej). These results demonstrate subcellular specificity of the binary complexes formed in tobacco epidermis, thus strengthening the credibility of the data.
DISCUSSION
Binary PPI Interaction Screens of the Arabidopsis Cell Cycle Regulators
Cell division in eukaryotes is performed by a network of functionally and structurally conserved core proteins (Murray, 1993). These proteins ultimately form complexes regulated by transcription, phosphorylation, subcellular translocation, and targeted degradation (Morgan, 1997). Plants are characterized by the presence of larger families of core cell cycle proteins, probably reflecting the complexity of cell cycle regulation within developmental contexts (de Jager et al., 2005). Therefore, an important goal of postgenomic plant cell cycle research is to define the function of cell cycle complexes by high-throughput analyses of PPIs and creation of interactome maps. Here, we generated such a comprehensive binary PPI network by applying two complementary approaches, BiFC and Y2H. The outcome of the analysis resulted in 357 interactions of which 293 were previously unknown. Interestingly, more interactions were detected with the BiFC assay than with the Y2H. Recently, Y2H and BiFC have been used to generate a high-quality binary PPI map of yeast in a high-throughput manner (Yu et al., 2008). In this work, both Y2H and BiFC detected a similar number of selected known interactions, although the BiFC assay resulted in a slightly higher number (5%) of false positives when tested on the random reference set of interactions. Although we cannot exclude the possibility that the PPIs detected by BiFC contain false positives, an additional explanation could be that the Y2H generated a considerable number of false negative interactions. Similarly, the Y2H assay used for protein interaction mapping in Caenorhabditis elegans displayed 45% of false negatives, probably because posttranslational modifications or particular localizations were suboptimal in yeast (Walhout et al., 2000). Alternatively, our Y2H screen was either very stringent because of the scoring criteria (requirement of both reporters being activated) or the Y2H sampling sensitivity was relatively low (Venkatesan et al., 2009). Moreover, the Y2H method applied in this work failed to pick up some of the previously detected Y2H interactions (see Supplemental Data Set 1C online). This might be explained by the use of a mating system, which in our hands fails to detect weak interactions. A simultaneous overproduction of core cell cycle regulators in yeast was reported previously to perturb cell growth and proliferation, resulting in growth inhibition (Akada et al., 1997). The BiFC assay, on the other hand, did not depend on cell survival and growth and detected interactions in the physiological environment. Nevertheless, synergetic application of experimental approaches to map PPIs increases the credibility of the results. Thus, we confirmed several of the previously unreported interactions between the transcriptionally anticorrelated pairs KRP1-CKS2 and KRP1-CYCA2;4 by FRET-FLIM. Although technically challenging, this method is applicable for in vivo measurements in plants (Russinova et al., 2004; Nougalli Tonaco et al., 2006), and it is thought to be independent of the fluorophore concentration (Wouters et al., 2001). Additionally, the stronger decrease in fluorescence lifetime at subnuclear speckles supplied high-resolution information on the subcellular distribution of the KRP1-CYCA2;4 complex.
It should be kept in mind that the PPI screen was performed in systems that do not provide spatio-temporal information of the complex formation and with protein levels exceeding the native conditions. Nevertheless, the binary PPI network confirmed most of the published PPIs (73%), strengthening the reliability of our results, whereas those not validated, but published, were mainly interactions between M phase–specific proteins, including, for example, CCS52 and CDKB2;1 (Kono et al., 2003; Fülöp et al., 2005). These complexes were experimentally detected in pull-down and coimmunoprecipitation assays, suggesting that these proteins may require additional factors, indispensable for the complex formation, but lacking in both yeast and nondividing tobacco epidermal cells. Alternatively, the conformation of the complex could account for some false negatives through steric hindrance. Interestingly, we found a subset of transcriptional positively correlated protein pairs that did not interact in our screen (see Supplemental Data Set 1C online). This observation might hint at a considerable number of higher-order interactions, as might be the case for at least the CCS52/CYC interaction. For instance, in mammalian cells, the APC-mediated degradation of cyclin A depends on the CKS of its CDK partner (Wolthuis et al., 2008).
The Network Connectivity Patterns
Visualization of the PPI data set with Cytoscape showed the connectivity of individual core cell cycle proteins. In agreement with previous predictions (Geisler-Lee et al., 2007), CDKA;1 was identified as a high-degree network node or a hub in our PPI network. Hub proteins are essential for mediating interactions among numerous, less connected, proteins and are involved in one or more vital biological processes. Therefore, a deletion of a hub protein is more likely to result in lethality (Jeong et al., 2001; He and Zhang, 2006; Zotenko et al., 2008). Indeed, the cdka;1 mutants are homozygous lethal (Iwakawa et al., 2006; Nowack et al., 2006). The high connectivity values for A- and B-type CDKs were due to their multiple interactions with different cyclin subunits at different cell cycle transitions. The PPI network revealed interactions between CDKA;1 and cyclins of the D, A2, A3, and B types, reflecting the activation of CDKA;1 from the G1/S until mid-M phases (Porceddu et al., 2001; Sorrell et al., 2001). The interaction between the CDKA;1 and A3-type cyclins during the S phase suggests that these complexes function in DNA replication and/or repair. Moreover, the A3-type cyclins are transcriptionally coregulated with several DNA repair-associated genes (Menges et al., 2005).
B1-type CDKs constitute the second most connected nodes in the PPI network, indicating that these proteins may be a plant-specific cell cycle hub. Although no lethal cdkb1 lines were reported, perhaps a double mutant with efficient inhibition of B1-type kinase activity would give more insight into the significance of these proteins in the cell cycle and/or plant development. Similarly, both Arabidopsis CKS proteins exhibit a high connectivity level. In mammals, double CKS depletion leads to embryonic lethality (Martinsson-Ahlzén et al., 2008), but it remains to be verified if plant CKS proteins play such an essential role in development.
Concurrently, the calculation of network parameters indicated that the global PPI network topology exhibited hubs (with high heterogeneity and centrality values) and differed from random networks generated with similar node and edge numbers. Calculation of network centrality revealed that the topologies of most subnetworks are star like, showing that these cell cycle–specific complexes require a central node (or nodes) with multiple interactions, as exemplified by the CDKB2;1 protein that binds several cyclins in M phase and by the CYCD4;2 that interacts with several KRPs in the S phase subcluster. The E2F/DP subcluster shows an opposite topology: the nodes are interconnected equally without a hub; thus, the subnetwork centrality is low. Even though the experimental network is tightly interconnected, it differs significantly from randomized data sets. Comparison of the density between cell cycle phase subnetworks revealed that the linkage between adjacent clusters (G1/S and G2/M) is higher than between clusters biologically separated by a cell cycle phase. The latter have a dispersed network topology with many disconnected components. Interestingly, there seems to be a crosstalk between G1 and G2 clusters. Additionally, many of these interactions were detected by both experimental methods. G1 and G2 subnetworks are relatively connected and dense, which is due to multiple interactions between B1-type CDKs and D-type cyclins, indicating that D-type cyclins operate at the G2/M transition, as reported before (Kono et al., 2003), or that B-type CDKs might function in the S phase, well before their expression peak during G2 and M. The latter hypothesis is supported by the observation that CDKB1;1 expression is regulated by the G1/S-specific E2F transcription factors (Boudolf et al., 2004).
Integrative Analysis of Gene Expression Patterns and the PPI Network
The integration of coexpression data with the PPI network led to the identification of several protein pairs for which the corresponding genes were expressed correlatively, substantiating the quality of our PPI data set. These results were in agreement with previous work in C. elegans and in yeast in which the correlation of the coexpression with interacting proteins was significantly high (Ge et al., 2001; Gunsalus et al., 2005). Complexes with the highest correlation values, such as CDKB/CYCB and RBR1-E2F-DP for the G2-to-M and G1-to-S transitions, respectively, imply that these cell cycle stages are strictly regulated at the transcriptional level. Interestingly, we found a strong positive correlation for the association of KRP3 with G2/M-specific molecules, indicative of a role for KRP3 in the G2-to-M transition, probably as an assembly factor rather than an inhibitor (LaBaer et al., 1997; Cheng et al., 1999; Chytil et al., 2004). Alternatively, KRP3 might prevent premature activation of the G2-to-M transition, ensuring that the M phase onset starts only when all S phase events are completed. KRP1, on the contrary, is highly anticorrelated with several of its interactors, particularly with G2/M phase regulators. Combined with previous published results (Weinl et al., 2005), these data substantiate the role of KRP1 in blocking entry into mitosis and/or exit from the cell cycle. Surprisingly, >50% of the positive PPI data set had PCC values without significant coexpression and interaction correlation. Although false positive interactions are not excluded, alternatively, the complex function might depend on just-in-time assembly rather than just-in-time synthesis (de Lichtenberg et al., 2005a).
The analysis of the binary PPI network sheds more light on specific cell cycle modules. The members of the RBR1-E2F-DP complex, which regulates the G1-to-S transition, were highly correlated among each other. The RBR1 protein bound directly to E2F and not to DP, but it also bound to CDKA;1, as previously shown by in vitro pull-down assays (Boniotti and Gutierrez, 2001), and multiple D-type cyclins. Thus, several CDKA;1/CYCD complexes might target RBR1 for phosphorylation. Interestingly, we observed that E2Fa, E2Fb, and DPb interact with CYCA2;2. In mammals, CDK2/CYCA phosphorylates E2F to enable G2 progression. Similarly, E2Fc proteolysis has been shown to depend on phosphorylation by CDKA1;1 bound to CYCA2;2 and CYCD2;1. CDKB can also phosphorylate the E2Fa protein from poplar (Populus trichocarpa) (Espinosa-Ruiz et al., 2004) and bind A-type cyclins in Arabidopsis, suggesting a function of CDKB in the early stages of the cell cycle. The G1/S complex CYCD4;2-CDKE;1 was highly anticorrelated, implying a negative role for CDKE;1 in the control of the G1-to-S transition. The CDKE;1 of Arabidopsis had primarily been detected as a cell cycle–related kinase, with a role in cell expansion and differentiation (Wang and Chen, 2004). So far, no cyclin partners for the plant CDKE;1 have been identified. In our interaction assays, CDKE;1 formed complexes with six D-type cyclins, suggesting integration of developmental and environmental signals with the cell cycle regulation. The G2/M complexes detected in this screen comprise B2-type CDKs, cyclins (A1-, A2-, and B-types), CCS52, and CKS2. The coexpression PCC analysis uncovered their positive correlation at the transcriptional level. The PPI screen also revealed general trends in cyclin binding specificity. The D1-, D2-, and D3-type cyclins displayed partly different binding partners than did the D4-, D5-, and D6-type cyclins. A-type cyclins have a broader range of interactors than B-type cyclins. A3-type cyclins differ from other A-type cyclins in the number and type of interactors, which might be explained by their broader window of expression during the cell cycle.
Subcellular Localization of the Binary Complexes
The subcellular localization data of the binary PPI network demonstrated that, as anticipated, proteins that interact reside in common compartments. A set of protein pairs interacted exclusively in one specific compartment, even though each of the binding partners had an overlapping localization in other compartments. Interestingly, several of the cyclins, when overproduced in the tobacco epidermis as a GFP fusion, localized to the MTs. In starfish (Asterina pectinifera) oocytes, CYCB1-bound Cdc2 regulates MT dynamics during mitosis (Ookata et al., 1995). In the plant cell suspension system, CYCB1 was cytoplasmic in the interphase and associated with condensed chromosomes during mitosis (Criqui et al., 2001). Nevertheless, when a nondegradable CYCB1 was overproduced in tobacco plants, the organization of phragmoplast MTs was impaired (Weingartner et al., 2004). Here, we demonstrated that CDKA;1 and B-type CDKs of Arabidopsis can bind B1- and A1-type cyclins and that some of these interactions occur on MTs. Microtubule localization in tobacco epidermal cells was never observed for any other CDK partners, suggesting that the cyclins might recruit the CDK/CYC complex to the MTs
In summary, the PPI network has revealed numerous previously unknown interactions and provides a framework for further in-depth analysis of the cell cycle machinery by validating the biological relevance of different complexes.
METHODS
Plasmid Construction
The BiFC constructs were obtained through recombinational Gateway cloning (Invitrogen). The GFP moieties were created by a PCR for N- and C-terminal parts of the fluorescent molecule using the following primer combinations: B2F-nGFP-F (5′-GGGGACAGCTTTCTTGTACAAAGTGGGGATGGTGAGCAAGGGCGAGGAGCTGTTCA-3′), B3R-nGFP+stop-R (5′-GGGGACAACTTTGTATAATAAAGTTGTTTAGGCCATGATATAGACGTTGTGGCTGTTGTA-3′), B2F-cGFP-F (5′-GGGGACAGCTTTCTTGTACAAAGTGGGGGACAAGCAGAAGAACGGCATCAAGGTGA-3′), and B3R-cGFP+stop-R (5′-GGGGACAACTTTGTATAATAAAGTTGTTTACTTGTACAGCTCGTCCATGCCGAGAGTG-3′). DNA sequences were generated that encoded the N terminus of the enhanced GFP, designated nGFP (1 to 465 bp, 1 to155 amino acids) and the C terminus, cGFP (466 to 717 bp, 156 to 239 amino acids) without linker sequences. The primers used to amplify this fragment contained a stop codon to use the GFP moieties at the C terminus of the fusion. All full-length ORFs (without stop codons) of the cell cycle proteins of interest were recombined into the pDONR221 entry vector (Invitrogen) by a BP reaction (see Supplemental Data Set 1G online). The MultiSite Gateway reaction resulted in translational fusions between the cell cycle ORFs and the moieties of GFP, driven by the CaMV 35S promoter. The expression clones were generated in the pH7m34GW and pK7m34GW destination vectors (for the ORF:nGFP and ORF:cGFP fusions, respectively; http://www.psb.ugent.be/gateway/index.php). Proteins fused with GFP were generated in the vector pK7FWG2 (http://www.psb.ugent.be/gateway/index.php).
For the Y2H assays, all analyzed core cell cycle full-length ORFs (containing stop codons) were recombined into the pDEST22 and pDEST32 vectors (Invitrogen) by an LR reaction, resulting in translational fusions between the ORFs and the GAL4 transcriptional activation and GAL4 DNA binding domains, respectively (see Supplemental Data Set 1G online).
Selected full-length ORFs (without stop codons) of the cell cycle proteins, used for the FRET assay, were fused C-terminally in-frame with the eCFP (in pK7CWG2 vector) and Venus YFP (in pK7m34GW). The expression of each fusion construct was driven by the CaMV p35S promoter (http://www.psb.ugent.be/gateway/index.php). All DNA stocks will be made available through the Arabidopsis Biological Resource Center (http://www.biosci.ohio-state.edu/∼plantbio/Facilities/abrc/abrchome.htm).
BiFC Screen
Wild-type tobacco (Nicotiana benthamiana) plants were grown under a normal light regime (14 h of light, 10 h of darkness) at 25°C and 70% relative humidity. All BiFC constructs were transferred into the Agrobacterium tumefaciens strain LBA4404 harboring the virulence plasmid VirG. The obtained Agrobacterium strains were used to infiltrate tobacco leaves, of which the transient expression was assayed according to Yang et al. (2000) with minor modifications. The transformed Agrobacterium strain harboring the constructs of interest was grown for 2 d in a shaking incubator (200 rpm) at 28°C in 5 mL of yeast extract broth medium, supplemented with appropriate antibiotics. After incubation, 2 mL of the bacterial culture was transferred to Eppendorf tubes and centrifuged (10 min, 7000 rpm). The pellets were washed twice with 1 mL of the infiltration buffer (50 mM MES, 2 mM Na3PO4, and 0.5% glucose, pH 5.6). The final pellet was resuspended in the infiltration buffer supplemented with 100 μM acetosyringone. The bacterial suspension was diluted with the same supplemented buffer to adjust the inoculum concentration to the final OD600 value (dilution series from 0.5 to 0.1). For coexpression experiments, 500 mL of each bacterial culture was mixed prior to the leaf infiltration, with the inoculum of each construct adjusted to the required final OD600. The inoculum was delivered to tobacco leaves by gentle pressure infiltration of the lower epidermis with a 1-mL syringe without needle. The infiltrated area of the leaf was delimited and labeled with an indelible pen. The plant was incubated under normal growing conditions and analyzed 3 to 5 d after infiltration. Five infiltrated tobacco leaf segments were analyzed per combination. Interactions were scored positive if at least 10 fluorescent cells per leaf segment were observed.
Y2H Screen
Plasmids encoding the baits (pDEST32) and preys (pDEST22) were transformed into the yeast strain PJ69-4α (MATα; trp1-901, leu2-3,112, ura3-52, his3-200, gal4Δ, gal80Δ, LYS2::GAL1-HIS3, GAL2-ADE2, met2GAL7-lacZ) and PJ69-4a (MATa; trp1-901, leu2-3,112, ura3-52, his3-200, gal4Δ, gal80Δ, LYS2TGAL1-HIS3, GAL2-ADE2, met2TGAL7-lacZ) by the LiAc method (Gietz et al., 1992). Transformed yeast cells were selected on synthetic dextrose (SD) plates without Leu (pDEST 32) or without Trp (pDEST22), respectively. Interactions between proteins were assayed by the mating method. All pDEST32 yeast cultures were inoculated in 200 μL SD without Trp in a 96-well microtiter plate (Falcon), while one pDEST22 yeast culture was inoculated in 50 mL SD medium without Leu. To scale up the yeast cultures, 20 μL of each culture grown for 2 d at 30°C were added to a microtiter plate containing 125 μL of YPD medium (10g/L bacto-yeast extract, 10g/L bacto-peptone, and 20% dextrose) and again grown for 24 h at 30°C. The YPD medium was replaced by SD medium without Leu and Trp. Diploid strains grown in a 96-well microtiter plate (NUNC) for 2 d at 30°C were diluted until OD600 = 0.2 and then added to a 96-well microtiter plate (Falcon) containing either 190 μL SD medium without Leu and Trp, but with His (as control) or SD medium without Leu, Trp, and His. The OD was measured after 2 d of incubation at 30°C. Interactions were scored positive when the ratio of the OD600 of SD medium without Leu, Trp, and His on that of the SD medium without Leu and Trp was exceeding 70%. For the LacZ test, 3 μL of the OD600 = 0.2 dilution was spotted on a Hybond-N+ membrane (GE Healthcare) on a YPD agar plate. After incubation for 2 d at 30°C, the membrane was snap frozen in liquid nitrogen and placed on two Whatmann papers soaked in 6 mL of Z buffer (16.1 g Na2HPO4·7H2O, 5.5 g NaH2PO4·H2O, 0.750 g KCl, and 0.246 g MgSO4·7H2O) with 11 μL β-mercaptoethanol and 100 μL 4% 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside. After incubation for 6 h at 37°C, colonies were scored visually for blue staining by comparison with positive and negative controls supplied with the ProQuest two-hybrid system kit (Invitrogen).
FRET-FLIM Analysis
As donor and acceptor fluorophores, the FRET pairs eCFP and Venus YFP were used. The cells containing the fluorescently tagged proteins were analyzed in vivo in transiently transfected tobacco cells (as described above). The donor fluorescence lifetime was determined by time-correlated single-photon counting (TCSPC) in tobacco epidermal cells. A short pulse of light was used to excite a sample and the intensity of the fluorescence signal was measured as a function of time. For donor fluorescence excitation, a pulsed picosecond diode laser (LDH series; PicoQuant) with an output wavelength of 440 nm at a frequency of 20 MHz along with a dedicated driver (PDL series; PicoQuant) was used. The excitation light was guided into a confocal laser scanning microscope (Olympus FV1000). Laser power was adjusted to give average photon counting rates not >104 to 105 photons/s (0.0001 to 0.001 photon counts per excitation event) to avoid pulse pileup. Images of 256 × 256 pixels were acquired with a ×63 C-Apochromat water immersion objective. Photons emitted by the sample were detected by a single photon avalanche diode (PDM series; PicoQuant). The data were acquired by the PicoHarp 300 TCSPC module (PicoQuant) working in the TTTR mode (time-tagged time-resolved). To calculate the fluorescence lifetime, the SymPhoTime software package (v4.7; PicoQuant) was used. Selected areas of the images (of n = 10 cells on average) corresponding to single nuclei were fitted by maximum likelihood estimation. Depending on the quality of a fit indicated by the value of χ2, a biexponential fitting model excluding IRF (the instrument response function) was applied. A model was rejected when χ2 exceeded a value of 1.0. Mean lifetimes τ for a series of control measurements were presented with a standard deviation and the FRET efficiency (E) was given for mean lifetimes of donor-acceptor pairs as well (see Supplemental Table 2 online). The efficiency of FRET was calculated using the lifetime of the donor in the presence (τDA) and absence of the acceptor probe (τD) according to the formula: E = 1 – (τDA/τD).
Confocal Microscopy and Image Analysis
Transfected tobacco leaves were assayed for fluorescence with a confocal microscope (Olympus FluoView FV1000) equipped with a ×63 water-corrected objective (numerical aperture of 1.2) and with a confocal microscope 100M with software package LSM510 (Zeiss) equipped with a ×63 water-corrected objective (numerical aperture 1.2). GFP fluorescence was imaged with a 488-nm laser excitation. Emission fluorescence was captured in the frame-scanning mode alternating GFP fluorescence via a 500-/550-nm band-pass emission filter.
Network Visualization
Cytoscape software (Shannon et al., 2003) was used to visualize protein–protein interaction networks to overlay it with transcriptional correlation and localization data. The Cytoscape files from this work are available as Supplemental Data Sets 2 to 8 online.
Network Parameter Calculations
The following parameters were calculated for the full network and the subnetworks: the number of connected components, network diameter, clustering coefficient (quantifying how close a network is to a clique, a fully connected set), centrality (referring to the extent to which a network revolves around a single node), and heterogeneity (the tendency of a network to contain hub-like nodes) (Watts and Strogatz, 1998; Dong and Horvath, 2007). Randomization of the network was performed on 1000 sets containing the same node and edge numbers as the experimental network. The random network parameters were calculated in the same way and compared with the values for the empirical data set.
Coexpression Analysis
The microarray compendium of Arabidopsis thaliana of 518 experiments (containing 19,937 genes/proteins) focused on cell cycle or plant growth and development was analyzed (see Supplemental Data Set 1E online). Coexpression between pairs was determined using the PCC. Distributions were created by randomly generating 100 networks consisting of 54 proteins (the same as those present in the experimental PPI network) and 348 pairs from Arabidopsis from within the collection of the tested interacting proteins. The transcript PCC was calculated for 256 (74%) of our binary PPI data set (Figure 3C; see Supplemental Data Set 1C online) because transcriptional data were not available for the remaining 92 (26%) pairs. The latter were not visualized in the transcript PCC network. The average PCC value was similar when calculated for the PPIs from the BiFC to those calculated from the Y2H assays (see Supplemental Figure 2 online). The Wilcoxon signed rank test was applied to verify the statistical significance of the shift between the distribution of the random networks and the experimental one (P value < 1.6E-9). The threshold selected for the positive correlations (0.6) is statistically significant with a P value < 2.2E-16.
Literature Search for Protein–Protein Interactions
The data were imported from publicly available databases, the Arabidopsis Information Resource (www.Arabidopsis.org), IntACT (http://www.ebi.ac.uk/intact/site/index.jsf), the General Repository for Interaction Data sets (BioGRID; http://www.thebiogrid.org/), the Biomolecular Interaction Network Database (www.bind.ca), the Database of Interacting Proteins (http://dip.doe-mbi.ucla.edu), and the Gold Reference data set constructor from Information on Protein complexes (http://rosalind.infj.ulst.ac.uk/GRIP). The data were extracted from PubMed (http://www.ncbi.nlm.nih.gov/sites/entrez?db=PubMed) as well.
Additional Data Online
A database of localization patterns can be found at http://www.psb.ugent.be/split-gfp/interactome.html.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Transcript PCC Value Distribution.
Supplemental Figure 2. Comparison of the Coexpression Correlation of Randomized Networks and the Experimental Binary Network.
Supplemental Figure 3. Distribution of Subcellular Localization Patterns of the Binary Complexes from the G1/S and the G2/M Modules.
Supplemental Table 1. A Random (Negative) Reference PPI Set for the BiFC Assay.
Supplemental Table 2. FLIM Analysis for the Selected Core Cell Cycle Proteins and Protein Pairs in Vivo in Tobacco Epidermis.
Supplemental Data Set 1A. The Arabidopsis Core Cell Cycle Proteins Analyzed in the PPI Screens.
Supplemental Data Set 1B. Tagging Combinations of nGFP and cGFP Detecting Arabidopsis Core Cell Cycle Protein Interactions in the BiFC Assay
Supplemental Data Set 1C. List of All Tested PPIs in the Binary Screens.
Supplemental Data Set 1D. Pairwise Yeast Two-Hybrid Interactions Tested between Arabidopsis Cell Cycle Proteins.
Supplemental Data Set 1E. List of Experiments Used to Build an Arabidopsis ATH1 Microarray Compendium of 518 Experiments Focused on Plant Growth and Development, Used to Calculate the Transcript Pearson Correlation Coefficients.
Supplemental Data Set 1F Network Parameters Calculated for the Cross-Linked Subnetworks Composed of Two Cell Cycle Phase-Specific Clusters.
Supplemental Data Set 1G. Cell Cycle ORF Primer Amplification Information.
Supplemental Data Set 2. Cytoscape File Corresponding to Figure 1B.
Supplemental Data Set 3. Cytoscape File Corresponding to Figure 3B.
Supplemental Data Set 4. Cytoscape File Corresponding to Figures 3C and 3D.
Supplemental Data Set 5. Cytoscape File Corresponding to Figures 5A to 5D.
Supplemental Data Set 6. Cytoscape File Corresponding to Figures 6C and 6D.
Supplemental Data Set 7. Cytoscape File Corresponding to Supplemental Figures 4A and 4B.
Supplemental Data Set 8. Cytoscape File Corresponding to Supplemental Data Set 1F.
Supplementary Material
Acknowledgments
We thank Volker Buschmann (PicoQuant) for his useful comments on the FLIM analysis, Danny Geelen, Els Van Der Scheuren, Raimundo Villarroel, Jan Zethof, Mike Boxem, and Marc Vidal for help at the initial stages of the project, Daniel Van Damme for critical reading of the manuscript and the constructive suggestions, Maria Duda and Oliwia Najder for technical support, and Martine De Cock for help with the manuscript preparation. This work was supported by grants from the European Union (SY-STEM, MRTN-GT-2004-005336), and the Interuniversity Attraction Poles Programme, initiated by the Belgium State Science Policy Office (IUAP VI/33). J.B. is indebted to the European Union-Human Resources and Mobility for an Early Stage Training (Grant MEST-CT-2004-514632) for a predoctoral fellowship and J.H to the Agency for Innovation by Science and Technology for a postdoctoral fellowship. L.D.V. is a postdoctoral fellow of the Research Foundation-Flanders.
References
- Akada R., Yamamoto J., Yamashita I. (1997). Screening and identification of yeast sequences that cause growth inhibition when overexpressed. Mol. Gen. Genet. 254: 267–274 [DOI] [PubMed] [Google Scholar]
- Aoki K., Ogata Y., Shibata D. (2007). Approaches for extracting practical information from gene co-expression networks in plant biology. Plant Cell Physiol. 48: 381–390 [DOI] [PubMed] [Google Scholar]
- Boniotti M.B., Gutierrez C. (2001). A cell-cycle-regulated kinase activity phosphorylates plant retinoblastoma protein and contains, in Arabidopsis, a CDKA/cyclin D complex. Plant J. 28: 341–350 [DOI] [PubMed] [Google Scholar]
- Boudolf V., et al. (2009). CDKB1;1 forms a functional complex with CYCA2;3 to suppress endocycle onset. Plant Physiol. 150: 1482–1493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boudolf V., Vlieghe K., Beemster G.T.S., Magyar Z., Torres Acosta J.A., Maes S., Van Der Schueren E., Inzé D., De Veylder L. (2004). The plant-specific cyclin-dependent kinase CDKB1;1 and transcription factor E2Fa-DPa control the balance of mitotically dividing and endoreduplicating cells in Arabidopsis. Plant Cell 16: 2683–2692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boutros R., Byrne J.A. (2005). D53 (TPD52L1) is a cell cycle-regulated protein maximally expressed at the G2-M transition in breast cancer cells. Exp. Cell Res. 310: 152–165 [DOI] [PubMed] [Google Scholar]
- Bryja V., Pacherník J., Vondráček J., Souček K., Čajánek L., Horvath V., Holubcová Z., Dvořák P., Hampl A. (2008). Lineage specific composition of cyclin D-CDK4/CDK6-p27 complexes reveals distinct functions of CDK4, CDK6 and individual D-type cyclins in differentiating cells of embryonic origin. Cell Prolif. 41: 875–893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burssens S., Himanen K., van de Cotte B., Beeckman T., Van Montagu M., Inzé D., Verbruggen N. (2000). Expression of cell cycle regulatory genes and morphological alterations in response to salt stress in Arabidopsis thaliana. Planta 211: 632–640 [DOI] [PubMed] [Google Scholar]
- Cheng M., Olivier P., Diehl J.A., Fero M., Roussel M.F., Roberts J.M., Sherr C.J. (1999). The p21Cip1 and p27Kip1 CDK 'inhibitors' are essential activators of cyclin D-dependent kinases in murine fibroblasts. EMBO J. 18: 1571–1583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chytil A., Waltner-Law M., West R., Friedman D., Aakre M., Barker D., Law B. (2004). Construction of a cyclin D1-Cdk2 fusion protein to model the biological functions of cyclin D1-Cdk2 complexes. J. Biol. Chem. 279: 47688–47698 [DOI] [PubMed] [Google Scholar]
- Criqui M.C., Weingartner M., Capron A., Parmentier Y., Shen W.-H., Heberle-Bors E., Bögre L., Genschik P. (2001). Sub-cellular localisation of GFP-tagged tobacco mitotic cyclins during the cell cycle and after spindle checkpoint activation. Plant J. 28: 569–581 [DOI] [PubMed] [Google Scholar]
- de Jager S.M., Maughan S., Dewitte W., Scofield S., Murray J.A.H. (2005). The developmental context of cell-cycle control in plants. Semin. Cell Dev. Biol. 16: 385–396 [DOI] [PubMed] [Google Scholar]
- de Lichtenberg U., Jensen L.J., Brunak S., Bork P. (2005a). Dynamic complex formation during the yeast cell cycle. Science 307: 724–727 [DOI] [PubMed] [Google Scholar]
- de Lichtenberg U., Jensen L.J., Fausbøll A., Jensen T.S., Bork P., Brunak S. (2005b). Comparison of computational methods for the identification of cell cycle-regulated genes. Bioinformatics 21: 1164–1171 [DOI] [PubMed] [Google Scholar]
- Dong J., Horvath S. (2007). Understanding network concepts in modules. BMC Syst. Biol. 1: 24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espinosa-Ruiz A., Saxena S., Schmidt J., Mellerowicz E., Miskolczi P., Bakó L., Bhalerao R.P. (2004). Differential stage-specific regulation of cyclin-dependent kinases during cambial dormancy in hybrid aspen. Plant J. 38: 603–615 [DOI] [PubMed] [Google Scholar]
- Fisher R.P., Morgan D.O. (1994). A novel cyclin associates with MO15/CDK7 to form the CDK-activating kinase. Cell 78: 713–724 [DOI] [PubMed] [Google Scholar]
- Fülöp K., Tarayre S., Kelemen Z., Horváth G., Kevei Z., Nikovics K., Bakó L., Brown S., Kondorosi A., Kondorosi E. (2005). Arabidopsis anaphase-promoting complexes: Multiple activators and wide range of substrates might keep APC perpetually busy. Cell Cycle 4: 1084–1092 [PubMed] [Google Scholar]
- Fung T.K., Poon R.Y.C. (2005). A roller coaster ride with the mitotic cyclins. Semin. Cell Dev. Biol. 16: 335–342 [DOI] [PubMed] [Google Scholar]
- Futcher B. (1999). Blast ahead. Nat. Genet. 23: 377–378 [DOI] [PubMed] [Google Scholar]
- Ge H., Liu Z., Church G.M., Vidal M. (2001). Correlation between transcriptome and interactome mapping data from Saccharomyces cerevisiae. Nat. Genet. 29: 482–486 [DOI] [PubMed] [Google Scholar]
- Geisler-Lee J., O'Toole N., Ammar R., Provart N.J., Millar A.H., Geisler M. (2007). A predicted interactome for Arabidopsis. Plant Physiol. 145: 317–329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gietz D., St. Jean A., Woods R.A., Schiestl R.H. (1992). Improved method for high efficiency transformation of intact yeast cells. Nucleic Acids Res. 20: 1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunsalus K.C., et al. (2005). Predictive models of molecular machines involved in Caenorhabditis elegans early embryogenesis. Nature 436: 861–865 [DOI] [PubMed] [Google Scholar]
- He X., Zhang J. (2006). Why do hubs tend to be essential in protein networks? PLoS Genet. 2: e88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humphrey T., Pearce A. (2005). Cell cycle molecules and mechanisms of the budding and fission yeasts. Cell Cycle Control: Mechanisms and Protocols, Methods in Molecular Biology, Vol. 296, Humphrey T., Brooks G., (Totowa, NJ: Humana Press: ), pp. 3–29 [DOI] [PubMed] [Google Scholar]
- Hunt T. (1991). Cyclins and their partners: from a simple idea to complicated reality. Semin. Cell Biol. 2: 213–222 [PubMed] [Google Scholar]
- Inzé D., De Veylder L. (2006). Cell cycle regulation in plant development. Annu. Rev. Genet. 40: 77–105 [DOI] [PubMed] [Google Scholar]
- Iwakawa H., Shinmyo A., Sekine M. (2006). Arabidopsis CDKA;1, cdc2 homologue, controls proliferation of generative cells in male gametogenesis. Plant J. 45: 819–831 [DOI] [PubMed] [Google Scholar]
- Jeong H., Mason S.P., Barabási A.-L., Oltvai Z.N. (2001). Lethality and centrality in protein networks. Nature 411: 41–42 [DOI] [PubMed] [Google Scholar]
- Joubès J., Chevalier C., Dudits D., Heberle-Bors E., Inzé D., Umeda M., Renaudin J.-P. (2000). CDK-related protein kinases in plants. Plant Mol. Biol. 43: 607–620 [DOI] [PubMed] [Google Scholar]
- Kaldis P. (1999). The cdk-activating kinase (CAK): From yeast to mammals. Cell. Mol. Life Sci. 55: 284–296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karimi M., Bleys A., Vanderhaeghen R., Hilson P. (2007). Building blocks for plant gene assembly. Plant Physiol. 145: 1183–1191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerppola T.K. (2008). Bimolecular fluorescence complementation: Visualization of molecular interactions in living cells. Fluorescent Proteins, Methods in Cell Biology, 2nd ed, Vol. 85, Sullivan K.F., (Amsterdam: Academic Press; ), pp. 431–470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitsios G., Alexiou K.G., Bush M., Shaw P., Doonan J.H. (2008). A cyclin-dependent protein kinase, CDKC2, colocalizes with and modulates the distribution of spliceosomal components in Arabidopsis. Plant J. 54: 220–235 [DOI] [PubMed] [Google Scholar]
- Kiyokawa H., Ray D. (2008). In vivo roles of CDC25 phosphatases: biological insight into the anti-cancer therapeutic targets. Anticancer. Agents Med. Chem. 8: 832–836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kono A., Umeda-Hara C., Lee J., Ito M., Ichimiya H., Umeda M. (2003). Arabidopsis D-type cyclin CYCD4;1 is a novel cyclin partner of B2-type cyclin-dependent kinase. Plant Physiol. 132: 1315–1321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosugi S., Ohashi Y. (2002). E2Ls, E2F-like repressors of Arabidopsis that bind to E2F sites in a monomeric form. J. Biol. Chem. 277: 16553–16558 [DOI] [PubMed] [Google Scholar]
- LaBaer J., Garrett M.D., Stevenson L.F., Slingerland J.M., Sandhu C., Chou H.S., Fattaey A., Harlow E. (1997). New functional activities of the p21 family of CDK inhibitors. Genes Dev. 11: 847–862 [DOI] [PubMed] [Google Scholar]
- Li M., Zhang P. (2009). The function of APC/CCdh1 in cell cycle and beyond. Cell Div. 4: 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mariconti L., Pellegrini B., Cantoni R., Stevens R., Bergounioux C., Cella R., Albani D. (2002). The E2F family of transcription factors from Arabidopsis thaliana. Novel and conserved components of the retinoblastoma/E2F pathway in plants. J. Biol. Chem. 277: 9911–9919 [DOI] [PubMed] [Google Scholar]
- Martinsson-Ahlzén H.-S., Liberal V., Grünenfelder B., Chaves S.R., Spruck C.H., Reed S.I. (2008). Cyclin-dependent kinase-associated proteins Cks1 and Cks2 are essential during early embryogenesis and for cell cycle progression in somatic cells. Mol. Cell. Biol. 28: 5698–5709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGowan C.H., Russell P. (1995). Cell cycle regulation of human WEE1. EMBO J. 14: 2166–2175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menges M., de Jager S.M., Gruissem W., Murray J.A.H. (2005). Global analysis of the core cell cycle regulators of Arabidopsis identifies novel genes, reveals multiple and highly specific profiles of expression and provides a coherent model for plant cell cycle control. Plant J. 41: 546–566 [DOI] [PubMed] [Google Scholar]
- Menges M., Hennig L., Gruissem W., Murray J.A.H. (2003). Genome-wide gene expression in an Arabidopsis cell suspension. Plant Mol. Biol. 53: 423–442 [DOI] [PubMed] [Google Scholar]
- Morgan D.O. (1997). Cyclin-dependent kinases: Engines, clocks, and microprocessors. Annu. Rev. Cell Dev. Biol. 13: 261–291 [DOI] [PubMed] [Google Scholar]
- Murray A.W. (1993). Turning on mitosis. Curr. Biol. 3: 291–293 [DOI] [PubMed] [Google Scholar]
- Nakai T., Kato K., Shinmyo A., Sekine M. (2006). Arabidopsis KRPs have distinct inhibitory activity toward cyclin D2-associated kinases, including plant-specific B-type cyclin-dependent kinase. FEBS Lett. 580: 336–340 [DOI] [PubMed] [Google Scholar]
- Nougalli Tonaco I.A., Borst J.W., de Vries S.C., Angenent G.C., Immink R.G.H. (2006). In vivo imaging of MADS-box transcription factor interactions. J. Exp. Bot. 57: 33–42 [DOI] [PubMed] [Google Scholar]
- Nowack M.K., Grini P.E., Jakoby M.J., Lafos M., Koncz C., Schnittger A. (2006). A positive signal from the fertilization of the egg cell sets off endosperm proliferation in angiosperm embryogenesis. Nat. Genet. 38: 63–67 [DOI] [PubMed] [Google Scholar]
- Nurse P. (1990). Universal control mechanism regulating onset of M-phase. Nature 344: 503–508 [DOI] [PubMed] [Google Scholar]
- Ookata K., Hisanaga S.-i., Bulinski J.C., Murofushi H., Aizawa H., Itoh T.J., Hotani H., Okumura E., Tachibana K., Kishimoto T. (1995). Cyclin B interaction with microtubule-associated protein 4 (MAP4) targets p34cdc2 kinase to microtubules and is a potential regulator of M-phase microtubule dynamics. J. Cell Biol. 128: 849–862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ormenese S., de Almeida Engler J., De Groodt R., De Veylder L., Inzé D., Jacqmard A. (2004). Analysis of the spatial expression pattern of seven Kip related proteins (KRPs) in the shoot apex of Arabidopsis thaliana. Ann. Bot. (Lond.) 93: 575–580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patra D., Dunphy W.G. (1998). Xe-p9, a Xenopus Suc1/Cks protein, is essential for the Cdc2-dependent phosphorylation of the anaphase-promoting complex at mitosis. Genes Dev. 12: 2549–2559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patra D., Wang S.X., Kumagai A., Dunphy W.G. (1999). The Xenopus Suc1/Cks protein promotes the phosphorylation of G2/M regulators. J. Biol. Chem. 274: 36839–36842 [DOI] [PubMed] [Google Scholar]
- Pines J. (1993). Cyclins and cyclin-dependent kinases: Take your partners. Trends Biochem. Sci. 18: 195–197 [DOI] [PubMed] [Google Scholar]
- Porceddu A., Stals H., Reichheld J.-P., Segers G., De Veylder L., De Pinho Barrôco R., Casteels P., Van Montagu M., Inzé D., Mironov V. (2001). A plant-specific cyclin-dependent kinase is involved in the control of G2/M progression in plants. J. Biol. Chem. 276: 36354–36360 [DOI] [PubMed] [Google Scholar]
- Russinova E., Borst J.W., Kwaaitaal M., Caño-Delgado A., Yin Y., Chory J., de Vries S.C. (2004). Heterodimerization and endocytosis of Arabidopsis brassinosteroid receptors BRI1 and AtSERK3 (BAK1). Plant Cell 16: 3216–3229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shakir R., Ngo N., Naresh K.N. (2008). Correlation of cyclin D1 transcript levels, transcript type and protein expression with proliferation and histology among mantle cell lymphoma. J. Clin. Pathol. 61: 920–927 [DOI] [PubMed] [Google Scholar]
- Shannon P., Markiel A., Ozier O., Baliga N.S., Wang J.T., Ramage D., Amin N., Schwikowski B., Ideker T. (2003). Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 13: 2498–2504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimotohno A., Umeda-Hara C., Bisova K., Uchimiya H., Umeda M. (2004). The plant specific kinase CDKF;1 is involved in activating phosphorylation in cyclin-dependent kinase-activating kinases in Arabidopsis. Plant Cell 16: 2954–2966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soellick T.-R., Uhrig J.F. (2001). Development of an optimized interaction-mating protocol for large-scale yeast two-hybrid analyses. Genome Biol. 2: research0052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorrell D.A., Menges M., Healy J.M.S., Deveaux Y., Amano X., Su Y., Nakagami H., Shinmyo A., Doonan J.H., Sekine M., Murray J.A.H. (2001). Cell cycle regulation of cyclin-dependent kinases in tobacco cultivar Bright Yellow-2 cells. Plant Physiol. 126: 1214–1223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stumpf M.P.H., Wiuf C., May R.M. (2005). Subnets of scale-free networks are not scale-free: Sampling properties of networks. Proc. Natl. Acad. Sci. USA 102: 4221–4224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ubersax J.A., Woodbury E.L., Quang P.N., Paraz M., Blethrow J.D., Shah K., Shokat K.M., Morgan D.O. (2003). Targets of the cyclin-dependent kinase Cdk1. Nature 425: 859–864 [DOI] [PubMed] [Google Scholar]
- Vandepoele K., Raes J., De Veylder L., Rouzé P., Rombauts S., Inzé D. (2002). Genome-wide analysis of core cell cycle genes in Arabidopsis. Plant Cell 14: 903–916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venkatesan K., et al. (2009). An empirical framework for binary interactome mapping. Nat. Methods 6: 83–90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walhout A.J.M., Sordella R., Lu X., Hartley J.L., Temple G.F., Brasch M.A., Thierry-Mieg N., Vidal M. (2000). Protein interaction mapping in C. elegans using proteins involved in vulval development. Science 287: 116–122 [DOI] [PubMed] [Google Scholar]
- Wang H., Zhou Y., Bird D.A., Fowke L.C. (2008). Functions, regulation and cellular localization of plant cyclin-dependent kinase inhibitors. J. Microsc. 231: 234–246 [DOI] [PubMed] [Google Scholar]
- Wang W., Chen X. (2004). HUA ENHANCER3 reveals a role for a cyclin-dependent protein kinase in the specification of floral organ identity in Arabidopsis. Development 131: 3147–3156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watts D.J., Strogatz S.H. (1998). Collective dynamics of 'small-world' networks. Nature 393: 440–442 [DOI] [PubMed] [Google Scholar]
- Weingartner M., Criqui M.-C., Mészáros T., Binarova P., Schmit A.-C., Helfer A., Derevier A., Erhardt M., Bögre L., Genschik P. (2004). Expression of a nondegradable cyclin B1 affects plant development and leads to endomitosis by inhibiting the formation of phragmoplast. Plant Cell 16: 643–657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinl C., Marquardt S., Kuijt S.J.H., Nowack M.K., Jakoby M.J., Hülskamp M., Schnittger A. (2005). Novel functions of plant cyclin-dependent kinase inhibitors, ICK1/KRP1, can act non-cell-autonomously and inhibit entry into mitosis. Plant Cell 17: 1704–1722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wittenberg C. (2005). Cyclin guides the way. Nature 434: 34–35 [DOI] [PubMed] [Google Scholar]
- Wolfe C.J., Kohane I.S., Butte A.J. (2005). Systematic survey reveals general applicability of “guilt-by-association” within gene coexpression networks. BMC Bioinformatics 6: 227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolthuis R., Clay-Farrace L., van Zon W., Yekezare M., Koop L., Ogink J., Medema R., Pines J. (2008). Cdc20 and Cks direct the spindle checkpoint-independent destruction of cyclin A. Mol. Cell 30: 290–302 [DOI] [PubMed] [Google Scholar]
- Wouters F.S., Verveer P.J., Bastiaens P.I.H. (2001). Imaging biochemistry inside cells. Trends Cell Biol. 11: 203–211 [DOI] [PubMed] [Google Scholar]
- Wydro M., Kozubek E., Lehmann P. (2006). Optimization of transient Agrobacterium-mediated gene expression system in leaves of Nicotiana benthamiana. Acta Biochim. Pol. 53: 289–298 [PubMed] [Google Scholar]
- Yamaguchi M., Fabian T., Sauter M., Bhalerao R.P., Schrader J., Sandberg G., Umeda M., Uchimiya H. (2000). Activation of CDK-activating kinase is dependent on interaction with H-type cyclins in plants. Plant J. 24: 11–20 [DOI] [PubMed] [Google Scholar]
- Yang Y., Li R., Qi M. (2000). In vivo analysis of plant promoters and transcription factors by agroinfiltration of tobacco leaves. Plant J. 22: 543–551 [DOI] [PubMed] [Google Scholar]
- Yu H., et al. (2008). High-quality binary protein interaction map of the yeast interactome network. Science 322: 104–110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zotenko E., Mestre J., O'Leary D.P., Przytycka T.M. (2008). Why do hubs in the yeast protein interaction network tend to be essential: reexamining the connection between the network topology and essentiality. PLOS Comput. Biol. 4: e1000140. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.