Abstract
The Central glycolytic genes Repressor (CggR) from Bacillus subtilis belongs to the SorC family of transcription factors that control major carbohydrate metabolic pathways. Recent studies have shown that CggR binds as a tetramer to its tandem operator DNA sequences and that the inducer metabolite, fructose 1,6-bisphosphate (FBP), reduces the binding cooperativity of the CggR/DNA complex. Here, we have determined the effect of FBP on the size, shape and stoichiometry of CggR complexes with full-length and half-site operator sequence by small-angle X-ray scattering, size-exclusion chromatography, fluorescence cross-correlation spectroscopy and noncovalent mass spectrometry (MS). Our results show that CggR forms a compact tetrameric assembly upon binding to either the full-length operator or two half-site DNAs and that FBP triggers a tetramer–dimer transition that leaves a single dimer on the half-site or two physically independent dimers on the full-length target. Although the binding of other phospho-sugars was evidenced by MS, only FBP was found to completely disrupt dimer–dimer contacts. We conclude that inducer-dependent dimer–dimer bridging interactions constitute the physical basis for CggR cooperative binding to DNA and the underlying repression mechanism. This work provides experimental evidences for a cooperativity-based regulation model that should apply to other SorC family members.
INTRODUCTION
The regulation of gene expression in all organisms from the simplest organisms to man involves modulation of the binding properties of protein/nucleic acid complexes, in most cases by small ligands, other protein partners or post-translational modifications. Besides changes in the affinity and stoichiometry of the complexes, transcription activation is often coupled to changes in cooperative interactions that dominate the assembly reaction (1–5). However, for most transcription factors the mechanisms underlying cooperativity are poorly understood. Structural information of the highest resolution possible is the key to elucidating the fine mechanisms of these regulatory events (6–10). However, given their modular nature and dynamic properties, atomic resolution structures of these proteins and their complexes with nucleic acids are often limited to particular domains or certain conformations and hence provide little information concerning the complex dynamic equilibria implicated in the control of their function. In such cases, the limited structural data must be complemented by information from other methods that determine the size, shape, composition and energetics of these complexes under a variety of conditions.
Metabolic pathways for the utilization of carbohydrates in bacteria are highly regulated at the level of gene expression (11–13). We have been interested in understanding the molecular mechanisms of transcriptional regulation that accompany the switch between glycolysis and gluconeogenesis in Bacillus subtilis. In this microorganism as well as other Gram-positive bacteria, two different enzymes, GapA and GapB, catalyze the inter-conversion of triose phosphates at a central control point in glycolysis (14). GapA is a NAD-dependent glyceraldehyde 3-phosphate dehydrogenase (GAPDH) that is produced and functions only under glycolytic conditions to produce 1,3-bisphosphoglycerate. GapB, on the other hand, is a NADP-dependent GAPDH that performs the reverse reaction under gluconeogenic conditions. At the level of transcription, the expression of the genes encoding GapA and four other key glycolytic enzymes is under the direct control of the Central glycolytic genes Repressor (CggR), the product of the first gene of the hexacistronic gapA operon (14–17).
CggR is a member of the SorC family of bacterial transcriptional repressors, characterized by an N-terminal DNA-binding domain (DBD) followed by a phospho-sugar binding (PSB) domain homologous to glucosamine-6-phosphate deaminases from the NagB family (18). Other characterized members of this family are the sorbitol operon regulator SorC from Klebesiella pneumoniae (19) and the deoxyribonucleoside repressor DeoR from B. subtilis (20,21). Within this family, CggR and its paralogs in other Gram-positive bacteria define a subclass of repressors with a 100-residue-long DBD predicted to contain a MarR-type winged-helix (wH) motif (17,18). Recently, a few crystal structures from SorC family members have been deposited in the Protein Data Bank, notably several liganded forms of the PSB domain from B. subtilis CggR (22) and the full-length structure of K. pneumoniae SorC (23). Identification of DNA operator sequences and effector ligands have been limited to B. subtilis DeoR (not structurally related to Escherichia coli DeoR) (24) and CggR (17). Recently, we have studied in detail CggR interactions with operator DNA using fluorescence anisotropy and other biochemical and biophysical approaches. These studies demonstrated that CggR interacts as a tetramer with the tandem DNA repeats forming the gapA operator. Each direct repeat constitutes the binding site for one CggR dimer, the dissociation constant (KD) being over a 100-fold lower for the 3′-repeat (right half-operator OR, KD < 0.5 nM) than for the 5′-repeat (left half-operator OL, KD = 50 nM) (25). We showed that this asymmetric binding is compensated by a highly cooperative mode of interaction on the full-length operator (OLR) and that the inducer metabolite, fructose 1,6-bisphosphate (FBP), has a strong negative effect on this interaction, the cooperative free energy dropping from 2.3 kcal/mol in the absence of FBP to 0.5 kcal/mol in the presence of 2 mM FBP (25). Besides the inhibition of repressor/operator binding cooperativity, FBP is also responsible of CggR conformational changes modifying the oligomerization and stability of the protein (26). However, no structural data has yet been provided on the CggR/operator complex and the molecular features responsible for its inducer-dependent cooperative assembly remain unknown.
In the present work, we used small-angle X-ray scattering (SAXS) in combination with size-exclusion chromatography (SEC), fluorescence cross-correlation spectroscopy (FCCS) and noncovalent mass spectrometry (MS) in order to characterize CggR/DNA complexes. SAXS is now a commonly used technique to obtain information on the size, shape, domain organization and interactions of biomacromolecules in solution. In addition to direct measurement of particle size parameters, SAXS data analysis also allows ab initio shape determination as well as rigid-body modeling of low-resolution structures, thanks to recently developed methods especially powerful for the characterization of macromolecular complexes [for review, see (27,28,29)]. In a limited but growing number of studies, SAXS has been employed to probe protein/nucleic acid interactions and determine solution structures of DNA complexes with proteins such as the tumor suppressor P53 (30,31), DNA polymerase (32), the basic helix-loop-helix leucine zipper domain from the human upstream stimulatory factor 1 (33) or proteins interacting with the herpes simplex virus 1 origin of replication (34). In all these studies, SAXS experiments have been combined with other biophysical methods such as analytical ultra-centrifugation, NMR, X-ray crystallography and electron microscopy.
Here, SAXS together with SEC, FCCS and MS experiments were performed with the CggR repressor bound to its full-length operator (OLR) DNA or the high-affinity right half-site (OR) under repressing or inducing condition, i.e. in the absence or in the presence of the inducer metabolite FBP. The ensemble of size and mass parameters retrieved from these experiments were then integrated to construct rigid-body models of the inducer-free and inducer-bound forms of the CggR/operator DNA complexes. Altogether our results demonstrate that CggR binds as a compact tetramer to its DNA target and that FBP binding abolishes dimer/dimer bridging interactions, allowing us to draw the schematic of a cooperativity-based regulation model for the repression/induction of gapA expression. Moreover, the relative high resolution of noncovalent mass spectra provided experimental support on inducer binding specificity and stoichiometry. This study illustrates how, in the absence of high-resolution 3D data on protein/DNA complexes, a multidisciplinary approach combining classical and state-of-the-art biophysical methods can be applied to elucidate the structural mechanisms underlying transcriptional regulation.
MATERIALS AND METHODS
Preparation of protein/DNA complexes
The CggR protein fused to an N-terminal His-tag was purified by affinity chromatography as described previously (25). The molecular weight (MW) of the purified protein monomer measured by MS was 38 780.3 ± 0.8 Da, in very good agreement with the MW calculated from the theoretical amino acid sequence (38 780.8 Da) (35).
The oligonucleotides corresponding to the full-length operator site (OLR, 45 bp, 5′-TGACGGGACGTTTTTTGTCATAGCGGGACATATAATGTCCAGCAA-3′) or the high-affinity right half-site operator (OR, 23 bp, 5′-TAGCGGGACATATAATGTCCAGC-3′) were purchased from Eurogentec (Figure 1a). The sense and anti-sense strands at 50 µM each were hybridized in TE buffer, 250 mM NaCl by heating to 90°C for 10 min and slowly cooling down to room temperature using a thermocycler. Complexes were formed by mixing the purified protein and DNA at a molar ratio of 4 : 1 (OLR) or 2 : 1 (OR) (protein/DNA) in a final volume of 500 µl, with ∼10% excess of protein. Protein/DNA complexes were purified by SEC on Superdex 200 HR 10/30 (GE Healthcare) pre-equilibrated with 20 mM Tris–HCl pH 8.0, 150 mM NaCl, 2 mM EDTA and 2 mM DTT. The protein/DNA complexes eluted in a single peak and the most concentrated fractions (500 µl) were pooled and eventually concentrated to a final protein concentration of 1 mg/ml as estimated by Bradford assay.
Figure 1.
The SEC and SAXS data of CggR/DNA complexes. (A) The B. subtilis gapA operator region targeted by the CggR repressor. The nucleotide sequence of the CggR-binding region identified by DNase I footprinting experiment (17) is shown, located 25-bp downstream of the transcription start (+1), and upstream of the translation initiation codon of cggR, the first coding sequence of the hexacistronic gapA operon (15). The full-length operator sequence OLR is composed of two tandem repeats (long arrows), OL and OR, each comprising a small palindrome (small black arrows) proposed to constitute the recognition motif for one CggR dimer (25). Indicated bellow is the length of the DNA fragments and the color code used in Figure 1 (B–D) for representing SEC and SAXS data obtained with the different CggR/DNA complexes in the presence/absence of the inducer metabolite FBP. (B) Elution profiles of 250 µl samples of purified CggR/DNA complexes (colored curves) at 1 mg/ml (protein concentration) and injected on Superdex 200 HR10/30 (GE Healthcare) at a flow rate of 0.5 ml/min in 150 mM NaCl, 20 mM Tris-HCl pH 8, 2 mM EDTA, 2 mM DTT, eventually supplemented with 0.5 mM FBP (red and green curves). Black curves, dsDNA alone corresponding to the full-length operator (OLR, 45 bp, dotted line) or the right half-site (OR, 23 bp, plain line). (C) Comparison of the normalized scattering profiles of CggR/DNA complexes in the presence and absence of FBP [identical samples and buffer condition as in (B)]. (D) Distance distribution function, P(r), computed from the experimental scattering data and normalized to the maximum value of unity using GNOM (39).
SEC
The SEC experiments were conducted at room temperature on Superdex 200 HR 10/30 using Akta purifier (GE Healthcare) in 20 mM Tris–HCl pH 8.0 containing 150 mM NaCl, 2 mM EDTA and 2 mM DTT (running buffer). Samples (250 µl) of purified CggR/DNA complexes at 1 mg/ml (protein concentration) were injected on the pre-equilibrated column at a flow rate of 0.5 ml/min and the elution profiles were continuously monitored with an online detector at a wavelength of 280 and 260 nm. For SEC experiments in the presence of FBP, samples were incubated with 2 mM FBP for 5 min before injection on the column equilibrated with the running buffer containing 0.5 mM FBP. Protein standards (purchased from GE Healthcare) used for column calibration under the same experimental conditions, and their corresponding Stokes radii, RS, were ovalbumin (30.5 Å), bovine serum albumin (35.5 Å), aldolase (48.1 Å), ferritin (61 Å) and thymoglobulin (85 Å). The void volume was determined from blue dextran and the bed volume from conductivity measurements. The partition coefficient, Kav = (Ve – V0)/(Vt – V0), was calculated, where Ve, Vt and V0 are the elution volumes of the protein samples, total bed volume and void volume, respectively (36). Calibration curves in the absence or in the presence of FBP were obtained by plotting the Stokes radius of the standard proteins versus Kav, and the Stokes radius of the CggR/DNA complexes were calculated from each curve although no significant difference was noticed between the two curves (data not shown).
SAXS
The SAXS data were collected on the EMBL X33 beamline with a linear gas detector on the storage ring DORIS III [Deutsches Elektronen-Synchrotron (DESY), Hamburg, Germany]. Samples containing protein/DNA complexes were prepared in 20 mM Tris–HCl pH 8.0, 150 mM NaCl, 2 mM EDTA, 2 mM DTT at a concentration of 1 mg/ml of protein as determined by Bradford assay. No radiation damage was noticed during data acquisition. All SAXS data were analyzed using the ATSAS package (37). Data were first processed using PRIMUS (38). Radii of gyration (Rg) were evaluated using the Guinier approximation (39) and the excluded volume Vp of the particles was computed from the Porod invariant (40). The pair-distance distribution functions P(r) were calculated from the entire scattering patterns using the indirect transform program GNOM (39) and manual adjustment of the particles maximum diameter (Dmax). Rg data were also computed from GNOM and these values were in excellent agreement with the results of the Guinier analysis.
Fluorescence labeling of DNA and cross-correlation spectroscopy
An oligonucleotide corresponding to the sense strand of the right half-operator (OR) site was purchased from Sigma Genosys with a 5′-reactive amine group. Labeling with Atto 647N succinimidyl ester dye (Invitrogen) was performed by adding a 10-fold molar excess of the dye to the oligonucleotide in 0.1 M Sodium Borate pH 9 buffer. The reaction was allowed to proceed at room temperature for 3 h with continuous agitation. The reaction was stopped by adding 10% Tris–HCl 1 M and after ethanol precipitation, the labeled oligonucleotide was further purified using reverse phase chromatography on a C2–C18 column. The anti-sense OR oligonucleotide was purchased 3′-labeled with fluorescein from Eurogentec as well the complementary unlabeled oligonucleotides. Hybridization of complementary strands was performed as described above, using a 10% molar excess of the non-labeled strand.
Two-photon, two-color FCCS (2γFCCS) was performed as described in Zorilla et al. (41) using a two-channel APD-based detector (ISS, Champaign, IL, USA). Excitation from a femtosecond pulsed infrared laser (MaiTai, Newport/Spectra Physics, Mountain View, CA, USA) was focused through a 63X 1.4NA oil immersion objective (Zeiss Apochromat) onto a 10-µl pegylated sample chamber containing the fluorescent sample of interest. The excitation wavelength used was 780 nm at a laser power of 10 mW on the back aperture of the objective. The power and wavelength were selected for optimal excitation of the two fluorophores used (Fluorescein and Atto647N) and the excitation power was chosen to give the best signal while avoiding saturation and photo bleaching effects (42). Infrared light was filtered from the detected light by using a 700-nm low-pass dichroic filter (Chroma Technology Corporation, Rockingham, VT). Emitted light was then split at 505 nm by another low-pass dichroic filter (Chroma). Light was then detected by two avalanche photodiodes (Perkin Elmer) and recorded at 2.4 MHz by a data acquisition card (ISS, Champaign, IL). FCCS measurements were performed at 20°C in 20 mM Tris-HCl pH 8.0, 150 mM NaCl, 2 mM EDTA, 2 mM DTT. The concentration of the labeled double-stranded DNA fragments was 60 nM that of the protein 300 nM in monomer units and FBP was added at 0.5 mM. Auto and cross-correlation functions were calculated from the photon streams of the two detection channels as previously described (41).
Noncovalent MS
Before MS experiments, CggR/DNA complexes were buffer exchanged against 150 mM ammonium acetate solution pH 8.0 (adjusted with ammonia) using seven successive concentration/dilution steps performed with centrifugal concentrators (Vivaspin, 30 kDa MW cutoff membranes, Sartorius, Göttingen, Germany). Protein concentration was determined by Bradford assay. FBP trisodium salt hydrate, fructose-6-phosphate (F6P) disodium salt hydrate and glucose-1,6-bisphosphate (GBP) tetra(cyclohexylammonium) salt hydrate were purchased from Sigma Aldrich (Steinheim, Germany).
Noncovalent MS experiments were performed on an electrospray time-of-flight mass spectrometer (LCT, Waters, Manchester, UK) equipped with an automated chip-based nanoelectrospray (nanoESI) source (Triversa Nanomate, Advion Biosciences, Ithaca, NY, USA). Mass spectra were recorded in the positive ion mode over the mass range m/z 500–12 000 after calibration with a solution of cesium iodide at 2 g/l in 1:1 water:isopropanol. Experiments were performed by diluting samples in 150 mM ammonium acetate buffer pH 8.0. Instrumental settings were carefully tuned in order to preserve the integrity of noncovalent assemblies in the gas phase while ensuring an optimal ion desolvation and transmission through the mass spectrometer. Particularly, the pressure (Pi) in the first pumping stage of the instrument was set to 7 mbar while the accelerating voltage applied on the sampling cone (Vc) was set to 200 V. Data analysis was performed with MassLynx 4.1 (Waters, Manchester, UK).
Molecular modeling
Since only the atomic structure of the CggR C-terminal domain has been determined experimentally (22), we constructed the N-terminal DBD of CggR (whCggR, residues 1–87) by homology modeling of the wH motif. Three different wH-containing proteins in complex with DNA available in the Protein Data Bank (PDB) were used as templates, from the diphtheria tox repressor DtxR (PDB ID: 1F5T) (43), the arginine repressor AhrC (PDB ID: 2P5L) (44) and the organic hyperoxide resistance protein regulator, OhrR from the MarR family (PDB ID: 1Z9C) (45). Automatic alignments retrieved from @TOM-2 (46) (http://atome.cbs.cnrs.fr/AT2/meta.html) were manually refined using ViTO (47) and models of whCggR dimers bound to the unmodified template DNA fragments were generated using MODELLER (http://salilab.org/modeller). The structure of a 23-bp DNA fragment having the OR sequence was then modeled using make-na server (http://structure.usc.edu/make-na/server.html) and substituted for the original DNA templates used for constructing the whCggR dimers. For this, the center of symmetry of the template palindromic sequence was manually aligned with that of the small palindrome forming the recognition motif for the CggR dimer as proposed by Zorilla et al. (25) (Figure 1A). The structure of the 45-bp full-length operator DNA (OLR) was modeled with the 1Z9C-derived whCggR dimer bound in tandem to the half-site palindromes.
Models of the full-length CggR/DNA complexes fitting the SAXS data were generated by rigid body refinement using the program SASREF (48). SASREF constructs molecular models from domains or subunits with known structure by rigid body movements and rotations. Starting from an arbitrary or prefixed initial configuration, the program employs simulated annealing to construct an interconnected model without steric overlaps fitting single data sets from the entire complex. Here, we used the different modeled structures of the whCggR/DNA complexes and the high-resolution structure of the C-terminal CggR regulatory domain (rdCggR, residues 89–340) determined by crystallography (PDB ID: 2OKG chain A) (22). For rigid-body modeling of the full-length CggR dimer or tetramer bound to DNA, the whCggR/OR or whCggR/OLR modeled structure was fixed and the rdCggR domains were free to move as independent units. The distance restraint between the C-terminus of the whCggR domains and the first residue of rdCggR units was fixed to different values (1, 2, 3, 4, 5, 6, 8, 10 or 12 Å) and no symmetry constraints were included in the final models. When possible, SASREF modeling was repeated until obtaining if possible 10 different models that fitted the scattering data with an experimental chi value (χe) < 1.5. The theoretical Stokes radius and radius of gyration of the SASREF atomic models were calculated using HYDROPRO (49).
RESULTS
Size parameters of the CggR/DNA complexes determined by SEC and SAXS
SEC, or analytical gel filtration, is a classical method for the characterization of diffusive properties of molecules related to their size and shape (36). From calibration curves with macromolecules of known diffusion coefficients, one can measure the Stokes radius (Rs), or hydrodynamic radius, which corresponds to the radius of a hard sphere that elutes at the same rate as the molecule of interest. A more extended molecule will, in general, have a larger Stokes radius compared to a more compact molecule of the same MW. Similarly, the radius of gyration (Rg), usually determined by SAXS, quantifies the spatial extent of the molecular structure. The hydrated volume (or Porod volume, Vp) and the maximum diameter (Dmax) of the diffusing particles are two other sized parameters that can be directly retrieved from scattering data.
SEC and SAXS were used here to determine the size parameters of CggR/DNA complexes with or without FBP (summarized in Table 1). Figure 1B shows the SEC profiles of CggR bound to a double-stranded (ds) oligonucleotide corresponding to the full-length operator sequence of the gapA operon (OLR, 45 bp), or the high-affinity right half-operator site (OR, 23 bp) (Figure 1A). As previously observed (26), the CggR protein alone showed a highly polydisperse elution profile indicating the presence of mainly high-order oligomers (data not shown). In contrast, no protein aggregate was observed in the presence of DNA and all the CggR/DNA complexes eluted in a single peak, with an eventual slower minor peak corresponding to unbound DNA.
Table 1.
Molecular sizes and mass of CggR/DNA complexes alone or with FBP
| Complex | Rs (nm) | Rg (nm) | Dmax (nm) | Vp (nm3) | Mass |
|---|---|---|---|---|---|
| CggR/OLR | 5.4 ± 0.2 | 4.46 ± 0.16 | 14 ± 1 | 202 ± 20 | 182 868 ± 9 |
| CggR/OR | 5.4 ± 0.2 | 4.23 ± 0.09 | 13 ± 1 | 227 ± 20 | 183 356 ± 5a |
| CggR/OLR + FBP | 5.8 ± 0.2 | 5.38 ± 0.11 | 17 ± 2 | 280 ± 30 | 183 900 ± 7a |
| CggR/OR + FBP | 4.6 ± 0.2 | 3.72 ± 0.10 | 11 ± 1 | 125 ± 10 | 92 350 ± 3a |
Stokes’ radius Rs determined by SEC; radius of gyration Rg, maximum diameter Dmax and excluded (Porod) volume Vp determined by SAXS; molecular mass determined by noncovalent nanoESI-MS. aMean value of the predominant species. Theoretical MW of CggR = 38 781 Da, OLR = 27 679 Da, OR = 14 087 Da, FBP = 340 Da.
In the absence of FBP, the CggR complexes with the full DNA target OLR or with the right half-operator OR eluted in the same volume (blue and magenta curves, respectively, in Figure 1B), corresponding to molecules having an Rs of 5.4 ± 0.2 nm. These complexes also produced nearly identical SAXS profiles (Figure 1C), from which equivalent size parameters were determined (Rg = 4.2–4.5 nm, Dmax = 13–14 nm, Vp = 200–230 nm3). The pair–distance distribution function P(r), which measures the distribution of pairwise interatomic distances within the molecule, demonstrated the strong similarity between the two complexes (Figure 1D). These results suggest that both inducer-free CggR/DNA complexes present a similar size and shape despite the 50% shorter length of the OR DNA fragment with respect to the full-length OLR target. They are consistent with a model in which two CggR/OR complexes interact together to form a macromolecular assembly that is structurally equivalent to the CggR/OLR complex.
Upon addition of 0.5 mM FBP, the SEC profiles show that the elution peaks corresponding to the free DNA fragments did not increase, indicating that at the rather high concentrations used for these experiments, the repressor protein remains bound to its operator sequences in the presence of inducer (red and green curves in Figure 1B). However, the hydrodynamic properties and the SAXS profiles of the CggR/OLR and CggR/OR complexes changed significantly. In the case of the full-length operator/repressor complex (red curves in Figure 1) addition of 0.5 mM FBP resulted in a slight increase of the determined Stokes radius (Rs = 5.8 ± 0.2 nm), radius of gyration (Rg = 5.38 ± 0.11 nm), maximal diameter (Dmax = 17 ± 2 nm) and excluded volume (Vp = 280 ± 30 nm3) as well as a significant change of the P(r) pattern. These results indicate that the CggR/OLR complex conserves the same stoichiometry but in a more extended conformation in the presence than in the absence of FBP. Indeed, the frictional coefficient [the ratio of Rg over Rs which provides information on the distribution of mass in a macromolecule, (36)] has a value of 0.928 for the FBP-bound CggR/OLR complex and 0.826 for the FBP-free form, respectively, indicating that the shape of the former deviates substantially from that of a globular molecule approximated to a hard sphere [Rg/Rs = (3/5)1/2 = 0.775].
In contrast, addition of the effector ligand to the CggR/OR complex (green curves in Figure 1) induced a large decrease of all the size parameters (Table 1): Rs dropped from 5.4 to 4.6 ± 0.2 nm, Rg from 4.23 ± 0.09 nm to 3.72 ± 0.10 nm and Dmax from 13 ± 1 nm to 11 ± 1 nm and the excluded volume of the particles was reduced by half. These observations suggest that the assembly of two CggR/OR complexes observed under repressing condition is disrupted upon addition of the inducing sugar.
Disruption of protein–protein contacts revealed by FCCS
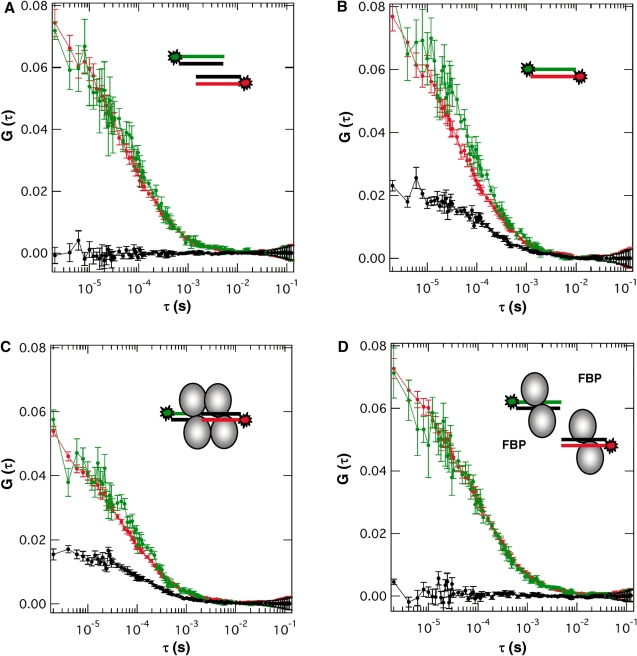
In order to further test our molecular interaction model for the CggR/OR complex, we employed 2γFCCS. The 2γFCCS is a very robust single-molecule-based method for detecting interactions between differently labeled molecules based on their concomitant diffusion through the very small two-photon excitation volume (<0.5 fl) (50,51). Here we performed 2γFCCS experiments with dsDNA fragments of the OR right half-operator labeled with either a red (Atto647N) or green (fluorescein) light emitting fluorophore. A mixture containing both fluorescent dsDNAs at 60 nM and no protein produced no significant cross-correlation signal (black curves in Figure 2), showing the absence of cross talk between the two detection channels and indicating that the labeled DNA strands were not cross hybridizing or aggregating (Figure 2A). Addition of CggR at 300 nm to this DNA mixture led to a strong correlation signal, providing evidence for a protein-dependent interaction between the two DNA species (Figure 2C). The maximum amplitude of the cross-correlation function [Gx(0)], which is related to the concentration of co-diffusing red and green dyes, was ∼30% of that of the autocorrelation functions retrieved from each detection channel [Gi(0)]. Given that the DNA probes are ∼80% labeled, this value of 30% (instead of a theoretical value of 50% if both probes were 100% labeled) indicates that most of the target oligonucleotides associate to form stable complexes with the CggR protein. As predicted by our molecular interaction model, this ternary complex was disrupted by the inducer metabolite, as visualized by the complete loss of cross correlation signal upon addition of FBP (Figure 2D). Meanwhile, the diffusion time through the observation volume of the labeled particles (132 ± 7 µs) remained significantly higher than that of the DNA fragments alone (78 ± 4 µs), indicating that, although, the DNA probes are no longer co-diffusing in the presence of FBP, they are not free but still associated to the protein, as observed above by SEC and SAXS. It can thus be concluded that the physical link that allows the self-association of the half-operator DNA molecules involves protein–protein contacts that are disrupted upon induction with FBP.
Figure 2.
Auto- and cross-correlation profiles recovered from FCCS measurements with Atto-647N-labeled or fluorescein-labeled oligonucleotides corresponding to the CggR OR half-site operator in the presence/absence of the repressor and the inducer metabolite FBP. Schematics of the labeled DNAs and protein present in the sample chambers are shown. Red, blue and black curves correspond to the autocorrelation traces recorded in the red (675 nm) and blue (525 nm) detection channels and the cross-correlation curve, respectively. (A) A mixture of the singly labeled dsDNA fragments showing the absence of cross-correlation signal. (B) A doubly labeled DNA hybrid serving as positive control of cross-correlation. The difference in the maximum amplitude of the auto- and cross-correlation function (G(0)) denotes the partial labeling and hybridization of the two labeled DNA strands. (C) Cross-correlation upon addition of the repressor protein to the singly labeled DNA mixture of panel (A), evidencing the CggR-mediated assembly of the DNA fragments. (D) Loss of cross correlation signal upon addition of the inducer metabolite to the protein/DNA mixture of panel (C), evidencing the FBP-induced disruption of the CggR/DNA ternary complex. Concentration was 60 nM for the labeled DNA fragments, 300 nM for CggR (monomer unit) and 0.5 mM for FBP.
Stoichiometry of the CggR/DNA/sugar complexes probed by noncovalent MS
The use of MS under conditions that preserve noncovalent complexes has previously been reported for the determination of protein oligomerization state and the study of protein/ligand interactions [for review, see (52)]. Recently, noncovalent MS has also encountered a growing success for the characterization of protein/oligonucleotide complexes (53–55). In the present study, we used noncovalent nanoESI-MS to determine the stoichiometry of the different CggR/DNA complexes as well as to retrieve information on the specificity and stoichiometry of binding of the effector ligand.
In the case of the OLR full-length operator, mass measurements confirmed the formation of a tetrameric 4:1 CggR/OLR complex that is preserved upon FBP addition (Table 1). The complex formed between CggR and the OR right half-operator was analyzed here in more details. The nanoESI mass spectrum recorded in the absence of FBP (Figure 3A) revealed the presence of an intense ion distribution in the mass range m/z 7000–9000 with a measured MW of 183 356 ± 5 Da corresponding to a macromolecular assembly containing four CggR subunits and two OR dsDNA oligonucleotides (4:2 CggR/OR complex, theoretical MW = 183 298 Da). An additional minor species was also detected in the mass range m/z 4000–6000 with measured MW (91 664 ± 5 Da) consistent with the presence of a small proportion of the dimeric protein bound to one OR dsDNA (2:1 CggR/OR complex, theoretical MW = 91 649 Da). Note that the MW determined for these protein/DNA complexes, or for the CggR protein alone under non-denaturing conditions (35) rules out the possibility that some carbohydrate ligand may have co-purified with the protein, in contrast to the protein preparation used for crystallization by Rezacova et al. (22).
Figure 3.
Noncovalent MS analysis of CggR interaction with the OR half-operator DNA and assessment of inducer binding specificity. The CggR/OR complex was diluted to 3 µM in 150 mM ammonium acetate buffer at pH 8.0 and analyzed either (A) alone or in presence of 30 µM (B) FBP, (C) F6P or (D) GBP. Chemical representations of the phospho-sugars are shown on the left. Insets on the right represent enlarged mass spectra showing the +20 charge state of the 2:1 CggR/OR complex and the +23 charge state of the 4:2 CggR/OR complex. Circled numbers correspond to the number of phospho-sugar molecules bound to each complex. Similar results were obtained when adding the phospho-sugars at 60 µM (data not shown).
The effect of FBP was then probed by incubating the CggR/OR complex (3 µM) in the presence of a 10-fold molar excess of the inducing sugar. This led to a complete and symmetrical dissociation of the tetrameric 4:2 CggR/OR complex into the dimeric 2:1 CggR/OR complex (Figure 3B). Under identical experimental and instrumental conditions, neither F6P nor GBP was able to fully dissociate the tetrameric 4:2 CggR/OR assembly (Figure 3C and D). Similar results were obtained using fructose-1-phosphate (F1P) or a 20-fold molar excess (60 µM) of these phospho-sugars (data not shown). These observations are in good agreement with our previous studies on CggR/DNA interactions by fluorescence anisotropy which showed that none of these phospho-sugars can prevent the cooperative binding of CggR to its target DNA as FBP does (18). It is also noteworthy that the marginal peaks detected in the mass range corresponding to the 2:1 CggR/OR complex are slightly more intense in the case of F6P than of GBP, which is also in good agreement with our previous findings indicating that F6P at high concentration (>5 mM) can indeed alter CggR/DNA-binding cooperativity (18). Unfortunately, the effect of the different sugars at high concentrations could not be tested here because of severe reduction in the signal-to-noise ratio of the mass spectra.
Interestingly, a close up on the most intense ionization peaks of the nanoESI mass spectra (Figure 3, right insets) revealed the presence of several molecular species with MW incrementing by exactly the mass of the added phospho-sugars. In case of FBP, the most populated state corresponded to the dimeric CggR/OR assembly with two inducer molecules (2:1:2 CggR/OR/FBP, MW = 92 350 ± 3 Da) but the complexes with three- or four-bound FBP molecules were also present in significant amount. Mass spectra obtained in the presence of F6P and GBP (Figure 3C and D) not only evidenced the binding of these sugar ligands to the dimeric CggR/OR complex, but also to the tetrameric 4:2 CggR/OR complex. Hence, these phospho-sugars could associate with the protein without disrupting the CggR tetramer. Moreover, at the concentration used for nanoESI MS experiments (30 µM), only one or two F6P or GBP adducts were visualized with the CggR/DNA complexes whereas up to four FBP molecules remain associated to the dimeric complex, i.e. two FBP per CggR monomer. Similarly, in the case of the CggR tetramer bound to the full-length operator, the most abundant species observed at 30 µM FBP corresponded to the 4:1:3 CggR/ORL/FBP complex (MW = 183 900 ± 7 Da, Table 1) but species with more than one sugar moiety per protein subunit were also detected with FBP and not with F1P (35). As discussed hereafter, these results could be explained by the presence in CggR of two distinct sugar-binding sites having different affinity and specificity for phosphorylated ligands.
Rigid-body models of CggR/DNA complexes
The ensemble of size and mass parameters characterizing the different CggR/DNA complexes are summarized in Table 1. Altogether, these data establish that (i) in the absence of FBP, two CggR dimers associate in a similar way with either the full-length OLR operator or two OR right half-operators in order to form a tetramer/DNA complex (4:1 CggR/OLR or 4:2 CggR/OR, respectively), and (ii) in the presence of FBP the 4:2 CggR/OR dissociates into a dimeric 2:1 CggR/OR complex while the tetrameric 4:1 CggR/OLR complex is preserved but adopts a more extended conformation. Based on this information, we attempted the construction of rigid-body models fitting the SAXS data of the different CggR/DNA complexes and using the program SASREF (48) as described in the experimental section.
Models of the CggR dimer bound to a 23-bp DNA fragment corresponding to the OR right half-operator were first generated with SASREF fitting the scattering data of the FBP-bound 2:1 CggR/OR complex. For this purpose, two monomer units from the rdCggR crystal structure (22) were automatically attached and rotated at the C-terminal ends of atomic models of the CggR wH DBD (whCggR). Three different models of the dimeric whCggR/OR complex were used, derived from PDB templates showing different position and orientation of the wH motifs on the DNA double helix. Satisfactory results were obtained only for the rigid-body models derived from OhrR (PDB ID: 1Z9C), which belongs to the same MarR superfamily as whCggR (45). These models consistently showed the two rdCggR domains as independent modules not directly interacting with each other (Figure 4A), although their relative distance and orientation could vary.
Figure 4.
SASREF rigid body models and their fit (magenta line) to experimental SAXS data (dots) of the CggR complexes with the half-site operator DNA OR (A and D) or the full-length operator OLR (B and C) and in the presence (A and B) or absence (C and D) of FBP. The 1Z9C-derived models of the dimeric or tetrameric N-terminal wH domain of CggR (whCggR) bound to the OR or OLR dsDNA are colored in dark blue, with the DNA shown as sticks and the whCggR dimers in surface representation. Red, cyan, yellow and green molecular surfaces represent the rdCggR that moved as independent units during rigid body modeling by SASREF (48). The experimental chi value (χe) of the fitting curves given by SASREF as well as the theoretical gyration radius (Rg) and Stokes’ radius (Rs) of the models calculated by HYDROPRO (49) are indicated in the bottom-left corner of the graphics. Shown here are representative examples of at least 10 different models generated automatically for each complex, combining one of the best fit and agreement with experimentally determined size parameters given in Table 1. Two orientations of the dimeric complex are shown in (A). Panels (C) and (D) purposely exemplify two different models that fit equally well the SAXS data of the tetrameric complex in the absence of FBP.
Modeling of the tetrameric CggR/DNA complexes was then pursued with four rdCggR monomers attached to the 1Z9C-derived whCggR dimer tandemly repeated onto a 45-bp OLR DNA. In the case of the CggR/OLR complex in the presence of FBP, the best fitting SASREF models always exhibited the four regulatory domains as non-interacting subunits, resulting in a fully extended structure (Figure 4B). In contrast, the different models obtained by fitting the scattering data of the FBP-free CggR/OLR or CggR/OR complexes presented a much more compact structure, as exemplified in Figure 4C and D. Although several solutions could be found that fit equally well the experimental data, a common feature was the bringing together of three or four regulatory subunits forming a compact central core that bridges the two whCggR dimers bound to the two half-operator sites. We conclude that such protein–protein interactions constitute the physical basis for the cooperative binding of the CggR tetramer to its tandem half-operator sites and that the inducer metabolite FBP disrupts these interactions.
DISCUSSION
An integrative approach for characterizing protein/DNA complexes
In this study, a combination of biophysical methods has been applied to characterize CggR/DNA complexes and elucidate the structural mechanism underlying the transcriptional regulation of central glycolytic genes in B. subtilis. Left unrevealed until the present study were the molecular forces responsible for the inducer-modulated cooperative formation of the repressor/operator complex. Here we provide experimental evidence showing that the cooperativity of the CggR/DNA assembly arises from dimer/dimer interactions linking the protein subunits bound at the two half-operator sites and that FBP disrupts these interactions. Based on this finding, we propose here below a regulation model which integrates not only the results of SEC, SAXS, FCCS and noncovalent MS experiments performed in the present work, but also information gathered from previous genetical, biochemical and other biophysical studies on this transcription factor since its identification (14,15,17,18,22,25,26,56). This combination of techniques provided an ensemble of complementary and sometimes overlapping data that together strengthen the conclusions and the proposed regulation model. For instance, by combining analytical centrifugation and time-resolved fluorescence anisotropy experiments, we had previously showed that the effector ligand FBP does not change the final stoichiometry, but rather the hydrodynamic properties of the CggR/OLR operator complex (25). This finding was confirmed and refined here through the precise determination of the complex MW by noncovalent MS and the estimation of the particles size and shape parameters by SEC and SAXS (Table 1). Recently, we also performed traveling-wave ion mobility MS experiments that evidenced further and quantified the conformational changes undergone by the CggR/OLR complex, revealing a 5% increase of its rotationally averaged collision cross section upon FBP binding (35).
Such an integrative approach presents an overwhelming advantage when attempts to obtain crystals of protein/DNA complexes remain unsuccessful, as this is the case for CggR and many other modular regulatory proteins. The conformational changes and domain motion on which rely most signal transduction processes are often responsible for the difficulties in solving 3D structures and capturing different active states of transcription factors. Although no detailed structural data are yet available for full-length CggR, alone or in complex with DNA, the low-resolution rigid-body models we obtained by fitting our SAXS data give a fairly good view of the repressed and induced form of the repressor/operator complex in solution. Further refinement of these models would require more information on the different protein/DNA complexes, including the precise position and relative orientation of the whCggR domains on both the OR and OL DNA targets, the actual symmetry of the complexes, the possible bending and/or distortion of the DNA double helix, the positioning and flexibility of the linker, etc. For this purpose, the combination of small-angle X-ray and neutron scattering data would be of particular interest (29).
Cooperativity-based regulation model
Together with previous results, the present study allows us to propose a structural mechanism of CggR-mediated regulation of the gapA operon (Figure 5). When B. subtilis cells are grown on a non-glycolytic carbon source, the intracellular concentration of FBP is low and the expression of the gapA operon is repressed through the cooperative binding of a compact CggR tetramer to the tandem OL and OR operator sequences. Given the position of these sequences (starting 25-bp downstream of the transcriptional start and ending 23-pb upstream of the start codon of cggR, the first gene of the gapA operon), the most likely mechanism by which CggR interferes with the transcriptional activity of the RNA polymerase is by a so-called ‘road-block’ mechanism. In absence of inducer, the CggR/operator DNA complex is stabilized by both the free energy of protein–DNA interactions and the coupled free energy of the CggR dimer–dimer interactions, leading to a very stable tetramer/DNA complex exhibiting a very low off-rate, and hence efficient repression of transcription (Figure 5A). Induction of the gapA operon occurs in the presence of glucose or other rapidly metabolized carbohydrates that activate glycolysis and raise the FBP intracellular concentration. Upon inducer binding, the interactions that maintain the CggR tetramer in the repression complex are disrupted, i.e. the coupling between protein–protein and protein–DNA interactions is abolished, and the CggR dimers bound to each half-operator behave as separate entities. This decreases the overall CggR/operator complex stability and hence increases the off-rate. This increased probability for CggR dissociation affords a larger window of opportunity for the RNA polymerase to transcribe through the first then the second operator half site (Figure 5B). According to this mechanism, the sequence of the operator half site plays a significant role. Indeed, the OL half-site, situated on the 5′-side of the operator where the transcribing RNA polymerase arrives, is of much lower affinity than the OR right half-site (25). Hence, the major rate limiting step for the polymerase under inducing conditions will be the dissociation of a CggR dimer from the OR right half-site, compared to waiting for the dissociation of the tetramer from the full-length operator in absence of inducing sugar.
Figure 5.
Structural mechanism for CggR-mediated regulation of the gapA operon. In the proposed model, transcriptional regulation by the CggR repressor and its inducer metabolite FBP is achieved by modulating the extent of cooperative interactions rather than simply DNA binding. (A) In the absence of glucose or other carbon sources producing high levels of FBP, CggR interacts as a dimer of dimers bridging the two half-sites of the gapA operator region. This tetrameric assembly can efficiently block the progression of the RNA polymerase, thereby leading to the arrest of transcription. (B) In the presence of glucose, the raise of FBP intracellular concentration provokes the disruption of dimer–dimer contacts through inducer binding to the low-affinity sugar-binding site of the CggR repressor. CggR dimers can still bind independently to each half-operator, however with much lower affinity for the left (5′) half-site than for the right (3′) half-site. This provides a higher opportunity for the transcribing RNA polymerase to read through the first then the second half-site, thereby leading to transcription of the downstream coding sequences.
The finding that CggR-mediated regulation of the gapA operon is based on the extent of cooperative protein–protein interactions rather than direct DNA binding may have a biological significance with respect to the adaptive response. Indeed, as soon as the concentration of FBP increases to a value signaling the presence of a preferred carbon source, disruption of the tetrameric CggR assembly will immediately permit read-through by the RNA polymerase even if repressor molecules can still bind to the gapA operator region. Hence transcription can take place even at relatively high repressor concentration. Conversely, when glycolytic nutrients are exhausted and FBP concentration drops, the regain of cooperativity will rapidly re-associate the regulatory subunits and maintain the repressive tetramer even at very low protein concentration. Such a cooperativity-based regulation mechanism might be of particular importance in the case of CggR, which represses the expression of its own gene and whose synthesis and stability is also dependent on differential mRNA processing and structural co-factor binding (26,56).
CggR is now by far the best characterized member of the SorC family of transcription factors. Its role in the regulation of essential glycolytic genes makes this repressor of particular interest for a global understanding of carbon and energy fluxes in the bacterial cells. Several lines of evidence suggest that the cooperativity-based regulation mechanism proposed for CggR applies also to other SorC-type regulators, even from different subclasses. The B. subtilis DeoR repressor, the only other member of this family for which the DNA-binding site has been identified (24), also interacts as a tetramer and in a cooperative manner with its operator DNA whose structural organization presents striking similarities with that of CggR (21,25). Recently, the sorbitol operon regulator SorC from K. pneumoniae was successfully crystallized in its full-length form and revealed a tetrameric structure that could be regarded as a dimer of two asymmetric dimers with the putative sugar-binding site located near the dimer–dimer interface (23). In this structure, the N-terminal DBDs protrude from each side of the tetrameric core, formed by the sugar-binding domains, and their position is compatible with DNA binding at two distinct sites separated by about two helix turns (∼70 Å). Based on these observations and the structural similarities between SorC and CggR, a transcriptional regulation mechanism was proposed for the sorbitol (sor) operon, in complete agreement with the one elaborated previously for the gapA operon and corroborated by the present experimental results. Regulation by a cooperativity-based rather than an affinity-based mechanism as proposed here could also account for the ability of SorC to act both as a repressor and an activator of the sor operon. In its compact tetrameric state, SorC could prevent transcription initiation or elongation, whereas in its induced dimeric state it could recruit the RNA polymerase and activate transcription. If so, the structural basis of this regulatory system would in some way resemble that recently elucidated for the genetic switch that governs the expression of restriction-modification genes (9).
Evidence for two FBP-binding sites in CggR
This work also brings further support to the peculiar feature of CggR to possess two binding sites for its effector sugar FBP (26,25). The relative high resolution of the noncovalent mass spectra obtained at 30 µM FBP revealed the possible liganding of two inducer molecules per CggR monomer unit whereas no more than one sugar adduct could be detected in case of F6P, GBP and F1P under identical conditions [Figure 3, (35)]. The fact that such distinct effects are observed between FBP and other phospho-sugars suggests that the multiple binding events observed with FBP results from specific association in solution, and not from any artifact of the technique. All the other tested sugars bind to a lower extent than FBP and without inducing the dissociation of the tetrameric CggR/OR assembly. Previous fluorescence anisotropy studies (18) as well as crystallographic studies and mobility shift assays (22) have shown that CggR can indeed interact with a variety of phosphorylated carbohydrates structurally related to FBP but with no inductor effect. Together these results corroborate our previous proposal for the existence of two different sugar-binding sites in CggR, one of high affinity for FBP where other phosphorylated compounds can bind without triggering the induction response, and the other of lower affinity where only FBP can bind efficiently and prevent CggR/DNA cooperative interaction (26). FBP bound at the high affinity site (KD ∼ 4–7 µm) was shown to play the role of a structural co-factor, protecting CggR against aggregation and proteolysis (26). Interestingly, a similar stabilizing effect was observed for other phosphorylated glycolytic intermediates (22). In the different liganded structures of the rdCggR recently reported (22), all the phosphorylated compounds (including FBP and F6P) co-crystallized at the conserved sugar-binding site that corresponds to the substrate binding site of the NagB enzymes from which rdCggR probably evolved (18). It can thus be inferred that under the conditions used for noncovalent MS, F6P and GBP bind at this conserved site only whereas FBP binds also at a secondary site, where it provokes the disruption of dimer–dimer interactions. As suggested by our previous studies (18), other phospho-sugars such as F6P could eventually bind also at this site, albeit with much lower affinity than FBP and therefore at concentrations that are unlikely to be reached in the bacterial cell. The location of this second, inducer-specific binding site remains unknown since in the FBP co-crystal structure (22), only one FBP molecule was observed per crystallographic dimer of rdCggR. In this structure, FBP is bound at the conserved sugar-binding site of one protein subunit, the binding site of the other subunit being occupied by a triose phosphate ligand that co-purified with the protein. It is thus unlikely that this structure represents the induced form of rdCggR.
FUNDING
Ministère de l’Education, de la Recherche et de la Technology (PhD grant to D.C. and C.A.); National Science Foundation (grant OISE 0710816 to M.L.F.). Funding for open access charge: Centre National de la Recherche Scientifique (CNRS), Institut National de la Santé et de la Recherche Médicale (INSERM).
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
We thank D. Svergun and his colleagues for assistance with SAXS data recording and analysis at the X33 beamline at European Molecular Biology Laboratory/Deutsches Elektronen-Synchrotron. We are also grateful to S. Arold for help in SAXS data analyses, E. Margeat for DNA labeling and J.L. Pons for homology modeling.
REFERENCES
- 1.Shea MA, Ackers GK. The OR control system of bacteriophage lambda. A physical-chemical model for gene regulation. J. Mol. Biol. 1985;181:211–230. doi: 10.1016/0022-2836(85)90086-5. [DOI] [PubMed] [Google Scholar]
- 2.Grillo AO, Brown MP, Royer CA. Probing the physical basis for trp repressor-operator recognition. J. Mol. Biol. 1999;287:539–554. doi: 10.1006/jmbi.1999.2625. [DOI] [PubMed] [Google Scholar]
- 3.Chahla M, Wooll J, Laue TM, Nguyen N, Senear DF. Role of protein-protein bridging interactions on cooperative assembly of DNA-bound CRP-CytR-CRP complex and regulation of the Escherichia coli CytR regulon. Biochemistry. 2003;42:3812–3825. doi: 10.1021/bi0271143. [DOI] [PubMed] [Google Scholar]
- 4.Beckett D. Linked equilibria in regulation of transcription initiation. Methods Cell Biol. 2008;84:25–52. doi: 10.1016/S0091-679X(07)84002-7. [DOI] [PubMed] [Google Scholar]
- 5.Connaghan KD, Heneghan AF, Miura MT, Bain DL. Na(+) and K(+) Allosterically regulate cooperative DNA binding by the human progesterone receptor. Biochemistry. 2010;49:422–431. doi: 10.1021/bi901525m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wilson DS, Guenther B, Desplan C, Kuriyan J. High resolution crystal structure of a paired (Pax) class cooperative homeodomain dimer on DNA. Cell. 1995;82:709–719. doi: 10.1016/0092-8674(95)90468-9. [DOI] [PubMed] [Google Scholar]
- 7.Schumacher MA, Miller MC, Grkovic S, Brown MH, Skurray RA, Brennan RG. Structural basis for cooperative DNA binding by two dimers of the multidrug-binding protein QacR. EMBO J. 2002;21:1210–1218. doi: 10.1093/emboj/21.5.1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kumarevel T, Nakano N, Ponnuraj K, Gopinath SC, Sakamoto K, Shinkai A, Kumar PK, Yokoyama S. Crystal structure of glutamine receptor protein from Sulfolobus tokodaii strain 7 in complex with its effector L-glutamine: implications of effector binding in molecular association and DNA binding. Nucleic Acids Res. 2008;36:4808–4820. doi: 10.1093/nar/gkn456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.McGeehan JE, Streeter SD, Thresh SJ, Ball N, Ravelli RB, Kneale GG. Structural analysis of the genetic switch that regulates the expression of restriction-modification genes. Nucleic Acids Res. 2008;36:4778–4787. doi: 10.1093/nar/gkn448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chung YS, Brendler T, Austin S, Guarne A. Structural insights into the cooperative binding of SeqA to a tandem GATC repeat. Nucleic Acids Res. 2009;37:3143–3152. doi: 10.1093/nar/gkp151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stulke J, Hillen W. Regulation of carbon catabolism in Bacillus species. Annu. Rev. Microbiol. 2000;54:849–880. doi: 10.1146/annurev.micro.54.1.849. [DOI] [PubMed] [Google Scholar]
- 12.Gorke B, Stulke J. Carbon catabolite repression in bacteria: many ways to make the most out of nutrients. Nat. Rev. Microbiol. 2008;6:613–624. doi: 10.1038/nrmicro1932. [DOI] [PubMed] [Google Scholar]
- 13.Fujita Y. Carbon catabolite control of the metabolic network in Bacillus subtilis. Biosci. Biotechnol. Biochem. 2009;73:245–259. doi: 10.1271/bbb.80479. [DOI] [PubMed] [Google Scholar]
- 14.Fillinger S, Boschi-Muller S, Azza S, Dervyn E, Branlant G, Aymerich S. Two glyceraldehyde-3-phosphate dehydrogenases with opposite physiological roles in a nonphotosynthetic bacterium. J. Biol. Chem. 2000;275:14031–14037. doi: 10.1074/jbc.275.19.14031. [DOI] [PubMed] [Google Scholar]
- 15.Ludwig H, Homuth G, Schmalisch M, Dyka FM, Hecker M, Stulke J. Transcription of glycolytic genes and operons in Bacillus subtilis: evidence for the presence of multiple levels of control of the gapA operon. Mol. Microbiol. 2001;41:409–422. doi: 10.1046/j.1365-2958.2001.02523.x. [DOI] [PubMed] [Google Scholar]
- 16.Ludwig H, Rebhan N, Blencke HM, Merzbacher M, Stulke J. Control of the glycolytic gapA operon by the catabolite control protein A in Bacillus subtilis: a novel mechanism of CcpA-mediated regulation. Mol. Microbiol. 2002;45:543–553. doi: 10.1046/j.1365-2958.2002.03034.x. [DOI] [PubMed] [Google Scholar]
- 17.Doan T, Aymerich S. Regulation of the central glycolytic genes in Bacillus subtilis: binding of the repressor CggR to its single DNA target sequence is modulated by fructose-1,6-bisphosphate. Mol. Microbiol. 2003;47:1709–1721. doi: 10.1046/j.1365-2958.2003.03404.x. [DOI] [PubMed] [Google Scholar]
- 18.Doan T, Martin L, Zorrilla S, Chaix D, Aymerich S, Labesse G, Declerck N. A phospho-sugar binding domain homologous to NagB enzymes regulates the activity of the central glycolytic genes repressor. Proteins. 2008;71:2038–2050. doi: 10.1002/prot.21883. [DOI] [PubMed] [Google Scholar]
- 19.Wohrl BM, Wehmeier UF, Lengeler JW. Positive and negative regulation of expression of the L-sorbose (sor) operon by SorC in Klebsiella pneumoniae. Mol. Gen. Genet. 1990;224:193–200. doi: 10.1007/BF00271552. [DOI] [PubMed] [Google Scholar]
- 20.Saxild HH, Andersen LN, Hammer K. Dra-nupC-pdp operon of Bacillus subtilis: nucleotide sequence, induction by deoxyribonucleosides, and transcriptional regulation by the deoR-encoded DeoR repressor protein. J. Bacteriol. 1996;178:424–434. doi: 10.1128/jb.178.2.424-434.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zeng X, Saxild HH, Switzer RL. Purification and characterization of the DeoR repressor of Bacillus subtilis. J. Bacteriol. 2000;182:1916–1922. doi: 10.1128/jb.182.7.1916-1922.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rezacova P, Kozisek M, Moy SF, Sieglova I, Joachimiak A, Machius M, Otwinowski Z. Crystal structures of the effector-binding domain of repressor Central glycolytic gene Regulator from Bacillus subtilis reveal ligand-induced structural changes upon binding of several glycolytic intermediates. Mol. Microbiol. 2008;69:895–910. doi: 10.1111/j.1365-2958.2008.06318.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.de Sanctis D, McVey CE, Enguita FJ, Carrondo MA. Crystal structure of the full-length sorbitol operon regulator SorC from Klebsiella pneumoniae: structural evidence for a novel transcriptional regulation mechanism. J. Mol. Biol. 2009;387:759–770. doi: 10.1016/j.jmb.2009.02.017. [DOI] [PubMed] [Google Scholar]
- 24.Zeng X, Saxild HH. Identification and characterization of a DeoR-specific operator sequence essential for induction of dra-nupC-pdp operon expression in Bacillus subtilis. J. Bacteriol. 1999;181:1719–1727. doi: 10.1128/jb.181.6.1719-1727.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zorrilla S, Doan T, Alfonso C, Margeat E, Ortega A, Rivas G, Aymerich S, Royer CA, Declerck N. Inducer-modulated cooperative binding of the tetrameric CggR repressor to operator DNA. Biophys. J. 2007;92:3215–3227. doi: 10.1529/biophysj.106.095109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zorrilla S, Chaix D, Ortega A, Alfonso C, Doan T, Margeat E, Rivas G, Aymerich S, Declerck N, Royer CA. Fructose-1,6-bisphosphate acts both as an inducer and as a structural cofactor of the central glycolytic genes repressor (CggR) Biochemistry. 2007;46:14996–15008. doi: 10.1021/bi701805e. [DOI] [PubMed] [Google Scholar]
- 27.Svergun DI, Koch MH. Advances in structure analysis using small-angle scattering in solution. Curr. Opin. Struct. Biol. 2002;12:654–660. doi: 10.1016/s0959-440x(02)00363-9. [DOI] [PubMed] [Google Scholar]
- 28.Koch MH, Vachette P, Svergun DI. Small-angle scattering: a view on the properties, structures and structural changes of biological macromolecules in solution. Q. Rev. Biophys. 2003;36:147–227. doi: 10.1017/s0033583503003871. [DOI] [PubMed] [Google Scholar]
- 29.Petoukhov MV, Svergun DI. Analysis of X-ray and neutron scattering from biomacromolecular solutions. Curr. Opin. Struct. Biol. 2007;17:562–571. doi: 10.1016/j.sbi.2007.06.009. [DOI] [PubMed] [Google Scholar]
- 30.Tidow H, Melero R, Mylonas E, Freund SM, Grossmann JG, Carazo JM, Svergun DI, Valle M, Fersht AR. Quaternary structures of tumor suppressor p53 and a specific p53 DNA complex. Proc. Natl Acad. Sci. USA. 2007;104:12324–12329. doi: 10.1073/pnas.0705069104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wells M, Tidow H, Rutherford TJ, Markwick P, Jensen MR, Mylonas E, Svergun DI, Blackledge M, Fersht AR. Structure of tumor suppressor p53 and its intrinsically disordered N-terminal transactivation domain. Proc. Natl Acad. Sci. USA. 2008;105:5762–5767. doi: 10.1073/pnas.0801353105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tang KH, Niebuhr M, Aulabaugh A, Tsai MD. Solution structures of 2 : 1 and 1 : 1 DNA polymerase-DNA complexes probed by ultracentrifugation and small-angle X-ray scattering. Nucleic Acids Res. 2008;36:849–860. doi: 10.1093/nar/gkm1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lamber EP, Wilmanns M, Svergun DI. Low resolution structural models of the basic helix-loop-helix leucine zipper domain of upstream stimulatory factor 1 and its complexes with DNA from small angle X-ray scattering data. Biophys. J. 2008;94:193–197. doi: 10.1529/biophysj.107.112243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Manolaridis I, Mumtsidu E, Konarev P, Makhov AM, Fullerton SW, Sinz A, Kalkhof S, McGeehan JE, Cary PD, Griffith JD, et al. Structural and biophysical characterization of the proteins interacting with the herpes simplex virus 1 origin of replication. J. Biol. Chem. 2009;284:16343–16353. doi: 10.1074/jbc.M806134200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Atmanene C, Chaix D, Bessin Y, Declerck N, Van Dorsselaer A, Sanglier-Cianferani S. Combination of Noncovalent Mass Spectrometry and Traveling Wave Ion Mobility Spectrometry Reveals Sugar-Induced Conformational Changes of Central Glycolytic Genes Repressor/110 DNA Complex. Anal. Chem. 2010;8:3597–3605. doi: 10.1021/ac902784n. [DOI] [PubMed] [Google Scholar]
- 36.Cantor CR, Schimmel PR. Techniques for the Study of Biological Structure and Function. New York: Freeman,W.H and Co; 1980. [Google Scholar]
- 37.Konarev PV, Petoukhov MV, Volkov VV, Svergun DI. ATSAS 2.1, a program package for small-angle scattering data analysis. J. Appl. Crystallogr. 2006;39:277–286. doi: 10.1107/S0021889812007662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Konarev PV, Volkov VV, Sokolova AV, Koch M.HJ, Svergun DI. PRIMUS: a Windows PC-based system for small-angle scattering data analysis. J. Appl. Crystallogr. 2003;36:1277–1282. [Google Scholar]
- 39.Svergun DI. Determination of the regularization parameter in indirect-transform methods using perceptual criteria. J. Appl. Crystallogr. 1992;25:495–503. [Google Scholar]
- 40.Porod G. General Theory, in Small-Angle X-ray Scattering. In: Glatter O, Kratky O, editors. London: Academic Press; 1982. pp. 17–51. [Google Scholar]
- 41.Zorrilla S, Ortega A, Chaix D, Alfonso C, Rivas G, Aymerich S, Lillo MP, Declerck N, Royer CA. Characterization of the control catabolite protein of gluconeogenic genes repressor by fluorescence cross-correlation spectroscopy and other biophysical approaches. Biophys. J. 2008;95:4403–4415. doi: 10.1529/biophysj.108.135863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schwille P, Meyer-Almes FJ, Rigler R. Dual-color fluorescence cross-correlation spectroscopy for multicomponent diffusional analysis in solution. Biophys. J. 1997;72:1878–1886. doi: 10.1016/S0006-3495(97)78833-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chen CS, White A, Love J, Murphy JR, Ringe D. Methyl groups of thymine bases are important for nucleic acid recognition by DtxR. Biochemistry. 2000;39:10397–10407. doi: 10.1021/bi0009284. [DOI] [PubMed] [Google Scholar]
- 44.Garnett JA, Marincs F, Baumberg S, Stockley PG, Phillips SE. Structure and function of the arginine repressor-operator complex from Bacillus subtilis. J. Mol. Biol. 2008;379:284–298. doi: 10.1016/j.jmb.2008.03.007. [DOI] [PubMed] [Google Scholar]
- 45.Hong M, Fuangthong M, Helmann JD, Brennan RG. Structure of an OhrR-ohrA operator complex reveals the DNA binding mechanism of the MarR family. Mol. Cell. 2005;20:131–141. doi: 10.1016/j.molcel.2005.09.013. [DOI] [PubMed] [Google Scholar]
- 46.Pons JL, Labesse G. @TOME-2: a new pipeline for comparative modeling of protein-ligand complexes. Nucleic Acids Res. 2009;37:W485–W491. doi: 10.1093/nar/gkp368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Catherinot V, Labesse G. ViTO: tool for refinement of protein sequence-structure alignments. Bioinformatics. 2004;20:3694–3696. doi: 10.1093/bioinformatics/bth429. [DOI] [PubMed] [Google Scholar]
- 48.Petoukhov MV, Svergun DI. Global rigid body modeling of macromolecular complexes against small-angle scattering data. Biophys. J. 2005;89:1237–1250. doi: 10.1529/biophysj.105.064154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Garcia De La Torre J, Huertas ML, Carrasco B. Calculation of hydrodynamic properties of globular proteins from their atomic-level structure. Biophys. J. 2000;78:719–730. doi: 10.1016/S0006-3495(00)76630-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Haustein E, Schwille P. Single-molecule spectroscopic methods. Curr. Opin. Struct. Biol. 2004;14:531–540. doi: 10.1016/j.sbi.2004.09.004. [DOI] [PubMed] [Google Scholar]
- 51.Zorrilla S, Lillo MP, Chaix D, Margeat E, Royer CA, Declerck N. Investigating transcriptional regulation by fluorescence spectroscopy, from traditional methods to state-of-the-art single-molecule approaches. Ann. NY Acad. Sci. 2008;1130:44–51. doi: 10.1196/annals.1430.023. [DOI] [PubMed] [Google Scholar]
- 52.Sanglier S, Atmanene C, Chevreux G, Dorsselaer AV. Nondenaturing mass spectrometry to study noncovalent protein/protein and protein/ligand complexes: technical aspects and application to the determination of binding stoichiometries. Methods Mol. Biol. 2008;484:217–243. doi: 10.1007/978-1-59745-398-1_15. [DOI] [PubMed] [Google Scholar]
- 53.Rusconi F, Guillonneau F, Praseuth D. Contributions of mass spectrometry in the study of nucleic acid-binding proteins and of nucleic acid-protein interactions. Mass Spectrom. Rev. 2002;21:305–348. doi: 10.1002/mas.10036. [DOI] [PubMed] [Google Scholar]
- 54.Barraud P, Golinelli-Pimpaneau B, Atmanene C, Sanglier S, Van Dorsselaer A, Droogmans L, Dardel F, Tisne C. Crystal structure of Thermus thermophilus tRNA m1A58 methyltransferase and biophysical characterization of its interaction with tRNA. J. Mol. Biol. 2008;377:535–550. doi: 10.1016/j.jmb.2008.01.041. [DOI] [PubMed] [Google Scholar]
- 55.Ritschel T, Atmanene C, Reuter K, Van Dorsselaer A, Sanglier-Cianferani S, Klebe G. An integrative approach combining noncovalent mass spectrometry, enzyme kinetics and X-ray crystallography to decipher Tgt protein-protein and protein-RNA interaction. J. Mol. Biol. 2009;393:833–847. doi: 10.1016/j.jmb.2009.07.040. [DOI] [PubMed] [Google Scholar]
- 56.Meinken C, Blencke HM, Ludwig H, Stulke J. Expression of the glycolytic gapA operon in Bacillus subtilis: differential syntheses of proteins encoded by the operon. Microbiology. 2003;149:751–761. doi: 10.1099/mic.0.26078-0. [DOI] [PubMed] [Google Scholar]