Abstract
Replication-factor C (RFC) is a protein complex that loads the processivity clamp PCNA onto DNA. Elg1 is a conserved protein with homology to the largest subunit of RFC, but its function remained enigmatic. Here, we show that yeast Elg1 interacts physically and genetically with PCNA, in a manner that depends on PCNA modification, and exhibits preferential affinity for SUMOylated PCNA. This interaction is mediated by three small ubiquitin-like modifier (SUMO)-interacting motifs and a PCNA-interacting protein box close to the N-terminus of Elg1. These motifs are important for the ability of Elg1 to maintain genomic stability. SUMOylated PCNA is known to recruit the helicase Srs2, and in the absence of Elg1, Srs2 and SUMOylated PCNA accumulate on chromatin. Strains carrying mutations in both ELG1 and SRS2 exhibit a synthetic fitness defect that depends on PCNA modification. Our results underscore the importance of Elg1, Srs2 and SUMOylated PCNA in the maintenance of genomic stability.
Keywords: genome stability, PCNA modification, post-replicational repair
Introduction
PCNA is a homotrimeric ring that encircles the double-stranded DNA (Krishna et al, 1994) and has an important function in DNA replication, enhancing the processivity of replicative DNA polymerases during DNA synthesis (Krishna et al, 1994; Eissenberg et al, 1997; Chilkova et al, 2007). In addition, PCNA constitutes a general platform for docking of DNA-processing enzymes involved in Okazaki fragment maturation, DNA repair, chromatin remodelling and sister chromatid cohesion (Warbrick, 2000; Majka and Burgers, 2004; Aroya and Kupiec, 2005; Lengronne et al, 2006; Moldovan et al, 2006).
In response to DNA damage, PCNA can be modified with ubiquitin at lysine 164, a process mediated by the E2/E3 pair Rad6 and Rad18 (Hoege et al, 2002). Mono-ubiquitination of PCNA was shown to enhance the binding of the translesion synthesis polymerases to PCNA in yeast and in human beings, resulting in an error-prone repair mechanism (Bienko et al, 2005; Parker et al, 2007; Acharya et al, 2008). Alternatively, PCNA can be further poly-ubiquitinated on the same lysine residue by a mechanism that additionally requires Ubc13-Mms2 (E2 heterodimer) and Rad5 (E3). Ubiquitin subunits in this chain are linked through the lysine 63 residue; the biochemical function of this poly-ubiquitin modification is still unknown; it coordinates a type of repair that is essentially error free (Blastyak et al, 2007; Branzei et al, 2008). Mutations in lysine 164 of PCNA, in RAD18 or in RAD5 cause high sensitivity to DNA damaging agents such as MMS and UV, emphasizing the importance of these modifications for DNA repair in living cells. Interestingly, the same residue (lysine 164) can be modified by another small protein named small ubiquitin-like modifier (SUMO). This modification takes place during S-phase (Hoege et al, 2002) or after high doses of DNA damage. An additional residue, lysine 127, can also be SUMOylated, but not ubiquitinated (Hoege et al, 2002). It was shown that the Srs2 helicase, which regulates homologous recombination, preferentially interacts with SUMOylated PCNA and that this interaction is harmful in the presence of DNA damage when PCNA cannot be ubiquitinated (Papouli et al, 2005; Pfander et al, 2005). However, it is not known whether other proteins also interact with SUMOylated PCNA and what is the function of this modification during ongoing replication. Recently, it was found that PCNA can also undergo polySUMOylation; the importance of this modification is still under investigation (Parker et al, 2008; Windecker and Ulrich, 2008).
A second way of regulating PCNA activity is by loading or unloading it from the DNA. This activity can be carried out by the replication-factor C (RFC) complex that binds PCNA, opens the homotrimeric ring and loads it on the template/primer junction in an ATP-dependent manner (Tsurimoto and Stillman, 1989, 1991). The clamp (PCNA) and clamp loader (RFC) are conserved throughout the evolutionary scale. Homologous proteins that share the same function and structure with RFC and PCNA were found in bacteria (Stukenberg et al, 1991), archaea (De Felice et al, 1999), yeast (Fien and Stillman, 1992) and other eukaryotes (Kelman and O'Donnell, 1995). In the last decade, three homologues of Rfc1, the large subunit of the clamp loader, were found in the yeast Saccharomyces cerevisiae. Each of these three proteins, Rad24, Ctf18 and Elg1, was found to bind to the small RFC subunits (Rfc2-5) forming RFC-like complexes. In contrast to Rfc1, these alternative large subunits are not essential for vegetative growth; however, they have an important function in maintaining genome stability (reviewed in Aroya and Kupiec, 2005).
ELG1 was found in several genome-wide screens in yeast (Scholes et al, 2001; Bellaoui et al, 2003; Ben-Aroya et al, 2003; Huang et al, 2003; Banerjee and Myung, 2004; Smolikov et al, 2004). The elg1 mutants exhibit increased recombination rates (Ben-Aroya et al, 2003; Ogiwara et al, 2007), chromosome loss (Ben-Aroya et al, 2003) and gross chromosomal rearrangements (Smith et al, 2004). They have elongated telomeres (Smolikov et al, 2004) and elevated levels of Ty transposition (Scholes et al, 2001). Thus, mutations in ELG1 cause increased levels of genomic instability. Orthologues of Elg1 were identified in many organisms, including human beings. hELG1 has been recently shown to have an important function in maintaining genome stability in S-phase (Sikdar et al, 2009). Targeted gene knockdown of hELG1 resulted in spontaneous foci formation of γ-H2AX, 53BP1 and phosphorylated ATM that usually mark chromosomal breaks, and to increased levels of recombination and chromosomal aberrations such as chromosomal fusions and inversions (Sikdar et al, 2009). hELG1 was also found to link DNA replication stalling with apoptosis (Ishii et al, 2005).
In this study, we have examined how Elg1 interacts with PCNA and we have identified the activity by which Elg1 contributes to genomic stability. We found that Elg1 preferentially interacts with SUMOylated PCNA through SUMO-interacting motifs (SIMs) and that this interaction affects the repair process chosen by the cells. The sensitivity of elg1 strains to DNA damage and the predicted function of Elg1 in DNA replication suggest that the alternative clamp loaders are responsible for the switch between DNA replication and DNA repair and for selecting the repair mechanism after PCNA modification.
Results
Deletion of ELG1 suppresses defects in the error-free branch of the post-replicative repair pathway
By genetic analysis, the Elg1 clamp loader complex has earlier been linked to the maintenance of genome stability. To gain insights into the precise function of Elg1 in the DNA damage response, we tested the DNA damage sensitivity of the Δelg1 mutant on its own and in combination with different DNA repair mutants.
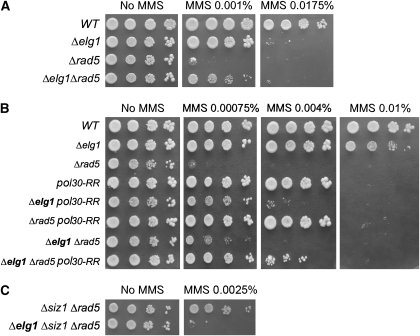
When chronically exposed to the DNA alkylating drug MMS, Δelg1 mutants displayed a mild sensitivity (Figure 1A). Surprisingly, the deletion of ELG1 clearly suppressed the (higher) sensitivity to MMS of rad5, a well-characterized mutant of the post-replicative repair pathway (Figure 1A). Rad5 is a large protein that contains a helicase domain and an E3 ubiquitin ligase domain (Chen et al, 2005; Gangavarapu et al, 2006). Supplementary Figure S1A shows that deletion of ELG1 suppresses a rad5 E3 ubiquitin ligase mutant (rad5-E3), but has an additive relation with the helicase mutant (rad5-h). We thus conclude that the suppressive effect of elg1 with respect to rad5 results from the poly-ubiquitination activity of this enzyme. A similar suppressive effect was obtained for strains deleted for MMS2 and UBC13, which encode the E2 poly-ubiquitin conjugation enzymes responsible for PCNA poly-ubiquitination and for the ubi-K63R mutant, which is unable to support PCNA poly-ubiquitination (Supplementary Figure S1A). This suppression was specific for the error-free (Rad5-dependent) pathway and not for the error-prone pathway, as shown by the additive interaction with the error-prone translesion polymerase ζ (Supplementary Figure S2).
Figure 1.
Genetic interactions between ELG1 and PCNA depend on PCNA modification. Serial dilutions of yeast cultures on minimal SD-complete plates with or without methylmethane sulphonate (MMS). Ten different MMS concentrations were used for each experiment; the informative ones are shown. (A) Deletion of ELG1 suppresses the sensitivity of Δrad5 to DNA damage. (B) elg1 suppression effect cannot be seen on a pol30-RR background. (C) elg1 suppression effect cannot be seen in the absence of the Siz1 SUMO ligase.
We thus conclude that in the presence of DNA damage, Elg1 activity becomes toxic if PCNA cannot be poly-ubiquitinated; deletion of the ELG1 gene under these circumstances suppresses the sensitivity to DNA damage. One possibility is that the toxic activity of the alternative clamp loader containing Elg1 depends on the modification that PCNA undergoes when poly-ubiquitination is inhibited. If this were the case, the suppression by elg1 should be abolished when PCNA cannot be modified. Indeed, in pol30-RR strains (PCNA mutants that cannot be modified neither by ubiquitin nor SUMO on lysine 127 and 164), deletion of ELG1 no longer suppresses the sensitivity to MMS of rad5 mutants: the triple rad5 pol30-RR elg1 mutant is not more resistant than the rad5 pol30-RR double mutant strain (Figure 1B; note that pol30-RR by itself partially suppresses the rad5 phenotype; Papouli et al, 2005). More specifically, the suppressive effect of elg1 was dependent on SIZ1, the E3 SUMO ligase that is responsible for the SUMOylation of lysine 164 of PCNA (Figure 1C; Supplementary Figure S1B). Similar results were obtained after acute MMS doses (Supplementary Figure S3). It can also be seen (Figure 1B; Supplementary Figure S1C) that in the absence of SUMOylation (pol30-RR or pol30-K164R), Elg1 activity is needed for DNA damage resistance. We thus conclude that the toxic effect caused by Elg1 in the absence of PCNA poly-ubiquitination depends on PCNA SUMOylation.
Physical interactions between Elg1 and PCNA
To prove that the genetic relations between elg1 mutants and modified PCNA result from direct physical contact between the proteins, we examined the interaction between full-length Elg1 and various PCNA mutants using a yeast-two-hybrid assay (James et al, 1996). Figure 2A shows that Elg1 binds PCNA. A similar level of binding was observed with versions of PCNA carrying mutations in lysine 127, lysine 164 or both (Figure 2A). The lack of effect of PCNA modifications could in principle be due to the fact that only a small fraction of PCNA is being modified and the two-hybrid assay is not sensitive enough to measure this change. In contrast, when we expressed prey constructs that contain either SUMO or ubiquitin fused to pol30-RR (a gift from H Ulrich; Parker et al, 2007) in the presence of Elg1 baits, the SUMO-PCNA fusion allows growth even under very stringent conditions (plates lacking adenine, Figure 2) indicating that indeed Elg1 binds with high affinity to the fusion SUMO-PCNA. Moreover, Figure 2A shows that Elg1 also binds to SUMO in a yeast-two-hybrid assay.
Figure 2.
Elg1 interacts physically with PCNA and SUMO. (A) Two-hybrid interaction between Elg1 fused to GAL4-binding domain (BD) and a number of constructs containing PCNA or SUMO fused to the GAL4-activating domain (AD). (B) Co-IP of an Elg1-13Myc protein with PCNA. After IP with IgG as a negative control or with anti-PCNA antibody, western blots were carried out with anti-PCNA or anti-myc antibodies. (C) Co-IP of an Elg1-13Myc protein with Histidine-tagged PCNA in ulp1-1 strains. The yeast culture was treated with 0.3% MMS for 1 h before lysis. The asterisk denotes a faint additional SUMOylation of PCNA at an unknown location that is sometimes detected.
Additional proof of the interaction between Elg1 and PCNA comes from immunoprecipitation experiments: Antibodies against PCNA were able to co-precipitate Myc-tagged versions of Elg1 under normal conditions (Figure 2B), confirming earlier observations obtained with overexpressed PCNA (Kanellis et al, 2003). Next, we precipitated Elg1 to find out if it interacts with modified PCNA (Figure 2C). These experiments were carried out in the presence of 0.3% MMS in ulp1-1 strains to maximize the SUMOylation signal (Hoege et al, 2002; Stelter and Ulrich, 2003). Elg1 co-precipitates with modified and unmodified PCNA. The same sample was probed with an anti-SUMO antibody proving that Elg1 interacts with SUMOylated PCNA (Figure 2C). It is important to notice that the SUMOylated PCNA is strongly enriched in the pellet compared with the whole cell extract (WCE), emphasizing that the Elg1-containing alternative clamp loader preferentially interacts with the modified form of PCNA. To find the SUMOylated form with which Elg1 interacts, we introduced ELG1-13MYC into strains with POL30 mutated at lysine 127, lysine 164 or both. We found that Elg1 preferentially precipitated with all forms of SUMOylated PCNA (Figure 2C).
N-terminus of Elg1 mediates the physical interaction with SUMOylated PCNA
To get better insights into the mechanism that enables Elg1 to interact with PCNA and in particular with SUMOylated PCNA, we carried out a deletion analysis of Elg1. We divided the Elg1 protein into a central AAA domain, bearing most of the homology to Rfc1 (Ben-Aroya et al, 2003), an N-terminus and a C-terminal domain. Figure 3 shows that both PCNA and SUMO bind exclusively to the N-terminus of Elg1 (Figure 3A and B). The fact that Elg1 binds PCNA through its N-terminus is surprising, as crystal structure studies have shown that the interactions of RFC with PCNA are mediated by the AAA domain of Rfc1 (Jeruzalmi et al, 2001). As expected, we found that the N-terminal domain of Elg1 preferentially binds to the SUMO-PCNA fusion, as seen with the full-length Elg1 protein (Supplementary Figure S4). When we examined by yeast two hybrid the interaction between the N-terminus of Elg1 and SUMO in a pol30-RR strain, this interaction was significantly reduced (data not shown). This means that the major interaction of Elg1 with SUMO is mediated by covalent modification of lysine 127 and 164 of PCNA (although other lysine residues on PCNA or other SUMOylated protein may also interact with Elg1).
Figure 3.
The N-terminus of Elg1 interacts with PCNA and SUMO. (A) Two-hybrid experiment with SUMO fused to the GAL4-activating domain (AD) and segments of ELG1 fused to the GAL4 DNA-binding domain (BD). (B) The same experiment as in (A) except that Pol30-RR is fused to AD. (C) Pull-down experiment with recombinant N-terminus of Elg1 fused to GST or with GST alone. The recombinant protein was mixed with lysates of yeast containing HIS-PCNA. Cells were treated with different amount of MMS as indicated. (D) Pull-down experiment with recombinant N-terminus of Elg1 fused to GST and yeast lysates (from cells treated with 0.3% MMS for 1 h) in which HIS-PCNA is mutated at the lysine that can undergo modification. (E) Quantitation of the relative amount of modified PCNA in the WCE and in the pellet (based on three experiments). (F) In vitro SUMOylation of PCNA was performed with purified Aos1p-Uba2p (E1), Ubc9 (E2), Siz1 (E3), PCNA, SUMO and ATP. The left panel shows the full reaction, as well as control reactions each missing one component. The right panel shows a pull-down experiment with recombinant N-terminus of Elg1 fused to GST or with GST and purified SUMOylated PCNA obtained in vitro.
Next, we carried out pull-down experiments with recombinant GST-Elg1(Nter) and lysates of yeast cells treated with different concentration of MMS. GST-Elg1(Nter) but not GST alone pulled down PCNA (Figure 3C). Importantly, the N-terminal domain of Elg1 preferentially interacted with SUMOylated PCNA (Figure 3C and D) and the amount of SUMOylated PCNA pulled down increased with increasing MMS concentration. Quantitation of the results (Figure 3E) shows a clear enrichment of SUMOylated PCNA (all forms) in the pellet compared with the WCE fraction. The higher level of SUMOylated PCNA in the presence of DNA damage may enhance the interaction between Elg1 and PCNA. To examine whether the interaction between the N-terminus of Elg1 and SUMOylated PCNA is direct or mediated by other cellular components, we examined the interactions in vitro. We performed an in vitro SUMOylation reaction of PCNA (see Materials and methods) followed by a pull-down experiment with GST or GST-Elg1(Nter) (Figure 3F). Again, a clear preferential interaction with SUMOylated PCNA could be detected. This experiment clearly shows that the interaction between Elg1 and SUMOylated PCNA is direct and does not depend on additional cellular factors.
SUMOylated PCNA accumulates in the chromatin fraction of Δelg1 strains
To better understand the effect of Elg1 on SUMOylated PCNA, we compared the amount of PCNA in the chromatin fraction of wild type versus Δelg1 strains. Figure 4A shows that in elg1 mutants, unmodified PCNA accumulates in the chromatin fraction (a ∼three-fold increase). It was earlier found that such an accumulation occurs during S-phase and that PCNA SUMOylation also occurs mainly in S-phase (Hoege et al, 2002; Parker et al, 2008). Thus, a possible explanation for the increased PCNA accumulation in elg1 mutants might be a prolonged S-phase. However, the difference in cell cycle distribution in wt and elg1 mutants is very small and cannot account for the effect seen (Supplementary Figure S5). Interestingly, the chromatin accumulation of SUMOylated PCNA was much more prominent than the unmodified form (Figure 4B and C). In contrast, ubiquitinated PCNA accumulated only to levels similar to those of the unmodified form (Supplementary Figure S6). In the absence of DNA damage, wt strains exhibit low levels of SUMOylated PCNA in the chromatin fraction; in contrast, elg1 mutants show a strong accumulation (Figure 4B and C). This result fits well with the preferential binding of Elg1 to SUMOylated PCNA in the yeast-two-hybrid assay (Figure 2A; without damage) and the pull-down assay (with damage; Figures 2 and 3) suggesting that the alternative clamp loader containing Elg1 might negatively regulate SUMOylated PCNA on chromatin.
Figure 4.
Higher levels of unmodified or SUMOylated PCNA in the chromatin of elg1 mutant cells. Wild-type or elg1 cells containing HIS-tagged PCNA were subjected to chromatin fractionation and Ni-NTA pull down after treatment without or with MMS for 60 min., followed by immunoblotting. (A) Unmodified or SUMOylated PCNA was visualized by western blotting using antibodies against SUMO or PCNA. PGK was used as a non-chromatin marker, whereas Acetylated Histone H4 (AcH4) was used as a chromatin marker. Whole cell extract (WCE), supernatant (Sup) and chromatin (CHR) fractions are shown. (B) Quantitation analysis of the amount of SUMOylated or unmodified PCNA of wt and elg1 mutants without MMS. The amount of PCNA or modified PCNA in the chromatin fraction was divided by the amount of AcH4 signal in the chromatin fraction. The average of four experiments is presented. (C) Quantitation analysis (as in B) of the samples treated with 0.3% MMS.
Genetic interactions between elg1 and srs2
In earlier publications, it was suggested that PCNA-SUMO and Srs2 work in the same pathway and that Srs2 preferentially binds to SUMOylated PCNA (Papouli et al, 2005; Pfander et al, 2005). In Figure 4, we show that in elg1 mutants SUMOylated PCNA accumulates in the chromatin fraction; as Srs2 preferentially interacts with SUMOylated PCNA, we expect to find an accumulation of Srs2 in the chromatin fraction in elg1 mutants, in which the alternative clamp loader is not active. Indeed, this prediction is confirmed (Figure 5A). This finding raises the question of whether Elg1 and Srs2 have similar modes of action. To address this question, we have examined the amount of SUMOylated PCNA in the chromatin fraction in srs2 mutants. Supplementary Figure S7 shows that deletion of the SRS2 gene does not lead to an accumulation of SUMOylated PCNA in the chromatin fraction. We also found no difference in the amount of Elg1 at the chromatin in srs2 mutants (data not shown). These results contradict the possibility that Elg1 and Srs2 compete on regulating the amount of SUMOylated PCNA on the chromatin. To get a better understanding of the mechanism by which these proteins control genomic stability, we examined the genetic interactions between elg1 and srs2. Notably, the double mutant srs2 elg1 is sick and grows poorly compared with the single mutants (Figure 5B). We, therefore, examined whether PCNA modification may affect this synthetic genetic interaction. Figure 5C shows that the pol30-RR allele improves the viability of elg1 srs2 strains. The same result was obtained in the presence of DNA damage: the high sensitivity of elg1 srs2 strains to MMS is suppressed when PCNA cannot be modified (Figure 5D). This effect did not result from the inability to ubiquitinate PCNA because a rad18 mutation (which prevents ubiquitination) could not suppress the growth defect or the sensitivity to MMS of an elg1 srs2 double mutant strain (Figure 5E). Thus, SUMOylation of PCNA in the absence of either protein is toxic, suggesting a model in which SUMOylated PCNA needs to be removed from the fork to allow survival. Recruitment of Srs2 promotes its clearance by allowing damage bypass or repair; alternatively, Elg1 may unload the modified PCNA molecules from the chromatin thus enabling other DNA repair mechanism to act. When both Elg1 and Srs2 are missing, SUMOylated PCNA stays at the fork and becomes toxic.
Figure 5.
Synthetic interactions of elg1 with Srs2 (A) Higher levels of Srs2 in the chromatin fraction of Δelg1 cells. Wild-type or elg1 cells containing Srs2-Myc were subjected to chromatin fractionation after treatment without or with 0.02% MMS for 60 min. PGK was used as a non-chromatin marker, whereas AcH4 was used as chromatin marker. (B) elg1 and srs2 show a synthetic fitness phenotype. Tetrad analysis of a diploid heterozygote srs2/+, +/elg1. (C) pol30-RR partially suppresses the synthetic sickness of elg1 srs2 mutants. Tetrad analysis of a diploid homozygote for elg1 (elg1/elg1) and heterozygote srs2/+, pol30-RR/+. (D) pol30-RR partially suppresses the sensitivity of elg1 srs2 to MMS. (E) rad18 (no ubiquitination of PCNA) does not suppresses the sensitivity of elg1 srs2 to MMS.
Three SIM motifs and a PCNA-interacting protein box enable the interaction of Elg1 with SUMOylated PCNA
To better understand the mechanism that enables Elg1 to interact with SUMOylated PCNA, we further defined, by deletion analysis, the regions of Elg1 that mediate the interaction with SUMO and PCNA. Two regions, between amino acids 67 and 94 and between residues 116 and 143, were important for the interactions with SUMO (Figure 6A), as deletion of each of them reduced the interaction. Careful examination of these regions uncovered three sequences similar to the earlier defined SIM, which enables proteins to interact with SUMO non-covalently (Minty et al, 2000; Song et al, 2004; Kerscher, 2007). This motif consists of a series of hydrophobic amino acids usually containing valine and isoleucine, followed by negatively charged amino acids. As shown in Figure 6C, each of the suspected regions contains a SIM motif. Next, we took the same approach to find the region of Elg1 that interacts with PCNA (using a version that cannot be modified by SUMO). Yeast-two-hybrid analysis defined a region located between amino acids 43 and 67 (Figure 6B). PCNA-binding motifs, termed PCNA-interacting protein (PIP) motifs, have been described for many proteins (Warbrick, 1998; Moldovan et al, 2007). Analysis of the Elg1 sequence in this region revealed a motif with general resemblance to the canonical PIP box (Figure 6D). Interestingly, the sequence found in the N-terminus of Elg1 is most similar to the PIP sequence of Rfc1 (Figure 6D; Bowman et al, 2004).
Figure 6.
SIM and PIP motifs mediate the interaction between Elg1, PCNA and SUMO. Interaction between different fragments of the N-terminus of ELG1 fused to GAL4 BD and (A) SUMO or (B) PCNA-RR fused to GAL4 AD. (C) The location of the PIP box (bright grey) and SIM motifs (dark grey) in the N-terminus of Elg1. (D) Sequence alignment between proteins that contain PIP boxes. (E) Elg1 interactions with SUMO and PCNA are reduced in SIM and PIP mutants, accordingly. Two-hybrid interactions between SUMO or PCNA-RR and the N-ter Elg1 carrying various mutations. The SIM alleles were I28A (SIM1), I93K (SIM2), II121, 122AA (SIM3) and combinations thereof; the PIP mutants were either SV57, 58AA (PIP57) or VV58, 59 (PIP58). (F) Pull down with the N-terminus of elg1 with or without mutation in the three SIM motifs. The yeast lysates were treated with 0.3% MMS for 1 h.
To examine whether the SIM and PIP motifs indeed contribute to the interactions with SUMO and PCNA, we mutated them (mutations are detailed in Supplementary Table S2) and tested the mutants in the yeast-two-hybrid assay. Mutation of each SIM motif reduced the ability to bind SUMO, and the motifs acted in an additive manner: the double SIM mutants showed a stronger effect and a triple SIM mutant showed no interaction at all (Figure 6E). According to Figure 6E, mutations in the PIP box cause a significant reduction in the interaction between the N-terminus of Elg1 and PCNA. In addition, mutations in either the PIP or the SIM motifs reduce the interaction with the fusion construct of SUMO-PCNA (Supplementary Figure S8).
Using pull-down experiments, we also found that mutations in the SIM motifs of a version of the amino terminus of Elg1 strongly reduced the interactions with PCNA and with SUMOylated PCNA (Figure 6F). In conclusion, these results uncover the motifs that enable Elg1 to interact with SUMOylated PCNA showing a novel mechanism that involves three SIM motifs and a PIP box.
We next examined by co-immunoprecipitation the interaction between PCNA and Elg1-containing site-specific mutations expressed from their endogenous promoters (Figure 7A). All strains exhibited similar levels of Elg1 protein (Figure 7B). Mutations in the three SIM motifs of Elg1 reduced the interaction with SUMOylated PCNA (Figure 7A). Mutations in the PIP motif had a minor effect on the interaction between Elg1 and PCNA. We, therefore, cannot exclude the possibility that there is more than one PCNA-interacting motif in Elg1, as was recently found in the human Pol η (Acharya et al, 2008) or that the interaction between Elg1 and PCNA is also mediated by the small subunits of the RFC complex.
Figure 7.
The SIMs and PIP motifs in ELG1 are important for its activity. (A) The interaction of Elg1 with SUMOylated PCNA is mediated by the SIM and PIP motifs. A Co-IP experiment with ELG1 mutants or wt cells tagged with 13-MYC or in strains in which ELG1 is not tagged (as a control). The SIM allele used is the triple mutant I28A, I93K, II121, 122AA; the PIP allele is SV57, 58AA, and the SIM-PIP (SP) allele is a combination of all the mutations. The yeast lysates were treated with 0.3% MMS for 1 h. (B) Western blot analysis to detect the protein amount of Elg1 mutants in vivo using anti-Myc antibody or anti-PGK antibody as loading control. (C, D) Sensitivity to MMS of elg1 mutants in RAD5 and rad5 mutant background, accordingly. (E) No effect on the sensitivity to MMS by elg1 mutants could be observed in a pol30-RR background.
Next, we wanted to examine whether the SIMs and PIP motifs at the N-terminus of Elg1 have biological significance. First, we measured the sensitivity to MMS of yeast strains mutated in these motifs. In this assay, we did not find an effect for the SIM motifs or the PIP motif separately; however, when we used a strain in which both SIM and PIP motifs were mutated, a clear sensitivity to MMS was observed (Figure 7C), suggesting alternative functions in PCNA binding. This sensitivity was somewhat lower than that of an elg1 null mutant, suggesting that other domains also contribute to the activity of Elg1. Figure 7D shows a clear suppression of rad5 by mutation of the SIM motif, and an even stronger suppression by the double SIM, PIP mutant. As in the case of the null mutant (Figure 1C), the suppression effect by the SIM, PIP mutant could not be observed in the pol30-RR background, in which PCNA cannot be SUMOylated (Figure 7E). From all the results presented, we conclude that the PIP and SIMs motifs mediate, in an additive manner, the physical interactions between Elg1 and SUMOylated PCNA. These interactions are responsible for most of the DNA damage sensitivity of rad5 mutants and are also responsible for the contribution of Elg1 to yeast resistance to DNA damage.
Discussion
In this work, we found that elg1 suppresses the sensitivity to DNA damage of rad5 mutants and other post-replication repair mutants such as ubc13 and mms2. We show that the suppression is due to the inability of these strains to poly-ubiquitinate PCNA (Figure 1; Supplementary Figure S1); under these circumstances, the presence of an active Elg1 protein is toxic. This toxicity required SUMOylation of PCNA, as no suppression effect could be seen on the background of siz1 or pol30-RR (which is mutated at the SUMOylation sites of PCNA; Figure 1B and C). We provide evidence for binding of PCNA by Elg1 and show that this interaction is mediated by the covalent modification of PCNA by SUMO. We base this conclusion on the two-hybrid assay results, the pull-down assay with the N-terminus of Elg1, the Co-IP experiments (Figures 2 and 3) and the in vitro assay with purified SUMOylated PCNA (Figure 3F). We have also identified three SIM motifs and a PIP box in the N-terminus of Elg1 that mediate the interaction (Figure 6). Moreover, our results show that in the absence of Elg1, SUMOylated PCNA accumulates in the chromatin fraction (Figure 4).
Taking into account the homology of ELG1 to the large subunit of the clamp loader/unloader complex RFC, the interaction of Elg1 with the small subunits of the RFC in vivo and in vitro (Bellaoui et al, 2003; Ben-Aroya et al, 2003; Kanellis et al, 2003; Bylund et al, 2006), the preferential genetic and physical interaction of Elg1 with SUMOylated PCNA and the accumulation of SUMOylated PCNA at the chromatin fraction when Elg1 is absent, the most reasonable model consistent with all the facts is that Elg1 participates in the unloading of SUMOylated PCNA. We cannot, however, rule out alternative models, such as one in which Elg1 is responsible for the recruitment of a de-SUMOylating enzyme (Stelter and Ulrich, 2003). Earlier publications suggested that PCNA modification can occur only after the homotrimer PCNA ring encircles the DNA (Parker et al, 2008). The switching-back mechanism, however, has not been explored yet. We propose a function for Elg1 in the clearance of SUMOylated PCNA, perhaps in parallel to mechanisms that cleave out the modification.
The exact function of each type of PCNA modification is being currently intensively investigated (Moldovan et al, 2007). For example, it was found that PCNA SUMOylation occurs mainly during S-phase, suggesting a function for SUMOylation of PCNA in the regulation of DNA replication (Hoege et al, 2002; Parker et al, 2008). Recently, Branzei et al (2008) showed that PCNA modification can control template switching at the DNA replication fork. If the replication fork stalls, the replicative DNA polymerase must be first evicted, and, after the completion of repair, a switch-back event should take place to enable processive continuity of replication (Ulrich, 2009). Recently, Zhuang et al (2008) suggested that the switching back from the TLS Pol η to the replicative Pol δ is not spontaneous and either de-modification of PCNA or unloading of the modified PCNA are needed for the continuity of the replication.
Other than Elg1, Srs2 is the only known protein that preferentially binds SUMOylated PCNA (Papouli et al, 2005; Pfander et al, 2005). Similarly to Elg1, Srs2 also contains SIM motifs that are essential for the interaction with SUMOylated PCNA (Le Breton et al, 2008). We found that in the absence of Elg1 not only SUMOylated PCNA, but also Srs2 accumulates in the chromatin fraction (Figures 4 and 5A). These results suggest a model in which SUMOylation of PCNA (by Siz1) allows recruitment of Srs2, which affects the repair mechanism chosen at the damage site. After repair is completed (or as a step in the repair process), Elg1 may take off SUMOylated PCNA and Srs2 to allow the continuity of replication. When Elg1 is not active, SUMOylated PCNA remains on the chromatin, leaving HR as the only alternative pathway for repair and viability This model is consistent with earlier finding that elg1 is synthetic lethal with genes involved in HR (Aroya and Kupiec, 2005).
The discovery of PCNA modification (Hoege et al, 2002) suggested a way in which competing PCNA-dependent DNA-affecting activities could be regulated. An important question that remains open is how the affinity of the various interactors may be affected by PCNA modification. We have identified a sequence in the Elg1 protein that binds PCNA and carries homology to the PIP motif (Xu et al, 2001; Moldovan et al, 2007). In addition, the affinity of Elg1 to SUMOylated PCNA is enhanced by three SIM motifs. The in vivo and in vitro interactions shown and the finding of the motifs in Elg1 that enable contact with SUMOylated PCNA strengthen the possibility that the interaction between Elg1 and SUMOylated PCNA is direct. Interestingly, the three SIM motifs are located in the N-terminus of Elg1 in a relatively small fragment (from AA 26 to AA 123) and the PIP box that enables the interaction with PCNA is embedded between them (Figure 6). Thus, our results present a possible mechanism of recruitment of Elg1 according to PCNA modification.
Increased affinity of a TLS polymerase to modified PCNA has been recently shown to be mediated both by PIP motifs and UBM/UBZ domains that enable proteins to interact with ubiquitin non-covalently (Bienko et al, 2005). Interestingly, in this case, pol η is ubiquitinated, raising the possibility that covalent modification of the polymerase may help regulate the non-covalent interaction with ubiquitinated PCNA. We have noticed that Elg1::MYC migrates as a double band (Figure 7B and see also Davidson and Brown, 2008). We are currently analysing the nature of this modification and its potential function in regulating interactions with other proteins.
Evidence for SUMOylation of PCNA was found in yeast, chicken DT40 cells (Arakawa et al, 2006) and Xenopus laevis egg extracts (Leach and Michael, 2005). Recently, it has been found that hELG1 has an important function in maintaining genomic stability during S-phase in human cells (Sikdar et al, 2009) and that the phenotypes of HeLa cells carrying siRNA directed against hELG1 are very similar to those of elg1 yeast cells. Sequences resembling SIM motifs can be found at the N-terminus of hELG1; their interactions with SUMOylated proteins and with SUMOylated PCNA will be the subject of future studies.
Materials and methods
Yeast strain and plasmids
A list of all strains used appears in Supplementary Table S1.
Plasmids
pGBU9 (pGBT9 in which TRP1 was replaced by URA3) was used in yeast two-hybrid experiments. JOY28 (Bellaoui et al, 2003) was the source of our ELG1::13MYC::KANMX strains. To construct the plasmids used in the pull-down assays, ELG1's N-terminus (AA 1–230) was cloned into pGEX3. Mutations in the PIP and SIM motifs in the pGEX3-NTD, a pGEM that contains ELG1::13MYC::KANMX and two-hybrid plasmids were created using PCR reaction with long primers that contain the relevant mutation as indicated in Supplementary Table S2, dissection with Dpn1 and transformation to bacteria.
Primer sequences used are available on request.
The plasmids carrying PCNA mutants and SUMO and ubiquitin fusions in pGAD424 are a generous gift from Helle Ulrich (Parker et al, 2007). The SUMO-PCNA 2 μ plasmid is kind gift from Hideo Shinagawa (Hishida et al, 2006).
DNA damage sensitivity
Serial 10-fold dilutions of logarithmic yeast cells were spotted on fresh SD-complete plates with or without MMS (Sigma) and incubated at 30°C for 3 days.
Two-hybrid assay
To detect two-hybrid interaction, yeast strain PJ69 (James et al, 1996) was co transformed with one LEU2-marked plasmid containing genes fused to the GAL4-activating domain (pACT or pGAD424) and one plasmid containing genes fused to the GAL4 DNA-binding domain (pGBU9). Yeast cultures were grown in SD-URA-LEU medium and spotted on SD-URA-LEU plates, SD-ADE plates and SD-HIS plates. Cells were incubated for 3 days at 30°C.
Cell lysis
Cells were lysed as described earlier (Hanna et al, 2001) with small modifications. 200 OD of logarithmic cells were washed and cells were resuspended in B60. The tubes were vortexed 12 times for 45 s, with 45 s intervals on ice.
Co-IP
Cell lysates were pre-treated with Invitrogen (Oregon, USA) Dynal beads and incubated with 20 μl of anti-Myc antibodies (Santa Cruz) or 7 μl anti-PCNA antibodies (generous gifts by Peter Burgers and Alain Verreault). After 2 h incubation at 4°C, 100 μl beads were added, and further incubated for an hour. Beads were collected, washed eight times and boiled with 50 μl sample buffer. Western blot analysis was performed using anti-PCNA, anti-SUMO (a gift from Phil Hieter) and anti-Myc antibodies. Ten per cent of yeast lysate (WCE) was taken for an Ni-NTA pull down (Ulrich, 2009) when HIS-PCNA modification was examined, to avoid non-specific bands in the western blot with polyclonal anti-PCNA.
GST pull down
Recombinant GST-Elg1 N-terminus (AA 1–230) with or without mutation in the PIP and SIMs motifs was overexpressed in bacteria by incubating overnight in the presence of IPTG at 16°C. A binding assay was carried out as described (Uzunova et al, 2007). Bacterial cells were lysed and incubated with glutation beads (Sigma) for 2 h at 4°C. After five washes with PBS+0.1% Triton, the beads were incubated for 2 h at 4°C with yeast cell lysate. The beads were collected, washed and boiled with 50 μl sample buffer. Ten percent of the yeast lysate (WCE) was used for Ni-NTA pull down (Ulrich, 2009) when HIS-PCNA modification was examined.
In vitro SUMOylation assay
E1 (Aos1/Uba2), E2 (Ubc9), E3 (Siz1) and SUMO (SMT3) were purified as described earlier by the Johnson laboratory (Johnson and Gupta, 2001); His-PCNA was purified as described for other substrates (Johnson and Gupta, 2001). The SUMOylation reaction was carried out in 40 μl of buffer containing 50 mM NaCl, 20 mM HEPES pH 7.5, 5 mM MgCl2, 1 mM DTT, 3 μg E1, 1500 ng E2, 4 μg E3, 1500 ng SUMO, 3 μg of PCNA and ATP to a final concentration of 2.5 mM. After 2 h at 30°C, the reaction mix was resuspended in 1 ml of binding buffer (Yunus and Lima, 2009) and subjected to GST pull down as described above.
Chromatin assay
Fractionation of soluble and chromatin-associated proteins was performed as described earlier (O'Shaughnessy et al, 2006), with a few modifications. Shortly, 7.5 × 106 cells/300 ml YPD medium were grown for 16 h to log phase, then treated with 0.02%, 0.3% of MMS (Sigma) or without MMS for 1 h. Cells were washed and incubated for 10 min at 37°C with 20 μg/μl of Zymolase T-100, next lysed with 0.25% Triton X-100. A total of 100 μl lysates were separated to supernatant and chromatin fractions by sucrose gradient. WCE, supernatant (Sup) and chromatin pellets (CHR) were subjected to SDS–PAGE western blot using anti-PCNA or anti-Myc (Santa Cruz), anti-acetylated histone H4 or anti-3-phosphoglycerate kinase (PGK) (Invitrogen) antibodies; 30–50 μg of total proteins was loaded in each lane. H4 served as a loading control as well as marker for chromatin fraction.
PCNA modifications were detected by denaturing Ni-NTA affinity chromatography and western blot analysis as described earlier, using PCNA- and SUMO-specific antibodies (Ulrich, 2009).
Supplementary Material
Acknowledgments
We thank Helle Ulrich for reagents, plasmids, strains and advice, Philip Hieter, Alain Verreault, Hideo Shinagawa, Amir Aharoni, Yearit Fridman, Giordano Liberi, Zhihao Zhuang and Peter Burgers for reagents and strains, Cristopher Lima and Erica S Johnson for plasmids, C̆eslovas Venclovas, Avital Parnas and Abdussalam Azem for their important advice and all members of the Kupiec laboratory for encouragement and ideas. This research was supported by grants from the German-Israeli bi-national Foundation (GIF) to MK and SJ and the Israel Science Foundation (ISF) and the Association for International Cancer Research (AICR) to MK.
Footnotes
The authors declare that they have no conflict of interest.
References
- Acharya N, Yoon JH, Gali H, Unk I, Haracska L, Johnson RE, Hurwitz J, Prakash L, Prakash S (2008) Roles of PCNA-binding and ubiquitin-binding domains in human DNA polymerase eta in translesion DNA synthesis. Proc Natl Acad Sci USA 105: 17724–17729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arakawa H, Moldovan GL, Saribasak H, Saribasak NN, Jentsch S, Buerstedde JM (2006) A role for PCNA ubiquitination in immunoglobulin hypermutation. PLoS Biol 4: e366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aroya SB, Kupiec M (2005) The Elg1 replication factor C-like complex: a novel guardian of genome stability. DNA Repair (Amst) 4: 409–417 [DOI] [PubMed] [Google Scholar]
- Banerjee S, Myung K (2004) Increased genome instability and telomere length in the elg1-deficient Saccharomyces cerevisiae mutant are regulated by S-phase checkpoints. Eukaryot Cell 3: 1557–1566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellaoui M, Chang M, Ou J, Xu H, Boone C, Brown GW (2003) Elg1 forms an alternative RFC complex important for DNA replication and genome integrity. EMBO J 22: 4304–4313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-Aroya S, Koren A, Liefshitz B, Steinlauf R, Kupiec M (2003) ELG1, a yeast gene required for genome stability, forms a complex related to replication factor C. Proc Natl Acad Sci USA 100: 9906–9911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bienko M, Green CM, Crosetto N, Rudolf F, Zapart G, Coull B, Kannouche P, Wider G, Peter M, Lehmann AR, Hofmann K, Dikic I (2005) Ubiquitin-binding domains in Y-family polymerases regulate translesion synthesis. Science 310: 1821–1824 [DOI] [PubMed] [Google Scholar]
- Blastyak A, Pinter L, Unk I, Prakash L, Prakash S, Haracska L (2007) Yeast Rad5 protein required for postreplication repair has a DNA helicase activity specific for replication fork regression. Mol Cell 28: 167–175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowman GD, O′Donnell M, Kuriyan J (2004) Structural analysis of a eukaryotic sliding DNA clamp-clamp loader complex. Nature 429: 724–730 [DOI] [PubMed] [Google Scholar]
- Branzei D, Vanoli F, Foiani M (2008) SUMOylation regulates Rad18-mediated template switch. Nature 456: 915–920 [DOI] [PubMed] [Google Scholar]
- Bylund GO, Majka J, Burgers PM (2006) Overproduction and purification of RFC-related clamp loaders and PCNA-related clamps from Saccharomyces cerevisiae. Methods Enzymol 409: 1–11 [DOI] [PubMed] [Google Scholar]
- Chen S, Davies AA, Sagan D, Ulrich HD (2005) The RING finger ATPase Rad5p of Saccharomyces cerevisiae contributes to DNA double-strand break repair in a ubiquitin-independent manner. Nucleic Acids Res 33: 5878–5886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chilkova O, Stenlund P, Isoz I, Stith CM, Grabowski P, Lundstrom EB, Burgers PM, Johansson E (2007) The eukaryotic leading and lagging strand DNA polymerases are loaded onto primer-ends via separate mechanisms but have comparable processivity in the presence of PCNA. Nucleic Acids Res 35: 6588–6597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson MB, Brown GW (2008) The N- and C-termini of Elg1 contribute to the maintenance of genome stability. DNA Repair (Amst) 7: 1221–1232 [DOI] [PubMed] [Google Scholar]
- De Felice M, Sensen CW, Charlebois RL, Rossi M, Pisani FM (1999) Two DNA polymerase sliding clamps from the thermophilic archaeon Sulfolobus solfataricus. J Mol Biol 291: 47–57 [DOI] [PubMed] [Google Scholar]
- Eissenberg JC, Ayyagari R, Gomes XV, Burgers PM (1997) Mutations in yeast proliferating cell nuclear antigen define distinct sites for interaction with DNA polymerase delta and DNA polymerase epsilon. Mol Cell Biol 17: 6367–6378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fien K, Stillman B (1992) Identification of replication factor C from Saccharomyces cerevisiae: a component of the leading-strand DNA replication complex. Mol Cell Biol 12: 155–163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gangavarapu V, Haracska L, Unk I, Johnson RE, Prakash S, Prakash L (2006) Mms2-Ubc13-dependent and -independent roles of Rad5 ubiquitin ligase in postreplication repair and translesion DNA synthesis in Saccharomyces cerevisiae. Mol Cell Biol 26: 7783–7790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanna JS, Kroll ES, Lundblad V, Spencer FA (2001) Saccharomyces cerevisiae CTF18 and CTF4 are required for sister chromatid cohesion. Mol Cell Biol 21: 3144–3158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hishida T, Ohya T, Kubota Y, Kamada Y, Shinagawa H (2006) Functional and physical interaction of yeast Mgs1 with PCNA: impact on RAD6-dependent DNA damage tolerance. Mol Cell Biol 26: 5509–5517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoege C, Pfander B, Moldovan GL, Pyrowolakis G, Jentsch S (2002) RAD6-dependent DNA repair is linked to modification of PCNA by ubiquitin and SUMO. Nature 419: 135–141 [DOI] [PubMed] [Google Scholar]
- Huang ME, Rio AG, Nicolas A, Kolodner RD (2003) A genomewide screen in Saccharomyces cerevisiae for genes that suppress the accumulation of mutations. Proc Natl Acad Sci USA 100: 11529–11534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishii H, Inageta T, Mimori K, Saito T, Sasaki H, Isobe M, Mori M, Croce CM, Huebner K, Ozawa K, Furukawa Y (2005) Frag1, a homolog of alternative replication factor C subunits, links replication stress surveillance with apoptosis. Proc Natl Acad Sci USA 102: 9655–9660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- James P, Halladay J, Craig EA (1996) Genomic libraries and a host strain designed for highly efficient two-hybrid selection in yeast. Genetics 144: 1425–1436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeruzalmi D, O′Donnell M, Kuriyan J (2001) Crystal structure of the processivity clamp loader gamma (gamma) complex of E. coli DNA polymerase III. Cell 106: 429–441 [DOI] [PubMed] [Google Scholar]
- Johnson ES, Gupta AA (2001) An E3-like factor that promotes SUMO conjugation to the yeast septins. Cell 106: 735–744 [DOI] [PubMed] [Google Scholar]
- Kanellis P, Agyei R, Durocher D (2003) Elg1 forms an alternative PCNA-interacting RFC complex required to maintain genome stability. Curr Biol 13: 1583–1595 [DOI] [PubMed] [Google Scholar]
- Kelman Z, O'Donnell M (1995) Structural and functional similarities of prokaryotic and eukaryotic DNA polymerase sliding clamps. Nucleic Acids Res 23: 3613–3620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerscher O (2007) SUMO junction-what's your function? New insights through SUMO-interacting motifs. EMBO Rep 8: 550–555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishna TS, Kong XP, Gary S, Burgers PM, Kuriyan J (1994) Crystal structure of the eukaryotic DNA polymerase processivity factor PCNA. Cell 79: 1233–1243 [DOI] [PubMed] [Google Scholar]
- Le Breton C, Dupaigne P, Robert T, Le Cam E, Gangloff S, Fabre F, Veaute X (2008) Srs2 removes deadly recombination intermediates independently of its interaction with SUMO-modified PCNA. Nucleic Acids Res 36: 4964–4974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leach CA, Michael WM (2005) Ubiquitin/SUMO modification of PCNA promotes replication fork progression in Xenopus laevis egg extracts. J Cell Biol 171: 947–954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lengronne A, McIntyre J, Katou Y, Kanoh Y, Hopfner KP, Shirahige K, Uhlmann F (2006) Establishment of sister chromatid cohesion at the S. cerevisiae replication fork. Mol Cell 23: 787–799 [DOI] [PubMed] [Google Scholar]
- Majka J, Burgers PM (2004) The PCNA-RFC families of DNA clamps and clamp loaders. Prog Nucleic Acid Res Mol Biol 78: 227–260 [DOI] [PubMed] [Google Scholar]
- Minty A, Dumont X, Kaghad M, Caput D (2000) Covalent modification of p73alpha by SUMO-1. Two-hybrid screening with p73 identifies novel SUMO-1-interacting proteins and a SUMO-1 interaction motif. J Biol Chem 275: 36316–36323 [DOI] [PubMed] [Google Scholar]
- Moldovan GL, Pfander B, Jentsch S (2006) PCNA controls establishment of sister chromatid cohesion during S phase. Mol Cell 23: 723–732 [DOI] [PubMed] [Google Scholar]
- Moldovan GL, Pfander B, Jentsch S (2007) PCNA, the maestro of the replication fork. Cell 129: 665–679 [DOI] [PubMed] [Google Scholar]
- O'Shaughnessy AM, Grenon M, Gilbert C, Toh GW, Green CM, Lowndes NF (2006) Multiple approaches to study S. cerevisiae Rad9 a prototypical checkpoint protein. Methods Enzymol 409: 131–150 [DOI] [PubMed] [Google Scholar]
- Ogiwara H, Ui A, Enomoto T, Seki M (2007) Role of Elg1 protein in double strand break repair. Nucleic Acids Res 35: 353–362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papouli E, Chen S, Davies AA, Huttner D, Krejci L, Sung P, Ulrich HD (2005) Crosstalk between SUMO and ubiquitin on PCNA is mediated by recruitment of the helicase Srs2p. Mol Cell 19: 123–133 [DOI] [PubMed] [Google Scholar]
- Parker JL, Bielen AB, Dikic I, Ulrich HD (2007) Contributions of ubiquitin- and PCNA-binding domains to the activity of Polymerase eta in Saccharomyces cerevisiae. Nucleic Acids Res 35: 881–889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker JL, Bucceri A, Davies AA, Heidrich K, Windecker H, Ulrich HD (2008) SUMO modification of PCNA is controlled by DNA. EMBO J 27: 2422–2431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfander B, Moldovan GL, Sacher M, Hoege C, Jentsch S (2005) SUMO-modified PCNA recruits Srs2 to prevent recombination during S phase. Nature 436: 428–433 [DOI] [PubMed] [Google Scholar]
- Scholes DT, Banerjee M, Bowen B, Curcio MJ (2001) Multiple regulators of Ty1 transposition in Saccharomyces cerevisiae have conserved roles in genome maintenance. Genetics 159: 1449–1465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sikdar N, Banerjee S, Lee KY, Wincovitch S, Pak E, Nakanishi K, Jasin M, Dutra A, Myung K (2009) DNA damage responses by human ELG1 in S phase are important to maintain genomic integrity. Cell Cycle 8: 3199–3207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith S, Hwang JY, Banerjee S, Majeed A, Gupta A, Myung K (2004) Mutator genes for suppression of gross chromosomal rearrangements identified by a genome-wide screening in Saccharomyces cerevisiae. Proc Natl Acad Sci USA 101: 9039–9044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smolikov S, Mazor Y, Krauskopf A (2004) ELG1, a regulator of genome stability, has a role in telomere length regulation and in silencing. Proc Natl Acad Sci USA 101: 1656–1661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song J, Durrin LK, Wilkinson TA, Krontiris TG, Chen Y (2004) Identification of a SUMO-binding motif that recognizes SUMO-modified proteins. Proc Natl Acad Sci USA 101: 14373–14378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stelter P, Ulrich HD (2003) Control of spontaneous and damage-induced mutagenesis by SUMO and ubiquitin conjugation. Nature 425: 188–191 [DOI] [PubMed] [Google Scholar]
- Stukenberg PT, Studwell-Vaughan PS, O′Donnell M (1991) Mechanism of the sliding beta-clamp of DNA polymerase III holoenzyme. J Biol Chem 266: 11328–11334 [PubMed] [Google Scholar]
- Tsurimoto T, Stillman B (1989) Purification of a cellular replication factor, RF-C, that is required for coordinated synthesis of leading and lagging strands during simian virus 40 DNA replication in vitro. Mol Cell Biol 9: 609–619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsurimoto T, Stillman B (1991) Replication factors required for SV40 DNA replication in vitro. I. DNA structure-specific recognition of a primer-template junction by eukaryotic DNA polymerases and their accessory proteins. J Biol Chem 266: 1950–1960 [PubMed] [Google Scholar]
- Ulrich HD (2009) Regulating post-translational modifications of the eukaryotic replication clamp PCNA. DNA Repair (Amst) 8: q461–469 [DOI] [PubMed] [Google Scholar]
- Uzunova K, Gottsche K, Miteva M, Weisshaar SR, Glanemann C, Schnellhardt M, Niessen M, Scheel H, Hofmann K, Johnson ES, Praefcke GJ, Dohmen RJ (2007) Ubiquitin-dependent proteolytic control of SUMO conjugates. J Biol Chem 282: 34167–34175 [DOI] [PubMed] [Google Scholar]
- Warbrick E (1998) PCNA binding through a conserved motif. Bioessays 20: 195–199 [DOI] [PubMed] [Google Scholar]
- Warbrick E (2000) The puzzle of PCNA's many partners. Bioessays 22: 997–1006 [DOI] [PubMed] [Google Scholar]
- Windecker H, Ulrich HD (2008) Architecture and assembly of poly-SUMO chains on PCNA in Saccharomyces cerevisiae. J Mol Biol 376: 221–231 [DOI] [PubMed] [Google Scholar]
- Xu H, Zhang P, Liu L, Lee MY (2001) A novel PCNA-binding motif identified by the panning of a random peptide display library. Biochemistry 40: 4512–4520 [DOI] [PubMed] [Google Scholar]
- Yunus AA, Lima CD (2009) Structure of the Siz/PIAS SUMO E3 ligase Siz1 and determinants required for SUMO modification of PCNA. Mol Cell 35: 669–682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhuang Z, Johnson RE, Haracska L, Prakash L, Prakash S, Benkovic SJ (2008) Regulation of polymerase exchange between Poleta and Poldelta by monoubiquitination of PCNA and the movement of DNA polymerase holoenzyme. Proc Natl Acad Sci USA 105: 5361–5366 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.