Abstract
La-related proteins (LARPs) belong to an evolutionarily conserved family of factors with predicted roles in RNA metabolism. Here, we have analyzed the cellular interactions and function of LARP4B, a thus far uncharacterized member of the LARP family. We show that LARP4B is a cytosolic protein that accumulates upon arsenite treatment in cellular stress granules. Biochemical experiments further uncovered an interaction of LARP4B with the cytosolic poly(A) binding protein 1 (PABPC1) and the receptor for activated C Kinase (RACK1), a component of the 40S ribosomal subunit. Under physiological conditions, LARP4B co-sedimented with polysomes in cellular extracts, suggesting a role in translation. In agreement with this notion, overexpression of LARP4B stimulated protein synthesis, whereas knockdown of the factor by RNA interference impaired translation of a large number of cellular mRNAs. In sum, we identified LARP4B as a stimulatory factor of translation. We speculate that LARP4B exerts its function by bridging mRNA factors of the 3′ end with initiating ribosomes.
Keywords: La-related protein 4B (LARP4B), PABPC1, RACK1, translation regulation, mRNP, stress granules
INTRODUCTION
The La-related proteins (LARPs) comprise an evolutionarily conserved group of factors with presumed roles in RNA metabolism (Bayfield et al. 2007; Bousquet-Antonelli and Deragon 2009). Thus far, seven different LARPs have been identified in humans (termed LARP1, 1B, 3, 4A, 4B, 6, and 7) based on their homology with the genuine La protein (also termed LARP3 according to the new nomenclature), the founding member of this family (Wolin and Cedervall 2002). The common structural hallmark of all LARPs is the La motif, which consists of ∼90 amino acids and is often adjacent to one or several RNA-recognition motifs (RRM). Crystal structure analysis of La revealed an elaborate winged helix–turn–helix fold for the La motif (Alfano et al. 2004; Dong et al. 2004). This motif in conjunction with the flanking RRM domain allows specific binding of the protein to 3′ oligo-uridylic acid stretches (Maraia and Bayfield 2006; Teplova et al. 2006; Kotik-Kogan et al. 2008) commonly found in primary transcripts generated by RNA polymerase III (Pol III) (Stefano 1984; Wolin and Cedervall 2002).
La is a highly abundant phosphoprotein predominantly localized in the nucleus. However, more recent studies indicated that this protein can shuttle between the nucleus and the cytoplasm and that the intracellular localization is influenced by its phosphorylation state (Intine et al. 2003; Fok et al. 2006). Several studies have linked La to specific functions in RNA metabolism. Upon binding to 3′ terminal oligo(U) stretches, it protects RNA polymerase III gene products from degradation by exonucleases (Copela et al. 2008). In addition, La was shown to be required for the normal maturation pathway of pre-tRNA and to take part in the quality control of a variety of noncoding RNAs (Intine et al. 2000; Chakshusmathi et al. 2003; Copela et al. 2006; Huang et al. 2006; Kadaba et al. 2006). Finally, La has been implicated in the translational control of a group of mRNAs containing a 5′ terminal oligopyrimidine (TOP) motif; however, its mode of action remains elusive (Cardinali et al. 2003).
Proteins homologous to human La are also present in the ciliate Euplotes aediculatus and in Tetrahymena thermophila (termed p43 and p65, respectively). Both proteins have been shown to associate in these organisms with the telomerase ribonucleoprotein and promote its assembly (Aigner et al. 2003; Aigner and Cech 2004; Prathapam et al. 2005; Stone et al. 2007). In contrast, the Saccharomyces cerevisiae homologs Sro9p and Slf1p are ribosome-associated factors and hence may act in translation (Sobel and Wolin 1999).
Recently, the other LARP family members have also been studied at the functional level. LARP7 has been shown to participate in the negative regulation of polymerase II transcribed genes through the 7SK RNP (Krueger et al. 2008; Markert et al. 2008; Barboric et al. 2009; Diribarne and Bensaude 2009). LARP7 binds by means of its La motif onto the uridylic acid-rich 3′ end of the 7SK RNA and allows the sequestration of the positive transcription factor pTEFb. As a consequence, the elongation phase of polymerase II is slowed down and transcription becomes inefficient. In contrast, LARP6 (also termed Acheron) interacts with Id transcription factors as well as CASK (calcium/calmodulin-dependent serine protein kinase) and has been linked to apoptosis (Valavanis et al. 2007; Wang et al. 2009; Weng et al. 2009). A regulatory effect on type I collagen mRNA translation by LARP6 has also been reported recently (Cai et al. 2010). Finally, evidence of a role of LARP1 in mRNA metabolism during development has been provided in Caenorhabditis elegans (Nykamp et al. 2008) and Drosophila melanogaster (Blagden et al. 2009).
In this study, we have employed biochemical strategies to gain insight into the function of the human LARP4B protein. This factor contains the La motif and an adjacent RRM-like domain (RRML) at its N-terminal region, whereas the C-terminal part lacks any known domains or sequence motifs (Bousquet-Antonelli and Deragon 2009). Our results suggest a stimulatory role of LARP4B in translation, through interactions with factors of the 5′ and 3′ ends of cellular mRNAs. Most mRNAs are translated via a cap-dependent initiation modus and hence require the recognition of the 5′ cap structure (m7GpppG/A) by the cap binding complex eIF4F (eukaryotic initiation factor 4F) (Sonenberg and Hinnebusch 2009). By means of this initiation factor, the 43S ribosomal subunit is recruited, and eventually 80S ribosome formation at the translation start site is enabled. In the active translation modus, mRNAs often circularize to allow efficient reinitiation events to occur after the translating ribosome has reached the end of the mRNA. This mRNA circularization is established by an interaction between poly(A) binding proteins (PABPs) associated with the 3′ end of nearly all mRNAs and eIF4G, a subunit of the 5′ cap binding complex eIF4F (Mangus et al. 2003; Gorgoni and Gray 2004; Kuhn and Wahle 2004). As a consequence, the 5′ and 3′ ends of mRNAs are brought into close proximity. This property is thought to underlie PABP's ability to stimulate mRNA binding to the 43S pre-initiation complex at least partly by enhancing eIF4F binding to the capped 5′ end of mRNA (Kahvejian et al. 2005). Forming the closed loop could facilitate reinitiation by post-termination ribosomes (Sonenberg and Hinnebusch 2009). However, the mechanism by which the mRNA 5′ cap and 3′ poly(A) tail synergize to stimulate translation is not fully understood.
Here we show that LARP4B is a cytoplasmic protein that co-sediments with polysomes and accumulates upon stress induction in stress granules. Biochemical studies further show that the protein interacts with two key factors of the translational machinery, namely, the cytoplasmic poly(A) binding protein (PABPC1) and the receptor for activated C Kinase (RACK1). Finally, in vivo experiments indicate that LARP4B positively affects the efficiency of translation of a large number of cellular mRNAs and hence acts as a general translation factor. By means of our results, we discuss here how LARP4B may exert its function in the context of the translation machinery.
RESULTS
Identification of PABPC1 and RACK1 as LARP4B-associated factors
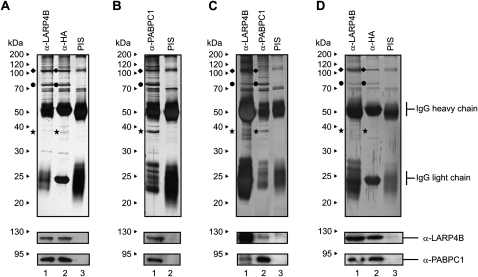
As a first approach to gain insight into the cellular interactions of LARP4B, we generated a stable cell line that allows the expression of HA-tagged LARP4B upon induction with tetracycline (FlpTRex-LARP4B; for details, see Materials and Methods). To identify proteins interacting with LARP4B, we performed immunoprecipitations using extracts from induced cells and antibodies that either directly recognize LARP4B (affinity-purified rabbit antiserum) or its N-terminal HA-tag (murine HA-tag monoclonal antibody). Both antibodies efficiently precipitated LARP4B (Fig. 1A, lanes 1,2, indicated with a rhombus) and two additional proteins of apparent molecular weights of 75 and 37 kDa (Fig. 1A, indicated by a dot and a star, respectively), which were not precipitated by a pre-immune serum (PIS) (Fig. 1A, lane 3). Analysis by mass spectrometry and Western blotting identified these proteins to be the poly(A) binding protein cytoplasmic 1 (PABPC1) and the receptor for activated C Kinase (RACK1). Our findings that an anti-PABPC1 antibody also co-precipitated LARP4B and RACK1 (Fig. 1B, lane 1) and that all interactions could also be detected in nontransfected cells (Fig. 1C, lanes 1,2) confirmed these results further.
FIGURE 1.
Identification of PABPC1 and RACK1 as interaction partners of LARP4B. (A) FlpTRex-LARP4B cells were induced with tetracycline and immunoprecipitated using antibodies against LARP4B (lane 1), HA (lane 2) or a pre-immune serum (PIS) (lane 3). (B) Immunoprecipitation of FlpTRex-LARP4B cell extract using anti-PABPC1 antibody (lane 1) or a PIS (lane 2). (C) Extract from nontransfected HEK293 cells was immunoprecipitated with antibodies against LARP4B (lane 1), PABPC1 (lane 2), or a PIS (lane 3). (D) Immunoprecipitation of RNase treated FlpTRex-LARP4B cell extract using antibodies against LARP4B (lane 1), HA (lane 2), or a PIS (lane 3). The gels were analyzed by mass spectrometry or Western blotting using anti-LARP4B and anti-PABPC1 antibodies (see lower panels). (◆) LARP4B protein; (●) PABPC1; (★) RACK1.
RACK1 and PABPC1 are well-known RNA/RNP-associated factors that bind to 40S ribosomal subunits and poly(A) tails of mRNAs, respectively (Mangus et al. 2003; Gorgoni and Gray 2004; Nilsson et al. 2004). However, RNase A treatment of extracts prior to immunoprecipitation with anti-LARP4B and anti-PABPC1 antibodies failed to affect the association between these proteins (Fig. 1D, lanes 1,2). Thus, it is very likely that LARP4B interacts with PABPC1 and RACK1 in an RNA-independent manner. These results received further support by experiments shown below and indicated that LARP4B is part of a protein complex associated with poly(A) mRNAs.
Two separate regions of LARP4B mediate binding to PABPC1 and RACK1
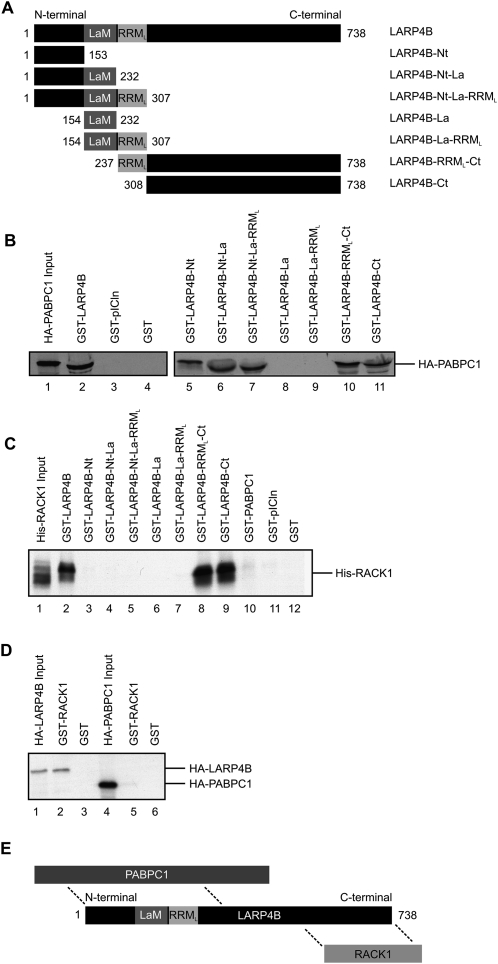
In vitro binding assays were performed to determine the regions of LARP4B involved in the identified interactions. For this, GST-fusion proteins were expressed in Escherichia coli encompassing either the N terminus (amino acids 1–153), the La motif (amino acids 154–232), the RRM-like domain (amino acids 237–307), the C terminus of LARP4B, or combinations thereof (see Fig. 2A for an overview of LARP4B truncations generated). The immobilized fusion proteins were incubated with in vitro translated and [35S]-methionine labeled PABPC1 or RACK1, respectively. Proteins that bound specifically were subsequently separated by denaturing gel electrophoresis and detected by autoradiography (Fig. 2B,C). Full-length LARP4B as well as all LARP4B fragments containing the N- and C-terminal parts of LARP4B were able to bind HA-PABPC1 (Fig. 2B, lanes 2,5–7,10,11). In contrast, no interaction was observed with GST alone (Fig. 2B, lane 4), an unrelated protein (Fig. 2B, GST-pICln, lane 3), or truncations of LARP4B containing only the La motif or the RRM-like domain (Fig. 2B, lanes 8,9). Thus, sequences flanking the putative RNA binding motifs mediated the interaction of LARP4B with PABPC1.
FIGURE 2.
Two separate segments of LARP4B mediate binding to PABPC1 and RACK1. (A) Schematic drawing of LARP4B fragments used for interaction mapping with PABPC1 and RACK1. (B) In vitro binding assay using the indicated immobilized GST-LARP4B fusions with in vitro translated [35S]-labeled HA-PABPC1. HA-PABPC1 was detected by autoradiography. (Lane 1) Five percent of labeled HA-PABPC1 input. (C) In vitro binding studies of in vitro translated [35S]-labeled His-RACK1 with indicated immobilized GST-LARP4B truncations. (Lane 1) Input, 5% of [35S]-labeled His-RACK1. (D) In vitro binding experiment of HA-LARP4B and HA-PABPC1 with immobilized GST-RACK1. (Lane 1) Five percent of [35S]-labeled HA-LARP4B; (lane 4) 5% of [35S]-labeled HA-PABPC1 input. (E) Schematic model of the interactions established in B and C.
In similar binding experiments, in vitro translated [35S]-labeled RACK1 was shown to interact with recombinant full-length LARP4B as well as with truncations containing only the C-terminal part (Fig. 2C, lanes 2,8,9; see lanes 11,12 as negative controls). In contrast, the N terminus of LARP4B and the RNA binding motifs were dispensable for this interaction (Fig. 2C, lanes 3–7). However, RACK1 and PABPC1 did not compete for binding to the C terminus of LARP4B, suggesting the presence of two different binding sites for both proteins (data not shown). Finally, we demonstrated that recombinant RACK1 interacted with [35S]-labeled LARP4B (Fig. 2D, lane 2), yet failed to interact with PABPC1 (Fig. 2D, lane 5). Thus, LARP4B provides a binding platform for PABPC1 and RACK1 via different binding modules. Their mode of interaction as determined by the binding experiments described above is schematically depicted in Figure 2E.
LARP4B is a cytoplasmic protein that accumulates in stress granules upon arsenite treatment
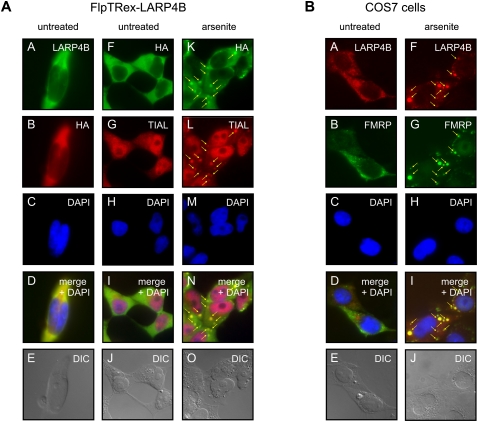
The identified interactions of LARP4B with PABPC1 and RACK1 pointed to a role in mRNA metabolism. To find support for this notion, we next analyzed the intracellular localization of LARP4B under normal growth as well as stress conditions. Immunofluorescence studies of both, endogenous LARP4B and HA-tagged LARP4B, overexpressed in the stable cell line revealed this protein to be predominantly cytoplasmic (Fig. 3A,B). A very similar localization has previously been reported for PABPC1 and RACK1 (Anderson and Kedersha 2008; Arimoto et al. 2008).
FIGURE 3.
LARP4B is a cytosolic protein and a component of SGs. (A) Tetracycline-induced FlpTRex-LARP4B cells were either mock-treated (panels A–J) or treated with arsenite (panels K–O). Immunofluorescence analyses of LARP4B were carried out using antibodies against HA-tag and TIAL as SG marker. (Panels A–E) Co-staining with anti-HA and anti-LARP4B antibodies. (Panels K–O) Stress-induced SG localization of TIAL and LARP4B. (Arrows) SGs. (B) Immunofluorescence studies in nontransfected COS7 cells using antibodies against LARP4B and FMRP as a SG marker protein. Cells were either mock-treated (panels A–E) or treated with arsenite (panels F–J).
Earlier studies had shown that PABPC1 translocates upon stress induction in discrete cytoplasmic foci termed stress granules (SGs) (Anderson and Kedersha 2008). These subcellular domains assemble in response to different forms of stress, such as heat shock, oxidative stress, or viral infections (Anderson and Kedersha 2008). It is believed that SGs serve under these conditions as storage pools for stalled, mRNA-bound translation initiation complexes formed upon polyribosome disassembly. In addition, a large number of mRNA binding proteins linked to translational silencing, mRNA stability, and regulation of mRNA metabolism are known components of SGs.
Given the interaction with PABPC1, we asked whether LARP4B also accumulates into SGs when stress is induced. For this purpose, FlpTRex-LARP4B as well as nontransfected COS7 cells were treated with arsenite, and SG formation was visualized by indirect immunofluorescence using antibodies against the TIA1 cytotoxic granule-associated RNA binding protein-like 1 (TIAL) and fragile X mental retardation protein (FMRP), two well-established markers for these domains (see Fig. 3A, panels G,L; 3B, panels B,G). As shown in Figure 3, LARP4B efficiently aggregated upon arsenite treatment in SGs. This re-distribution was observed in FlpTRex-LARP4B cells (Fig. 3A, panels K–O) as well as in untransfected COS7 cells (Fig. 3B, panels F–J) and lead to a complete co-localization with TIAL and FMRP (Fig. 3A, panel N; 3B, panel I). Hence, these data identified LARP4B as a new factor associated with SGs and raised the possibility that LARP4B might be a component of RNP complexes acting in translation.
LARP4B associates with polysomes via an interaction with the 40S subunit
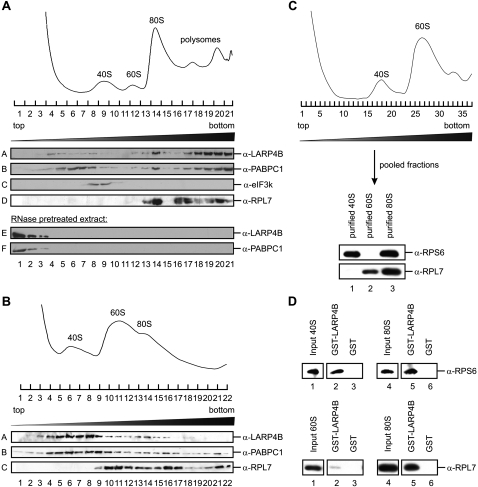
We next performed gradient centrifugation experiments to determine whether LARP4B associates with the translational machinery in cellular extracts. Fractionation of cytosolic extract in a linear 15%–45% sucrose gradient allowed the separation of ribosomal 40S and 60S subunits, 80S ribosomes, and polysomes (see A260 profile in Fig. 4A). The position of these ribosomal complexes could be independently confirmed by Western blotting using antibodies against eukaryotic initiation factor 3k (eIF3k; marker for 43S) and ribosomal protein L7 (RPL7; marker for 60S, 80S, and polysomes). A nearly perfect co-sedimentation of LARP4B and PABPC1 with RPL7 was observed in regions of the gradient corresponding to 80S ribosomes and polysomes (Fig. 4A, cf. panels A, B, and D). Varying amounts of LARP4B and PABPC1 were also identified in subribosomal fractions (Fig. 4A, panels A,B).
FIGURE 4.
LARP4B associates with the 80S complex and polysomes. (A) A 15%–45% sucrose gradient of cycloheximide-treated HeLa cell extract. Shown are the Western blot analyses of individual gradient fractions with antibodies against LARP4B, PABPC1, eIF3k, and RPL7 (panels A–D). (E,F) Sedimentations of LARP4B and PABPC1 in extract that has been treated with RNase A prior to centrifugation. (B) A 10%–30% sucrose gradient of EDTA-treated HeLa cell extract. The individual gradient fractions were analyzed by Western blotting using antibodies against LARP4B, PABPC1, and RPL7 (panels A–C). (Upper part) The A260 RNA profile of the gradient. (C) Purification of 40S and 60S ribosomal subunits. Ribosomal subunits were dissociated using puromycin and separated on a 10%–30% sucrose gradient. (Upper panel) A260 RNA profile. Purified ribosomal subunits were analyzed by Western blotting using anti-RPS6 and anti-RPL7 antibodies as markers for 40S and 60S subunits, respectively. (D) In vitro binding assays of purified 40S and 60S ribosomal subunits to immobilized GST-LARP4B. (Lanes 3,6) GST was used as a negative control. (Lanes 2,5) Bound ribosomal complexes were detected by Western blot analyses with antibodies against RPS6 and RPL7, respectively.
To test whether LARP4B is, indeed, a component of ribosomal complexes, we pretreated HEK293 cell extract with RNase A prior to sedimentation analysis. This resulted in a quantitative shift of LARP4B and PABPC1 to the top fractions of the gradient, reinforcing that both proteins are part of large RNPs (Fig. 4A, panels E,F). In addition, we analyzed sedimentation of LARP4B in extracts that had been treated with EDTA prior to gradient centrifugation leading to the dissociation of ribosomal complexes into the 40S and 60S subunits. Under these conditions, LARP4B as well as PABPC1 shifted almost quantitatively from the ribosomal/polysomal fractions to the fractions containing 40S complexes (Fig. 4B, panels A,B; cf. also the 60S marker RPL7 in panel C).
To analyze this interaction further, we purified 40S and 60S ribosomal subunits by gradient centrifugation of puromycin-treated 80S complexes (Fig. 4C; for more details, see Materials and Methods). The purified subunits were then used for in vitro binding experiments with recombinant immobilized GST-LARP4B (Fig. 4D). In accordance with the data shown in Figure 4B, the 40S ribosomal subunit was bound as efficient by LARP4B as the 80S ribosome (Fig. 4D, upper part, lanes 2,5 [inputs are depicted in lanes 1,4]), whereas almost no interaction with the 60S subunit could be detected (Fig. 4D, lower part, lanes 2,5). Moreover, no binding was observed to GST alone, showing the specificity of this interaction (Fig. 4D, lanes 3,6). Taken together, our experiments strongly suggest that LARP4B associates with actively translating ribosomes. This interaction appears to be mediated, at least in part, by binding to the small ribosomal subunit.
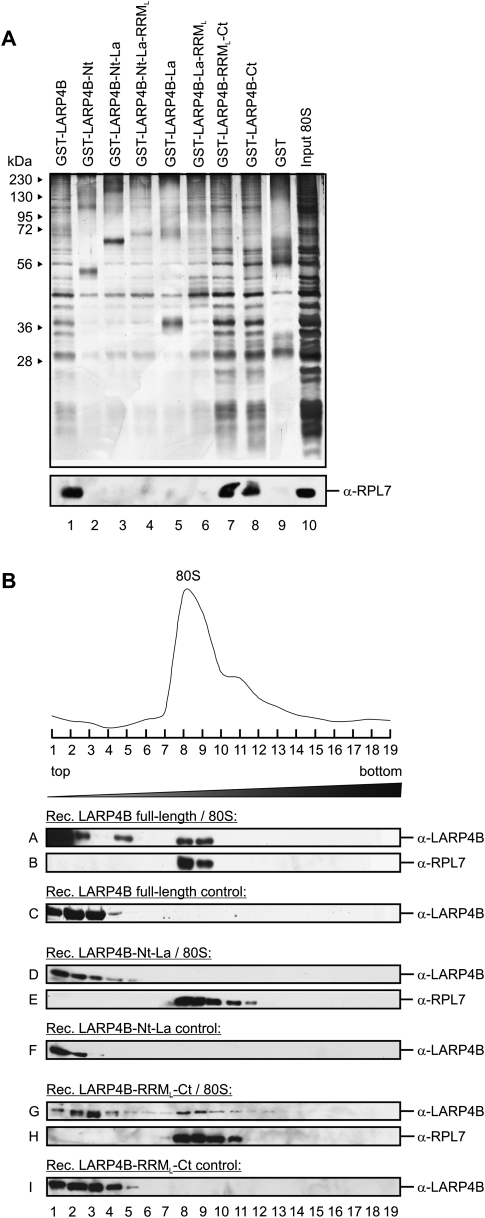
The C-terminal part of LARP4B mediates the interaction with ribosomes
Based on the findings described above, we next mapped the sequence element within LARP4B that is responsible for its interaction with ribosomes. To address this issue, in vitro binding assays were performed using isolated 80S ribosomes and GST-fusions of LARP4B introduced in Figure 2A. As shown in Figure 5, GST-LARP4B and all truncations containing the C terminus bound specifically to ribosomes (see Fig. 5A, lanes 1,7,8 [upper panel for bound ribosomes visualized by silver staining; lower panel for the corresponding Western blot analysis using an antibody against the ribosomal protein RPL7]). In contrast, no binding was observed to truncations of LARP4B containing only the La motif, the RRM-like domain, or the N-terminal part (Fig. 5A, lanes 2–6). To confirm this interaction by an independent experiment, we incubated full-length LARP4B or fragments thereof containing either the N or the C terminus with isolated 80S ribosomes and separated the mixture by sucrose gradient centrifugation (Fig. 5B). Western blot analyses of the individual gradient fractions with anti-LARP4B and anti-RPL7 antibodies confirmed our finding that only full-length LARP4B or the C terminus of LARP4B is able to bind ribosomal complexes and hence co-sedimented with 80S ribosomes (Fig. 5B, panels A,B,G,H). The N terminus alone, however, failed to interact with the purified ribosomes and therefore stayed at the top of the gradient (Fig. 5B, panels D,E), as it was the case in the absence of ribosomes (Fig. 5B, panels C,F,I). Thus, LARP4B binds ribosomal complexes through its C-terminal domain.
FIGURE 5.
The association of LARP4B with 80S ribosomal complexes is mediated via its C terminus. (A) In vitro interaction of immobilized GST-LARP4B truncations with purified 80S complexes. Interactions were analyzed by silver staining (upper part) and Western blotting using anti-RPL7 antibody (lower part). (B) Determination of the ribosome binding site of LARP4B. Mixtures of the indicated LARP4B truncations and purified ribosomes were separated by a 5%–45% sucrose gradient. Shown are the A260 RNA profile (upper part) and Western blot analyses of individual gradient fractions with antibodies against LARP4B (A,D,G) and RPL7 (B,E,H). (C,F,I) As a control, migration of the LARP4B truncations in the absence of 80S complexes is shown.
LARP4B affects translation of a broad spectrum of mRNAs in vivo
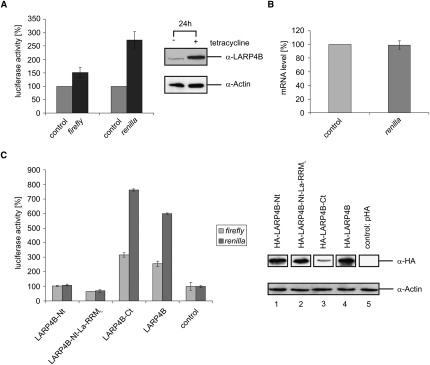
The presence of LARP4B in polysomal fractions suggested a role in translation. We hence reasoned that overexpression of LARP4B enhances its effect on translation, whereas its silencing results in the adverse effect. Two different experimental strategies were employed to test this hypothesis. In a first approach, we tested how overexpression of LARP4B affects expression of reporter plasmids encoding either firefly (pGL3 vector) or Renilla luciferase (pRL-TK vector). For this, both plasmids were co-transfected into either tetracycline-induced or uninduced FlpTRex-LARP4B cells. Expression of both luciferase variants was then determined 24 h later. The firefly and Renilla luciferase activity was analyzed using a luminometer and the dual luciferase reporter assay system (for details, see Materials and Methods). As shown in Figure 6A, LARP4B overexpression caused an increase in the enzymatic activity of firefly and Renilla luciferase. Since the amount of luciferase mRNA was not altered by LARP4B overexpression, the protein is likely to exert its effect at the translational, rather than transcriptional level (Fig. 6B). In addition, we mapped the region within LARP4B that is required for the detected stimulation of translation. As shown in Figure 6C, all transfected truncations lacking the C terminus of LARP4B failed to affect translation. In contrast, the fragment containing the C-terminal region strongly stimulated translation of the appropriated reporters. Thus, the same domain of LARP4B necessary for its interaction with PABPC1 and RACK1, as well as with the ribosome, is responsible for the identified up-regulation of translation.
FIGURE 6.
Overexpression of LARP4B enhances translation in vivo. (A) Luciferase activity in tetracycline-induced FlpTRex-LARP4B cells (black bars) and noninduced control cells (gray bars) after 24 h of cell growth. Shown are the activity of firefly luciferase expressed from transfected pGL3 vector and of Renilla luciferase expressed from co-transfected pRL-TK vector. Tetracycline-induced overexpression of HA-LARP4B was analyzed by Western blotting using anti-LARP4B and anti-Actin antibodies. Presented data were normalized to global protein level by Bradford measurement. (B) Renilla luciferase mRNA level in tetracycline-induced FlpTRex-LARP4B cells measured by qRT-PCR (dark gray bar). (C) Influence on luciferase activity of the indicated pHA/LARP4B constructs transfected in HEK293 cells. Shown are the firefly (pGL3; gray bars) and Renilla luciferase activities (pRL-TK; dark gray bars) 48 h post-transfection. As a control, empty pHA plasmid was transfected (see two bars on the right). Transfection efficiency was determined by Western blot analysis using anti-HA and anti-Actin antibodies. Extracts were normalized by Bradford measurement.
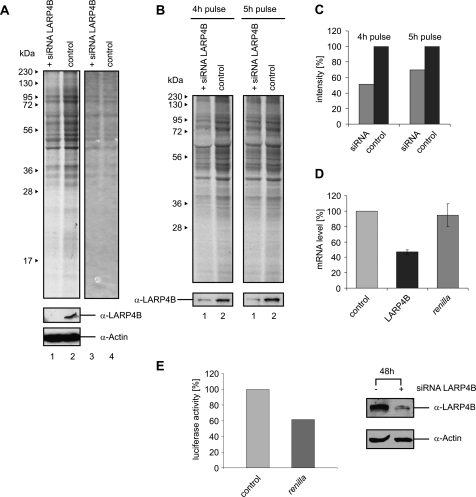
Encouraged by the results above, we tested in a second approach whether the inhibition of LARP4B expression would lead to a reduction in translation. For this, we knocked down the LARP4B protein levels in HEK293 cells by RNA interference (see Western blot in Fig. 7A, lower panel). To assess the de novo protein synthesis under these conditions, these cells as well as mock-treated control cells were transferred to medium containing [35S]-methionine to label newly synthesized proteins. After 5 h of growth, cells were lysed and proteins separated by SDS-PAGE. Autoradiography uncovered a reduction of de novo translation of a broad spectrum of cellular proteins in LARP4B-deficient cells (Fig. 7A, cf. lanes 1 and 2; see also Fig. 7B,C for further support by a kinetic investigation of translation). QRT-PCR analysis further revealed that the level of reporter mRNA was not altered under these conditions, illustrating that the observed effect is due to alterations in translation (Fig. 7D; note that the mRNA level of LARP4B was strongly reduced due to the siRNA knockdown). Furthermore, we could also detect the reduction of luciferase reporter synthesis when the LARP4B protein level was decreased by RNA interference, as shown in Figure 7E. Taken together, our data suggest LARP4B to act as a general stimulatory factor of translation, associated in poly(A)-mRNA-bound mRNP complexes.
FIGURE 7.
Knockdown of LARP4B decreases translation levels in vivo. (A) HEK293 cells were transfected with LARP4B siRNA and incubated for 48 h at 37°C. After addition of medium containing [35S]-methionine and subsequent incubation for 5 h, cells were harvested and the extracts separated by SDS-PAGE. The gel was analyzed by autoradiography (lanes 1,2) and Coomassie staining (lanes 3,4). Western blots using anti-LARP4B and anti-Actin antibodies were performed to analyze the efficiency of RNAi-induced LARP4B knockdown. (B) Comparison of the global translation level for different time points of [35S]-methionine labeling. (C) Translation level after LARP4B knockdown and 4 h or 5 h of labeling with [35S]-methionine. (D) QRT-PCR analysis of mRNA levels of LARP4B (black bar) and Renilla luciferase (dark gray bar) in HEK293 cells treated with siRNA against LARP4B. (E) Influence of LARP4B siRNA transfection on Renilla luciferase activity. Shown is the activity of Renilla luciferase (pRL-TK; dark gray bar) 48 h post-transfection.
DISCUSSION
Within the family of mammalian La-related proteins, the members 3 (the genuine La) and 7 represent the so far best characterized at the functional level (Wolin and Cedervall 2002; Krueger et al. 2008; Markert et al. 2008). Both proteins have shown to be located predominantly in the nucleus and to determine the fate and function of various Pol III RNA transcripts by targeting their U-rich 3′ terminus. In this report, we have determined the cellular interactions and function of LARP4B. In contrast to La and LARP7, LARP4B localizes in the cytoplasm and exerts a role in mRNA metabolism. Several lines of evidence obtained in vitro and in vivo support our conclusion. By means of immunoprecipitations from cellular extracts, we initially established interactions of LARP4B with PABPC1 and RACK1, two well-known components of the cellular translation system. These interactions were subsequently confirmed by in vitro binding experiments using recombinant as well as in vitro translated proteins. By means of this approach we identified N- and C-terminal sequence elements of LARP4B as binding sites for PABPC1. The interaction of RACK1, in contrast, was only mediated by a region within the C terminus of LARP4B that appeared to be different from the PABPC1 interaction module. Of note, neither the sequences merely encompassing the La motif and the RRM-like domain nor bridging RNA were required for the uncovered protein–protein interactions. Thus, formation of a ternary protein complex is possible in vitro and most likely also in vivo. Because the in vitro binding experiments presented in this study were performed using unpurified radioactively labeled proteins translated in the reticulocyte system, it cannot be precluded that these interactions may be partially indirect, mediated by factors present in the lysate. However, considering the efficiency of these interactions as well as the fact that all proteins can be coimmunoprecipitated from cellular extracts, such a scenario seems to be unlikely.
Our data are in agreement with the recent findings of Angenstein et al. (2002), who discovered that RACK1, PABPC1, and LARP4B are components of ribosome-associated mRNP complexes in rat brain extracts. Interestingly, evidence of a role of LARP1 in mRNA metabolism during development has been provided in C. elegans (Nykamp et al. 2008) and D. melanogaster (Blagden et al. 2009), whereas PABPC1 has also been described as a binding partner of dLARP1. In addition, the two yeast La motif-containing proteins Sro9p and Slf1p were found in close association with translating ribosomes (Sobel and Wolin 1999). Hence, these data raise the possibility that several members of the LARP family, including LARP1 and 4B, are involved in cytoplasmic mRNA metabolism.
A detailed sedimentation analysis revealed an association of LARP4B with 80S ribosomes and polysomes. It is tempting to speculate that LARP4B is recruited to these complexes upon binding to PABPC1 and RACK1, as their association with mRNPs and ribosomes has been well established. Consistently, PABPC1 exhibited a sedimentation profile very similar to LARP4B.
Upon dissociation of ribosomes into their subunits by EDTA treatment of cellular extracts, LARP4B remained mainly associated with the 40S subunit, suggesting the presence of a direct binding partner on this entity. Whether this interaction is mediated via RACK1, a known 40S-associated factor, is currently unclear. However, our findings that the interactions with both, RACK1 and ribosomes, are mediated via the C-terminal part of LARP4B are consistent with this idea (Pfaffl et al. 2002). Alternatively and not mutually exclusive, the La and/or the RRM-like domain of LARP4B may specifically bind the ribosomal RNA of the 40S subunit.
In accordance with our biochemical data obtained in vitro, several in vivo experiments pointed to a role of LARP4B in translation regulation. One hint for this connection in vivo came from our finding that LARP4B accumulates in SGs when cellular stress is induced. Notably, PABPC1 as one major component of poly(A) mRNAs likewise has been identified in SGs under comparable conditions (Anderson and Kedersha 2008). As SGs serve as cytoplasmic storage sites for mRNPs under conditions of inhibited translation (Anderson and Kedersha 2008), the accumulation of LARP4B in this subcellular domains is in line with our conclusion of a tight association with the translational machinery.
Direct evidence for a role of LARP4B in translation was obtained from a series of in vivo experiments showing that, dependent on the cellular level of LARP4B, translation efficiency was significantly altered. Overexpression of LARP4B stimulated the expression of two different luciferase gene reporters, whereas RNA-interference-induced knockdown of LARP4B resulted in the adverse effect. Importantly, neither overexpression nor down-regulation of LARP4B affected the mRNA level, as indicated by our qRT-PCR experiments. Because we observed alteration of protein synthesis of a large number of proteins in response to altered cellular LARP4B levels, the protein is likely to exert a general rather than specific role in translation. Moreover, we identified the C-terminal part of LARP4B to be primarily responsible for the detected stimulatory effect.
The biochemical and functional data of LARP4B presented in this study suggest a possible mode of action of LARP4B in translation. Assuming that LARP4B interacts with mRNA-associated PABPC1 and RACK1 simultaneously, it may form a bridge between the 3′ end of mRNAs and the initiating ribosome. This process would lead to mRNA circularization, possibly in an analogous way as it has been described for PABPC1 and eIF4G, the scaffold protein of the cap-binding complex. Whether such a model entirely explains the general stimulatory role of LARP4B in translation requires additional functional studies, which are currently in progress.
MATERIALS AND METHODS
Cloning, recombinant protein expression, and antibodies
For in vitro translation of [35S]-labeled proteins, the respective cDNAs were cloned into the pHA vector (a derivative of pcDNA3.1, allowing generation of N-terminally tagged HA proteins; Invitrogen). For expression in E. coli BL21, we cloned the cDNA of LARP4B and truncations thereof into pGex6.1 (GE Healthcare). The luciferase expression vectors pGL3 and pRL-TK were purchased from Promega (Nakano et al. 2005).
PGex6.1-LARP4B or pGex6.1-LARP4B deletion mutants were transformed in E. coli BL21 (DE3) and grown to an OD600 of 0.6–0.8. After induction with 1 mM IPTG for 3 h at 15°C, bacteria were pelleted and resuspended in 700 mM NaCl, 50 mM Tris-HCl (pH 7.5), 5 mM EDTA, 5 mM EGTA, and 0.01% (v/v) Igepal CA630 (P500) supplemented with a mix of different protease-inhibitors. The clarified lysates were incubated with Glutathione-Sepharose (GE Healthcare) and washed extensively to remove unbound proteins (same buffer as described above). GST-fusion proteins were then either eluted with 20 mM glutathione or cleaved on the column using PreScission protease (4 U/μL; GE Healthcare). The proteins were subsequently dialyzed against phosphate-buffered saline (PBS) and used for the indicated assays. In vitro translated proteins were produced using the TnT-T7 quick coupled transcription/translation system (Promega). Antibodies against LARP4B were raised by injection of full-length human proteins into rabbits and affinity-purified on columns with the respective covalently linked antigen. Commercial primary antibodies were purchased from Abcam (rabbit polyclonal PABPC1 antibody; rabbit polyclonal TIAL antibody) and Sigma (mouse monoclonal HA antibody; mouse monoclonal β-Actin antibody).
Generation of stable cell lines and cell culture
To create stable cell lines (FlpTRex-LARP4B; HEK293 cells stably expressing LARP4B) with the Flp-In T-Rex core kit (Invitrogen), we generated a pcHA-LARP4B/FRT/TO plasmid, which contains LARP4B under control of the CMV/TetO2 promoter, the binding/cleavage site for Flp recombinase (FRT-site) as well as the hygromycin resistance (O'Gorman et al. 1991). POG44 vector encoding Flp recombinase and pcHA-LARP4B/FRT/TO were co-transfected in Flp-In T-Rex host cell lines. Cells were afterward selected by using medium containing hygromycin, picked, and allowed to grow separately. To test whether the plasmids were incorporated, cells were analyzed for their zeocin sensitivity and their lack of β-galactosidase activity. Positive FlpTRex-LARP4B cells were grown in DMEM containing 10% FCS and 1% blasticidin/hygromycin. HEK293 as well as COS7 cells were maintained in DMEM/10% FCS with 1% penicillin/streptomycin added.
Immunological procedures
HEK293 cells or induced FlpTRex-LARP4B cells were harvested in lysis buffer (20 mM Tris-HCl at pH 7.5, 100 mM NaCl, 0.5 mM EDTA, 0.5 mM PMSF, 0.5% [w/v] NP-40), incubated on ice for 20 min, and cell debris were pelleted. For RNase sensitivity studies, the supernatant was incubated with 2 μL of RNase A (20 μg/μL) for 30 min at room temperature. For immunoprecipitations the indicated antibodies were immobilized on protein-G–Sepharose in PBS containing 0.01% (w/v) NP-40 and incubated with cellular extracts for 90 min at 4°C. After extensive washing of the beads, bound proteins were separated on a 12% SDS-PAGE. Western blot analyses were performed as described previously (Otter et al. 2007), using appropriate antibodies. Immunocytochemistry and stress treatment were carried out as depicted in Linder et al. (2008).
Purification of 80S ribosomes and gradient centrifugations
Twelve liters of HeLa S3 cells (5 × 105/mL density) grown in suspension were pelleted, washed with PBS, and lysed in 90 mL of buffer containing 20 mM HEPES (pH 7.6), 6 mM Mg(OAc)2, 30 mM NH4Cl, 4 mM β-mercaptoethanol, and 1% (w/v) Triton X-100 for 30 min at 4°C. Ribosomes were pelleted by centrifugation of the cellular extract through a 1 M sucrose cushion using a 45Ti rotor (Beckman Coulter). Pellets were resuspended and subsequently subjected to a 10%–30% sucrose gradient centrifugation (SW32Ti, 22,000 rpm, 12 h, 4°C). The 80S-containing fractions were pooled and pelleted (45Ti, 40,000 rpm, 4 h, 4°C). To determine interactions of LARP4B truncations with ribosomes, the indicated recombinant proteins and purified 80S ribosomes were incubated in PBS containing 0.01% (w/v) NP-40 for 20 min at 30°C and subsequently processed in a 5%–45% sucrose gradient (SW60Ti, 34,500 rpm, 105 min, 4°C). Gradient fractions were separated by SDS-PAGE, and the indicated proteins were detected by Western blotting.
Purification of ribosomal subunits
HeLa S3 cells grown in suspension to a density of 4.5 × 105/mL were lysed in buffer containing 20 mM Tris (pH 7.5), 10 mM KCl, 5 mM MgCl2, and 2 mM DTT by douncing. The lysate was cleared by centrifugation (JA25.50, 23,000 rpm, 30 min, 4°C), and the KCl concentration was adjusted to 100 mM. Starting with this crude extract, purification of ribosomal subunits was performed as described previously (Pisarev et al. 2007). The conceived 60S-containing fractions were loaded onto a second gradient.
Polysomal gradient centrifugation and EDTA treatment
To stabilize polysomes, cells were treated with cycloheximide for 20 min (CHX; 5 mg/mL, 1:100). Hereafter, the cells were washed with ice-cold PBS containing 10 μg/mL CHX, pelleted by centrifugation (3 min, 2000 rpm, 4°C), and lysed in buffer containing 100 mM KCl, 20 mM Tris (pH 7.5), 5 mM MgCl2, 2% (v/v) CHX (5 mg/mL), 0.5% (w/v) NP-40, a 1:1000 mix of protease inhibitors, and 1 mM DTT. After incubation on ice for 10 min, cell debris were pelleted by centrifugation (10,000 rpm, 10 min, 4°C). The supernatant was then either directly separated by centrifugation in a 15%–45% sucrose gradient (SW41Ti, 38,000 rpm, 4 h, 4°C) or incubated with 25 mM EDTA for 30 min on ice to dissociate 80S complexes. The EDTA-treated extract was separated on a 10%–30% sucrose gradient (SW60Ti, 34,500 rpm, 4 h, 4°C). Gradient fractions were separated by SDS-PAGE and analyzed by Western blotting.
In vitro binding studies
In vitro binding assays with [35S]-labeled proteins and recombinant proteins expressed in E. coli were performed as described previously (Otter et al. 2007; Markert et al. 2008). For interaction studies of ribosomes with LARP4B, purified 80S and immobilized GST-LARP4B fusions were incubated (1:1 ratio) in PBS containing 0.01% (w/v) NP-40 for 90 min at 4°C. The beads were then washed with PBS/0.01% NP-40. Interaction studies with 40S and 60S subunits were performed using buffer containing 20 mM Tris (pH 7.5), 100 mM KCl, 2 mM MgCl2, 2 mM DTT, and 1 mg/mL BSA. After incubation for 90 min at 4°C, beads were washed with the buffer described above supplemented with 300 mM KCl. Proteins were separated by SDS-PAGE and visualized by silver staining or Western blotting as described (Otter et al. 2007).
Analysis of translation in vivo
FlpTRex-LARP4B cells were induced with 1 μg/mL tetracycline, and luciferase expression vectors (pGL3 and pRL-TK) were co-transfected using Nanofectin (PAA). Twenty-four hours later, cell extracts were prepared and luciferase activity was determined by using a standard chemoluminescence detection procedure and the dual luciferase reporter assay system according to the manufacturer's instruction (Promega). LARP4B truncations cloned in pHA were transfected in HEK293 cells using Nanofectin. PGL3 and pRL-TK were co-transfected as reporter vectors. Luciferase activity was measured as described above.
For RNAi experiments, HEK293 cells grown in DMEM supplemented with 10% (v/v) FCS were transfected with the indicated siRNA using Lipofectamine 2000 (Invitrogen; LARP4B siRNA was purchased from Santa Cruz Biotechnology, Inc.). Forty-eight hours post-transfection, the cells were washed with PBS before they were transferred into DMEM containing 10% (v/v) FCS, antibiotics, and [35S]-methionine (100 μCi, 10-cm Petri dishes). After incubation for 3, 4, or 5 h at 37°C, cell extracts were prepared and the degree of silencing of LARP4B was assessed by Western blotting. The de novo protein synthesis was analyzed by autoradiography (Thoma et al. 2004).
Reverse transcription and real-time PCR (qRT-PCR)
Luciferase expression vector pRF was transfected in induced FlpTRex-LARP4B cells. Twenty-four hours post-transfection, cells were harvested and RNA was phenol-extracted. In parallel, LARP4B siRNA and pRF were co-transfected in HEK293 cells and the cells treated likewise after 48 h. The amplifications of the target genes were performed by reverse transcription (RT) using Superscript II reverse transcriptase (Invitrogen). QRT-PCR was performed with the Stratagene MX 3000P cycler and ThermoABsolut QPCR SYBR Green mix as indicated by the manufacturer (Thermo Scientific). The following primers were used for the amplifications of LARP4B, RPLP0 (ribosomal protein large subunit P0), and Renilla luciferase mRNAs:
LARP4B-1, 5′-GAAGAATTCCTTTGGCTACCG-3′;
LARP4B-2, 5′-AGACTGTGTCTGGCTGCTTG-3′;
RPLP0-1, 5′-TTCCAGGCTTTAGGTATCAC-3′;
RPLP0-2, 5′-ACTTCAGGGTTGTAGATG-3′;
pRF(Renilla luciferase)-1, 5′-GCCTCGTGAAATCCCGTTAGTAAAAGG-3′; and
pRF(Renilla luciferase)-2, 5′-GGAAACTTCTTGGCACCTTCAACAATAGC-3′.
Threshold values (ct) were calculated, and all reactions were run in triplicate. LARP4B and Renilla ct values were normalized to RPLP0. The evaluation was performed using the ΔΔct method (Pfaffl et al. 2002).
Sample preparation for MS analysis
Protein bands of interest were excised from the SDS-PAAG and further processed according to a modified protocol of Shevchenko et al. (1996) (Winkler et al. 2007). Tryptic digestion was performed overnight at 37°C. The resulting peptides were extracted twice with 0.1% (v/v) trifluoroacetic acid (TFA) for 15 min and a third time by application of 0.05% (v/v) TFA and 50% (v/v) acetonitrile (MeCN).
LC-MS/MS analysis and data interpretation
Nano-LC-MS/MS analyses were accomplished on an LTQ XL with ETD (Thermo Scientific) or a 4000 Q TRAP (Applied Biosystems) coupled to an inert Ultimate 3000 (Dionex) or an inert Famos/Switchos/Ultimate nano-HPLC-system, respectively, as described in Chari et al. (2008) and references therein.
ACKNOWLEDGMENTS
This work was supported by grants to U.F. (FOR855) and the RVZ-Network program. We thank the members of our laboratory for helpful comments on the manuscript as well as S. Knaup, D. Meier, G. Stoll, M. Küspert, F. Amelingmeier, N. Neuenkirchen, and members of the Institute of Human Genetics at the University of Wuerzburg for technical help.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.2146910.
REFERENCES
- Aigner S, Cech TR 2004. The Euplotes telomerase subunit p43 stimulates enzymatic activity and processivity in vitro. RNA 10: 1108–1118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aigner S, Postberg J, Lipps HJ, Cech TR 2003. The Euplotes La motif protein p43 has properties of a telomerase-specific subunit. Biochemistry 42: 5736–5747 [DOI] [PubMed] [Google Scholar]
- Alfano C, Sanfelice D, Babon J, Kelly G, Jacks A, Curry S, Conte MR 2004. Structural analysis of cooperative RNA binding by the La motif and central RRM domain of human La protein. Nat Struct Mol Biol 11: 323–329 [DOI] [PubMed] [Google Scholar]
- Anderson P, Kedersha N 2008. Stress granules: The Tao of RNA triage. Trends Biochem Sci 33: 141–150 [DOI] [PubMed] [Google Scholar]
- Angenstein F, Evans AM, Settlage RE, Moran ST, Ling SC, Klintsova AY, Shabanowitz J, Hunt DF, Greenough WT 2002. A receptor for activated C kinase is part of messenger ribonucleoprotein complexes associated with poly(A)-mRNAs in neurons. J Neurosci 22: 8827–8837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arimoto K, Fukuda H, Imajoh-Ohmi S, Saito H, Takekawa M 2008. Formation of stress granules inhibits apoptosis by suppressing stress-responsive MAPK pathways. Nat Cell Biol 10: 1324–1332 [DOI] [PubMed] [Google Scholar]
- Barboric M, Lenasi T, Chen H, Johansen EB, Guo S, Peterlin BM 2009. 7SK snRNP/P-TEFb couples transcription elongation with alternative splicing and is essential for vertebrate development. Proc Natl Acad Sci 106: 7798–7803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayfield MA, Kaiser TE, Intine RV, Maraia RJ 2007. Conservation of a masked nuclear export activity of La proteins and its effects on tRNA maturation. Mol Cell Biol 27: 3303–3312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blagden SP, Gatt MK, Archambault V, Lada K, Ichihara K, Lilley KS, Inoue YH, Glover DM 2009. Drosophila Larp associates with poly(A)-binding protein and is required for male fertility and syncytial embryo development. Dev Biol 334: 186–197 [DOI] [PubMed] [Google Scholar]
- Bousquet-Antonelli C, Deragon JM 2009. A comprehensive analysis of the La-motif protein superfamily. RNA 15: 750–764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai L, Fritz D, Stefanovic L, Stefanovic B 2010. Binding of LARP6 to the conserved 5′ stem–loop regulates translation of mRNAs encoding type I collagen. J Mol Biol 395: 309–326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardinali B, Carissimi C, Gravina P, Pierandrei-Amaldi P 2003. La protein is associated with terminal oligopyrimidine mRNAs in actively translating polysomes. J Biol Chem 278: 35145–35151 [DOI] [PubMed] [Google Scholar]
- Chakshusmathi G, Kim SD, Rubinson DA, Wolin SL 2003. A La protein requirement for efficient pre-tRNA folding. EMBO J 22: 6562–6572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chari A, Golas MM, Klingenhager M, Neuenkirchen N, Sander B, Englbrecht C, Sickmann A, Stark H, Fischer U 2008. An assembly chaperone collaborates with the SMN complex to generate spliceosomal SnRNPs. Cell 135: 497–509 [DOI] [PubMed] [Google Scholar]
- Copela LA, Chakshusmathi G, Sherrer RL, Wolin SL 2006. The La protein functions redundantly with tRNA modification enzymes to ensure tRNA structural stability. RNA 12: 644–654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Copela LA, Fernandez CF, Sherrer RL, Wolin SL 2008. Competition between the Rex1 exonuclease and the La protein affects both Trf4p-mediated RNA quality control and pre-tRNA maturation. RNA 14: 1214–1227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diribarne G, Bensaude O 2009. 7SK RNA, a noncoding RNA regulating P-TEFb, a general transcription factor. RNA Biol 6: 122–128 [DOI] [PubMed] [Google Scholar]
- Dong G, Chakshusmathi G, Wolin SL, Reinisch KM 2004. Structure of the La motif: A winged helix domain mediates RNA binding via a conserved aromatic patch. EMBO J 23: 1000–1007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fok V, Friend K, Steitz JA 2006. Epstein-Barr virus noncoding RNAs are confined to the nucleus, whereas their partner, the human La protein, undergoes nucleocytoplasmic shuttling. J Cell Biol 173: 319–325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorgoni B, Gray NK 2004. The roles of cytoplasmic poly(A)-binding proteins in regulating gene expression: A developmental perspective. Brief Funct Genomics Proteomics 3: 125–141 [DOI] [PubMed] [Google Scholar]
- Huang Y, Bayfield MA, Intine RV, Maraia RJ 2006. Separate RNA-binding surfaces on the multifunctional La protein mediate distinguishable activities in tRNA maturation. Nat Struct Mol Biol 13: 611–618 [DOI] [PubMed] [Google Scholar]
- Intine RV, Sakulich AL, Koduru SB, Huang Y, Pierstorff E, Goodier JL, Phan L, Maraia RJ 2000. Control of transfer RNA maturation by phosphorylation of the human La antigen on serine 366. Mol Cell 6: 339–348 [DOI] [PubMed] [Google Scholar]
- Intine RV, Tenenbaum SA, Sakulich AL, Keene JD, Maraia RJ 2003. Differential phosphorylation and subcellular localization of La RNPs associated with precursor tRNAs and translation-related mRNAs. Mol Cell 12: 1301–1307 [DOI] [PubMed] [Google Scholar]
- Kadaba S, Wang X, Anderson JT 2006. Nuclear RNA surveillance in Saccharomyces cerevisiae: Trf4p-dependent polyadenylation of nascent hypomethylated tRNA and an aberrant form of 5S rRNA. RNA 12: 508–521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kahvejian A, Svitkin YV, Sukarieh R, M'Boutchou MN, Sonenberg N 2005. Mammalian poly(A)-binding protein is a eukaryotic translation initiation factor, which acts via multiple mechanisms. Genes Dev 19: 104–113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotik-Kogan O, Valentine ER, Sanfelice D, Conte MR, Curry S 2008. Structural analysis reveals conformational plasticity in the recognition of RNA 3′ ends by the human La protein. Structure 16: 852–862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krueger BJ, Jeronimo C, Roy BB, Bouchard A, Barrandon C, Byers SA, Searcey CE, Cooper JJ, Bensaude O, Cohen EA, et al. 2008. LARP7 is a stable component of the 7SK snRNP while P-TEFb, HEXIM1, and hnRNP A1 are reversibly associated. Nucleic Acids Res 36: 2219–2229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhn U, Wahle E 2004. Structure and function of poly(A) binding proteins. Biochim Biophys Acta 1678: 67–84 [DOI] [PubMed] [Google Scholar]
- Linder B, Plottner O, Kroiss M, Hartmann E, Laggerbauer B, Meister G, Keidel E, Fischer U 2008. Tdrd3 is a novel stress granule-associated protein interacting with the fragile-X syndrome protein FMRP. Hum Mol Genet 17: 3236–3246 [DOI] [PubMed] [Google Scholar]
- Mangus DA, Evans MC, Jacobson A 2003. Poly(A)-binding proteins: Multifunctional scaffolds for the post-transcriptional control of gene expression. Genome Biol 4: 223 doi: 10.1186/gb-2003-4-7-223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maraia RJ, Bayfield MA 2006. The La protein–RNA complex surfaces. Mol Cell 21: 149–152 [DOI] [PubMed] [Google Scholar]
- Markert A, Grimm M, Martinez J, Wiesner J, Meyerhans A, Meyuhas O, Sickmann A, Fischer U 2008. The La-related protein LARP7 is a component of the 7SK ribonucleoprotein and affects transcription of cellular and viral polymerase II genes. EMBO Rep 9: 569–575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakano S, Shinde A, Ito H, Kusaka H 2005. Messenger RNA degradation may be inhibited in sporadic inclusion body myositis. Neurology 65: 420–425 [DOI] [PubMed] [Google Scholar]
- Nilsson J, Sengupta J, Frank J, Nissen P 2004. Regulation of eukaryotic translation by the RACK1 protein: A platform for signaling molecules on the ribosome. EMBO Rep 5: 1137–1141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nykamp K, Lee MH, Kimble J 2008. C. elegans La-related protein, LARP-1, localizes to germline P bodies and attenuates Ras-MAPK signaling during oogenesis. RNA 14: 1378–1389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Gorman S, Fox DT, Wahl GM 1991. Recombinase-mediated gene activation and site-specific integration in mammalian cells. Science 251: 1351–1355 [DOI] [PubMed] [Google Scholar]
- Otter S, Grimmler M, Neuenkirchen N, Chari A, Sickmann A, Fischer U 2007. A comprehensive interaction map of the human survival of motor neuron (SMN) complex. J Biol Chem 282: 5825–5833 [DOI] [PubMed] [Google Scholar]
- Pfaffl MW, Georgieva TM, Georgiev IP, Ontsouka E, Hageleit M, Blum JW 2002. Real-time RT-PCR quantification of insulin-like growth factor (IGF)-1, IGF-1 receptor, IGF-2, IGF-2 receptor, insulin receptor, growth hormone receptor, IGF-binding proteins 1, 2, and 3 in the bovine species. Domest Anim Endocrinol 22: 91–102 [DOI] [PubMed] [Google Scholar]
- Pisarev AV, Unbehaun A, Hellen CU, Pestova TV 2007. Assembly and analysis of eukaryotic translation initiation complexes. Methods Enzymol 430: 147–177 [DOI] [PubMed] [Google Scholar]
- Prathapam R, Witkin KL, O'Connor CM, Collins K 2005. A telomerase holoenzyme protein enhances telomerase RNA assembly with telomerase reverse transcriptase. Nat Struct Mol Biol 12: 252–257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shevchenko A, Wilm M, Vorm O, Mann M 1996. Mass spectrometric sequencing of proteins silver-stained polyacrylamide gels. Anal Chem 68: 850–858 [DOI] [PubMed] [Google Scholar]
- Sobel SG, Wolin SL 1999. Two yeast La motif-containing proteins are RNA-binding proteins that associate with polyribosomes. Mol Biol Cell 10: 3849–3862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonenberg N, Hinnebusch AG 2009. Regulation of translation initiation in eukaryotes: Mechanisms and biological targets. Cell 136: 731–745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stefano JE 1984. Purified lupus antigen La recognizes an oligouridylate stretch common to the 3′ termini of RNA polymerase III transcripts. Cell 36: 145–154 [DOI] [PubMed] [Google Scholar]
- Stone MD, Mihalusova M, O'Connor CM, Prathapam R, Collins K, Zhuang X 2007. Stepwise protein-mediated RNA folding directs assembly of telomerase ribonucleoprotein. Nature 446: 458–461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teplova M, Yuan YR, Phan AT, Malinina L, Ilin S, Teplov A, Patel DJ 2006. Structural basis for recognition and sequestration of UUU(OH) 3′ temini of nascent RNA polymerase III transcripts by La, a rheumatic disease autoantigen. Mol Cell 21: 75–85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thoma C, Ostareck-Lederer A, Hentze MW 2004. A poly(A) tail-responsive in vitro system for cap- or IRES-driven translation from HeLa cells. Methods Mol Biol 257: 171–180 [DOI] [PubMed] [Google Scholar]
- Valavanis C, Wang Z, Sun D, Vaine M, Schwartz LM 2007. Acheron, a novel member of the Lupus Antigen family, is induced during the programmed cell death of skeletal muscles in the moth Manduca sexta. Gene 393: 101–109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Glenn H, Brown C, Valavanis C, Liu JX, Seth A, Thomas JE, Karlstrom RO, Schwartz LM 2009. Regulation of muscle differentiation and survival by Acheron. Mech Dev 126: 700–709 [DOI] [PubMed] [Google Scholar]
- Weng H, Kim C, Valavanis C, Wang Z, Schwartz LM 2009. Acheron, a novel LA antigen family member, binds to CASK and forms a complex with Id transcription factors. Cell Mol Biol Lett 14: 273–287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winkler C, Denker K, Wortelkamp S, Sickmann A 2007. Silver- and Coomassie-staining protocols: Detection limits and compatibility with ESI MS. Electrophoresis 28: 2095–2099 [DOI] [PubMed] [Google Scholar]
- Wolin SL, Cedervall T 2002. The La protein. Annu Rev Biochem 71: 375–403 [DOI] [PubMed] [Google Scholar]