Abstract
To understand the mechanisms for structural diversification of Pseudomonas-derived toluene-catabolic (TOL) plasmids, the complete sequence of a self-transmissible plasmid pDK1 with a size of 128,921 bp from Pseudomonas putida HS1 was determined. Comparative analysis revealed that (i) pDK1 consisted of a 75.6-kb IncP-7 plasmid backbone and 53.2-kb accessory gene segments that were bounded by transposon-associated regions, (ii) the genes for conjugative transfer of pDK1 were highly similar to those of MOBH group of mobilizable plasmids, and (iii) the toluene-catabolic (xyl) gene clusters of pDK1 were derived through homologous recombination, transposition, and site-specific recombination from the xyl gene clusters homologous to another TOL plasmid, pWW53. The minireplicons of pDK1 and its related IncP-7 plasmids, pWW53 and pCAR1, that contain replication and partition genes were maintained in all of six Pseudomonas strains tested, but not in alpha- or betaproteobacterial strains. The recipient host range of conjugative transfer of pDK1 was, however, limited to two Pseudomonas strains. These results indicate that IncP-7 plasmids are essentially narrow-host-range and self-transmissible plasmids that encode MOBH group-related transfer functions and that the host range of IncP-7-specified conjugative transfer was, unlike the situation in other well-known plasmids, narrower than that of its replication.
Bacterial genes for the utilization of recalcitrant environmental pollutants such as herbicides, pesticides, and petroleum and other industrial waste compounds are often found on plasmids and chromosomally specified integrative and conjugative elements (ICEs) (57). Although the origins of such catabolic genes still remain unknown, it seems most likely that, once established, the catabolic gene modules spread between plasmids and chromosomes through intracellular movements of insertion sequence (IS)-flanked composite transposons (68) and class II (Tn3-related) transposons (61, 63) and intercellular conjugative transfers of plasmids and ICEs (36, 65). The genes associated with the degradation of man-made xenobiotic compounds (e.g., atrazine, 2,4-dichlorophenoxyacetate, and haloacetates) have predominantly been found on broad-host-range and incompatibility group P-1 (IncP-1) plasmids, whereas the genes responsible for the degradation of natural aromatic hydrocarbons (e.g., phenol, naphthalene, and toluene/xylenes) via the meta-cleavage catabolic pathways are mainly located on IncP-2, IncP-7, and IncP-9 plasmids, which have been found exclusively in Pseudomonas species (38).
Studies of the archetypal 119-kb and IncP-9 toluene/xylene-catabolic (TOL) plasmid pWW0 from Pseudomonas putida mt-2 have greatly contributed to the detailed clarification of genetic and biochemical mechanisms for the aerobic degradation of toluene/xylenes (72). The pWW0-specified xyl genes are organized as four transcriptional units within the class II transposons, Tn4651 and Tn4653 (62): (i) the upper pathway operon (xylXYZLTEGFJQKIH) for the conversion of toluene and xylenes to benzoate and its methyl derivatives, respectively; (ii) the meta pathway operon (xylXYZLTEGFJQKIH) for the subsequent conversion to tricarboxylic acid (TCA) cycle intermediates; (iii) and the two transcriptional regulator genes, xylS and xylR (42). After the discovery of pWW0, various TOL plasmids carrying the xyl gene clusters very similar to those on pWW0 were discovered in soil bacteria from geographically distant areas around the world (5, 22, 23, 26, 50, 71). Such TOL plasmids differ in sizes, incompatibilities, and the numbers and relative orientations of xyl gene clusters, implying the plausible movement and other rearrangements of xyl genes between and within plasmids (5, 70). The rearrangement of catabolic gene clusters is considered to be an important step for the host cell to improve its performance of the preexisting catabolic function(s) (8, 43, 75). Our major aim is to clarify the underlying mechanisms of the recombinations which generate diverse catabolic gene clusters. Comparison of the pWW0 sequence and its related catabolic genes from different Pseudomonas strains suggested that the recombination between IS copies on two different replicons led to the establishment of the pWW0-type configuration of the xyl gene cluster (70). Our recent studies of an IncP-7 TOL plasmid, pWW53, which possesses two meta operons (meta 1 and meta 2) and three xylS genes (two are functional but one is truncated) revealed that (i) pWW53 acquired all of its xyl gene clusters through transposition of class II transposon Tn4660 and (ii) the site-specific recombination mediated by a transposon-encoded resolvase gave rise to an additional transposon (Tn4656) that carried part of the xyl gene cluster on pWW53 (60, 73). These studies implicated transposition-related genes in the rearrangements and dissemination of xyl gene clusters.
Another TOL plasmid pDK1 was discovered in a 3-methylbenzoate-utilizing P. putida strain, HS1, that was isolated in Minnesota (26). The restriction maps of regions covering the xyl genes and other available nucleotide sequences of pDK1 showed remarkable similarity to those of pWW53 (1-3, 41, 51, 67). However, the relative orientation of the xyl upper and meta operons on pDK1 differs from that of the upper and meta 2 operons in pWW53 (see Fig. S1 in the supplemental material). Furthermore, one of the two meta operons (meta 1) of pWW53 is absent from pDK1 (1), and pDK1 is self-transmissible, in contrast to pWW53 (26). To elucidate the evolutionary relationship between pWW53 and pDK1, we have determined the complete sequence of pDK1. Comparative analysis of the two plasmids strongly suggests their diversification from an ancestral IncP-7 TOL plasmid through both homology-dependent and site-specific recombination events. We also investigated in this study the host range of IncP-7 plasmids.
MATERIALS AND METHODS
Bacterial strains, plasmids, and culture conditions.
Bacterial strains and plasmids used in this study are listed in Table 1. Escherichia coli strains were grown at 37°C in Luria-Bertani (LB) broth (48), and Pseudomonas, Sphingomonas, and Burkholderia strains were grown at 30°C in LB broth diluted 1/3 (1/3-LB broth) (74). Solid media were prepared by the addition of 1.5% agar. Antibiotics were added at final concentrations of 10 μg/ml for gentamicin (Gm), 20 μg/ml for tetracycline (Tc), 50 μg/ml for kanamycin (Km), 100 μg/ml for rifampin (Rif), and 20 μg/ml for nalidixic acid (Nal). M9 minimal agar (48) supplemented with 5 mM 3-methylbenzoate (3MB) as a sole source of carbon and energy was used to select the Pseudomonas strains carrying the pDK1 derivatives with an intact xyl meta pathway operon.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Relevant characteristic(s)a | Reference or source |
|---|---|---|
| Strains | ||
| E. coli | ||
| S17-1 | recA pro thi hsdR; chromosomally integrated RP4-2-Tc::Mu-Km::Tn7; Tpr Smr | 55 |
| HB101 | recA13hsdS20(rB− mB−) ara-14proA2 lacI1galK2 rpsL20xyl-5mtl-1supE44 | 48 |
| DH5α | λ− φ80dlacZΔM15 Δ(lacZYA-argF)U169recA1endA1 hsdR17(rK− mK+) supE44thi-1gyrArelA1 | 48 |
| DH5α (λpir) | DH5α derivative, λpir lysogen | Laboratory collection |
| P. putida | ||
| HS1 | Soil isolate, carrying pDK1; TOL+ 3MB+ | 26 |
| HS1C | HS1 derivative cured of pDK1 | This study |
| HS1CR | Spontaneous rifampin-resistant mutant of HS1C; Rifr | This study |
| HS1CG | HS1C::TnMod-OGm; Gmr | This study |
| KT2440 | Type strain | ATCC 47054 |
| KT2440G2 | KT2440::mini-Tn7-TGm; Gmr | This study |
| F1 | Soil isolate | 13 |
| F1G | F1::TnMod-OGm; Gmr | This study |
| P. fluorescens | ||
| Pf-5 | Type strain | ATCC BAA477 |
| Pf-5S | Spontaneous streptomycin-resistant mutant of Pf-5; Smr | This study |
| Pf-5G | Pf-5::mini-Tn7-TGm; Gmr | This study |
| P. aeruginosa | ||
| KG2512 | P. aeruginosa PAO1 aph; Km-sensitive strain | N. Gotoh, Kyoto Pharmaceutical University |
| KG2512G | KG2512::mini-Tn7-TGm; Gmr | This study |
| P. resinovorans | ||
| CA10dm4RG | pCAR1-free derivative of P. resinovorans CA10 with chromosomal copy of TnMod-OGm; Rifr Gmr | 54 |
| S. paucimobilis | ||
| IAM12578G | IAM12578::TnMod-OGm; Gmr | 32 |
| B. multivorans | ||
| ATCC 17616G | ATCC 17616::TnMod-OGm; Gmr | 32 |
| Plasmids | ||
| pEX18Tc | pUC replicon carrying sacB; Mob+ Tcr | 21 |
| pK18mob | pMB1 replicon; Mob+ Kmr | 49 |
| pJP5608 | R6K replicon; Mob+ Tcr | 40 |
| pUC4K | pUC replicon; Km resistance gene cassette donor; Apr Kmr | 66 |
| pUCARori004ΔparA | pUC19 carrying the repA-oriV-parW-parA-parS-parB region of pCAR1; parA::kan; Apr Kmr; unstable in P. putida due to inactivation of parA | 54 |
| pUCARori004 | pUC19 carrying the repA-oriV-parW-parA-parS-parB region of pCAR1 | 54 |
| pRK2013 | ColEI replicon, helper plasmid for RK2 oriT-mediated conjugative transfer; Tra+ Kmr | 12 |
| pTNS3 | R6K replicon carrying tnsABCD genes of Tn7; Apr | 6 |
| pUC18-mini-Tn7T-Gm | pUC replicon carrying mini-Tn7::aacC1; Apr Gmr | 6 |
| pTnMod-OGm | pUC replicon carrying mini-Tn5::aacC1 (= TnMod-OGm); Apr Gmr | 11 |
| pDK1 | IncP-7 replicon; Tra+ TOL+ 3MB+ | 26 |
| pDK1K | pDK1xylU::kan; Tra+ 3MB+ Kmr | This study |
| pDK2 | RK2::Tn4663; Tcr TOL+ | 51 |
| pEXxylUKm | pEX18Tc carrying the xylUWC region of pDK1 with the kan gene in xylU; Mob+ Tcr | This study |
| pJPxylY | pJP5608 carrying the xylYZ region of pDK1; Mob+ Tcr | This study |
| pDK1K::pJP | pDK1K derivative with the pJPxylY insertion into the xylY region; Mob+ 3MB+ Kmr Tcr | This study |
| pHY118 | pK18mob carrying the repA-oriV-parW-parA-parS-parB region of pWW53; Mob+ Kmr | 73 |
| pMM67 | pK18mob carrying the repA-oriV-parW-parA-parS-parB region of pCAR1; Mob+ Kmr | This study |
| pMM68 | pK18mob carrying the repA-oriV-parW-parA-parS-parB region of pDK1; Mob+ Kmr | This study |
Abbreviations are as follows: Km, kanamycin; Ap, ampicillin; Tc, tetracycline; Gm, gentamicin; Tra+, self-transmissible; Mob+, mobilizable; TOL+, degradation of toluene/xylenes; 3MB+, degradation of 3-methylbenzoate.
The details of construction of antibiotic-resistant mutants of Pseudomonas strains are described in the supplemental material. Curing of pDK1 from P. putida HS1 generated HS1C. For this purpose, HS1 was electroporated with a Km-resistant (Kmr) and unstable IncP-7 mini-pCAR1 plasmid, pUCARori004Δpar (54), to obtain the Kmr transformants. The resulting transformant lacking the catabolic activity of 3MB was next cultivated overnight in LB broth without any antibiotics so as to facilitate the spontaneous loss of the unstable pUCARori004Δpar replicon. One of the Km-sensitive derivatives without any plasmids was HS1C.
Conjugation experiments.
Filter matings on LB agar plates used for the construction of plasmids and liquid matings in 1/3-LB broth for the transfer of pDK1 derivatives were performed at 30°C for approximately 12 h (69). In liquid matings, approximately a 1:5 ratio of donor and recipient cells in log phase was used.
Standard DNA manipulation and construction of plasmids.
CaCl2-induced competent cells of E. coli were subjected to heat shock transformation and used for DNA cloning (48). Electrocompetent cells of Pseudomonas strains were prepared according to the protocol by Choi et al. (7). PCR was carried out using KOD-plus high-fidelity DNA polymerase (Toyobo, Osaka, Japan). Extraction of DNA fragments from agarose gel was performed using a QiaEX II bead kit (Qiagen, Valencia, CA) according to the manufacturer's protocol. Standard alkaline lysis with phenol-chloroform purification (48) and a LaboPass minikit (Hokkaido System Sciences, Sapporo, Japan) were used for the large- and small-scale preparations, respectively, of plasmids from E. coli. The DNA sample prepared by the large-scale preparation was used for shotgun sequencing analysis.
pDK1K, a derivative of pDK1 with an insert of the Kmr gene in the xylU gene, and a cointegrate of pDK1K and pJP5608 (40), designated pDK1K::pJP, were constructed according to the procedures described in the supplemental material. A mini-pDK1 plasmid, pMM68, was constructed by cloning of the 6,870-bp repA-oriV-parWASB-containing HindIII fragment of pDK1K::pJP into the HindIII site of pK18mob (49), while a mini-pCAR1 plasmid, pMM67, was constructed by cloning the HindIII fragment containing repA-oriV-parWASB region from pUCARori004 (54) into the HindIII site of pK18mob.
Sequencing and annotation.
The pDK1K::pJP DNA purified from DH5α (λpir) by the large-scale preparation was partially digested with Sau3AI, shotgun cloned into the BamHI site of pUC18, and sequenced. The sequencing reaction was carried out using a BigDye terminator kit, version 3 (Applied Biosystems, Foster City, CA), and the sequencing determination was performed with ABI Prism model 310 and 3730 sequencers (Applied Biosystems). The PCR-based gap-closing process was performed using wild-type pDK1 DNA as a template. Annotation and sequence comparison were carried out using GLIMMER 3 (10), BLAST (16), GenomeMatcher (39), and GENETYX-MAC, version 13 (Genetyx, Tokyo). The Web-based analysis software programs Jpred3 (http://www.compbio.dundee.ac.uk/∼www-jpred/) (9) and SOSUI (http://bp.nuap.nagoya-u.ac.jp/sosui/) (20) were used for the prediction of protein secondary structures and the identification of the signal peptide and transmembrane helix, respectively. The putative transcriptional promoter and Rho-independent terminator were predicted using BPROM and FindTerm, respectively (Softberry).
Nucleotide sequence accession number.
Nucleotide sequence of pDK1 was submitted under DDBJ/EMBL/GenBank accession number AB434906. In the annotation of the pDK1 genome, a gene name was not given for a protein with ambiguous function, and such a gene was designated ofn (orf with no name).
RESULTS AND DISCUSSION
Strategy of pDK1 sequencing.
The pDK1 DNA samples prepared from the original P. putida host strain (HS1) were always contaminated with a large amount of chromosomal DNA. To overcome this, we attempted to prepare high-purity plasmid DNA from the E. coli cells. For this purpose, a pDK1 derivative, pDK1K::pJP, which carried the RP4-derived oriT (oriTRP4) and R6K-derived oriV regions, was first constructed in the HS1 background (see the supplemental material). The former region was expected to allow the conjugative transfer of pDK1K::pJP to E. coli in the presence of transfer genes of RP4, and the latter region allows the autonomous replication of pDK1K::pJP in the E. coli strain with the pir gene that encodes the R6K-specific replication protein. The triparental mating led to the successful conjugative transfer and autonomous replication of pDK1K::pJP in E. coli DΗ5α (λpir). Shotgun sequencing of pDK1K::pJP prepared from DΗ5α (λpir) generated five contigs with five gaps. The gap closing was performed by PCR-based walking approaches to obtain the complete sequence of pDK1K::pJP.
Overview of pDK1 genome.
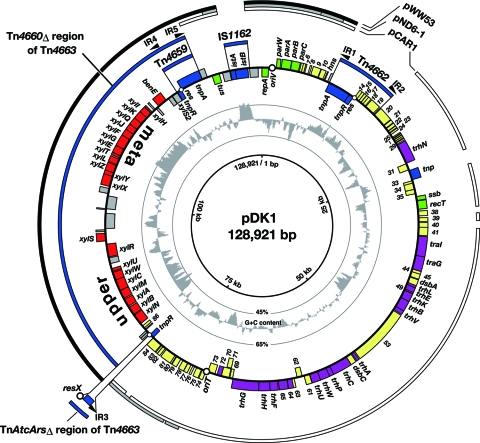
In silico elimination of the artificially integrated DNA regions (Kmr gene and pJP5608) from the pDK1K::pJP sequence indicated that pDK1 was 128,921 bp in size (accession number AB434906) with an average G+C content of 55.9%. Although five sequences of the pDK1 xyl gene clusters (25,644 bp in total) had already been deposited by several other groups (GenBank accession numbers AF134348, L02642, AF019635, M65205, and L02358), our subsequent analysis employed only the pDK1 sequence determined in this study. pDK1 carried 114 probable coding sequences (CDSs) (see Table S1 in the supplemental material). Among them, 22 were predicted to be involved in catabolism and transport of toluene/xylenes (xyl and ben), 6 are involved in replication and partition (rep, par, and tus), 19 are involved in conjugative DNA transfer (trh, tra, ofn64, ofn65, and ofn72), 7 are involved in transposition and site-specific recombination (tnp and ist), 8 are involved in DNA-associated functions (ssb, recT, ofn41, ofn61, ofn80, ofn81, ofn83, and ofn84), and 51 are involved in other known or unknown functions. Seventeen CDSs were predicted to code for membrane-associated proteins and were mostly located within the conjugative DNA transfer region. Most of the pDK1-specified gene products showed high (>83%) identities to those of published IncP-7 plasmids: the self-transmissible and carbazole/dioxin-catabolic plasmid pCAR1 (31), the nontransmissible TOL plasmid pWW53 (73), and the nontransmissible naphthalene-catabolic plasmid pND6-1 (30) (see Table S1).
The pairwise BLASTn analysis revealed that two regions of pDK1, totaling 49.3 kb (the 9.1-kb region from coordinates 1 to 9110 and the 40.2-kb region from 80702 to 120926) showed high (>98%) identities to the DNA regions on pWW53 (Fig. 1). The first carried the genes for replication and partition and also shared high degrees of sequence identity with the corresponding regions on pCAR1 and pND6-1. The second, 40.2-kb, region carried part (here referred to as Tn4660Δ) of the toluene-catabolic transposon Tn4660 from pWW53 (Fig. 1 and 2). In the pWW53 genome, Tn4660 is inserted within a putative nuclease gene equivalent to ORF59 of pCAR1 and is flanked by direct repeats (Fig. 2) (73). Furthermore, one extremity of Tn4660 is occupied by a nested class II transposon Tn4659. The Tn4659-proximal end of Tn4660 and the truncated ORF59-like gene on pWW53 were also present in the pDK1 genome, suggesting that pWW53 and pDK1 originated from a common ancestral IncP-7 plasmid that had received a Tn4660-like ancestral transposon into the same position (Fig. 2). The other extremity of Tn4660, which contains the xyl meta 1 region, was absent from pDK1. Instead, part of a class II transposon, TnAtcArs (64), was present next to the xyl upper operon in pDK1. The complex structure of the Tn4660Δ region in pDK1 suggests its structural rearrangement(s) after insertion of the Tn4660-like transposon into the ancestral plasmid of pDK1 (see below).
FIG. 1.
Circular map of pDK1. CDSs are shown as boxes inside or outside the circle with gene names or ofn numbers. Color indicates proposed functions of gene products: green, replication, partition, and DNA-processing; blue, transposition and site-specific recombination; purple, conjugative DNA transfer; red, degradation of toluene/xylenes; yellow, other known or unknown functions; and gray, gene remnants. Transposon-derived segments are represented by blue arcs. Terminal IRs and res regions of class II transposons are indicated as filled triangles and open circles, respectively. The segments highly homologous to those in pWW53, pND6-1, and pCAR1 (revealed by pairwise BLASTn analysis) are indicated by black, gray, and white arcs, respectively.
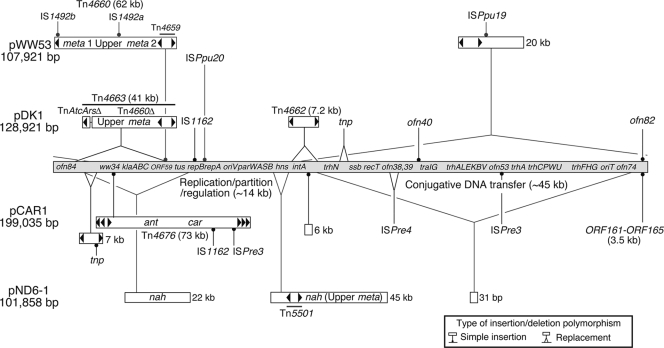
FIG. 2.
Schematic representation of insertions and deletions in pDK1, pWW53, pCAR1, and pND6-1. A proposed IncP-7 backbone is represented by a gray bar with landmark genes. pDK1- and pWW53-specific inserts are shown above the backbone, while pCAR1- and pND6-1-specific inserts are shown below the backbone. Genes for the proteins with ambiguous or hypothetical functions in pDK1, pWW53, and pCAR1 are designated ofn, ww, and ORF, respectively. Filled arrowheads indicate terminal IRs of class II transposons. The replication/partition/regulation region (the 14-kb segment from tus to hns) and conjugative DNA transfer region (the 45-kb segment from trhN to ofn74) are indicated.
In contrast to the sequence similarity in the two regions described above, pDK1 differed from pWW53 in the presence of a 63.8-kb region (from inverted repeat 2 [IR2] to ofn84, coordinates 16311 to 80150) that carries the genes for conjugative DNA transfer (Fig. 1; see also Table S1 in the supplemental material). The gene organization in this region is the same as that of pCAR1 except for insertion/deletion polymorphisms at four sites (Fig. 2). The transfer-related gene products of pDK1 show high (83 to 99%) identities to the corresponding products of pCAR1 but show low (<40%) identities to those of the other reported plasmids. The conservation of the regions for DNA transfer, as well as for replication and partition, between pDK1 and pCAR1 strongly suggests (i) that IncP-7 plasmids originally had a common self-transmissible backbone of at least 75 kb (Fig. 2) and (ii) that nontransmissible IncP-7 plasmids such as pWW53 and pND6-1 have lost their DNA transfer-related genes during their evolution.
Features of replication/partition function region.
The 15.5 kb of pDK1 from coordinates 122283 to 8760 contain genes and gene remnants associated with replication (repA, repB, and tus), partition (parW, parA, and parB), and transcriptional regulation (hns) (Fig. 3). The gene synteny in this region was conserved in three other IncP-7 plasmids, pDK1, pWW53, and pCAR1, but not in pND6-1; the previously intact hns and tus gene regions in the ancestral form of pND6-1 must have been replaced by large gene clusters for the catabolism of naphthalene (Fig. 2). The genes necessary for the stable maintenance of pCAR1 have been suggested to be repA, parW, parA, and parB (54). The replication initiator protein (RepA) of pDK1 was identical to the RepA proteins of pWW53, pND6-1, and the partially sequenced TOL plasmid pL6.5 (59) and 98.9% identical to the RepA of pCAR1. The RepA proteins of these IncP-7 plasmids were predicted to show structural similarity to replication initiation proteins encoded by iteron-containing plasmids, such as RepA of pPS10 (GenBank accession number CAA41700) and RepE of F plasmid (BAA97915). The copy numbers and relative positions of repeat sequences in the oriV regions are the same in pDK1, pCAR1, pWW53, pND6-1, and pL6.5.
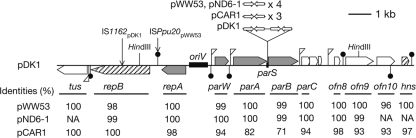
FIG. 3.
Gene organization in replication/partition region of the IncP-7 plasmid. Depicted is the pDK1 segment from coordinates 22283 to 8760. Coding sequences are represented by pentagons. Arrows inserted between parA and parB indicate the 17-bp palindromic motifs. Gene products required for stable maintenance of the pCAR1 replicon in P. putida (54) are indicated in gray. The repB and hns genes in pDK1 (hatched pentagons) are inactivated by IS1162 insertion and nonsense mutations, respectively (see text for details). The identities of gene products of other IncP-7 plasmids with those of pDK1 are shown below the gene names. To calculate the identities for RepB and H-NS, their respective wild-type proteins of pDK1 are predicted by the in silico removal of mutations (the insertion mutation of IS1162 in repB and the nonsense mutation at the 38th codon in hns in pDK1). NA, not available due to lack of gene homologs in pND6-1. Putative promoters and Rho-independent terminators are indicated by flags and pins, respectively. Two HindIII sites used for construction of mini-IncP-7 replicons, pMM67 (mini-pCAR1), pMM68 (mini-pDK1), and pHY118 (mini-pWW53) (73), are depicted.
Most plasmid partition systems so far characterized are comprised of three components: (i) a centromere, (ii) a DNA-binding protein that specifically recognizes the centromere, and (iii) a partition ATPase that moves the complex of centromere and the binding protein (14). These components correspond to sopC, SopB, and SopA, respectively, in the F-plasmid (SopAF and SopBF) partition system (33). The ParA protein of pDK1 (ParApDK1) had an ATP-binding motif typical of a partition ATPase and showed structural similarity to SopA, while the ParB protein (ParBpDK1) showed structural similarity to SopB. The ParW proteins encoded by all the sequenced IncP-7 plasmids possessed transmembrane helix motifs at the N-terminal portions, but their detailed function(s) still remains unclear. The ParW, ParA, and ParB proteins of pDK1 were 98 to 100% identical to the respective proteins of pWW53, pDN6-1, and pL6.5 but exhibited relatively low identities to the respective proteins of pCAR1, with 94% identity for ParW, 82% for ParA, and 71% for ParB (Fig. 3). This suggests that pCAR1 is phylogenetically distant from the four other IncP-7 plasmids. General similarity of ParApDK1 and ParBpDK1 to SopAF and SopBF, respectively, implied the presence of a sopC-like region in pDK1. The sopC region contains 16-bp palindromic sequence repeats where SopB binds (33). pDK1 and other IncP-7 plasmids had, between their parA and parB genes, the conserved 17-bp palindrome sequence, 5′-TGGTGCTCgGAGCACCA-3′ (g, symmetric axis), the copy numbers of which were different among the plasmids: two in pDK1, three in pCAR1, and four in pWW53, pND6-1, and pL6.5 (Fig. 3). This 17-bp motif was not found in the rest of the pDK1 genome. We hypothesized that this 17-bp motif region functions as the centromere designated parS. The purified ParB protein of pDK1 was, indeed, able to specifically bind to the parS region (our unpublished data).
Features of DNA transfer-associated genes.
Conjugative systems in many plasmids from Gram-negative bacteria are comprised of three components: the mating-pair formation (Mpf) apparatus which spans inner and outer membranes and is responsible for the synthesis of the conjugative pilus; the relaxosome, which is a complex of proteins that process the DNA at the origin of transfer (oriT), and the coupling protein which connects mating-pair formation apparatus and relaxosome (28). Mobilizable plasmids (MOBs) have recently been classified into six families (MOBF, MOBH, MOBQ, MOBC, MOBp, and MOBV) on the basis of the phylogeny of relaxase proteins in the relaxosome components (15). The probable relaxase protein (TraI) of pDK1 (TraIpDK1) showed 96% identity to that of pCAR1, and both TraI relaxases more resembled, in their predicted secondary structures, the relaxase from R27 (a representative of MOBH group) than the relaxases from other MOB groups (data not shown). TraIpDK1 possessed both the three-H motif (HQ-X2-PASE-X-HHH-X3-GG-X3-H-X-L) and the HD hydrolase motif (HD-X-DK), which are uniquely conserved in the MOBH relaxases (15). Other plausible relaxosome components of pDK1 were Ofn61 and Ofn65. Ofn61 was 26 and 28% identical to the TraO primase of plasmid pIPO2 (MOBP group) (58) and Orf220 of Rts1, an IncT plasmid, respectively, while Ofn65, exhibiting 44% identity to R0204 of R27 (R0204R27), was a conserved hypothetical protein with a Zn-binding primase-helicase domain (Pfam accession number 8273).
The coupling protein of pDK1, designated TraG, contained an ATPase (Walker A and B) motif and showed 99, 38, and 27% identities, respectively, to the pCAR1, Rts1, and R27 homologs. The Mpf components, which probably constituted the F-plasmid-type retractile pilus, were also shared by pDK1, pCAR1, Rts1, and R27 (see Table S2 in the supplemental material). The Mpf components of pDK1 showed 83 to 99% identities to the pCAR1 homologs, 25 to 46% identities to the Rts1 homologues, and 21 to 33% identities to the R27 homologs. Taking into consideration the conservation of Mpf-related components among the MOBH plasmids, the annotation of mpf genes of R27 was applied to those of IncP-7 plasmids in this paper. Thus, the genes referred to as trhF, traF, and traD in the pCAR1 paper (31) correspond to trhP, trhF, and traG, respectively, of pDK1.
A total of 48 kb of pDK1 (from trhN to ofn74) was highly conserved in pCAR1 (Fig. 1) except for the following insertion/deletion polymorphisms: (i) the presence in pDK1 of the IS200/IS605 family transposase gene and the ofn40 region that are absent from pCAR1, (ii) the replacement of the ofn38 region of pDK1 by the ISPre4 region in pCAR1, and (iii) the interruption of the ofn53 region of pDK1 by insertion of ISPre3 on pCAR1 (Fig. 2). In spite of these polymorphisms, pDK1 and pCAR1 possessed their self-transmissibility, indicating that the genes essential for conjugative transfer remain intact in both plasmids.
Analysis with the tBLASTx program (16) revealed that homologs of ofn71, ofn72, ofn73, and ofn74 were also present in Rts1 as a cluster with a few additional CDSs, suggesting evolution of the cluster as a module; ofn71, ofn72, ofn73, and ofn74 in pDK1 corresponded to orf246, orf248, orf250, and orf252, respectively, in Rts1 (orf246Rts1, orf248Rts1, orf250Rts1, and orf252Rts1, respectively). The oriT site of Rts1 has been experimentally determined to be located between orf250Rts1 and orf252Rts1 (34), and the oriT site of pDK1 was assumed to be located between ofn73 and ofn74 on the basis of conservation of oriT-flanking genes (with orf250 corresponding to ofn73 and orf252 corresponding to ofn74). Although repeated sequences and possible stem-loop motifs were present in the probable oriT regions of IncP-7 plasmids, no significant sequence similarities in the oriT regions were detected between the IncP-7 plasmids and other, better-characterized MOBH group elements such as R27 (29) and SXT (4). Further experimental approaches are required to elucidate the location of the oriT site of pDK1.
Transposons and IS elements.
pDK1 carries three, apparently intact, class II transposons (Tn4659, Tn4662, and Tn4663) and an insertion sequence IS1162 (Fig. 1 and 2). Tn4662 and Tn4663 were designated in this study according to the criteria proposed by Roberts et al. (45), whereas IS1162 and Tn4659 have previously been reported (56, 73). Tn4663 (40.8 kb) had two Tn21-like 38-bp terminal inverted repeats (IR3 and IR5) at the ends and contained all the xyl genes on pDK1. Shaw and Williams (51) reported that an approximately 40-kb segment of pDK1 including the entire xyl gene was able to translocate to RP4 to generate pDK2, leading to the suggestion that the 40-kb segment might be an active catabolic transposon. To determine whether this segment corresponded to Tn4663, the borders of the pDK1-derived segment in pDK2 were sequenced. Our analysis of pDK2 revealed that the entire Tn4663 unit was, indeed, inserted downstream of the tetR gene (RP4 coordinate 13245; GenBank accession no. NC_001621) with a 5-bp duplication of RP4 sequence.
An intact class II transposon usually encodes the transpositional cointegration system (the transposase [tnpA product] and its target sequences, i.e., terminal IRs) as well as the cointegrate resolution system (the resolvase [tnpR product] and its target sequence [res region]) (25). Tn4663 encoded two resolution systems; one was encoded within the nested Tn4659 region, and the other was encoded in the 0.7-kb res-tnpR-IR cluster, designated resX-tnpRTnAtcArs-IR3 (where tnpRTnAtcArs is tnpR in TnAtcArs), which was located at the Tn4659-distal end. The nucleotide sequence of resX-tnpRTnAtcArs-IR3 was 98% identical to that in the corresponding cluster of an arsenate resistance transposon, TnAtcArs, from Acidithiobacillus caldusin (64). The tnpR and tnpA genes in TnAtcArs are divergently transcribed and are physically separated by a 10-kb arsenate resistance (ars) operon. The similarity of the resX-tnpRTnAtcArs-IR3 region to the original TnAtcArs form suggests that resX-tnpRTnAtcArs-IR3 is a remnant of an intact TnAtcArs-like transposon that might be formed by deletion of the ars-tnpA-covering portion.
Transposon Tn4662 was very closely related to Tn5501-Tn5502 (27) and Tn5503 (18) in terms of gene content and sequence similarity. Tn4662 differed from Tn5503 on Rms149, an IncP-6 Pseudomonas plasmid, only in the repeat number of the 7-bp sequence (5′-CCCAGAG-3′) in ofn19; the repeat number was 16 in Tn4662 and 9 in the corresponding gene of Tn5503. A deletion derivative of Tn4662 is also located on pND6-1 (30). Although IS1162 was present on pDK1 and pCAR1, the positions of IS1162 were different between the two plasmids. The presence of identical or very closely related transposable elements on pDK1, pND6-1, pCAR1, and Rms149 suggests that these plasmids have evolved in phylogenetically related host strains.
Rearrangements of toluene-catabolic gene clusters.
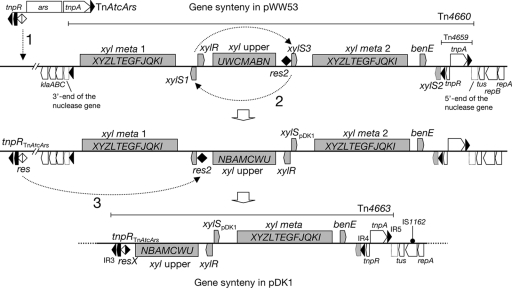
pDK1 has been reported to carry an xyl meta operon that was functionally homologous to the pWW53 meta 2 operon (51). The complete sequence of pDK1 revealed that both the xyl upper and meta operons of pDK1 were more similar to those from pWW53 than to those from pWW0 and that the meta operon of pDK1 was more similar to the meta 2 operon than to the meta 1 operon of pWW53 (the identities of gene products are shown in Fig. S2 in supplemental material), confirming the previous report (51). On the basis of sequence similarity between the xylS gene of pDK1 (xylSpDK1) and two intact xylS genes on pWW53 (xylS1 and xylS3 [xylS1pWW53 and xylS3pWW53, respectively]), Assinder et al. (1) proposed that xylSpDK1 could be a hybrid gene of xylS1pWW53 and xylS3pWW53 (the 5′ portion of xylSpDK1 from xylS1pWW53 and the 3′ portion from xylS3pWW53). They also suggested that the current organization of xyl operons in pDK1, wherein the upper operon is located on the opposite strand relative to the meta operon, could have originated from an ancestral form, in which both the upper and meta operons are located on the same DNA strand, by homology-dependent intramolecular inversion between two copies of xylS genes (1). If the xyl gene cluster of pDK1 had originated from the pWW53-type xyl gene cluster, pDK1 must have lost one meta operon as well as one copy of the xylS gene. The loss of such a gene cluster can be explained most simply by the following mechanism.
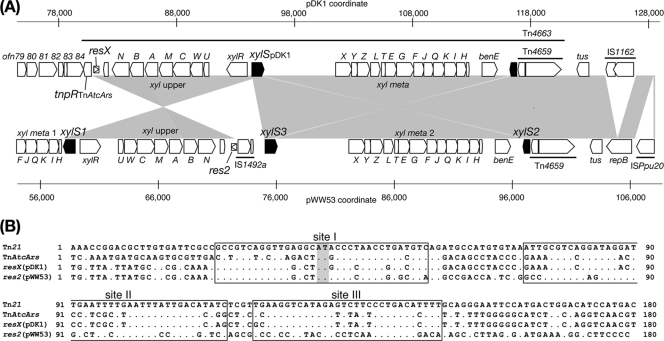
pDK1 and pWW53 share two highly conserved segments. One is the 45.5-kb segment of pDK1 that starts at IR1 (coordinate 9110) and ends at the xylS region (coordinate 95348), and the other is the 14.4-kb segment of pDK1 that starts at the resX region (coordinate 80871) and ends in the xylS region (coordinate 95251) (Fig. 1 and 4A). In pWW53, the 14.4-kb segment starts from xylS1 (coordinate 57656) and ends in res2 (coordinate 72064), which is a remnant of Tn21-like transposon and is functional as a target site of the pWW53-encoded resolvase (Fig. 4A) (73). The BLAST searches against databases revealed the closest similarities of the 45.5- and 14.5-kb regions of pDK1 to those from pWW53 (see Table S1 and Fig. S2 in the supplemental material). These structural features and the sequence similarity between the two plasmids are consistent with the previous hypothesis (1) that the current xyl upper operon-containing region of pDK1 originated from the opposite strand of pWW53 by the intramolecular inversion of the segment comprised of xylS1, the upper operon, and xylS3 (Fig. 5).
FIG. 4.
Comparison of pDK1 and pWW53. (A) Similarity in xyl-containing regions of pDK1 and pWW53. Results of BLAST analysis are shown. Pentagons indicate coding sequences. The two xylS genes (xylS1 and xylS3) and a truncated xylS gene (xylS2) are represented by filled pentagons. (B) Sequence comparison among Tn21-related res regions. Dots indicate nucleotides identical to those in Tn21. Boxed are the three binding sites of TnpR in the Tn21 res region (46) and putatively in the res regions in other Tn21-related transposons. The center of site I, where the staggered-cut and subsequent strand exchange takes place, is shaded.
FIG. 5.
Proposed model for generation of a pDK1-type xyl gene cluster from a pWW53-type cluster: event 1, transposition of TnAtcArs into the pDK1 ancestor; event 2, homology-dependent intracellular inversion between xylS1 and xylS3; event 3, TnpR-mediated site-specific resolution between res2 and the res region of the TnAtcArs-like transposon. The order of events 1 and 2 is unclear. See text for details.
The biological role of TnpR and the res region is to resolve a cointegrate that is formed in the transposition process of class II transposons. TnpR catalyzes the DNA-cleavage and -rejoining reaction between two directly repeated res regions such that the DNA segment flanked by two res regions is deleted, leaving one res region on a replicon (19). Thus, the loss of the meta 1 operon and one xylS gene can be explained by the TnpR-mediated intramolecular site-specific recombination between the two res regions. The res regions of class II transposons are generally comprised of three subsites (sites I, II, and III) to each of which the dimer form of TnpR binds, and crossing-over takes place at the center of site I (44). The resX region of pDK1 (resXpDK1) showed high similarity to the res2 region of pWW53 (res2pWW53) and the res region of TnAtcArs (resTnAtcArs) (Fig. 4B). Among the 180-bp res regions shown in Fig. 4B, resXpDK1 and res2pWW53 share the first 41 bp up to the center of the putative site I sequences, while resXpDK1 and resTnAtcArs share the rest of the 180-bp regions except for two mismatches in the site I sequences. This similarity in the three res regions is thus consistent with the site-specific recombination event between res2 and the res region in the TnAtcArs-like transposon. It is possible that the transposition of the TnAtcArs-like transposon downstream of conjugative DNA transfer genes happened to give rise to two directly repeated res sites, and subsequently the meta 1-xylS region, part of the plasmid backbone, and part of the TnAtcArs-like transposon were eliminated from the pDK1 ancestor by the TnpRTnAtcArs-mediated resolution reaction (Fig. 5).
It has been pointed out that the res regions of class II transposons can be hot spots of recombination that facilitate rearrangement and shuffling of replicons through intermolecular and intramolecular recombinations (24). Our analyses of two IncP-7 TOL plasmids have revealed two different examples where two related res regions present on a plasmid have led to rearrangements. One is the case of pWW53, in which two related res regions (res2 and Tn4658 res) with inverse orientations caused the inversion of pWW53, leading to the generation of another class II transposon, Tn4656 (73). The other case is described in this paper: the transient formation of directly repeated res regions on a plasmid resulted in the deletion of the segment flanked by the two res regions (Fig. 5). Interestingly, the rearrangement of pDK1 also generated a toluene-catabolic transposon, Tn4663, which moved to RP4 to form pDK2 (51). These observations suggest that the resolution system contributes to the formation of class II transposons with novel genetic contexts.
The host ranges of IncP-7 plasmids.
The complete sequence determination of pDK1 has provided insights for genetic organization of the ancestral IncP-7 plasmid (Fig. 2). However, available information for the IncP-7 group was still limited with regard to basic plasmid functions and the range of its potential hosts (47, 52, 53). To obtain the consensus information on host ranges of IncP-7 plasmids, minireplicons of three IncP-7 plasmids (pHY118 for pWW53, pMM67 for pCAR1, and pMM68 for pDK1) that carried the repA-oriV-parWASB region (Fig. 3) were introduced from E. coli S17-1 into five plasmid-free Pseudomonas strains (i.e., the derivatives of Pseudomonas resinovorans CA10 [designated CA10dm4G], P. putida F1 [F1G], P. putida KT2440 [KT2440G2], Pseudomonas aeruginosa PAO1 [KG2512G], and P. fluorescens Pf-5 [Pf-5G]) as well as the pDK1-free derivative of HS1 (HS1CG) through the RP4-specified conjugation machinery (the oriTRP4 site on the three minireplicons and all of the trans-acting transfer-related genes from the S17-1 chromosome). We additionally included Sphingomonas paucimobilis IAM12578G (an alphaproteobacterial strain) and Burkholderia multivorans ATCC 17616G (a betaproteobacterial strain) as recipients. The three minireplicons were transferred into all six Pseudomonas strains at frequencies of 10−5 to 10−2 transconjugants per donor cell but not to the S. paucimobilis or B. multivorans strain. These results indicate that IncP-7 plasmids are able to replicate at least in the Pseudomonas strains examined but not in alpha- or betaproteobacteria.
Conjugative transfer of pDK1 was next investigated using pDK1K, a pDK1 derivative having a Kmr gene in xylU. Among the six plasmid-free Pseudomonas strains that were employed as the recipient in the pMM68 transfer (see above), pDK1K was transferred from P. putida HS1CR to P. putida HS1CG and Pseudomonas fluorescens Pf-5G at frequencies of 4.4 × 10−4 and 2.5 × 10−3, respectively, per donor cell. When P. fluorescens Pf-5S was used as a donor strain, the plasmid was transferred to Pf-5G and HS1CG at frequencies of 3.0 × 10−5 and 2.3 × 10−7, respectively, per donor cell. No transfer of pDK1K was detected (<10−8 per donor cell) to P. putida F1G, P. putida KT2440G2, P. resinovorans CA10dm4G, or P. aeruginosa PAO1G. This indicates that pDK1 retains self-transmissibility but with a limited recipient range even in P. putida strains. This property contrasted with the replication ability of pMM68 in the six Pseudomonas strains. The limited recipient range of conjugative transfer is also associated with pCAR1 (53), and it is therefore most likely that the host range of self-transmissible IncP-7 plasmids is, in general, restricted by the conjugation system rather than the replication system.
Conclusions.
The primary purpose of this study was to clarify the evolutionary relationship between the two TOL plasmids pDK1 and pWW53. Analysis of the pDK1 genome revealed the evolutionary history of catabolic gene clusters on this plasmid and the conservation of genes for basic plasmid functions among IncP-7 plasmids. We additionally performed experimental analyses to obtain insights for the host range of the IncP-7 group. Four major findings concerning the nature of IncP-7 plasmids from this study are summarized as follows. First, pDK1 and pWW53 shared 49.3 kb that cover the plasmid backbone region and the catabolic transposon region, and this clearly demonstrated that the two plasmids have originated from an ancestral IncP-7 TOL plasmid. Second, generation of the xyl gene cluster on pDK1 was most simply explained by the three sequential recombination events: (i) homology-dependent inversion between two copies of an xylS gene, (ii) insertion of a TnAtsArs-like transposon, and (iii) the TnpRTnAtsArs-mediated site-specific deletion between directly repeated res regions (Fig. 5). Third, the limited replication host ranges of pDK1 and pCAR1 and the conservation of MOBH group-related DNA transfer genes in pDK1 and pCAR1 suggest that the IncP-7 plasmids are narrow-host-range and self-transmissible plasmids specialized to Pseudomonas strains (52). Lastly, for several Pseudomonas strains in which the mini-pDK1 plasmid could replicate, no conjugative transfer of pDK1 was detected. This result contrasted with the paradigm that the host range of transfer is broader than that of replication in other self-transmissible plasmids (17).
Several TOL plasmids other than pWW53 contain two homologous or higher-similarity meta pathway operons (37, 50), and there is a report indicating that the xyl-related catabolic genes are located on the chromosomes (35). However, the relationship of these catabolic genes to mobile genetic elements is still unclear. Sequence analyses of such catabolic gene clusters and their flanking regions will provide many clues leading to a better understanding of how functional gene clusters are assembled and disseminate in nature.
Supplementary Material
Acknowledgments
We are grateful to P. A. Williams for P. putida HS1 and pDK2 and for critical reading and improvement of the manuscript. We thank H. Nojiri for his gift of pUCARori004, pUCARori004ΔparA, and unpublished data. We also thank the National BioResource Project of National Institute of Genetics (Mishima, Japan) for providing pJP5608 and N. Gotoh for providing P. aeruginosa KG2512. We appreciate Y. Ohtsubo for useful discussion and technical advice and K. Osada for technical assistance.
This work was supported by Grants-in-Aid from the Ministry of Education, Culture, Sports, Science and Technology, Japan. M.M. was supported by a Research Fellowship of the Japan Society for the Promotion of Science for Young Scientist.
Footnotes
Published ahead of print on 25 June 2010.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Assinder, S. J., P. De Marco, D. J. Osborne, C. L. Poh, L. E. Shaw, M. K. Winson, and P. A. Williams. 1993. A comparison of the multiple alleles of xylS carried by TOL plasmids pWW53 and pDK1 and its implications for their evolutionary relationship. J. Gen. Microbiol. 139:557-568. [DOI] [PubMed] [Google Scholar]
- 2.Assinder, S. J., P. de Marco, J. R. Sayers, L. E. Shaw, M. K. Winson, and P. A. Williams. 1992. Identical resolvases are encoded by Pseudomonas TOL plasmids pWW53 and pDK1. Nucleic Acids Res. 20:5476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Benjamin, R. C., J. A. Voss, and D. A. Kunz. 1991. Nucleotide sequence of xylE from the TOL pDK1 plasmid and structural comparison with isofunctional catechol-2,3-dioxygenase genes from TOL, pWW0 and NAH7. J. Bacteriol. 173:2724-2728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ceccarelli, D., A. Daccord, M. Rene, and V. Burrus. 2008. Identification of the origin of transfer (oriT) and a new gene required for mobilization of the SXT/R391 family of integrating conjugative elements. J. Bacteriol. 190:5328-5338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chatfield, L. K., and P. A. Williams. 1986. Naturally occurring TOL plasmids in Pseudomonas strains carry either two homologous or two nonhomologous catechol 2,3-oxygenase genes. J. Bacteriol. 168:878-885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Choi, K. H., J. B. Gaynor, K. G. White, C. Lopez, C. M. Bosio, R. R. Karkhoff-Schweizer, and H. P. Schweizer. 2005. A Tn7-based broad-range bacterial cloning and expression system. Nat. Methods 2:443-448. [DOI] [PubMed] [Google Scholar]
- 7.Choi, K. H., A. Kumar, and H. P. Schweizer. 2006. A 10-min method for preparation of highly electrocompetent Pseudomonas aeruginosa cells: application for DNA fragment transfer between chromosomes and plasmid transformation. J. Microbiol. Methods 64:391-397. [DOI] [PubMed] [Google Scholar]
- 8.Clement, P., D. H. Pieper, and B. Gonzalez. 2001. Molecular characterization of a deletion/duplication rearrangement in tfd genes from Ralstonia eutropha JMP134(pJP4) that improves growth on 3-chlorobenzoic acid but abolishes growth on 2,4-dichlorophenoxyacetic acid. Microbiology 147:2141-2148. [DOI] [PubMed] [Google Scholar]
- 9.Cuff, J. A., M. E. Clamp, A. S. Siddiqui, M. Finlay, and G. J. Barton. 1998. JPred: a consensus secondary structure prediction server. Bioinformatics 14:892-893. [DOI] [PubMed] [Google Scholar]
- 10.Delcher, A. L., D. Harmon, S. Kasif, O. White, and S. L. Salzberg. 1999. Improved microbial gene identification with GLIMMER. Nucleic Acids Res. 27:4636-4641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dennis, J. J., and G. J. Zylstra. 1998. Plasposons: modular self-cloning minitransposon derivatives for rapid genetic analysis of gram-negative bacterial genomes. Appl. Environ. Microbiol. 64:2710-2715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Figurski, D. H., and D. R. Helinski. 1979. Replication of an origin-containing derivative of plasmid RK2 dependent on a plasmid function provided in trans. Proc. Natl. Acad. Sci. U. S. A. 76:1648-1652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Finette, B. A., V. Subramanian, and D. T. Gibson. 1984. Isolation and characterization of Pseudomonas putida PpF1 mutants defective in the toluene dioxygenase enzyme system. J. Bacteriol. 160:1003-1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Funnell, B. E., and R. A. Slavcev. 2004. Partition systems of bacterial plasmids, p. 81-103. In B. E. Funnell and G. J. Phillips (ed.), Plasmid biology. ASM Press, Washington, DC.
- 15.Garcillan-Barcia, M. P., M. V. Francia, and F. de la Cruz. 2009. The diversity of conjugative relaxases and its application in plasmid classification. FEMS Microbiol. Rev. 33:657-687. [DOI] [PubMed] [Google Scholar]
- 16.Gertz, E. M., Y. K. Yu, R. Agarwala, A. A. Schaffer, and S. F. Altschul. 2006. Composition-based statistics and translated nucleotide searches: improving the TBLASTN module of BLAST. BMC Biol. 4:41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Guiney, D. G. 1982. Host range of conjugation and replication functions of the Escherichia coli sex plasmid Flac. Comparison with the broad host-range plasmid RK2. J. Mol. Biol. 162:699-703. [DOI] [PubMed] [Google Scholar]
- 18.Haines, A. S., K. Jones, M. Cheung, and C. M. Thomas. 2005. The IncP-6 plasmid Rms149 consists of a small mobilizable backbone with multiple large insertions. J. Bacteriol. 187:4728-4738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hallet, B., V. Vanhooff, and F. Cornet. 2004. DNA site-specific resolution systems, p. 145-180. In B. E. Funnell and G. J. Phillips (ed.), Plasmid biology. ASM Press, Washington, DC.
- 20.Hirokawa, T., S. Boon-Chieng, and S. Mitaku. 1998. SOSUI: classification and secondary structure prediction system for membrane proteins. Bioinformatics 14:378-379. [DOI] [PubMed] [Google Scholar]
- 21.Hoang, T. T., R. R. Karkhoff-Schweizer, A. J. Kutchma, and H. P. Schweizer. 1998. A broad-host-range Flp-FRT recombination system for site-specific excision of chromosomally located DNA sequences: application for isolation of unmarked Pseudomonas aeruginosa mutants. Gene 212:77-86. [DOI] [PubMed] [Google Scholar]
- 22.Keil, H., S. Keil, R. W. Pickup, and P. A. Williams. 1985. Evolutionary conservation of genes coding for meta pathway enzymes within TOL plasmids pWW0 and pWW53. J. Bacteriol. 164:887-895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Keil, H., M. R. Lebens, and P. A. Williams. 1985. TOL plasmid pWW15 contains two nonhomologous, independently regulated catechol 2,3-oxygenase genes. J. Bacteriol. 163:248-255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kholodii, G. 2001. The shuffling function of resolvases. Gene 269:121-130. [DOI] [PubMed] [Google Scholar]
- 25.Kleckner, N. 1981. Transposable elements in prokaryotes. Annu. Rev. Genet. 15:341-404. [DOI] [PubMed] [Google Scholar]
- 26.Kunz, D. A., and P. J. Chapman. 1981. Isolation and characterization of spontaneously occurring TOL plasmid mutants of Pseudomonas putida HS1. J. Bacteriol. 146:952-964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lauf, U., C. Muller, and H. Herrmann. 1998. The transposable elements resident on the plasmids of Pseudomonas putida strain H, Tn5501 and Tn5502, are cryptic transposons of the Tn3 family. Mol. Gen. Genet. 259:674-678. [DOI] [PubMed] [Google Scholar]
- 28.Lawley, T., B. M. Wilkins, and L. S. Frost. 2004. Conjugation in gram-negative bacteria, p. 203-226. In B. E. Funnell and G. J. Phillips (ed.), Plasmid biology. ASM press, Washington, DC.
- 29.Lawley, T. D., M. W. Gilmour, J. E. Gunton, L. J. Standeven, and D. E. Taylor. 2002. Functional and mutational analysis of conjugative transfer region 1 (Tra1) from the IncHI1 plasmid R27. J. Bacteriol. 184:2173-2180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li, W., J. Shi, X. Wang, Y. Han, W. Tong, L. Ma, B. Liu, and B. Cai. 2004. Complete nucleotide sequence and organization of the naphthalene catabolic plasmid pND6-1 from Pseudomonas sp. strain ND6. Gene 336:231-240. [DOI] [PubMed] [Google Scholar]
- 31.Maeda, K., H. Nojiri, M. Shintani, T. Yoshida, H. Habe, and T. Omori. 2003. Complete nucleotide sequence of carbazole/dioxin-degrading plasmid pCAR1 in Pseudomonas resinovorans strain CA10 indicates its mosaicity and the presence of large catabolic transposon Tn4676. J. Mol. Biol. 326:21-33. [DOI] [PubMed] [Google Scholar]
- 32.Miyazaki, R., Y. Sato, M. Ito, Y. Ohtsubo, Y. Nagata, and M. Tsuda. 2006. Complete nucleotide sequence of an exogenously isolated plasmid, pLB1, involved in gamma-hexachlorocyclohexane degradation. Appl. Environ. Microbiol. 72:6923-6933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mori, H., Y. Mori, C. Ichinose, H. Niki, T. Ogura, A. Kato, and S. Hiraga. 1989. Purification and characterization of SopA and SopB proteins essential for F plasmid partitioning. J. Biol. Chem. 264:15535-15541. [PubMed] [Google Scholar]
- 34.Murata, T., M. Ohnishi, T. Ara, J. Kaneko, C. G. Han, Y. F. Li, K. Takashima, H. Nojima, K. Nakayama, A. Kaji, Y. Kamio, T. Miki, H. Mori, E. Ohtsubo, Y. Terawaki, and T. Hayashi. 2002. Complete nucleotide sequence of plasmid Rts1: implications for evolution of large plasmid genomes. J. Bacteriol. 184:3194-3202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Noh, S. J., Y. Kim, K. H. Min, T. B. Karegoudar, and C. K. Kim. 2000. Cloning and nucleotide sequence analysis of xylE gene responsible for meta-cleavage of 4-chlorocatechol from Pseudomonas sp. S-47. Mol. Cells 10:475-479. [PubMed] [Google Scholar]
- 36.Nojiri, H., M. Shintani, and T. Omori. 2004. Divergence of mobile genetic elements involved in the distribution of xenobiotic-catabolic capacity. Appl. Microbiol. Biotechnol. 64:154-174. [DOI] [PubMed] [Google Scholar]
- 37.O'Donnell, K. J., and P. A. Williams. 1991. Duplication of both xyl catabolic operons on TOL plasmid pWW15. J. Gen. Microbiol. 137:2831-2838. [DOI] [PubMed] [Google Scholar]
- 38.Ogawa, N., A. M. Chackrabarty, and O. Zaborina. 2004. Degradative plasmid, p. 341-392. In B. E. Funnell and G. J. Phillips (ed.), Plasmid biology. ASM Press, Washington, DC.
- 39.Ohtsubo, Y., W. Ikeda-Ohtsubo, Y. Nagata, and M. Tsuda. 2008. GenomeMatcher: a graphical user interface for DNA sequence comparison. BMC Bioinformatics 9:376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Penfold, R. J., and J. M. Pemberton. 1992. An improved suicide vector for construction of chromosomal insertion mutations in bacteria. Gene 118:145-146. [DOI] [PubMed] [Google Scholar]
- 41.Platt, A., V. Shingler, S. C. Taylor, and P. A. Williams. 1995. The 4-hydroxy-2-oxovalerate aldolase and acetaldehyde dehydrogenase (acylating) encoded by the nahM and nahO genes of the naphthalene catabolic plasmid pWW60-22 provide further evidence of conservation of meta-cleavage pathway gene sequences. Microbiology 141:2223-2233. [DOI] [PubMed] [Google Scholar]
- 42.Ramos, J. L., S. Marques, and K. N. Timmis. 1997. Transcriptional control of the Pseudomonas TOL plasmid catabolic operons is achieved through an interplay of host factors and plasmid-encoded regulators. Annu. Rev. Microbiol. 51:341-373. [DOI] [PubMed] [Google Scholar]
- 43.Reams, A. B., and E. L. Neidle. 2003. Genome plasticity in Acinetobacter: new degradative capabilities acquired by the spontaneous amplification of large chromosomal segments. Mol. Microbiol. 47:1291-1304. [DOI] [PubMed] [Google Scholar]
- 44.Reed, R. R. 1981. Transposon-mediated site-specific recombination: a defined in vitro system. Cell 25:713-719. [DOI] [PubMed] [Google Scholar]
- 45.Roberts, A. P., M. Chandler, P. Courvalin, G. Guedon, P. Mullany, T. Pembroke, J. I. Rood, C. J. Smith, A. O. Summers, M. Tsuda, and D. E. Berg. 2008. Revised nomenclature for transposable genetic elements. Plasmid 60:167-173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rogowsky, P., S. E. Halford, and R. Schmitt. 1985. Definition of three resolvase binding sites at the res loci of Tn21 and Tn1721. EMBO J. 4:2135-2141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sagai, H., V. Krcmery, K. Hasuda, S. Iyobe, and H. Knothe. 1975. R factor-mediated resistance to aminoglycoside antibiotics in Pseudomonas aeruginosa. Jpn. J. Microbiol. 19:427-432. [DOI] [PubMed] [Google Scholar]
- 48.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 49.Schafer, A., A. Tauch, W. Jager, J. Kalinowski, G. Thierbach, and A. Puhler. 1994. Small mobilizable multi-purpose cloning vectors derived from the Escherichia coli plasmids pK18 and pK19: selection of defined deletions in the chromosome of Corynebacterium glutamicum. Gene 145:69-73. [DOI] [PubMed] [Google Scholar]
- 50.Sentchilo, V. S., A. N. Perebituk, A. J. Zehnder, and J. R. van der Meer. 2000. Molecular diversity of plasmids bearing genes that encode toluene and xylene metabolism in Pseudomonas strains isolated from different contaminated sites in Belarus. Appl. Environ. Microbiol. 66:2842-2852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Shaw, L. E., and P. A. Williams. 1988. Physical and functional mapping of two cointegrate plasmids derived from RP4 and TOL plasmid pDK1. J. Gen. Microbiol. 134:2463-2474. [DOI] [PubMed] [Google Scholar]
- 52.Shintani, M., N. Fukushima, M. Tezuka, H. Yamane, and H. Nojiri. 2008. Conjugative transfer of the IncP-7 carbazole degradative plasmid, pCAR1, in river water samples. Biotechnol. Lett. 30:117-122. [DOI] [PubMed] [Google Scholar]
- 53.Shintani, M., H. Habe, M. Tsuda, T. Omori, H. Yamane, and H. Nojiri. 2005. Recipient range of IncP-7 conjugative plasmid pCAR2 from Pseudomonas putida HS01 is broader than from other Pseudomonas strains. Biotechnol. Lett. 27:1847-1853. [DOI] [PubMed] [Google Scholar]
- 54.Shintani, M., H. Yano, H. Habe, T. Omori, H. Yamane, M. Tsuda, and H. Nojiri. 2006. Characterization of the replication, maintenance, and transfer features of the IncP-7 plasmid pCAR1, which carries genes involved in carbazole and dioxin degradation. Appl. Environ. Microbiol. 72:3206-3216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Simon, R., U. Priefer, and A. Puhler. 1983. A broad host range mobilization systems for in vivo genetic engineering: transposon mutagenesis in gram negative bacteria. Biotechnology (NY) 1:784-791. [Google Scholar]
- 56.Solinas, F., A. M. Marconi, M. Ruzzi, and E. Zennaro. 1995. Characterization and sequence of a novel insertion sequence, IS1162, from Pseudomonas fluorescens. Gene 155:77-82. [DOI] [PubMed] [Google Scholar]
- 57.Springael, D., and E. M. Top. 2004. Horizontal gene transfer and microbial adaptation to xenobiotics: new types of mobile genetic elements and lessons from ecological studies. Trends Microbiol. 12:53-58. [DOI] [PubMed] [Google Scholar]
- 58.Tauch, A., S. Schneiker, W. Selbitschka, A. Puhler, L. S. van Overbeek, K. Smalla, C. M. Thomas, M. J. Bailey, L. J. Forney, A. Weightman, P. Ceglowski, T. Pembroke, E. Tietze, G. Schroder, E. Lanka, and J. D. van Elsas. 2002. The complete nucleotide sequence and environmental distribution of the cryptic, conjugative, broad-host-range plasmid pIPO2 isolated from bacteria of the wheat rhizosphere. Microbiology 148:1637-1653. [DOI] [PubMed] [Google Scholar]
- 59.Thomas, C. M., and A. S. Haines. 2004. Plasmids of the genus Pseudomonas, p. 197-231. In J. L. Ramos (ed.), Pseudomonas, vol. 1. Kluwer Academic/Plenum Publishers, New York, NY. [Google Scholar]
- 60.Tsuda, M., and H. Genka. 2001. Identification and characterization of Tn4656, a novel class II transposon carrying a set of toluene-degrading genes from TOL plasmid pWW53. J. Bacteriol. 183:6215-6224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Tsuda, M., and T. Iino. 1987. Genetic analysis of a transposon carrying toluene degrading genes on a TOL plasmid pWW0. Mol. Gen. Genet. 210:270-276. [DOI] [PubMed] [Google Scholar]
- 62.Tsuda, M., and T. Iino. 1988. Identification and characterization of Tn4653, a transposon covering the toluene transposon Tn4651 on TOL plasmid pWW0. Mol. Gen. Genet. 213:72-77. [DOI] [PubMed] [Google Scholar]
- 63.Tsuda, M., and T. Iino. 1990. Naphthalene degrading genes on plasmid NAH7 are on a defective transposon. Mol. Gen. Genet. 223:33-39. [DOI] [PubMed] [Google Scholar]
- 64.Tuffin, I. M., P. de Groot, S. M. Deane, and D. E. Rawlings. 2005. An unusual Tn21-like transposon containing an ars operon is present in highly arsenic-resistant strains of the biomining bacterium Acidithiobacillus caldus. Microbiology 151:3027-3039. [DOI] [PubMed] [Google Scholar]
- 65.van der Meer, J. R., R. Ravatn, and V. Sentchilo. 2001. The clc element of Pseudomonas sp. strain B13 and other mobile degradative elements employing phage-like integrases. Arch. Microbiol. 175:79-85. [DOI] [PubMed] [Google Scholar]
- 66.Vieira, J., and J. Messing. 1982. The pUC plasmids, an M13mp7-derived system for insertion mutagenesis and sequencing with synthetic universal primers. Gene 19:259-268. [DOI] [PubMed] [Google Scholar]
- 67.Voss, J. A., H. Khedairy, R. F. Baker, and R. C. Benjamin. 1990. Molecular cloning of the xylL-xylE region from the P. putida TOL plasmid, pDK1. SAAS Bull. Biochem. Biotechnol. 3:54-57. [PubMed] [Google Scholar]
- 68.Weightman, A. J., A. W. Topping, K. E. Hill, L. L. Lee, K. Sakai, J. H. Slater, and A. W. Thomas. 2002. Transposition of DEH, a broad-host-range transposon flanked by ISPpu12, in Pseudomonas putida is associated with genomic rearrangements and dehalogenase gene silencing. J. Bacteriol. 184:6581-6591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Willetts, N. 1988. Conjugation, p. 49-77. In J. Grinsted and P. M. Bennett (ed.), Method in microbiology. Academic Press, London, United Kingdom.
- 70.Williams, P. A., R. M. Jones, and G. J. Zylstra. 2004. Genomics of catabolic plasmids, p. 165-195. In J. L. Ramos (ed.), Pseudomonas, vol. 1. Kluwer Academic/Plenum Publishers, New York, NY. [Google Scholar]
- 71.Williams, P. A., and M. J. Worsey. 1976. Ubiquity of plasmids in coding for toluene and xylene metabolism in soil bacteria: evidence for the existence of new TOL plasmids. J. Bacteriol. 125:818-828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Worsey, M. J., F. C. Franklin, and P. A. Williams. 1978. Regulation of the degradative pathway enzymes coded for by the TOL plasmid (pWWO) from Pseudomonas putida mt-2. J. Bacteriol. 134:757-764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Yano, H., C. E. Garruto, M. Sota, Y. Ohtsubo, Y. Nagata, G. J. Zylstra, P. A. Williams, and M. Tsuda. 2007. Complete sequence determination combined with analysis of transposition/site-specific recombination events to explain genetic organization of IncP-7 TOL plasmid pWW53 and related mobile genetic elements. J. Mol. Biol. 369:11-26. [DOI] [PubMed] [Google Scholar]
- 74.Yuhara, S., H. Komatsu, H. Goto, Y. Ohtsubo, Y. Nagata, and M. Tsuda. 2008. Pleiotropic roles of iron-responsive transcriptional regulator Fur in Burkholderia multivorans. Microbiology 154:1763-1774. [DOI] [PubMed] [Google Scholar]
- 75.Zhong, S., A. Khodursky, D. E. Dykhuizen, and A. M. Dean. 2004. Evolutionary genomics of ecological specialization. Proc. Natl. Acad. Sci. U. S. A. 101:11719-11724. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.