Abstract
In this critical review we highlight recent advances in the use of peptide- and protein-related materials as smart building blocks in nanotechnology. Peptides and proteins can be very practical for new material synthesis and device fabrications. For example, peptides and proteins have superior specificity for target binding as seen in the antibody recognition and this biological recognition function can be used to assemble them into specific structures and shapes in large scale, as observed in the S-layer protein assembly. Collagens are assembled from triple helix peptides in micron-size with precise recognition between peptides and these biological assemblies can undergo smart structural change with pH, ionic strength, temperature, electric/magnetic fields. In addition, assemblies of peptides can template complex 3D crystallization processes with catalytic function, thus enabling to grow various materials in physiological conditions at low temperature in aqueous solution. The biomimetic growth of nanomaterials in aqueous solution is extremely useful when they are applied to therapeutics and medical imaging in vivo since these nanomaterials will be well dispersed in bodies. Peptides also play significant roles in signal transduction pathways in cells. For example, neuropeptides are used as neurotransmitters between synapses and these peptides bind receptors on the surface of cells to cascade the signal transduction. These versatile functions of peptides are extremely practical and here we discuss them with examples of relevant applications such as nanoreactors, sensors, electronics, and stimulus-responsive materials. It should be noted that peptide/protein assemblies can be applied to build up micron-scale materials that still feature excellent nano-scale ensembles, which essentially bridges the nano-world and the micro-world (86 references).
I. Introduction
Nanotechnology is revolutionizing a variety of technological applications by unveiling the exciting physicochemical properties of nanometric building blocks as compared to bulk materials.1–3 While downscaling the size of functional building blocks has been a major endeavor for this cutting-edge technology, in order to further broaden the range of applications a simple and straightforward methodology is necessary to develop complex material building blocks, difficult to synthesize in conventional growth methods, and organize them into large-scale assemblies that process smart functionality for nano-devices.
The complexity in material development includes 3D shape control, size control, crystalline-structural control, functionality integration, and catalytic low temperature growth. The key strategy to construct complex assemblies of nanoscale building blocks is to program the assembly so that each building block binds at a specific location on the substrate or device geometry, and this process can be reconfigured by triggering the assembly-disassembly command. The molecular recognition with the on-off switching capability such as protein-receptor interactions can be extremely useful to program the assembly of building blocks. Moreover, if an external stimulus triggers any physical change of the building blocks such as volume, optical property, conductivity, or binding affinity, it may be applied in reconfigurable photonic/electric devices and smart reusable sensing systems.
Despite the great interest in this approach, the current stage of smart self-assembly nanotechnology is far behind this goal and it is difficult to even incorporate one or two of these functions into nanoscale building blocks. When multiple functions are integrated into a single device, the final response of the material is difficult to predict because these outputs as physical responses might interfere with each other or because the combination of these functions might lead to another unexpected functions. If this is the case, why not start by understating existing smart functional building blocks in nature and mimic them? Life already possesses a unique set of nanomachines that replicate and adapt to the environment with complex smart functions as a consequence of millenniums of evolution. For example, organisms such as diatoms and marine sponges use templates of self-assembled peptides, proteins, and organic–inorganic suprastructures to control nucleation and the amorphous-to-crystal phase transformation, which leads to the generation of crystals with unusual shapes and complexity in controlled size, orientation, composition, and hierarchical organization from nanoscale to microscale.4–7 S-layer proteins, defensive layers on bacteria surfaces, are also shown to organize their assembled structure on membranes by molecular recognition of proteins in addition to the robust nucleation.8,9 Mimicking these functions of peptides and proteins and integrating them with nano-scale device building blocks provide the opportunity to self-assemble novel complex device configurations.10,11
In this review we highlight recent advances in the use of protein-related materials as smart building blocks in nanotechnology. In the cellular factory, proteins are workforces that perform broad functions via perfectly orchestrated yet intricate metabolic pathways. For instance, while antibodies act as security agents that specifically recognize alien elements to the organism, membrane proteins are the postal service to communicate with the environment, and enzymes are engineers that regulate every aspect of the cell machinery. Moreover, structural proteins such as collagen are powerful building blocks that sustain the cell shape and function. This outstanding degree of organization and efficiency to accomplish a task could have significant merit to use proteins and protein-related materials to solve nanotechnology-related issues, and this approach is comprised in the young yet exciting field of bionanotechnology.12 The problem of the application of peptides and proteins in nanotechnology is that sometimes it is difficult to predict their functions in a non-biological environment. However, an advantage is that these functions are reproducible and less likely to fail in appropriate conditions since biological systems have already optimized each function or the integrated functions during millenniums of evolution.
Traditionally, DNA molecules have received more attention than peptides for its application in bionanotechnology. However, peptides and proteins can be very practical for new material synthesis and device fabrications due to the following features. For example, although peptides and proteins do not have the versatile base-pair rule to program DNA assemblies,13 proteins also have superior specificity for target binding with complex molecular recognition mechanism. Peptides can also use the recognition function to assemble into specific shapes in large scale, as observed in the S-layer protein assembly. This peptide function can be useful for developing nanoreactors, sensors, and electronics. In addition, assemblies of peptides can template complex 3D crystallization processes with catalytic function, enabling to grow various materials in physiological conditions at low temperature and in aqueous solution. Nanoreactors and sensors are the fields of application of this peptide function. The biomimetic growth of nano-materials in aqueous solution is extremely useful when they are applied to therapeutics and medical imaging in vivo since these nanomaterials will be well dispersed in bodies without aggregating them at undesired locations. Peptides also play significant roles in signal transduction in cells.14 For example, neuropeptides are used as neurotransmitters between synapses and these peptides bind receptors on the surface of cells to cascade the signal transduction. This peptide function can be applied in electronics and sensors. Since neuropeptides need to be cleared out from synapses efficiently so that these synapses can be ready to function again, this assembly/disassembly switching can be applied to smart functional materials such as reconfigurable photonic/electronic devices where building blocks can be disassembled and reconfigured in different device configurations on command. The robust assembly nature of peptides is very powerful in the applications of nanoreactors, electronics, and sensor fabrication. For example, collagens are assembled from triple helix peptides in micron-size with precise recognition between peptides. By mimicking this assembly process, it is possible to build up micron-scale materials that still feature excellent nano-scale ensembles, which essentially bridges the nano-world and the micro-world. The conformation change of peptides and proteins can be applied for smart responsive materials. For example, protein assembly can undergo structural change with variations in pH, ionic strength, temperature, electric/magnetic fields. If this function is incorporated, these building blocks can be applied to actuators and responsive photonic films whose physical property depends on the structural change induced by these external inputs. Finally, it should be noted that the advancement of genetic engineering is advantageous to further generate precise peptide functionalization for bionanotechnology. For example, the biotinylation at a specific amino acid residue is extremely difficult with non-genetic methods due to the limitation of the chemical protections on preserved groups.
In this tutorial article, after a brief classification of peptidic materials with their degree of structural complexity, the versatile functions of peptides are discussed with examples of relevant applications such as nanoreactors, sensors, electronics, and stimulus-responsive materials. The use of proteins in nanobiotechnology such as in drug delivery and nanomedicine has already been extensively reviewed and will not be the subject in the present review.15–17 By the same reason, bio-nanotechnology based on the use of nucleic acids, viruses, and cells is also not discussed in this article.13,18,19
II. Peptides, proteins and their supramolecular structures
Small peptides consist of short amino acid sequences that have less intricate functionality than proteins. While these oligomeric polymers may not perform highly specialized tasks compared to proteins, they can be synthesized more easily with desired amino acid sequences by well-established chemical and genetic engineering procedures, and therefore they are versatile components of the bionanotechnology toolbox.20 When peptides are designed to be folded in desired conformations (α-helix, β-sheet, etc.), these three-dimensional building blocks may yield supramolecular structures via self-assembly.17,21 Moreover, the 3D spatial distribution of chemical moieties can be controlled by changing the peptide conformation via tuning the physicochemical properties of the environment such as pH, temperature and salt content, which makes peptides versatile smart materials for the de novo design of nanoreactors.22,23
Proteins, including antibodies, enzymes and structural proteins possess long amino acid sequences with complex 3D structures and usually perform highly specialized tasks in cells. The complex folding steps that lead to the native conformation of proteins are difficult to replicate in vitro, and hence it is difficult to design proteins de novo with tailored functions. Therefore, the availability of a given protein relies on its extraction from natural sources or its production in a micro-organism via genetic engineering. The latter approach also paves the way to modify the physicochemical properties of proteins to possess a particular function by expressing a specific plasmid DNA vector in the bacterial host.24,25 Regarding their functions, the high specificity and strength of the antibody-antigen interaction can be applied to target nanoscale building blocks onto desired locations for the construction of complex nano-architectures, while enzymes can be viewed as valuable nanoreactors when these proteins catalyze synthetic reactions of interests and also template the products such as metals and semiconductors in controlled structures.
The function of peptides and proteins to yield supra-molecular architectures with controlled shapes and physicochemical properties is a significant feature for their application in bionanotechnology. For instance, α-helical motifs of peptides can self-assemble into diverse structures from micrometer-long fibers to star-burst arrangements that work as polynanoreactors.26 The aggregation of peptides into amyloid fibers in Parkinson’s disease inspired the self-assembly of peptide nanotubes, which posses exceptional robustness and electrical conductivity along the axis of the tube.27,28 Glycylglycine bolaamphiphile peptides could be assembled into peptide nanotubes in a pH-dependent manner,29 which are useful templates for the fabrication of nanowire building blocks in nanosensors.30,31 Metalorganic protein frameworks were assembled into one-dimensional protein bundles, which mimics collagen assembly in biomineralization processes, by using the strong interaction between streptavidin-conjugated and biotinylated organometallic monomers.32 Recently, the streptavidin-biotin interaction was also exploited to obtain large-scale 3D peptide scaffolds where metal nanoparticles that conjugate streptavidin are incorporated in the cage-like unit cells made from biotylated peptides.33 Also, genetically engineered bacterial proteins showed great potential for the fabrication of nanotubes.34
III. Nanoreactors
Biomineralizing organisms have evolved the capability of synthesizing complex inorganic structures at the nano- and micro-scale aimed by a battery of peptides and enzymes, which allow synthesis of materials in mild conditions, selection of the polymorph, and its programmed assembly to yield exquisite structures as observed in starfish and sea urchins.4,6,35,36 This natural crystal growth process has inspired the search for de novo peptide sequences that could catalyze the synthesis of materials of technological relevance via molecular biology tools such as phage display.37–39 In this approach, DNA fragments encoding random peptide sequences are inserted into bacteriophages so that the peptides are expressed on the capsids and the affinity of those peptides for the material of interest is tested by applying stringent selection protocols. The resulting peptides optimized from naturally observed mineralizing sequences not only catalyze the material synthesis in mild conditions but also direct the growth of a particular crystalline structure, shape and size depending on the experimental conditions.40 Although the mechanism behind this bio-inspired mineralization processes remains elusive, the amino acid composition and the spatial arrangement of chemical moieties dictated by the tertiary structure are known to be important for these peptides to function as biomolecular nanoreactors, nanoscale catalytic templates assembled from peptides and proteins. For example, certain conformations of Pt-binding peptides that can specifically recognize the (110) face of Pt crystals have a 3D structure resembling the grooves of the crystalline structure.41 While the crystallinity can be relatively well controlled by the biomimetic material growth, the size and the shape are more difficult to be regulated. But recently the development of these morphology controls with biomimetic approach showed significant improvement. For example, when immobilized on biomolecular nanotubes, peptides with high affinity for Cu can grow monodisperse Cu nanoparticles and the size of Cu nanoparticles on the nanotube is directly related to the conformation of the peptide, dictated by the pH of the growth solution (Fig. 1).42 The shape of Ag nanocrystals was also optimized by peptides that can slow down or catalyze the growth on certain crystalline faces over others due to characteristic amino acid sequences.40,43 Similarly, fluorescent metal nanoclusters could be obtained by the biomimetic route.44 These experiments pave the way for the programmed 3D supramolecular assembly of peptides to tailor nanoreactors for the synthesis of technologically relevant materials. For example, a 3D nanostructure self-assembled from leucine zipper sequences could template highly monodisperse silver nanoparticles as Ag-binding sulfide moieties were inserted onto the interior cavity (Fig. 2a).23 In another approach, bolaamphiphile peptides are assembled into nanoring structures whose cavity has high affinity for heavy metals, and these peptide nanorings are capable of growing Ga2O3 nanocrystals catalytically (Fig. 2b).45 The constraint size of the growth space of nanostructures along with the presence of high affinity groups, and a hydrophobic confinement are essential for the outstanding function of these ring-shaped nanoreactors.
Fig. 1.
(a) A proposed structure of the Cu nanocrystal–HG12 peptide complex on the template nanotube. The conformation change of peptides influences the nucleation and the growth rate to control the Cu nanoparticle domains on bionanotubes. (b) Cu nanocrystals grown on the bionanotube at pH 6; (top-left) TEM image. (top-center) Electron-diffraction pattern. (top-right) Size distribution. (inset) The TEM image in higher magnification. (bottom). (c) Cu nanocrystals grown on the bionanotube at pH 8; (bottom-left) TEM image. (bottom-center) Electron-diffraction pattern. (bottom-right) Size distribution. (Inset) The TEM image in higher magnification. (Scale bar = 100 nm.)
Fig. 2.
Peptide and protein nanoreactors; (a) Illustration of mono-disperse silver nanoparticle growth inside the cavity of a peptide polynanoreactor; (b) TEM image of peptide nanorings assembled from bolaamphiphile precursor grow β-Ga2O3 inside their cavity; (c) TEM image of silver nanoparticles synthesized inside the genetically engineered protein cage apoferritin; (d) TEM image and SAED pattern of highly crystalline ZnO nanoshells grown at room temperature by using the enzyme urease as a biocatalytic template for the reaction. (a,d reproduced with permission from ref. 23 and 50, copyright Wiley-VCH Verlag GmbH & Co. KGaA; b, c, reprinted with permission from ref. 45 and 24, copyright 2007, 2004 American Chemical Society).
While the programmed assembly of peptides to build nanoreactors is a challenging task, in eukaryotic organisms the protein ferritin naturally stores iron as crystallites in a cavity of 7 nm formed by the assembly of 24 subunits. This crystallization process in a monodisperse biotemplate has been used to synthesize several materials such as iron and cobalt oxide in uniform size inside apoferritin.46 Furthermore, the inner cavity could be genetically engineered to display peptide sequences optimized to grow desired materials, thus making apoferritin a versatile nanoreactor for the synthesis of nanocrystals of diverse origins (Fig. 2c).24 All peptide/protein-based nanoreactors introduced here are designed to have a catalytic cavity that has optimum physicochemical properties for the synthesis of the target materials. However, an emerging field in bionanotechnology is to apply enzymes that already possess superior catalytic activity as nanoreactors to control the crystal growth kinetics in benign conditions such as aqueous solution and room temperature. In this approach, the enzyme-solvent interface plays an important role in the nucleation of the material of interest. Moreover, the local production of a concentration gradient of precursors around the enzyme surface favors the growth of the crystal under kinetic control.47 This is a key feature of enzyme nanoreactors, which is a different mineralization mechanism as compared to previous examples based in the use of peptides. For example, the enzyme peroxidase can be a nanoreactor for the synthesis of water-soluble polyaniline, a conductive polymer.48 Silicateins, a class of enzymes with a central role in biomineralization processes, were harnessed to synthesize oxide semiconductors such as titanium dioxide at room temperature by catalyzing the sol–gel route to these materials.49 Similarly, urease was found to work as an ideal nanoreactor to synthesize ZnO nanoshells at room temperature by the exquisite pH modulation mechanism around the enzyme-solvent interface (Fig. 2d),50 and recently this enzyme could regulate another semiconductor crystallization process by templating monodisperse Ag2S nanocrystals in aqueous solution.51 In material sciences, the growth of defect-free crystals always remains as a challenge because the high temperature processing leads to aggregations and defects due to local thermal stresses. However, the biomimetic approaches could have better prospect on the reduction of defects because the crystalline growth can be processed in low temperature.52
IV. Sensors
While peptides and proteins have been applied to sensors as simply recognition units, peptides and proteins are playing more active roles as useful building blocks in the design of sensors. One of the best-known examples of such a device is the utilization of protein nanopores as nanosensors (Fig. 3).53–55 In nanopore-based sensors, ionic current blockade occurs as a single molecule is translocated though the channel protein, and this ionic current blockade contains information about the identity, concentration, structure and dynamics of the target molecule. This pre-based sensing unit has been extended to the biological protein pore α-hemolysin and other porins since these protein pores present a ca. 1 nm diameter that makes them ideal candidates for the detection of single-stranded DNA. Indeed, it has been shown that ss-DNA could be detected label-free in a sequence-specific manner via translocation in nanopores, and this approach shows great promise for the development of new economical DNA sequencing devices.55,56
Fig. 3.
Protein nanopore sensors detect a current decay as single stranded DNA molecules are translocated trough the pore. (reprinted by permission from Macmillan Publishers Ltd: Nature Biotech., ref. 55, copyright 2008).
In another approach, self-assembly of peptides can be used to trigger an optical signal for biosensing. For example, nanoparticle enzyme sensors can be generated by immobilizing a peptide sequence that is the substrate of an enzyme on gold nanoparticles, and the enzyme activity induces the assembly or the disassembly of the nanoparticles, thus resulting in a variation in the gold plasmon resonance that yields a color change of the solution.57 Nanoparticle-based sensor systems containing biotinylated ATP molecules were also demonstrated to detect target peptides as the kinase reaction induced by the peptide could drive the crosslinking of gold nanoparticles functionalized with the peptide substrate and streptavidin.58 A similar procedure has also been applied to the detection of phosphatases.59 To detect proteases, the peptide-modified nanoparticles were first self-assembled into aggregates that yielded blue emission, and the proteolytic action of the enzyme could break up the aggregates to change the color of the solution to red.60 Similarly, gold nanoparticles were functionalized with peptides with high affinity for heavy metal and the presence of the metal ions could aggregate these nanoparticles to induce changes in the intensity and position of the plasmon absorbance peak, depending on the types of ions.61
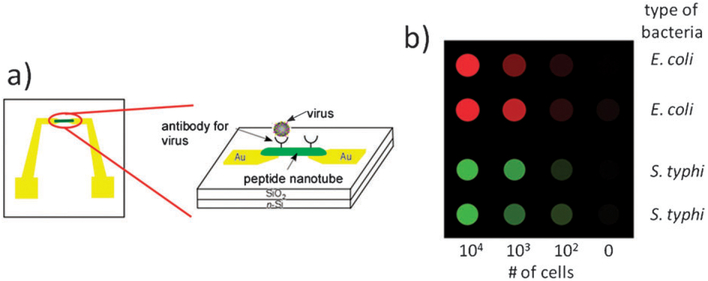
While the integration of peptides and proteins with optical probes is one of the most popular systems in biomolecular sensor applications, the integration of peptides and their derivatives in electrochemical sensors is emerging as a new sensor format. For example, conductive peptide nanotubes immobilized on electrodes could be used for the electrochemical detection of redox species, similar to the inorganic nanotube-based sensing systems.62 This amperometric biosensor could detect glucose or ethanol by monitoring the concentration of either hydrogen peroxide or NADH generated by glucose oxidase and ethanol dehydrogenase, respectively. Electrodes modified with nanofibers assembled from ionic complementary peptides could also improve the biocompatibility of the electrode surface without compromising the electrochemical transduction.63 In another approach, non-conductive peptide nanotubes were modified with antibodies to develop sensitive sensors for viruses (Fig. 4a).31 For this sensor, the antibody nanotubes were directed to the gap between two electrodes by positive dielectrophoresis, which enables large-scale assembly of various nanotubes on the platform for multiplexed sensing. For the transduction mechanism of this sensor, the interaction between the antibody and the low permittivity virus results in an increase of the impedance at the gap between the electrodes at high frequency and the binding event between the nanotube and virus can be monitored by measuring the variation of the capacitance of the solution. Recently this concept has been extended to the multiplexed detection of bacteria with a fully reusable pathogen biochip.64 In this approach an array of 36 pairs of interdigitated transducers was used as the detection platform. On each transducer, peptide nanotubes agglutinate bacteria, and the increased mass of the nanotube-pathogen adducts results in their fast precipitation, therefore inducing a fast increase of the impedance with time. For the reusability, the nanotube transducers were not firmly bound on the electrodes so that the residual bacteria could be washed off with the nanotubes. The prototype of this peptide nanotube multisensor could detect E. coli and S. typhimurium cells in the range from 102 to 104 cells, and the chip could be reused many times without tedious cleaning protocols with harsh chemicals (Fig. 4b).64
Fig. 4.
Impedimetric pathogen biosensors assembled from peptide nanotubes; (a) antibody modified nanotubes concentrate virus at the gap between two electrodes and the impedance at high frequency increases; (b) peptide nanotube biochips for the multiplexed detection of bacterial cells via agglutination on an array of impedimetric transducers.
V. Electronics
Downscaling the size of circuit components is a key factor for the development of more powerful electronic devices. Although biological-inorganic assemblies were considered to be inadequate for the fabrication of electronic devices, a growing number of examples indicate that biomolecules can be practical building blocks for electrical circuits. One example is to use metallo-proteins from natural sources as molecular wires that can mediate efficient electron transfers by tuning their redox energy levels with external electric fields. (Fig. 5)65 For example, the protein azurin from P. aeruginosa is capable of transfer electrons from cytochrome C551 to nitrite reductase with an unusually high redox potential at 628 nm absorption.66 By assembling azurin proteins between source and drain electrodes as grafted monolayers and depositing a silver gate on the back of p-type Si substrate in the FET configuration with electron-beam lithography, diode-like or transistor-like devices could be fabricated in two- and three-electrode settings.67,68 The redox sites were correctly oriented to allow the electron hopping along these monolayers, and then electrons could be transported between donor (reduced CuI protein) and acceptor (adjacent oxidized CuII protein). In this device configuration, the gate voltage, Vg, controls the redox state of azurin (e.g., higher Vg, more reduction) and the bias between source and drain is the in-plane driving source for the intermolecular electron transfer rate.65 As Vg was adjusted as the numbers of reduced proteins and oxidized proteins were present in equal amounts, the electron transfer was observed to become maximal.
Fig. 5.
Transport mechanism of the protein FET. The monolayer of the blue-copper protein azurin, containing the central Cu ion as redox site, connecting two arrow-shaped Cr/Au electrodes on a SiO2 substrate. An Ag back-electrode acts as the gate. The transport is based on sequential electron hopping between one reduced azurin (blue copper ion in the inset) to an adjacent oxidized one (red ion in the inset). The vertical gate can influence the oxidation state of the redox site to induce the field effect. (reproduced with permission from ref. 65, copyright Wiley-VCH Verlag GmbH & Co. KGaA).
Another example is to incorporate metal-protein complexes in the metal-oxide semiconductor (MOS) device configuration by assembling proteins onto targeted areas on electric substrates. It should be noted that floating gates assembled from nanoparticles have a significant advantage in smaller operating voltages, better endurance, and faster writing speeds by reducing charge loss however, for this improved nanoparticle-based floating gate memory these nanoparticles need to have precise size, shape, in-plane density in the MOS layers.69 Therefore, a protein cage, ferritin, is the perfect template to generate this type of nanoparticle-assembled memory. Recently, CoO3 nanoparticles, p-type wide band-gap semiconductors, were synthesized in apoferritin and embedded in the MOS stacked structure for charge storage nodes of floating gate memory (Fig. 6).70 The tunneling SiO2 layer above the MOS-FET channel was coated by positively-charged poly(ethyleneimine) (PEI) and negatively-charged ferritin was immobilized on the PEI surface. After protein shells were removed by sintering, the resulting floating gate memory could be operated up to 1 × 105 cycles of program/erase operation and the charges were stored for a practical length of time.69 This excellent charge capacity, long charge retention, and stress resistance are due to the superb charge confinement to the metal–Co potential well.71 Instead of ferritin protein, a mutated ring protein, tryptophan attenuation protein (TRAP), could also be used as building blocks for MOS capacitor after Au nanoparticles were incorporated in the cavity of the protein.72 In this fabrication, the targeted immobilization of proteins on Ti surface was controlled by Ti-binding peptides displayed on the TRAP via mutation. These nanoparticles were assembled between gates by peptide recognition toward Ti. The binding of biological molecules could be even more specificity for ligands and recently an even more precise assembly method of nanoparticles and nanowires was proposed by using antibody-protein recognition.73,74 In this biomimetic assembly methodology, multiple nanowires, incorporating various antibodies, were selectively immobilized on different locations on electric substrates where areas were labeled by complementary proteins. When the concentrations of proteins on the substrate were optimized, complex electric device geometries could be fabricated with high yields (Fig. 7).75,76
Fig. 6.
Assembly of ferritin-Co3O4 composites between source and drain electrodes (left) and the cross sectional TEM of the device showing Co3O4 nanoparticles embedded in the control SiO2 layer after proteins are sintered (right). (Reproduced by permission of The Royal Society of Chemistry).
Fig. 7.
Scheme to assemble two different antibody nanotubes, anti-mouse IgG-coated nanotube and anti-human IgG-coated nanotube, into the cross-bar geometry by biomolecular recognition (left). AFM image of the two antibody nanotubes assembled in the cross-bar geometry (right). Scale bar = 200 nm.
Previous two examples are based on 2D assembly of peptide/protein nanostructures. The 3D assembly such as vertical growth of peptide nanotubes will further broaden their electronic applications to field emission, solar cells, and batteries. Recently, peptide nanotubes were assembled into vertical forests from diphenylalanine peptide monomers by solvent evaporation and vapor deposition techniques, respectively. For the solvent evaporation technique the rapid evaporation of highly volatile fluorinated alcohol solvent induces a super-saturation state of diphenylalanine peptides near substrate so that these peptide monomers nucleate and stack vertically from substrate to the liquid–air interface to produce 3D peptide nanotube arrays in high density77 while for the vapor deposition technique low molecular weight peptides are vaporized and these gaseous peptides are deposited on a cooled substrate (Fig. 8a).78 By this method peptide nanotubes in the diameter of 50–300 nm were assembled with a density of 4 × 108 nanotubes/cm2 and their dimensions such as the nanotube length could be controlled up to 40 μm by the deposition time (Fig. 8b). For the application of ultracapacitor, these peptide nanotubes were assembled vertically on carbon electrodes and the resulting peptide nanotube-modified electrodes showed higher double-layer capacitance as compared to the same electrode configuration but modified by vertically-aligned carbon nanotubes. The peptide nanotube forest was also applied as super-hydrophobic coating on glass substrates and their contact angle could reach to 140° as the longer peptides with the larger number of phenylalanine residues were used for the vertical assembly.
Fig. 8.
(a) Schematic of the peptide vapor deposition technique. Low-molecular weight diphenylalanine peptides are vaporized from a bottom substrate heated at 220 °C in the form of a cyclic structure and deposited on the upper substrate at 80 °C to form ordered vertically aligned peptide nanotubes. (b) Side view SEM image of vertically aligned peptide nanotubes on a carbon substrate. (reprinted by permission from Macmillan Publishers Ltd: Nature Nanotech., ref. 78, copyright 2009).
VI. Stimulus responsive materials
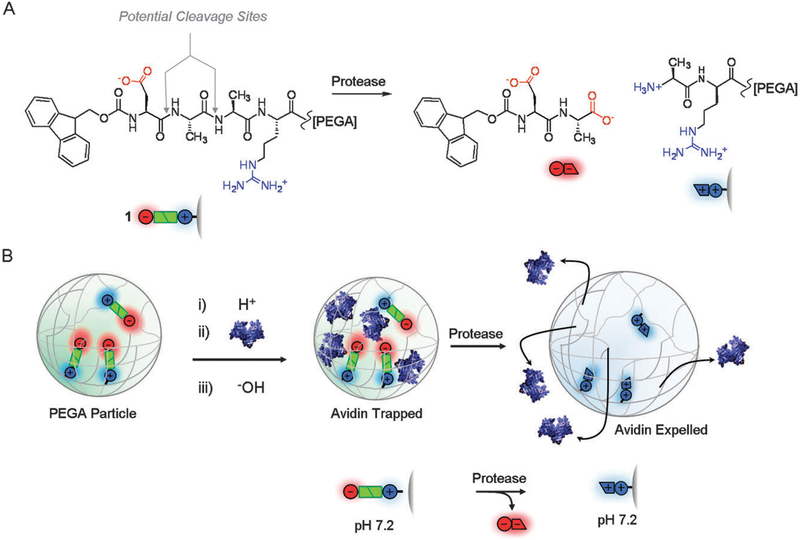
Stimulus responsive nanomaterials are defined as solid structures that undergo the change with external stimulus such as pH, ionic strength, temperature, electric/magnetic fields, and photon. To trigger this function by peptides, the use of their conformation change is the most popular approach. The basic idea for this approach is that peptides incorporated in nanomaterials assemblies or matrices alter the spacing or the volume by swelling or shrinking induced by the conformation change. For this reason, some people called this peptide as a peptide actuator. For example, elastin-like protein (ELP) aggregates can be swelled by ligand binding of proteins or peptides that isothermally trigger the phase transition of the appended ELPs (Fig. 9).79 The amino hydrogel particles functionalized by the peptide actuator could also induce swelling and release incorporated proteins as enzymes cleave these peptides and the charge–charge repulsion of cationic peptide fragments increases the particle volume (Fig. 10).80 The peptide conformation change with pH was also applied in material synthesis where the domain size and the crystalline structure of semiconductors on substrates can be controlled since the charge and hydrophobic distribution on surfaces is sensitively tuned by the peptide conformation via the actuation process (Fig. 1).42 However, in nature, this function is already developed with more complexity such as camouflaging. For example, cephalopods, Hawaiian bobtail squid, can manipulate the color of skin by the external stimulus of photon.81 The photonic structure of the skin consists of multilayer stacks of refractive tissues, reflectin that function as diffraction grading (Fig. 11(a) and (b)).82,83 This squid alternates the refractive index dielectrics of skin by changing the spacing between high refractive index platelets or the thickness of the platelets, which ultimately change the light interference to control the absorption and the reflection. The hurdle for the application of these reflectin proteins to biomimetic devices is to process them into films, which are insoluble in most organic solvents. This barrier was overcome by using an ionic liquid solvent to cast thin films with a flow-coating technique.84 Recently, this function was further developed to simpler biopolymer-based responsive photonic crystals and full color pixels (Fig. 11(c)).85,86 In this system, the hydrophilic layer of block-copolymers swells by the contact with water and the spacing of glassy hydrophobic layers change the index of refraction. This changing of the periodicity of the biomimetic structure displays tunability form the ultraviolet-visible to near-infrared regions.85 Electrochemical stimulus was also applied to induce rapid change in color of biopolymer pixels across the visible spectrum.86 Many organisms triggers color changes by chemical secretion by the sympathetic nerve system, and the application of electrochemical reactions for changes of refractive index is truly biomimetic.
Fig. 9.
Schematic representation of ELP fusion protein actuators. When apoCaM (a) binds Ca2+, the attached ELPs are assembled into meso–microscale particles (b). Chelation of the bound Ca2+ by EDTA reverses the LCST transition to the apoCaM–ELP monomers. Legend: CaM, cyan; ELPs, orange; Ca2+, gray. (reprinted with permission from ref. 79, copyright 2008 American Chemical Society).
Fig. 10.
(A) The actuator peptide, consisting of negative and positive residues, is designed to release positively charged parts by enzymatic reactions. (B) The scheme of the actuation motion. The positive charges generated by enzymatic cleavage of the bond between alanine residues swells protein particles via charge–charge repulsion. The enzymatic cleavage also leads to diffuse cationic proteins through the polymer pores as payload release. (reproduced by permission of The Royal Society of Chemistry).
Fig. 11.
(a) Squid, Loligo pealeii, adjusts the spacing and thickness of multilayers of proteins to control refractive indexes. (b) TEM image of the squid iridophores, protein refractive platelets. Scale bar = 1 μm. (c) Reflectin-mimetic biopolymer platelets that change the spacing by electrochemical reactions (d3 < d2 < d1). (a, b, reproduced with kind permission from Springer Science; c, reproduced with permission from ref. 86, copyright Wiley-VCH Verlag GmbH & Co. KGaA).
VII. Summary
Peptides and proteins can be designed to yield functional assemblies with molecular recognition. Molecular recognition can be used to assemble more complex structures as compared to traditional molecular self-assembly or it can bind them onto locations where complementary ligands are present. As external stimulus changes their conformation, their recognition function is modified and generates new recognition patterns, which can be applied as smart responsive materials and flexible sensor systems. In fact, peptides often have combined functions, which make them exciting building blocks for novel applications. Here we only introduced their applications in nanoreactors, sensors, electronics, and stimulus-responsive materials, however their broader applications are expected to be explored in near future. The high reproducibility of their assemblies and functionalities are very advantageous to their practical developments.
Acknowledgements
This research was supported by the U.S. Department of Energy, Office of Basic Energy Sciences, Division of Materials Sciences and Engineering under Award No. DE-FG-02–01ER45935. Hunter College infrastructure is supported by the National Institutes of Health, the RCMI program (G12-RR003037–245476), and NSF MRI shared instrument grant (ID. 0521709). R.R. acknowledges a postdoctoral fellowship from the Spanish Ministerio de Innovación y Ciencia and Fundación Española para la Ciencia y la Tecnología.
Biographies
Roberto de la Rica

After completing his PhD in the Institute of Microelectronics of Barcelona, Spain in the field of biosensors, Dr de la Rica joined Prof. Matsui’s lab as a research associate. There he developed new hybrid technologies combining nanolithography and bio-inspired concepts for semiconductor fabrication and nanosensor development. He is currently working in supramolecular biomimetics in the Supra-molecular Chemistry and Technology Group at the MESA + Institute, University of Twente.
Hiroshi Matsui

Hiroshi Matsui received his MS in Materials Science and Engineering from Stanford University and PhD in Chemistry from Purdue University in 1996 after completion of his BS in Chemistry at Sophia University. He worked at Columbia University as a postdoctoral associate in Chemistry Department for two years, and currently he is a Professor in Chemistry Department at City University of New York—Hunter College. His research focuses on self-assemblies of biological molecules and their applications in biosensors, photovoltaics, medical imaging, tissue engineering, genetic engineering and cancer research.
Footnotes
Part of the peptide- and protein-based materials themed issue.
References
- 1.Cheng MMC, Cuda G, Bunimovich YL, Gaspari M, Heath JR, Hill HD, Mirkin CA, Nijdam AJ, Terracciano R, Thundat T and Ferrari M, Curr. Opin. Chem. Biol, 2006, 10, 11. [DOI] [PubMed] [Google Scholar]
- 2.Lu W and Lieber CM, Nat. Mater, 2007, 6, 841. [DOI] [PubMed] [Google Scholar]
- 3.Sun YG and Rogers JA, Adv. Mater, 2007, 19, 1897. [Google Scholar]
- 4.Aizenberg J, Weaver JC, Thanawala MS, Sundar VC, Morse DE and Fratzl P, Science, 2005, 309, 275. [DOI] [PubMed] [Google Scholar]
- 5.Aizenberg J, Muller DA, Grazul JL and Hamann DR, Science, 2003, 299, 1205. [DOI] [PubMed] [Google Scholar]
- 6.Meldrum FC and Colfen H, Chem. Rev, 2008, 108, 4332. [DOI] [PubMed] [Google Scholar]
- 7.Losic D, Mitchell JG and Voelcker NH, Adv. Mater, 2009, 21, 2947. [Google Scholar]
- 8.Sleytr UB, Huber C, Ilk N, Pum D, Schuster B and Egelseer EM, FEMS Microbiol. Lett, 2007, 267, 131. [DOI] [PubMed] [Google Scholar]
- 9.Tang JL, Ebner A, Badelt-Lichtblau H, Vollenkle C, Rankl C, Kraxberger B, Leitner M, Wildling L, Gruber HJ, Sleytr UB, Ilk N and Hinterdorfer P, Nano Lett, 2008, 8, 4312. [DOI] [PubMed] [Google Scholar]
- 10.Gao X and Matsui H, Adv. Mater, 2005, 17, 2037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dickerson MB, Sandhage KH and Naik RR, Chem. Rev, 2008, 108, 4935. [DOI] [PubMed] [Google Scholar]
- 12.Gazit E, FEBS J, 2007, 274, 317. [DOI] [PubMed] [Google Scholar]
- 13.Lin C, Liu Y and Yan H, Biochemistry, 2009, 48, 1663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nassel DR, Invertebrate Neuroscience, 2009, 9, 57. [DOI] [PubMed] [Google Scholar]
- 15.Prato M, Kostarelos K and Bianco A, Acc. Chem. Res, 2008, 41, 60. [DOI] [PubMed] [Google Scholar]
- 16.Jain KK, in The Role of Nanobiotechnology in Drug Discovery, Berlin, 2009. [DOI] [PubMed] [Google Scholar]
- 17.Mart RJ, Osborne RD, Stevens MM and Ulijn RV, Soft Matter, 2006, 2, 822. [DOI] [PubMed] [Google Scholar]
- 18.Young M, Willits D, Uchida M and Douglas T, Annu. Rev. Phytopathol, 2008, 46, 361. [DOI] [PubMed] [Google Scholar]
- 19.Gao JH and Xu B, Nano Today, 2009, 4, 37. [Google Scholar]
- 20.Tamerler C and Sarikaya M, ACS Nano, 2009, 3, 1606. [DOI] [PubMed] [Google Scholar]
- 21.Zhao XJ and Zhang SG, Chem. Soc. Rev, 2006, 35, 1105.17057839 [Google Scholar]
- 22.Sarikaya M, Tamerler C, Jen AKY, Schulten K and Baneyx F, Nat. Mater, 2003, 2, 577. [DOI] [PubMed] [Google Scholar]
- 23.Ryadnov MG, Angew. Chem., Int. Ed, 2007, 46, 969. [DOI] [PubMed] [Google Scholar]
- 24.Kramer RM, Li C, Carter DC, Stone MO and Naik RR, J. Am. Chem. Soc, 2004, 126, 13282. [DOI] [PubMed] [Google Scholar]
- 25.Bai H, Xu K, Xu Y and Matsui H, Angew. Chem., Int. Ed, 2007, 46, 3319. [DOI] [PubMed] [Google Scholar]
- 26.Ryadnov MG, Biochem. Soc. Trans, 2007, 35, 487. [DOI] [PubMed] [Google Scholar]
- 27.Sedman VL, Allen S, Chen XY, Roberts CJ and Tendler SJB, Langmuir, 2009, 25, 7256. [DOI] [PubMed] [Google Scholar]
- 28.Castillo J, Tanzi S, Dimaki M and Svendsen W, Electrophoresis, 2008, 29, 5026. [DOI] [PubMed] [Google Scholar]
- 29.Matsui H and Gologan B, J. Phys. Chem. B, 2000, 104, 3383. [Google Scholar]
- 30.Djalali R, Chen Y-F and Matsui H, J. Am. Chem. Soc, 2003, 125, 5873. [DOI] [PubMed] [Google Scholar]
- 31.de la Rica R, Mendoza E, Lechuga M and Matsui H, Angew. Chem., Int. Ed, 2008, 47, 9752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Burazerovic S, Gradinaru J, Pierron J and Ward TR, Angew. Chem., Int. Ed, 2007, 46, 5510. [DOI] [PubMed] [Google Scholar]
- 33.Kaur P, Maeda Y, Mutter AC, Matsunaga T, Xu Y and Matsui H, submitted. [DOI] [PMC free article] [PubMed]
- 34.Ballister ER, Lai AH, Zuckermann RN, Cheng Y and Mougous JD, Proc. Natl. Acad. Sci. U. S. A, 2008, 105, 3733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Belcher AM, Wu XH, Christensen RJ, Hansma PK, Stucky GD and Morse DE, Nature, 1996, 381, 56. [Google Scholar]
- 36.Tsuji T, Onuma K, Yamamoto A, Iijima M and Shiba K, Proc. Natl. Acad. Sci. U. S. A, 2008, 105, 16866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tamerler C and Sarikaya M, Philos. Trans. R. Soc. London, Ser. A, 2009, 367, 1705. [DOI] [PubMed] [Google Scholar]
- 38.Dickerson MB, Naik RR, Stone MO, Cai Y and Sandhage KH, Chem. Commun, 2004, 1776. [DOI] [PubMed] [Google Scholar]
- 39.Sewell SL and Wright DW, Chem. Mater, 2006, 18, 3108. [Google Scholar]
- 40.Yu LT, Banerjee IA and Matsui H, J. Am. Chem. Soc, 2003, 125, 14837. [DOI] [PubMed] [Google Scholar]
- 41.Oren EE, Tamerler C and Sarikaya M, Nano Lett, 2005, 5, 415. [DOI] [PubMed] [Google Scholar]
- 42.Banerjee IA, Yu LT and Matsui H, Proc. Natl. Acad. Sci. U. S. A, 2003, 100, 14678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Naik RR, Stringer SJ, Agarwal G, Jones SE and Stone MO, Nat. Mater, 2002, 1, 169. [DOI] [PubMed] [Google Scholar]
- 44.Pat. Br., 11/786,190, 2007; US Pat., 0009427, 2010.
- 45.Lee S-Y, Gao X and Matsui H, J. Am. Chem. Soc, 2007, 129, 2954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hosein HA, Strongin DR, Allen M and Douglas T, Langmuir, 2004, 20, 10283. [DOI] [PubMed] [Google Scholar]
- 47.Kisailus D, Schwenzer B, Gomm J, Weaver JC and Morse DE, J. Am. Chem. Soc, 2006, 128, 10276. [DOI] [PubMed] [Google Scholar]
- 48.Liu W, Kumar J, Tripathy S, Senecal KJ and Samuelson L, J. Am. Chem. Soc, 1999, 121, 71. [Google Scholar]
- 49.Brutchey RL and Morse DE, Chem. Rev, 2008, 108, 4915. [DOI] [PubMed] [Google Scholar]
- 50.de la Rica R and Matsui H, Angew. Chem., Int. Ed, 2008, 47, 5415. [DOI] [PubMed] [Google Scholar]
- 51.Pejoux C, de la Rica R and Matsui H, Small, 2010, 6, 999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Nuraje N, Su K, Haboosheh A, Samson J, Manning EP, Yang N-L and Matsui H, Adv. Mater, 2006, 18, 807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Butler TZ, Pavlenok M, Derrington IM, Niederweis M and Gundlach JH, Proc. Natl. Acad. Sci. U. S. A, 2008, 105, 20647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Meller A, Nivon L, Brandin E, Golovchenko J and Branton D, Proc. Natl. Acad. Sci. U. S. A, 2000, 97, 1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Branton D, Deamer DW, Marziali A, Bayley H, Benner SA, Butler T, Di Ventra M, Garaj S, Hibbs A, Huang XH, Jovanovich SB, Krstic PS, Lindsay S, Ling XSS, Mastrangelo CH, Meller A, Oliver JS, Pershin YV, Ramsey JM, Riehn R, Soni GV, Tabard-Cossa V, Wanunu M, Wiggin M and Schloss JA, Nat. Biotechnol, 2008, 26, 1146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Clarke J, Wu HC, Jayasinghe L, Patel A, Reid S and Bayley H, Nat. Nanotechnol, 2009, 4, 265. [DOI] [PubMed] [Google Scholar]
- 57.Ghadiali JE and Stevens MM, Adv. Mater, 2008, 20, 4359. [Google Scholar]
- 58.Wang ZX, Levy R, Fernig DG and Brust M, J. Am. Chem. Soc, 2006, 128, 2214. [DOI] [PubMed] [Google Scholar]
- 59.Choi Y, Ho NH and Tung CH, Angew. Chem., Int. Ed, 2007, 46, 707. [DOI] [PubMed] [Google Scholar]
- 60.Laromaine A, Koh LL, Murugesan M, Ulijn RV and Stevens MM, J. Am. Chem. Soc, 2007, 129, 4156. [DOI] [PubMed] [Google Scholar]
- 61.Slocik JM, Zabinski JS, Phillips DM and Naik RR, Small, 2008, 4, 548. [DOI] [PubMed] [Google Scholar]
- 62.Yemini M, Reches M, Gazit E and Rishpon J, Anal. Chem, 2005, 77, 5155. [DOI] [PubMed] [Google Scholar]
- 63.Yang H, Fung SY, Pritzker M and Chen P, Langmuir, 2009, 25, 7773. [DOI] [PubMed] [Google Scholar]
- 64.de la Rica R, Pejoux C, Fernandez-Sanchez C, Baldi A and Matsui H, Small, 2010, 6, 1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Maruccio G, Biasco A, Visconti P, Bramanti A, Pompa PP, Calabi F, Cingolani R, Rinaldi R, Corni S, Di Felice R, Molinari E, Verbeet MR and Canters GW, Adv. Mater, 2005, 17, 816. [Google Scholar]
- 66.Vijgenboom E, Busch JE and Canters GW, Microbiology, 1997, 143, 2853. [DOI] [PubMed] [Google Scholar]
- 67.Maruccio G, Visconti P, D’Amico S, Calogiuri P, D’Amone E, Cingolani R and Rinaldi R, Microelectron. Eng, 2003, 67–68, 838. [Google Scholar]
- 68.Rinaldi R and Cingolani R, Phys. E, 2004, 21, 45. [Google Scholar]
- 69.Miura A, Uraoka Y, Fuyuki T, Yoshii S and Yamashita I, J. Appl. Phys, 2008, 103, 074503. [Google Scholar]
- 70.Yamashita I, J. Mater. Chem, 2008, 18, 3813. [Google Scholar]
- 71.Miura A, Tsukamoto R, Yoshii S, Yamashita I, Uraoka Y and Fuyuki T, Nanotechnology, 2008, 19, 255201. [DOI] [PubMed] [Google Scholar]
- 72.Heddle JG, Fujiwara I, Yamadaki H, Yoshii S, Nishio K, Addy C, Yamashita I and Tame JRH, Small, 2007, 3, 1950. [DOI] [PubMed] [Google Scholar]
- 73.Nuraje N, Banerjee IA, MacCuspie RI, Yu L and Matsui H, J. Am. Chem. Soc, 2004, 126, 8088. [DOI] [PubMed] [Google Scholar]
- 74.Zhao Z, Banerjee IA and Matsui H, J. Am. Chem. Soc, 2005, 127, 8930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Zhao Z and Matsui H, Small, 2007, 3, 1390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Yang L, Nuraje N, Bai H and Matsui H, J. Pept. Sci, 2008, 14, 203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Reches M and Gazit E, Nat. Nanotechnol, 2006, 1, 195. [DOI] [PubMed] [Google Scholar]
- 78.Adler-Abramovich L, Aronov D, Beker P, Yevnin M, Stempler S, Buzhansky L, Rosenman G and Gazit E, Nat. Nanotechnol, 2009, 4, 849. [DOI] [PubMed] [Google Scholar]
- 79.Kim B and Chilkoti A, J. Am. Chem. Soc, 2008, 130, 17867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Thornton PD, Mart RJ, Webb SJ and Ulijn RV, Soft Matter, 2008, 4, 821. [DOI] [PubMed] [Google Scholar]
- 81.M. L and Hanlon RT, Cell Tissue Res, 2007, 329, 179. [DOI] [PubMed] [Google Scholar]
- 82.Hanlon RT, Cooper KM, Budelmann BU and Pappas TC, Cell Tissue Res, 1990, 259, 3. [DOI] [PubMed] [Google Scholar]
- 83.Cooper KM, Hanlon RT and Budelmann BU, Cell Tissue Res, 1990, 259, 15. [DOI] [PubMed] [Google Scholar]
- 84.Kramer RM, Crookes-Goodson WJ and Naik RR, Nat. Mater, 2007, 6, 533. [DOI] [PubMed] [Google Scholar]
- 85.Kang Y, Walish JJ, Gorishnyy T and Thomas EL, Nat. Mater, 2007, 6, 957. [DOI] [PubMed] [Google Scholar]
- 86.Walish JJ, Kang Y, Mickiewicz RA and Thomas EL, Adv. Mater, 2009, 21, 3078. [Google Scholar]