The Putzig (Pzg) protein is associated with the NURF nucleosome remodeling complex, thereby promoting Notch target gene expression. Our findings suggest a novel Pzg-NURF complex that is responsible for the epigenetic regulation of Notch target genes.
Abstract
Drosophila putzig was identified as a member of the TRF2–DREF complex that is involved in core promoter selection. Additionally, putzig regulates Notch signaling, however independently of DREF. Here, we show that Putzig associates with the NURF complex. Loss of any NURF component including the NURF-specific subunit Nurf 301 impedes binding of Putzig to Notch target genes, suggesting that NURF recruits Putzig to these sites. Accordingly, Putzig can be copurified with any NURF member. Moreover, Nurf 301 mutants show reduced Notch target gene activity and enhance Notch mutant phenotypes. These data suggest a novel Putzig–NURF chromatin complex required for epigenetic activation of Notch targets.
INTRODUCTION
Putzig (Pzg) is a component of a large multiprotein complex that includes the TATA-box-binding-protein–related factor 2 (TRF2) and the DNA-replication related element (DRE) binding factor DREF (Hochheimer et al., 2002). The TRF2–DREF complex has been associated with the transcriptional regulation of replication-related genes that contain DREF binding sites (Hochheimer et al., 2002). Accordingly, Pzg acts as a positive regulator of cell cycle and replication-related genes (Hochheimer et al., 2002; Kugler and Nagel, 2007). In addition to this, we could show that Pzg is also required for Notch target gene activation in a DREF-independent manner (Kugler and Nagel, 2007). Presumably, Pzg functions at the level of chromatin activation, because the open chromatin structure typical of active Notch target genes is no longer detectable in a pzg mutant background (Kugler and Nagel, 2007).
The TRF2–DREF complex consists of more than a dozen of proteins and the biochemical function of most of them remains still elusive. Interestingly, it also contains three members of the nucleosome remodeling factor (NURF), imitation switch (ISWI), Nurf 55 and Nurf 38 (Hochheimer et al., 2002). NURF is a multisubunit complex that has been associated with chromatin activation and repression (Badenhorst et al., 2002, 2005; Kwon et al., 2008). NURF triggers nucleosome sliding thereby provoking changes in the dynamic properties of the chromatin (Tsukiyama and Wu, 1995; Hamiche et al., 1999; Kang et al., 2002; reviewed in Bouazoune and Brehm, 2006). The subunit ISWI is a member of the SWI2/SNF ATPase family and is thought to provide energy for nucleosome remodeling (Tsukiyama and Wu, 1995). Nurf 38 encodes an inorganic pyrophosphatase, which catalyzes the incorporation of nucleotides into a growing nucleic acid chain during transcription, replication, and DNA repair mechanisms (Gdula et al., 1998). Nurf 55 harbors WD-40 repeats, which allow interaction with other proteins and protein complexes (Neer and Smith, 1996; Martinez-Balbas et al., 1998). The fourth and largest subunit Nurf 301 is specific to the NURF complex, whereas all other members are shared with other chromatin modifying complexes (Xiao et al., 2001; Barak et al., 2003). Accordingly, Nurf 301 is not a component of the TRF2–DREF complex. Nurf 301 exhibits a number of protein motifs that typify transcription factors and other chromatin modifying proteins (Xiao et al., 2001). In addition, the N-terminal region of Nurf 301 shows homology to the DNA-binding protein HMGA (high mobility group A) implying that Nurf 301 mediates the contact with the DNA or provides a platform to recruit other transcription factors (Reeves and Beckerbauer, 2001; Xiao et al., 2001). In this context it has already been shown that Nurf 301 is required for the transcriptional activation for example of homeotic genes and notably of Ecdyson-receptor (EcR) and Wingless target genes (Badenhorst et al., 2002; Badenhorst et al., 2005; Song et al., 2009).
The DREF independence of Pzg during the activation of Notch target genes raised the possibility that it may instead involve the NURF complex for chromatin activation. Here, we provide evidence for a functional interplay between Pzg and the NURF complex with regard to Notch target gene activation. Coimmunoprecipitations revealed that Pzg is present in protein complexes containing the known NURF subunits. Moreover, Pzg binding on Notch target genes is neither detectable in mutants of the NURF-specific subunit Nurf301, nor in mutants affecting other subunits of NURF. In addition, Nurf301 is required for Notch target gene expression, which is impaired in Nurf301 mutant cell clones. Consistent with this, Nurf301 mutants enhance the Notch mutant wing phenotype, strongly arguing for an involvement of the NURF complex in Pzg-mediated epigenetic Notch target gene activation.
MATERIALS AND METHODS
Genetics, Fly Strains, and Documentation of Phenotypes
To generate Nurf301 mutant clones using the Flp/FRT system, the Nurf3012 null mutant allele was recombined with FRT80B and selected according to its gentamicin resistance and failure of complementation with other Nurf alleles. Flies of the genotype ywhsflp; Nurf3012FRT80B/FRT80BM(3)67cubi-GFP (Janody et al., 2004, gift of J. Treisman, NYU Medical Center, NY) were exposed to a heat shock (37°C for 30 min) for 48–72 h after egg laying (AEL), and wing imaginal discs were prepared 120–144 h after AEL.
The following genotypes (fly stocks) were used for the experiments: Gal4/UAS system: enGFP-Gal4 (Neufeld and Edgar, 1998), Omb-Gal4/FM7a (Lecuit et al., 1996), ptc-Gal4::UAS-pzg-RNAi (Kugler and Nagel, 2007), SD-Gal4 (Roy et al., 1997; gift of K. Irvine, Rutgers, The State University of New York), UAS-Nurf301-RNAi (VDRC 24740), UAS-Nurf55-RNAi (VDRC 26455), UAS-Nurf38-RNAi (VDRC 3200) all obtained from Vienna Stock Center, UAS-pzg-RNAi (Kugler and Nagel, 2007); mutants: Df(1)N5419/FM7c, N55e11 (gift of A. Preiss, University of Hohenheim, Germany), Iswi2/CyoGFP (Deuring et al., 2000; gift of P. Badenhorst, University of Birmingham, United Kingdom); Nurf381/CyO (BL12206); Nurf3012/TM6GFP (Badenhorst et al., 2005); and reporter lines: m8-lacZ (Lecourtois and Schweisguth, 1995) and vg-BElacZ (Kim et al., 1996). Generally, flies were raised at 25°C on standard fly food with the exception of the Nurf301-RNAi induction assays, which were performed at 25 and at 29°C.
Adult wings were embedded in Euparal (Roth, Karlsruhe, Germany), and at least 30 wings of each genotype were measured. Wing size was determined using ImageJ software for pixel measurements (http://rsb.info.nih.gov/ij/). The wing area was encircled with the Polygon tool. The Segmented Line tool was used to reconstruct the length of wing notches. To test statistical significance, p values were calculated according to Student's t test (http://www.physics.csbsju.edu/stats/t-test.html). Pictures were taken with Normarski optics on a Zeiss Axiophot (Carl Zeiss, Jena, Germany).
Immunhistochemistry, Immunoprecipitation, and Chromatin Immunoprecipitation
Wing imaginal discs were prepared from crawling third instar larvae of the respective genotype. The following antibodies were used: mouse anti-β-galactosidase (1:100; JIE7), mouse anti-Cut (1:20; 2B10), and mouse anti-Wg (1:25; 4D4), all obtained from the Developmental Studies Hybridoma Bank, University of Iowa (NICHD contract NO1-HD-7-3262), rabbit anti-cleaved caspase 3 (1:200, NEB Cell Signaling Technology, Beverly, MA), rat anti-Ci 2A1 at 1:2 dilutions (Motzny and Holmgren, 1995). Pictures of stained discs were taken with a Bio-Rad MRC1024 confocal microscope (Hercules, CA) on a Zeiss Axiophot.
Immunoprecipitations were performed according to Nagel et al. (2005) by using protein extracts from 100 first instar larvae. For precipitations we used guinea pig anti-Pzg antibodies (1:250; Kugler and Nagel, 2007) and rabbit anti-MOF antibodies (Matyunina et al., 2008; gift of John Lucchesi, Emory University, Atlanta, GA). For detection, rabbit anti-ISWI (Tsukiyama and Wu, 1995), rabbit anti-Nurf301 (Kwon et al., 2008), rabbit anti-Nurf55 (Martinez-Balbas et al., 1998), rabbit anti-Nurf38 (Gdula et al., 1998), and mouse anti-Notch intra (1:10; C17.9C6, DSHB) were used; all three Nurf antibodies were a gift from Hua Xiao (National Cancer Institute, Bethesda, MD).
Chromatin immunoprecipitations (ChIPs) were done with 300 first instar larvae each, using the ChIP Assay Kit from Upstate Biotechnology (Lake Placid, NY; Kugler and Nagel, 2007). The following primer sets were used: m8UP (5′-CAACAACCAAGGCGACCCGGCACGA) and m8LP (5′-TTTTTGAAAAATTTTGTATTCGGCTTGTTGCTG) for amplification of Enhancer of split m8 [E(spl)m8] 5′ regulatory region and vgdvUP (5′-GGGTGAATTCCGCAACTCAATGTTGGCTT) and vgdvLP (5′-TATTTGGATCCGTTTAACTTTAGGTTTCGGGACTGG) for amplification of the d/v regulatory region of the vestigial (vg) locus.
The following primer pairs for control reactions: Enhancer of split m8 (3′) control: m83primUP (5′-AACAAGGGGTTAAGTGGCAGGAAG GTAAGG) and m83primLP (5′-C-AGTGTTGCATGCGCCAGTGTTAGGA); roX-1: UP (5′-GTCGAATTCGAAAAACACATTTACTAACAAATAA-3′) and LP (5′-GTCGAATTCCCCAAAGAAATCCACATAACAT-3′); open reading frame (ORF) vg primer: vg ORF UP (5′-TTGTACTCCTCCTCGGGCGTC-3′) and vg ORF LP (5′-GTGTGTGCGAGTGCGGGTG-3′); and ORF m8 primer: m8 ORF UP (5′-CACCAAGACCCAGATCTACCAG-3′) and m8 ORF LP (5′-CGCTGTGATATCCGGAGGA-3′).
RESULTS
Pzg Does Not Directly Bind to the DNA of Notch Target Genes
We recently showed by cross-linking chromatin immunoprecipitation (XChIP) that Pzg is bound to the regulatory regions of Notch target genes, however, in a DREF-independent manner (Kugler and Nagel, 2007). Because pzg encodes a Zn-finger protein, we investigated at first whether Pzg binds directly to Notch target gene promoters. We performed electromobility shift analyses with the sequences precipitated by Pzg in the XChIP, however, found no hints for a direct DNA binding of Pzg to these sites (data not shown). From this we conclude that Pzg requires at least one further partner for binding to Notch target gene promoters.
Pzg's Presence at Notch Target Genes Depends on the Presence of the NURF Complex
Pzg depletion is correlated with a reduction of open chromatin structure at different Notch target genes, assuming an important role of Pzg in chromatin activation at these Notch target sites (Kugler and Nagel, 2007). Because Dref mutant cells do not show this effect, Pzg must influence Notch signaling in a TRF2–DREF-independent manner (Kugler and Nagel, 2007). Three proteins of the TRF2–DREF complex are also components of the nucleosome remodeling factor NURF that has been involved in chromatin activation (reviewed in Bouazoune and Brehm, 2006). Hence, Pzg might act together with NURF in the context of Notch target gene activation. To investigate a possible association of Pzg and NURF in more detail, we first performed XChIPs with anti-Pzg antibodies in mutants of all four components of the NURF complex. This includes the NURF-specific subunit Nurf 301, which is not part of the TRF2–DREF complex (Xiao et al., 2001; Hochheimer et al., 2002; Barak et al., 2003). Indeed, no Pzg protein was bound to the promoters of the Notch target genes vg and E(spl)m8 in the absence of ISWI, Nurf 38, Nurf 55, or Nurf 301, whereas it was present in the wild type at these sites (Figure 1). This demonstrates that the NURF complex as a whole is involved in the Pzg-mediated epigenetic activation of Notch target genes. To further confirm our conclusion we compared the binding of Pzg to the vg and E(spl)m8 loci on wild-type and Nurf301 mutant salivary gland chromosomes. Consistent with the data gained in the XChIP, binding of Pzg is impaired at these loci in the absence of Nurf301 in support of an important role for NURF in targeting Pzg to these sites (Supplemental Figure S1, A and B).
Figure 1.
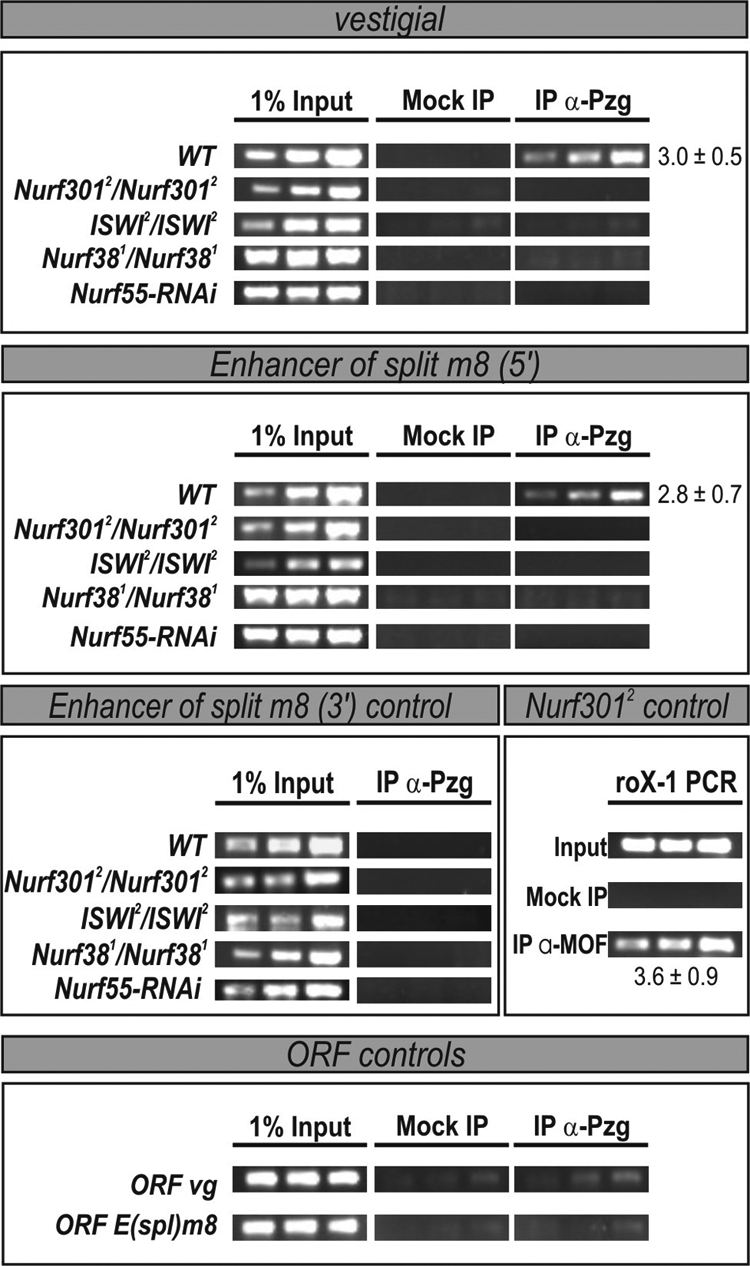
NURF complex is required for Pzg binding at Notch target genes. XChIP analyses were performed on chromatin isolated from either wild-type or Nurf3012, Iswi2, and Nurf381 homozygous first instar larvae (as indicated). In the case of Nurf55, da-Gal4::UAS Nurf55-RNAi first instars were used. Anti-Pzg antibodies as well as guinea pig preimmune sera (mock control) and anti-MOF antibodies (positive control) were used for precipitations. Sequences from the vestigial-boundary enhancer (vg) and the Enhancer-of-split m8 [E(spl)m8] promoter (m85′) were amplified. Samples of the 31st, 33rd and 35th amplification cycle are shown. Relative enrichment was estimated for the 33rd PCR cycle sample from the ratio between Pzg immunoprecipitations and mock controls. Mean values and standard deviations of at least three independent experiments were calculated. PCR-amplification of the E(spl)m8 3′UTR as well as a region in the ORF of vg and E(spl)m8 is shown as unrelated control. As a positive control MOF binding was monitored at the roX-1 locus in Nurf3012 mutant chromatin.
Pzg Associates with NURF Complex Members In Vivo.
The failure of Pzg to bind to Notch target genes in NURF mutants—notably in the absence of the NURF specific subunit Nurf301—suggested an association of Pzg with NURF itself. To this end, we tested whether Pzg coprecipitated with NURF complex members using protein extracts from first instar larvae and Pzg antibodies. As expected for members of the TRF2–DREF complex, the three NURF components ISWI, Nurf 38, and Nurf 55 were found in a complex with Pzg (Figure 2). However, we also found Nurf 301 coprecipitating with Pzg, indicating that Pzg associates with the NURF complex as well (Figure 2). These data provide evidence that Pzg is present in at least two different protein complexes, TRF2–DREF and NURF.
Figure 2.

Pzg is associated with the whole NURF complex. Proteins immunoprecipitated (IP) from wild-type larval protein extracts by anti-Putzig antibodies were probed for Nurf 301, ISWI, Nurf 55, and Nurf 38 as indicated. The input column shows 25% of the extract used for the IP. IPs with guinea pig preimmune sera served as negative control (mock). Specificity of the used α-Pzg antibody was shown with the intracellular domain of Notch, which could not be copurified.
NURF Promotes Expression of Notch Target Genes.
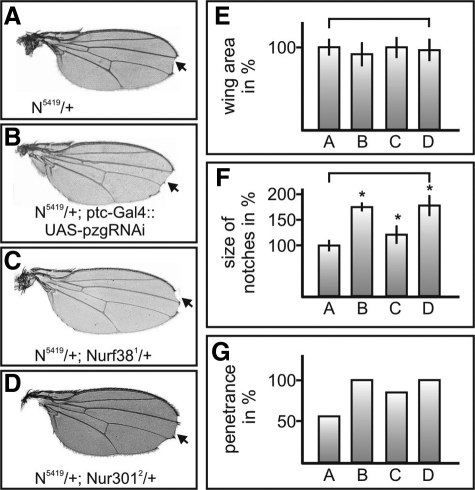
The observation that Pzg associates with the NURF complex suggests that the influence of Pzg on Notch target genes involves chromatin activation by NURF. To further investigate this possibility, we first looked for genetic interactions between Notch and the NURF complex. To this end, we used the sensitized genetic background of heterozygous N5419/+ mutant animals and analyzed the haplo-insufficient Notch mutant wing phenotype, which is characterized by wing notches and thickening of the third longitudinal vein (Figure 3A).
Figure 3.
NURF genetically interacts with Notch. (A–D) The Notch mutant wing phenotype (marked by arrows) is enhanced by pzg-RNAi as well as by one mutant allele of different NURF complex members including the large subunit Nurf301. The genotypes were as followed: (A) N5419/+, (B) N5419/+; ptc-Gal4::UAS-pzgRNAi, (C) N5419/+; Nurf381/+, and (D) N5419/+; Nurf3012/+. (E–G) The phenotypes from A through D were statistically analyzed. The labels A–D correspond to the genotypes shown in Figure 3, A–D. Mean values of at least 30 wings of each genotype were considered using N5419/+ as reference (100%). Error bars, SD. *p < 0.0005 by Student's t test; brackets indicate phenotypes compared. (E) Quantification of the wing sizes. The wing sizes of the tested combinations do not statistically differ. (F) The length of the notches varies between the different combinations. A strong increase can be observed when Nurf301 and pzg activity is reduced, whereas in combination with Nurf38 the effect is not as strong. As the overall size of the wings is comparable, changes in the size of the notches are not due to a modified wing size. (G) Penetrance of wing notches was determined for the respective genotypes. Both, Nurf381 and Nurf3012 mutants increased the penetrance of this phenotype.
Indeed, we noted an enhancement of the wing defects if the dose of either NURF gene, Nurf38, Nurf55, or Nurf301 was reduced (Figure 3, A–G). Notably, decreasing the activity of Nurf301 in a Nurf3012 heterozygous mutant background strongly increased both, the penetrance and severity of wing margin defects (Figure 3, A and D). Similar results were obtained using another N allele, N55e11, confirming specificity of the genetic interaction data (Supplemental Figure S1, C–G). Because Nurf 301 is obligatory in the assembly of the NURF complex (Xiao et al., 2001), the sensitivity of Notch toward the presence of Nurf 301 indicates that the NURF complex functions a positive regulator of Notch signaling in normal wing development (Figure 3, A–G).
To validate these genetic interaction results we analyzed also the expression of the well-defined Notch target genes cut (ct), Enhancer of split m8 [E(spl)m8], wingless (wg), and vestigial (vg) in a Nurf301 mutant background. In a first approach, we addressed this issue by using a Nurf301-RNAi line under UAS control, allowing us to down-regulate the activity of Nurf301 specifically in the wing anlagen. In addition, we took advantage of the temperature sensitivity of the system: strong Nurf301 reduction was induced by raising the flies at 29°C, whereas intermediate levels were obtained at 25°C. However, the expression levels of all four target genes tested were affected, to a different degree: The expression of Cut, E(spl)m8-lacZ and vg-lacZ was strongly reduced or even abolished in Nurf301 RNAi–treated cells at both temperature settings, whereas that of Wg was affected only at 29 but not at 25°C (Supplemental Figure S2). Because induction of RNAi reduces gene activity but does not necessarily abolish it, we wondered whether residual Nurf301 activity supported the expression of the tested genes notwithstanding the RNAi treatment. To test this, we looked at cells that completely lacked Nurf301 activity by inducing mutant cell clones using the null allele Nurf3012 (Badenhorst et al., 2005).
In a first attempt we failed to generate Nurf3012 mutant clones, most probably because these cells have a growth disadvantage. Therefore, we generated Nurf3012 mutant clones in a Minute mutant background, which allows growth of disadvantaged cells. Here, Nurf3012 mutant clones exhibit a conspicuous reduction of the respective Notch target gene expression (Figure 4, A–C″). To exclude cell death as artificial trigger of the observed effects, levels of activated caspase 3 were examined but were unchanged in the mutant cells (Figure 4, D–D″).
Figure 4.
Notch target gene activity is reduced in Nurf3012 mutant cell clones. (A–C″) Nurf3012 mutant clones in the wing imaginal disk marked by the absence of green fluorescent protein (GFP; outlined) reveal an autonomous reduction of the Notch target gene activity, including Cut (red in A and A″), vg-lacZ (red in B, B″), and Wg (red in C and C″). (D–D″) No increase of apoptosis, detected by activated Caspase 3, was observed in Nurf3012 mutant cells (loss of GFP) in comparison to the surrounding Minute GFP-positive cells. (E–E″) Nurf301 activity is not required for the expression of Ci (red in E and E″).
To ensure that the Nurf3012 mutation does not generally impede expression of developmentally regulated genes, we monitored Ci, which is an integral component of the Hedgehog signaling pathway known to be unaffected in a pzg-RNAi mutant background (Kugler and Nagel, 2007). As predicted, Nurf3012 mutant cells have no visible effect on the expression of Ci, ruling out a global requirement of Nurf for the transcription of developmentally regulated genes (Figure 4, E–E″).
Together, these results suggest that the NURF chromatin-remodelling complex is required for the activation of Notch target genes. To further confirm this conclusion, we analyzed the effects of a local down-regulation of Nurf38 and Nurf55 by RNAi on vg and on E(spl)m8 expression using the Gal4/UAS system. Again, a strong reduction of Notch target gene activity was observed in the affected cells (Supplemental Figure S3). Together, these data indicate that Pzg interacts with NURF to promote transcription of Notch target genes.
DISCUSSION
Our work shows that Pzg is associated with at least two different types of protein complexes that are involved in transcriptional activation: the TRF2–DREF complex and the NURF complex. Interestingly, these two complexes share several members apart from Pzg despite their different roles in core promoter selection versus nucleosome sliding and chromatin activation (Martinez-Balbas et al., 1998; Hochheimer et al., 2002; Narlikar et al., 2002). However, the specific role for Pzg in the promotion of Notch target gene transcription involves NURF and not the TRF2–DREF complex. Notably, NURF also promotes efficient expression of a subset of Wingless target genes (Song et al., 2009). In this case, a direct interaction between ISWI and Armadillo, the major transcriptional coactivator of Wingless targets, was shown. We have no indication however, that pzg is involved in the regulation of wg, suggesting that the NURF complex recruits Pzg only onto specific promotors. Furthermore, the NURF subunit Nurf 301 contacts the Ecdysone receptor (EcR), thereby modulating the activity of ecdysone signaling during the larval and pupal stages of Drosophila development (Badenhorst et al., 2005). How is NURF recruited to Notch target sites? Notch target gene activation involves a ternary complex containing the DNA-binding protein Suppressor of Hairless [Su(H)], intracellular Notch, and Mastermind, plus other more general coactivators (reviewed in Bray, 2006; Kopan and Ilagan, 2009). We have no indication of a direct contact of Pzg to either Notch or Su(H), tested by coimmunoprecipitations as well as yeast two-hybrid assays (Figure 2 and data not shown). However, we cannot exclude contacts between the other components, notably Mastermind or ISWI. Mastermind has been shown to interact with several chromatin modifying proteins, for example, with the histone acetyltransferase p300 or with cyclin-dependent kinase 8 (Fryer et al., 2004; Hansson et al., 2009).
Several studies in Drosophila and vertebrates have shown that many Notch-responsive target genes are regulated by combinatorial signal inputs, which need the Notch ternary complex and additional cooperators bound to sites nearby. In contrast to cofactors within the transactivation complex, these other factors do not physically interact with the Notch ternary complex but instead synergize during transcriptional activation at Notch target gene promoters (e.g., Fu and Noll, 1997; Furriols and Bray, 2001; Lee et al., 2007). It is conceivable, that a Pzg-NURF complex is likewise needed in conjunction with the Notch transactivator complex for full Notch target gene expression.
Gene Regulation Complexes Share Different Subunits
It is well established, that chromatin modification complexes share several components. For example, ISWI is not only contained in NURF and TRF2–DREF complexes but also in chromatin-remodeling and assembly factor (CHRAC) and ATP-utilizing chromatin-remodeling and assembly factor (ACF) in Drosophila, where it serves to increase the accessibility of nucleosomal DNA (Hochheimer et al., 2002; reviewed in Narlikar et al., 2002). Nurf 55, also known as CAF-1, forms a stable complex with Drosophila Myb and E2F2/RBf and regulates the transcription of several developmentally important genes (Martinez-Balbas et al., 1998; Lewis et al., 2004). Like ISWI and Nurf 55, also Nurf 38 is present in the TRF2–DREF complex (Hochheimer et al., 2002). Pzg is contained within the TRF2–DREF and within the NURF complex serving the activation of proliferation related genes and N target genes, respectively (Hochheimer et al., 2002; Kugler and Nagel, 2007). Not all NURF complexes, however, require pzg, for example, as during the activation of Wg target genes. Sharing components raises the question, how specificity of the different complexes is achieved. Obviously, specificity is mediated either by unique subunits or by certain combinations of shared subunits. These subunits may specifically modulate the activity of the ATPase subunit or, more likely, may help to target the remodeling complexes to particular promoters. Two members of the NURF complex, ISWI and Nurf 301, have been shown to directly target transcription factors (Badenhorst et al., 2005; Song et al., 2009). It is tempting to speculate, that Pzg might be a specific cofactor needed to realize some of the operation spectrum of NURF, notably during the epigenetic regulation of Notch target genes.
Supplementary Material
ACKNOWLEDGMENTS
We are indebted to I. Wech for her technical assistance. We thank the Bloomington Stock Center (Indiana University), Developmental Studies Hybridoma Bank (University of Iowa), P. Badenhorst, J. Lucchesi, H. Xiao, and J. Treisman for fly stocks and antibodies. We thank A. Preiss for invaluable suggestions and discussions and for critical reading of the manuscript. This work was supported by a Deutsche Forschungsgemeinschaft grant NA 427/2-1 to A.C.N.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E10-03-0212) on August 4, 2010.
REFERENCES
- Badenhorst P., Voas M., Rebay I., Wu C. Biological functions of the ISWI chromatin remodeling complex NURF. Genes Dev. 2002;16:3186–3198. doi: 10.1101/gad.1032202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badenhorst P., Xiao H., Cherbas L., Kwon S. Y., Voas M., Rebay I., Cherbas P., Wu C. The Drosophila nucleosome remodeling factor NURF is required for Ecdysteroid signaling and metamorphosis. Genes Dev. 2005;19:2540–2545. doi: 10.1101/gad.1342605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barak O., Lazzaro M. A., Lane W. S., Speicher D. W., Picketts D. J., Shiekhattar R. Isolation of the human NURF: a regulator of engrailed expression. EMBO J. 2003;22:6089–6100. doi: 10.1093/emboj/cdg582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouazoune K., Brehm A. ATP-dependent chromatin remodeling complexes in Drosophila. Chromosome Res. 2006;14:433–449. doi: 10.1007/s10577-006-1067-0. [DOI] [PubMed] [Google Scholar]
- Bray S. J. Notch signalling: a simple pathway becomes complex. Nat. Rev. Mol. Cell Biol. 2006;7:678–689. doi: 10.1038/nrm2009. [DOI] [PubMed] [Google Scholar]
- Deuring R., et al. The ISWI chromatin-remodeling protein is required for gene expression and the maintenance of high order chromatin structure in vivo. Mol. Cell. 2000;5:355–365. doi: 10.1016/s1097-2765(00)80430-x. [DOI] [PubMed] [Google Scholar]
- Fryer C. H., White J. B., Jones K. A. Mastermind recruits CycD: CDK8 to phosphorylate the Notch ICD and coordinate activation with turnover. Mol. Cell. 2004;16:509–520. doi: 10.1016/j.molcel.2004.10.014. [DOI] [PubMed] [Google Scholar]
- Fu W., Noll M. The Pax2 homolog sparkling is required for development of cone and pigment cells in the Drosophila eye. Genes Dev. 1997;11:2066–2078. doi: 10.1101/gad.11.16.2066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furriols M., Bray S. A model Notch response element detects Suppressor of Hairless-dependent molecular switch. Curr. Biol. 2001;11:60–64. doi: 10.1016/s0960-9822(00)00044-0. [DOI] [PubMed] [Google Scholar]
- Gdula D. A., Sandaltzopoulos R., Tsukiyama T., Ossipow V., Wu C. Inorganic pyrophosphatase is a component of the Drosophila nucleosome remodeling factor complex. Genes Dev. 1998;12:3206–3216. doi: 10.1101/gad.12.20.3206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamiche A., Sandaltzopoulos R., Gdula D. A., Wu C. ATP-dependent histone octamer sliding mediated by the chromatin remodeling complex NURF. Cell. 1999;97:833–842. doi: 10.1016/s0092-8674(00)80796-5. [DOI] [PubMed] [Google Scholar]
- Hansson M. L., Popko-Scibor A. E., Saint Just Ribeiro M., Dancy B. M., Lindberg M. J., Cole P. A., Wallberg A. E. The transcriptional coactivator MAM1 regulates p300 autoacetylation and HAT activity. Nucleic Acids Res. 2009;37:2996–3006. doi: 10.1093/nar/gkp163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hochheimer A., Zhou S., Zheng S., Holmes M. C., Tjian R. TRF2 associates with DREF and directs promoter-selective gene expression in Drosophila. Nature. 2002;420:439–445. doi: 10.1038/nature01167. [DOI] [PubMed] [Google Scholar]
- Janody F., Lee J. D., Jahren N., Hazelett D. J., Benlali A., Miura G. I., Draskovic I., Treisman J. E. A mosaic genetic screen reveals distinct roles for trithorax and polycomb group genes in Drosophila eye development. Genetics. 2004;166:187–200. doi: 10.1534/genetics.166.1.187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang J. G., Hamiche A., Wu C. Gal4 directs nucleosome sliding induced by NURF. EMBO J. 2002;21:1406–1413. doi: 10.1093/emboj/21.6.1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J., Sebring A., Esch J. J., Kraus M. E., Vorwerk K., Magee J., Carroll S. B. Integration of positional signals and regulation of wing formation and identity by Drosophila vestigial gene. Nature. 1996;382:133–138. doi: 10.1038/382133a0. [DOI] [PubMed] [Google Scholar]
- Kopan R., Ilagan M. X. The canonical Notch signaling pathway: unfolding the activation mechanism. Cell. 2009;137:216–233. doi: 10.1016/j.cell.2009.03.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kugler S. J., Nagel A. C. putzig is required for cell proliferation and regulates Notch activity in Drosophila. Mol. Biol. Cell. 2007;18:3733–3740. doi: 10.1091/mbc.E07-03-0263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon S. Y., Xiao H., Glover B. P., Tjian R., Wu C., Badenhorst P. The nuclesosome remodeling factor (NURF) regulates genes involved in Drosophila innate immunity. Dev. Biol. 2008;316:538–547. doi: 10.1016/j.ydbio.2008.01.033. [DOI] [PubMed] [Google Scholar]
- Lecourtois M., Schweisguth F. The neurogenic Suppressor of Hairless DNA-binding protein mediates the transcriptional activation of the Enhancer of split genes triggered by Notch signaling. Genes Dev. 1995;9:2598–2608. doi: 10.1101/gad.9.21.2598. [DOI] [PubMed] [Google Scholar]
- Lecuit T., Brook W. J., Ng M., Celleja M., Sun H., Cohen S. M. Two distinct mechanisms for long-range patterning by Decapentaplegic in the Drosophila wing. Nature. 1996;381:387–393. doi: 10.1038/381387a0. [DOI] [PubMed] [Google Scholar]
- Lee J., Basak J. M., Demehri S., Kopan R. Bi-compartmental communication contributes to the opposite behaviour of Notch1-deficient hair follicle and epidermal keratinocytes. Development. 2007;134:2795–2806. doi: 10.1242/dev.02868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis P. W., Beall E. L., Fleischer T. C., Georlette D., Link A. J., Botchan M. R. Identification of a Drosophila Myb-E2F2/RBF transcriptional repressor complex. Genes Dev. 2004;18:2929–2940. doi: 10.1101/gad.1255204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Balbas M. A., Tsukiyama T., Wisniewski J., Wu C. Drosophila NURF-55, a WD repeat protein involved in histone metabolism. Proc. Natl. Acad. Sci. USA. 1998;95:132–137. doi: 10.1073/pnas.95.1.132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matyunina L. V., Bowen N. J., McDonald J. F. LTR retrotransposons and the evolution of dosage compensation in Drosophila. BMC Mol. Biol. 2008;9:55. doi: 10.1186/1471-2199-9-55. Jun 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motzny C. K., Holmgren R. The Drosophila cubitus interruptus protein and its role in the wingless and hedgehog signal transduction pathways. Mech. Dev. 1995;52:137–150. doi: 10.1016/0925-4773(95)00397-j. [DOI] [PubMed] [Google Scholar]
- Nagel A. C., Krejci A., Tenin G., Bravo-Patino A., Bray S., Maier D., Preiss. A. Hairless mediated repression of Notch target genes requires combined activity of Groucho and CtBP co-repressors. Mol. Cell. Biol. 2005;25:10433–10441. doi: 10.1128/MCB.25.23.10433-10441.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narlikar G. J., Fan H-Y., Kingston R. E. Cooperation between complexes that regulate chromatin structure and transcription. Cell. 2002;108:475–487. doi: 10.1016/s0092-8674(02)00654-2. [DOI] [PubMed] [Google Scholar]
- Neer E. J., Smith T. F. G protein heterodimers: new structures propel new questions. Cell. 1996;84:175–178. doi: 10.1016/s0092-8674(00)80969-1. [DOI] [PubMed] [Google Scholar]
- Neufeld T. P., Edgar B. A. Connections between growth and the cell cycle. Curr. Opin. Cell Biol. 1998;10:784–790. doi: 10.1016/s0955-0674(98)80122-1. [DOI] [PubMed] [Google Scholar]
- Reeves R., Beckerbauer L. HMGI/Y proteins: flexible regulators of transcription and chromatin structure. Biochem. Biophys. Acta. 2001;1519:13–29. doi: 10.1016/s0167-4781(01)00215-9. [DOI] [PubMed] [Google Scholar]
- Roy S., Shashidhara L. S., Vigay Raghavan K. Muscles in the Drosophila second thoracic segment are patterned independently of autonomous homeotic gene function. Curr. Biol. 1997;7:222–227. doi: 10.1016/s0960-9822(06)00117-5. [DOI] [PubMed] [Google Scholar]
- Song H., Spichiger-Haeusermann C., Basler K. The ISWI-containing NURF complex regulates the output of the canonical Wingless pathway. EMBO Rep. 2009;10:1140–1146. doi: 10.1038/embor.2009.157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsukiyama T., Wu C. ISWI, a member of the SWI2/SNF2 ATPase family, encodes the 140kDa subunit of the nucleosome remodeling factor. Cell. 1995;83:1011–1020. doi: 10.1016/0092-8674(95)90217-1. [DOI] [PubMed] [Google Scholar]
- Xiao H., Sandaltzopoulos R., Wang H. M., Hamiche A., Renallo R., Lee K. M., Fu D., Wu C. Dual functions of the largest NURF subunit NURF301 in nucleosome sliding and transcription factor interactions. Mol. Cell. 2001;8:531–543. doi: 10.1016/s1097-2765(01)00345-8. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.