Abstract
Plasma membrane monoamine transporter (PMAT) is a polyspecific organic cation transporter in the solute carrier 29 (SLC29) family. Previous studies suggested that the major substrate recognition site is located within TM1-6 and PMAT interaction with organic cations may involve aromatic residues. In this study, we analyzed the role of tyrosine and tryptophan residues located within TM1-6 with the goal to identify potential residues involved in substrate recognition and translocation. The six Tyr and one Trp residues in this region were each mutated to alanine followed by analysis of these mutants’ membrane localization and transport activity towards 1-methyl-4-phenylpyridinium (MPP+), serotonin (5-HT) and dopamine. Two mutants, Y85A and Y112A, showed normal cell surface expression but lost their transport activity towards organic cations. At position Y85, aromatic substitution with Phe or Trp fully restored organic cation transport activity. Interestingly, at position Y112, Phe substitution is not allowed. Trp substitution at Y112 partially restored transport activity towards 5-HT and dopamine, but severely impaired MPP+ transport. Detailed kinetic analysis revealed that Trp substitution at Y85 and Y112 affected apparent binding affinity (Km) and maximal transport velocity (Vmax) in a substrate-dependent manner. Together these data suggest that Y85 and Y112 are important molecular determinants for PMAT function, and Y112 is indispensable for optimal interaction with organic cation substrates. Our analysis also suggested the involvement of transmembrane domains 1 and 2 in forming the substrate permeation pathway of PMAT.
The solute carrier 29 (SLC29) family consists of membrane transporters that mediate cellular uptake of important nutrients, signaling molecules and therapeutic drugs (1, 2). There are four SLC29 isoforms, SLC29A1-4, in the human genome. SLC29A1 and SLC29A2 encode the classic equilibrative nucleoside transporters 1 and 2 (ENT1 and ENT2), which transport purine and pyrimidine nucleosides and their structural analogs (1, 2). ENT1 and ENT2 play important regulatory roles in nucleic acid synthesis and purinergic signaling pathways, and are also important determinants of cellular response to nucleoside drugs used in anticancer, antiviral and immunosuppressive therapies (1, 2). The third member, ENT3 (SLC29A3), also transports nucleosides and nucleoside analogs, and mainly functions as an intracellular transporter (3, 4). The fourth member, SLC29A4, was cloned and characterized in our laboratory as the plasma membrane monoamine transporter (PMAT) (5). PMAT is different from ENT1-3 in that it does not typically transport nucleosides and nucleoside analogs, but rather functions as a polyspecific transporter for a wide range of small hydrophilic organic cations (5, 6). Representative PMAT substrates include the neurotoxin 1-methyl-4-phenylpyridinium (MPP+) and the monoamine neurotransmitters (e.g. serotonin, dopamine), which are also transported by organic cation transporters (OCTs) in the SLC22 family (7–10). In humans, PMAT mRNA is most strongly expressed in brain, and transcripts are also found in other organs such as the kidney, heart, and small intestine (5, 11, 12). Immunolocalization and biochemical studies suggested that PMAT may play important roles in brain monoamine clearance as well as tissue-specific disposition of organic cations in vivo (13–15).
The human and rodent PMAT proteins consist of about 530 amino acid residues and are predicted to possess an 11 transmembrane domain (TM) (5, 13). PMAT (ENT4) is unique in the SLC29 family in that while other SLC29 members (i.e. ENT1-3) are nucleoside transporters, PMAT functions as a polyspecific transporter for structurally diverse organic cations. Despite their distinct substrate specificity, PMAT and ENTs share a similar 11 TM membrane topology and previous studies have shown the major substrate recognition sites for PMAT and ENT1 reside in their N-terminal half containing TM1-6 (16–18). Of note, we have previously shown that transplanting the TM1-6 in PMAT into ENT1 converted ENT1 from a nucleoside transporter to an organic cation transporter (16). Based on these data, we hypothesized that PMAT and ENTs share a similar protein structure, and their distinct substrate specificity is determined by a unique set of residues that line the substrate binding and permeation pathway. Identification of such residues will provide novel insights into the structure-function relationships of transporters in the SLC29 family.
Using a series of structurally diverse organic cations, we previously probed the structural requirement of PMAT substrates and inhibitors (6). Our analysis revealed that a positive charge and a hydrophobic moiety are the two basic molecular features for transporter-substrate/inhibitor interaction. Interestingly, high-affinity PMAT substrates and inhibitors all possess at least one aromatic ring, whereas cations without a planar aromatic moiety are either low-affinity substrates or do not interact with PMAT (6). These results suggested that negatively charged residues and aromatic residues may be involved in transporter-substrate recognition through electrostatic interaction and π-π stacking. Using site-directed mutagenesis we previously identified a negatively charged residue on TM5, E206, that functions as a charge sensor and is critical for the cation selectivity of PMAT (16).
In this study, we aim to test the hypothesis that aromatic residues in TM1-6 are involved in substrate recognition and transport. We particularly focused on tyrosine and tryptophan residues because such residues have been implicated in interacting with organic cation substrates (e.g. MPP+) in the OCTs (19, 20). The tyrosine and tryptophan residues in TM1-6 were first mutated to alanine to identify the ones that are essential for PMAT activity. Confocal microscopy and membrane protein biotinylation were used to determine cell surface expression. Additional mutations were carried out at critical positions to further probe the structural-functional relationship of PMAT.
EXPERIMENTAL PROCEDURES
Sequence Alignment and Analysis
Sequences of mammalian ENTs were obtained from GenBank™ with the following accession numbers: hENT1 (AF_079117), rENT1 (NM_031684), mENT1 (AF_218255), macaques ENT1 (AB_168783), canine ENT1 (NM_001003367), cattle ENT1 (NM_001034398), hENT2 (AF_034102), rENT2 (NM_031738), mENT2 (AF_183397), cattle ENT2 (NM_001103269), rabbit ENT2 (AF_323951), hENT3 (AF_326987), rENT3 (AY_273196), mENT3 (AF_326986), cattle ENT3 (NM_001080223), hPMAT (AY_485959), rPMAT (NP_001099381), mPMAT (NM_146257). ClustalW2 software was used for sequence analysis and multiple alignments.
Generation of Mutant Constructs
To facilitate the determination of membrane localization, mutants were constructed using yellow fluorescence protein (YFP)-tagged wild type (WT) human PMAT as template. YFP was tagged at the N-termini of the WT and mutant PMAT transporters, and our previous studies have shown that the YFP tagging had no effect on substrate selectivity and kinetic behaviors of the transporter (16). The WT human PMAT was previously sub-cloned into the YFP vector pEYFP-C1 (Clontech, Palo Alto, CA) (5). Mutants were generated by site-directed mutagenesis using the QuickChange kit (Stratagene, La Jolla, CA) according to the manufacturer’s protocol. The sequence of each mutant was confirmed by direct DNA sequencing in the Department of Biochemistry at the University of Washington.
Stable Expression in MDCK Cells
YFP-tagged mutant constructs were transfected into MDCK cells using Lipofectamine 2000 transfection reagent (Invitrogen, Carlsbad, CA). Stably transfected cell lines were obtained by culturing cells in minimal essential medium containing 10% fetal bovine serum and G418 (1000 μg/ml). Empty pEYFP-C1 vector was transfected into MDCK cells to obtain the control cell line. After 2–3 weeks of drug selection, fluorescence-positive cells were purified by a FACS Vantage SE sorter (BD Biosciences, San Jose, CA) at the Cell Analysis Center at the University of Washington, Health Sciences Center. The sorted cells were cultured and maintained in minimal essential medium containing G418 (200 μg/ml).
Confocal Fluorescence Microscopy
To determine the cellular localization of YFP-tagged mutant transporters, ~2×105 cells were grown on top of microscope cover glass in 6-well plates (Falcon) for 2–3 days until confluent. Cells were mounted onto microscope glass slides with Fluoromount-G (Electron Microscopy Sciences, Hatfield, PA) and visualized with a Leica SP1 confocal microscope equipped with an argon laser as the light source at the Keck Microscopy Facility at the University of Washington. Images were captured by excitation at 488 nm and emission at 515 nm.
Functional Characterization in MDCK cells
Stably transfected MDCK cells were plated in 24-well plates and allowed to grow for 2–3 days until confluent. Growth medium was aspirated and each well was rinsed once with Krebs-Ringer-Henseleit (KRH) buffer (5.6 mM glucose, 125 mM NaCl, 4.8 mM KCl, 1.2 mM KH2PO4, 1.2 mM CaCl2, 1.2 mM MgSO4, 25 mM HEPES, pH 7.4) and preincubated in the same buffer for 15 min at 37°C. Transport assays were performed at 37°C by incubating cells in KRH buffer containing a 3H-labeled ligand. [3H] MPP+ (85 Ci/mmol) and [3H] uridine (30 Ci/mmol) were obtained from American Radiolabeled Chemicals, Inc. (St. Louis, MO). [3H] 5-HT (5-hydroxy-[1,2-3H] tryptamine creatinine sulfate, 28.1 Ci/mmol) and [3H] dopamine (3,4-dihydroxy-[2,5,6-3H] phenylethylamine, 51.3 Ci/mmol) were from PerkinElmer Life Sciences, Inc. All other chemicals were obtained from Sigma (St. Louis, MO). For transport studies using 3H-labeled nucleoside uridine, 0.5 μM NBMPR was added to the transport buffer to suppress endogenous nucleoside uptake activities. For uridine inhibition studies, cells were incubated with 3H-labeled ligands in the presence of 1 mM uridine and transport assays were performed at 37°C. Uptake was terminated by washing the cells three times with ice-cold KRH buffer. Cells were then solubilized with 0.5 ml of 1N NaOH and neutralized with 0.5 ml of 1N HCl. Radioactivity in the cell lysate was quantified by liquid scintillation counting. Protein concentration in each well was measured using BCA protein assay kit (Pierce) and the uptake in each well was normalized to its protein content. In all studies, cells transfected with an empty vector were served as a background control. Transporter-specific uptake was calculated by subtracting the background uptake in vector-transfected cells.
Isolation of Plasma Membrane Proteins by Cell Surface Biotinylation
Stably transfected MDCK cells were plated onto 60 mm plates and cultured until confluent. Cells were washed twice with 3 ml of ice-cold PBS/CM (138 mM NaCl, 2.7 mM KCl, 8 mM Na2HPO4, 1.5 mM KH2PO4, 0.1 mM CaCl2, 1 mM MgCl2, pH 8.0). Biotinylation was carried out on ice by incubation with 1 ml of ice-cold PBS/CM containing a membrane-impermeable biotinylation reagent Sulfo-NHS-SS-biotin (0.5 mg/ml) (Pierce, Rockford, IL). After two successive 20 min incubations at 4°C with freshly prepared NHS-SS-biotin and gentle shaking, cells were briefly rinsed with 3 ml of PBS/CM containing 100 mM glycine. Cells were further incubated at 4°C with the same solution for 20 min to ensure complete quenching of the unreacted NHS-SS-biotin. Cells were then solubilized on ice by incubating in 1 ml of lysis buffer containing 20 mM Tris, 150 mM NaCl, 1 mM EDTA, 1% Triton X-100, 1 mM phenylmethyl-sulfonyl fluoride and Protease Inhibitors Cocktail (Roche) for 1 h with occasional vortexing. Protein concentrations were measured from the supernatant lysate and fifty microliters of UltraLink Immobilized NeutrAvidin protein (Pierce) was then added to the supernatant for the isolation of membrane proteins. Membrane proteins were subjected to Western blot using a mouse monoclonal anti-yellow fluorescent protein antibody (JL-8) (BD Biosciences) at 1:1000 dilution, followed by horseradish peroxidase-conjugated goat anti-mouse IgG (1:20,000 dilution). The chemiluminescent signals in the Western blots were detected by using SuperSignal West Pico Chemiluminescent Substrate (Pierce) followed by exposure of the blots to x-ray films. Band intensity was quantified by densitometry using the ImageQuant software (Molecular Dynamics). As reported previously (16), double or multiple protein bands around the expected molecular size (~ 75 kDa) were observed for the YFP tagged PMAT proteins, which could be due to differential glycosylation of PMAT.
Helical Wheel Analysis
The helical wheels were generated using the Helix Wheel program on the EXPASY molecular biology server and subsequently transposed onto a helical wheel template. The transmembrane domain is assumed to be a standard α–helix (3.6 residues/helical turn). Each residue in TM is plotted every 100° around the center of a circle. The projection of the positions of the residues was shown on a plane perpendicular to the helical axis. Hydrophobicity and hydrophilicity are assigned according to the consensus scale of Eisenberg et al. (21).
Data Analysis
For all uptake experiments, data were expressed as the mean ± S.D. from three independent experiments (n=3) with different cell passages. For each experiment, uptake was carried out in triplicates in three different wells on the same plate. Where applicable, p values were obtained through Student’s t-test. For Michaelis-Menten studies, data were fit to the equation V= Vmax [S]/(Km+[S]) using Kaleidagraph Version 3.6 (Synergy Software, Reading, PA), where V is the transport rate and [S] is the substrate concentration. Kinetic parameters were determined by nonlinear least-squares regression fitting as described previously (5, 6).
RESULTS
Analysis of Tyr and Trp Residues in TM1-6 by Alanine Substitution
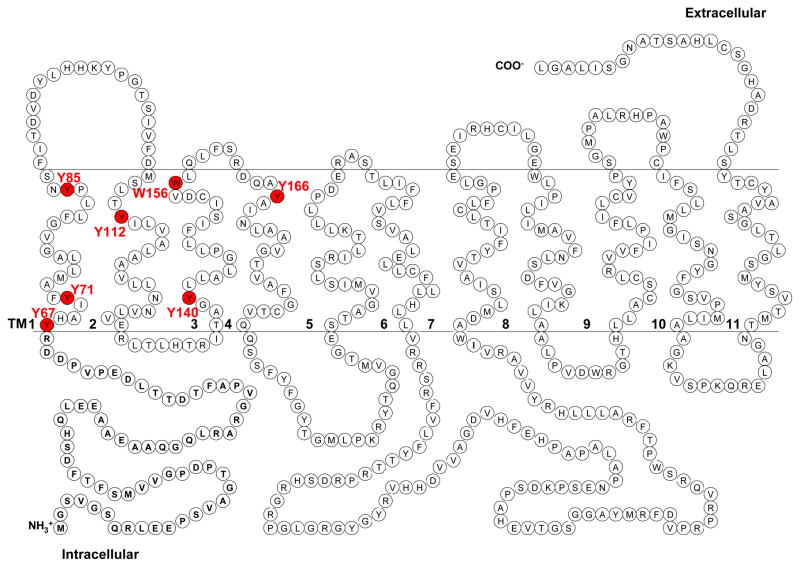
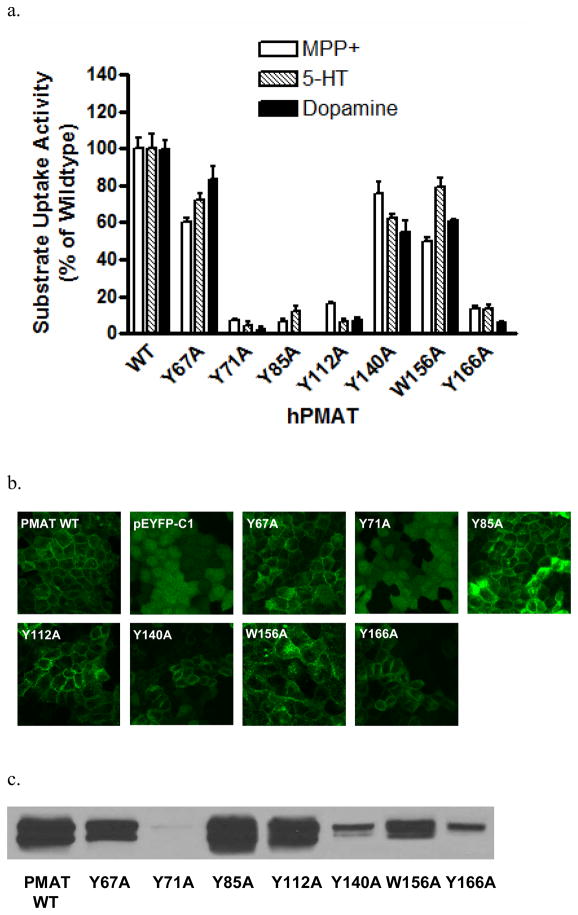
PMAT functions as a polyspecific organic cation transporter and it has been shown that all high-affinity substrates and inhibitors of PMAT possess at least one aromatic ring in their structures (6). We thus hypothesized that aromatic residues, especially Tyr and Trp, in TM1-6 may be involved in substrate recognition and/or translocation. There are six tyrosine (Y67, Y71, Y85, Y112, Y140, and Y166) and one tryptophan (W156) residues on the predicted TM1-4 (Fig. 1). No Tyr and Trp were found on TM5-6. All seven residues are completely conserved among human, rat and mouse PMAT proteins. Alanine substitution was used to test the functional significance of these residues in PMAT function. YFP-tagged mutants Y67A, Y71A, Y85A, Y112A, Y140A, W156A, and Y166A were constructed and stably expressed in MDCK cells. The function of each alanine mutant was analyzed by uptake studies using the prototype PMAT substrates MPP+, serotonin (5-HT) and dopamine. As shown in Fig. 2a, mutation of Y67, Y140, and W156 to alanine largely retained the transport activities for all substrates tested, suggesting that aromatic residues at these positions are not essential for transporter function. In contrast, mutation of Y71, Y85, Y112, and Y166 to alanine resulted in loss of transport activities towards all three substrates tested. The loss in the uptake activity of these alanine mutants could be due to impairment in transport function or alteration of cell surface expression levels of the transporter. To differentiate these mechanisms, cellular localization of the alanine mutants was visualized by confocal microscopy (Fig. 2b) and their plasma membrane YFP-fusion protein expression levels were detected by cell surface biotinylation followed by Western blot analysis (Fig. 2c). The three functional mutants (Y67A, Y140A and W156A) are localized on plasma membrane. In contrast, mutants Y71A, which lost organic cation transport activity, exhibited diffused fluorescence throughout the cytoplasm (Fig. 2b) with no detectable protein on plasma membrane (Fig. 2c). Mutant Y166 showed some plasma membrane localization but the cell surface expression level was only about one-tenth of that of the wild type (WT) PMAT. These data suggest that the loss of organic cation transport activity in cells expressing Y71A and Y166A was mainly due to problems in PMAT protein stability and/or membrane trafficking. Importantly, two mutants, Y85A and Y112A, showed normal plasma membrane expression but lost uptake activities towards organic cations (Fig. 2), suggesting that substitution of Tyr with Ala at these two positions affected the catalytic activity of the transporter. Y85 and Y112 were predicted to be located within TM1 and 2 respectively. These potentially important aromatic residues were chosen for more detailed analysis.
Figure 1.
Proposed secondary structure of PMAT. Positions of candidate aromatic residues selected for mutagenesis analysis are highlighted in red.
Figure 2.
(a) Uptake of 3H-labeled MPP+ (1 μM), 5-HT (10 μM) and dopamine (10 μM) by WT PMAT and various alanine mutants in stably transfected MDCK cells. Uptake was performed at 37°C for 4 min. Substrate uptake was corrected by subtracting nonspecific uptake in vector-transfected cells. Values are expressed as percentage of uptake in cells expressing WT PMAT. Each value represents the mean ± S.D. from three independent experiments (n=3) with different cell passages. For each experiment, uptake was carried out in triplicates in three different wells on the same plate. (b) Confocal imaging of cellular localization of WT PMAT, pEYFP-C1, and various PMAT alanine mutants in stably transfected MDCK cells. (c) Plasma membrane expression of WT PMAT and various PMAT alanine mutants detected by biotinylation followed by Western blot with an anti-yellow fluorescent protein monoclonal antibody.
Effect of Side Chain Substitution at Y85
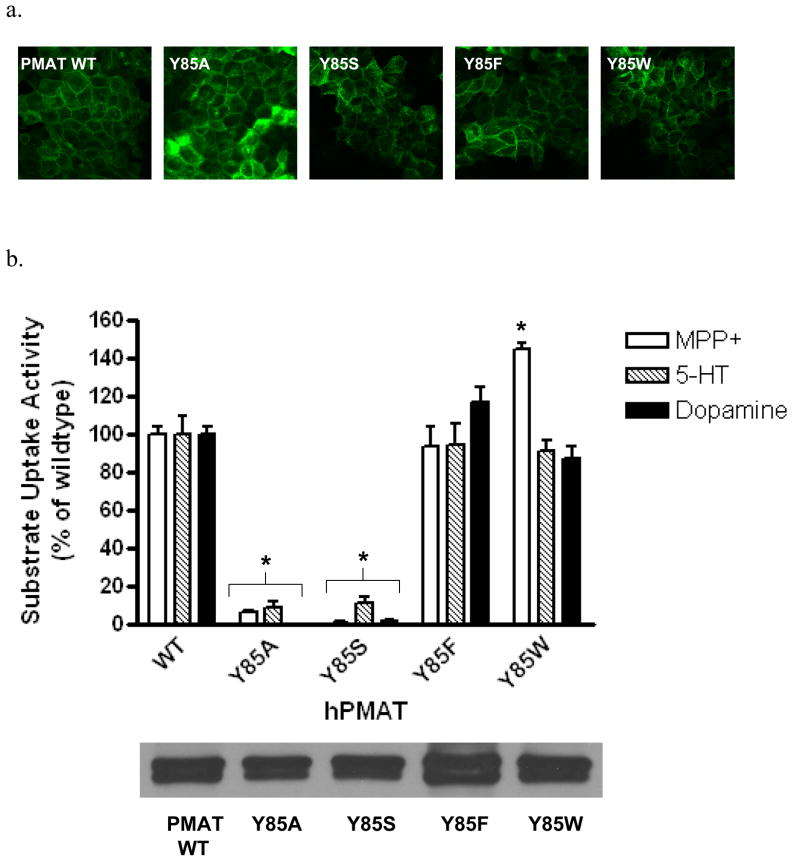
To investigate whether an aromatic side chain is essential for PMAT function at position 85, we next mutated Tyr to Trp and Phe. Tyrosine was also changed to Ser, a residue that retains a hydroxyl group but lacks an aromatic feature. Y85S, Y85F and Y85W were constructed and stably expressed in MDCK cells. All mutants exhibited plasma membrane expression patterns similar to that of WT PMAT (Fig. 3a). Cell surface biotinylation followed by Western blot analysis revealed that the membrane expression levels of the YFP-tagged proteins in all Y85 mutant cell lines are comparable with WT PMAT cell line (Fig. 3b lower panel). Functional assays showed that substitution of Y85 with non-aromatic residues (Y85A, Y85S) resulted in almost complete loss of activities toward all substrates tested (Fig. 3b upper panel). In contrast, substitution of Y85with aromatic moiety-containing residues (Y85F, Y85W) fully restored transporter activity (Fig. 3b upper panel). Interestingly, compared to WT PMAT, mutant Y85W showed similar activity towards 5-HT and dopamine, but significantly higher transport activity towards MPP+ (Fig. 3b upper panel).
Figure 3.
(a) Confocal imaging of cellular localization of WT PMAT and various Y85 mutants in stably transfected MDCK cells. (b) Upper panel, uptake of 3H-labeled MPP+ (1 μM), 5-HT (10 μM) and dopamine (10 μM) by WT PMAT and various Y85 mutants in stably transfected MDCK cells. Uptake was performed at 37°C for 4 min. Substrate uptake was corrected by subtracting nonspecific uptake in vector-transfected cells. Values are expressed as percentage of uptake in cells expressing WT PMAT. Each value represents the mean ± S.D. from three independent experiments (n=3) with different cell passages. For each experiment, uptake was carried out in triplicates in three different wells on the same plate. *, p<0.01, vs WT values. Lower panel, plasma membrane expression of WT PMAT and various Y85 mutants detected by biotinylation followed by Western blot with an anti-yellow fluorescent protein monoclonal antibody.
Effect of Side Chain Substitution at Y112
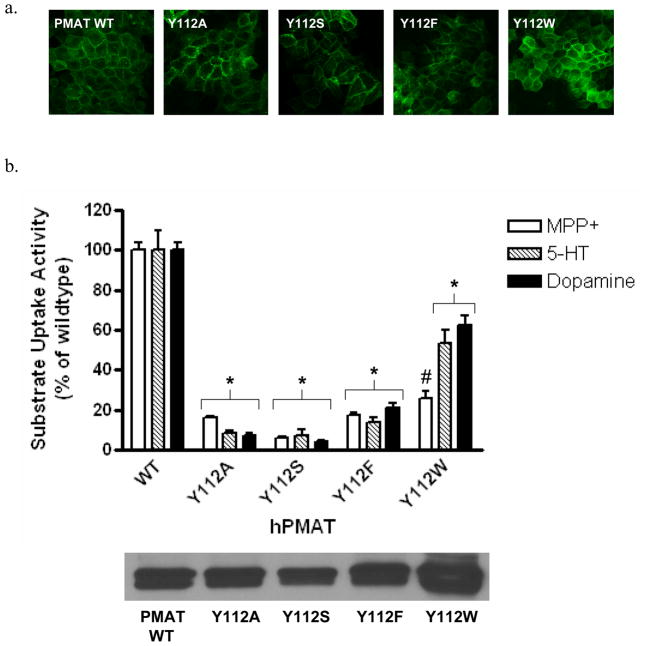
Y112S, Y112F and Y112W were constructed and stably expressed in MDCK cells. All mutants exhibited plasma membrane localization (Fig. 4a). The cell surface expressions of Y112A, Y112S and Y112F were comparable to WT PMAT, while Y112W showed slightly higher (~ 1.7 fold) expression (Fig. 4b lower panel). Functional assays showed that substitution of Y112 with non-aromatic residues (Y112A, Y112S) resulted in about 90% loss of transporter activity towards all organic cations tested (Fig. 4b upper panel). Mutant Y112F only showed less than 20% activity towards all tested substrates, suggesting that substitution of Tyr with Phe is not allowed at position 112. Interestingly, Trp substitution at Y112 appeared to have differential impact on transport activity towards MPP+, 5-HT and dopamine (Fig. 4b upper panel). Compared to cells expressing WT PMAT, cells expressing Y112W retained 50–60% activity for 5-HT and dopamine. However, much less (<20%) activity was retained for MPP+. These data suggested that Y112 in TM2 plays a specific role in interacting with organic cation substrates and the effect of Trp substitution at this position is substrate-dependent.
Figure 4.
(a) Confocal imaging of cellular localization of WT PMAT and various Y112 mutants in stably transfected MDCK cells. (b) Upper panel, uptake of 3H-labeled MPP+ (1 μM), 5-HT (10 μM) and dopamine (10 μM) by WT PMAT and various Y112 mutants in stably transfected MDCK cells. Uptake was performed at 37°C for 4 min. Substrate uptake was corrected by subtracting nonspecific uptake in vector-transfected cells. Values are expressed as percentage of uptake in cells expressing WT PMAT. Each value represents the mean ± S.D. from three independent experiments (n=3) with different cell passages. For each experiment, uptake was carried out in triplicates in three different wells on the same plate. *, p<0.01, vs WT values. #, p<0.01, vs Y112W-mediated 5-HT and dopamine uptake values. Lower panel, plasma membrane expression of WT PMAT and various Y112 mutants detected by biotinylation followed by Western blot with an anti-yellow fluorescent protein monoclonal antibody.
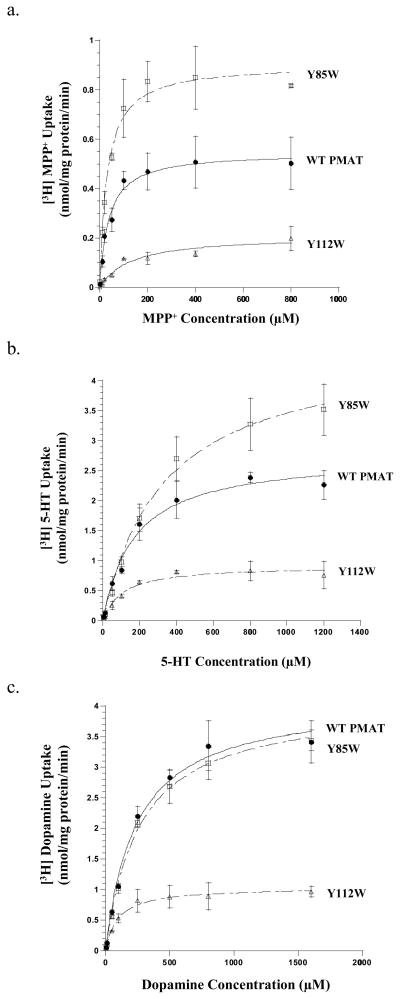
Transport kinetics of Y85W and Y112W
While Trp substitution is allowed at position Y85 and partially permitted at position Y112, the impacts of these side chain changes on transport activity appeared to be substrate-dependent (Fig. 3b and Fig. 4b). To confirm these data and to understand the kinetic mechanisms underlying such observations, we performed detailed kinetic analysis on WT PMAT, Y85W and Y112W (Fig. 5). The resulting kinetic parameters (summarized in Table 1) clearly demonstrated that the Tyr to Trp substitution at both positions differentially influenced the transport kinetics of MPP+, 5-HT and dopamine. For MPP+, Y85W substitution produced a 1.6-fold increase in maximal velocity (Vmax), with little effect on apparent binding affinity (Km), resulting in about 2-fold increase in transport efficiency (Vmax/Km) towards MPP+ (Fig. 5a and Table 1). For 5-HT, Y85W substitution caused a similar degree of increase in both the Km and the Vmax, leading to an unaltered transport efficiency towards this substrate (Fig. 5b and Table 1). For dopamine, Y85W substitution had no effects on either the Km or the Vmax (Fig. 5c and Table 1). At position Y112, Trp substitution produced a marked (2.6- to 4-fold) reduction in Vmax for all three tested substrates, suggesting a universal impairment in the transporter’s turnover rate (Table 1). Remarkably, the effect on Km is largely substrate-dependent. For MPP+, a 2.6-fold reduction in Vmax is accompanied by a significant increase (3.5 folds) in Km, resulting in a substantial (7 folds) reduction in MPP+ transport efficiency (Vmax/Km) (Fig. 5a and Table 1). For 5-HT and dopamine, the reduction in Vmax was, however, compensated by a simultaneous decrease in Km, resulting in transport efficiencies that were still 60–70% of the WT PMAT (Fig. 5b,c and Table 1). These data are consistent with the results from single concentration uptake studies performed at a low substrate concentration (Fig. 4b), and further demonstrate the importance of Y112 in recognizing and transporting MPP+. Taken together, these results confirmed that Trp substitution at Y85 and Y112 influenced substrate binding and/or transport in a substrate-dependent manner, and these two residues may play specific roles in interacting with the substrates.
Figure 5.
Concentration-dependent transport of (a) MPP+, (b) 5-HT, and (c) dopamine by WT PMAT, Y85W and Y112W. Vector-, WT PMAT-, Y85W-, and Y112W-transfected MDCK cells were incubated with varying concentrations of the substrates for 1 min at 37°C. Specific uptake was calculated by subtracting the uptake values in vector-transfected cells. WT PMAT (●), Y85W (□), and Y112W (Δ) concentration-dependent uptake were shown. Each value represents the mean ± S.D. from three independent experiments (n=3) with different cell passages. For each experiment, uptake was carried out in triplicates in three different wells on the same plate.
Table 1.
Transport kinetics of PMAT wild type, mutants Y85W and Y112W.
| MPP+ |
Serotonin |
Dopamine |
|||||||
|---|---|---|---|---|---|---|---|---|---|
|
Km (μM) |
Vmax (nmol/mg protein/min) |
Vmax/Km |
Km (μM) |
Vmax (nmol/mg protein/min) |
Vmax/Km |
Km (μM) |
Vmax (nmol/mg protein/min) |
Vmax/Km | |
| Wild Type | 42.2 ± 5.0 | 0.60 ± 0.04 | 0.014 ± 0.002 | 167.5 ± 18.9 | 2.80 ± 0.24 | 0.017 ± 0.002 | 244.4 ± 16.2 | 4.12 ± 0.35 | 0.017 ± 0.002 |
| Y85W | 31.1 ± 6.9 | 0.94 ± 0.10** | 0.030 ± 0.007* | 308.2 ± 50.8* | 4.08 ± 0.30** | 0.013 ± 0.002 | 242.6 ± 30.4 | 3.87 ± 0.14 | 0.016 ± 0.002 |
| Y112W | 146.8 ± 36.3** | 0.23 ± 0.02** | 0.002 ± 0.0004** | 103.9 ± 13.2** | 1.04 ± 0.13** | 0.010 ± 0.002* | 88.0 ± 6.8** | 1.03 ± 0.09** | 0.012 ± 0.001* |
All experiments were performed in triplicate in three different wells on the same plate and repeated three times. Values were expressed as mean ± S.D. from parameters obtained from three independent experiments (n=3) with different cell passages.
p<0.05,
p<0.01 vs WT PMAT values.
Uridine Uptake and Interaction with PMAT Mutants
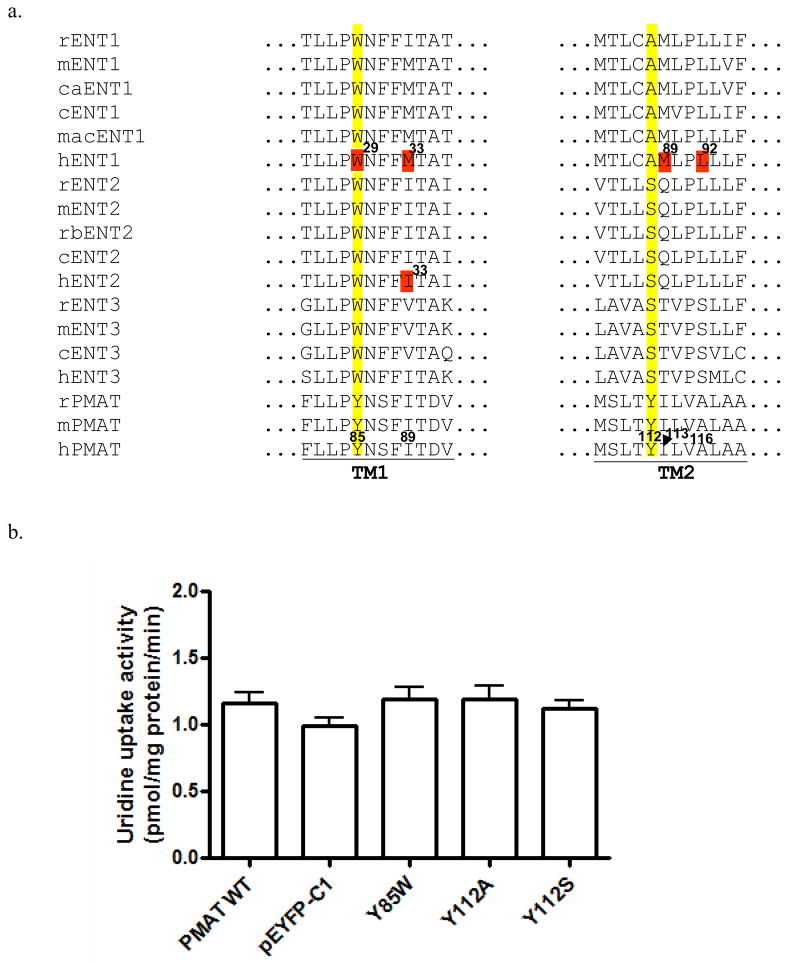
Sequence alignments showed that at the equivalent positions of Y85, the Trp residue was conserved in all ENT1-3 members from various species (Fig. 6a). At the equivalent position of Y112 in PMAT, the Tyr residue is respectively substituted by an Ala in ENT1 transporters and a Ser in ENT2 and ENT3 isoforms (Fig. 6a). We thus tested whether mutating Y85 and Y112 to their corresponding residues in ENT1 or ENT2 would lead to a gain of transport activity towards uridine, a prototypical substrate for the ENTs. Like WT PMAT, Y85W, Y112A and Y112S mutants showed no statistically significant transport activity for uridine (Fig. 6b). In addition, no significant inhibitory effect of uridine (1 mM) was observed for Y85W-mediated organic cations transport (data not shown). These data suggest that single substitution with an ENT1/2 equivalent residue at these positions is not sufficient to enhance PMAT interaction with the nucleosides.
Figure 6.
(a) Multiple sequence alignment of PMATs with mammalian ENTs in TM1 and TM2 regions. Y85 and Y112 in PMATs and their corresponding residues on ENTs are numbered and highlighted in yellow. Functionally important residues previously identified in ENTs are numbered and highlighted in red. Corresponding residues in human PMAT are numbered. (r: rat; m: mouse; ca: canine; c: cattle; mac: macaques; h: human; rb: rabbit) (b) Uptake of uridine in the presence of NBMPR by various PMAT mutants in stably transfected MDCK cells. Values are expressed as pmol/mg protein/min. Each value represents the mean ± S.D. from three independent experiments (n=3) with different cell passages. For each experiment, uptake was carried out in triplicates in three different wells on the same plate. There was no statistically difference in uridine transport for WT and mutant PMAT proteins as compared to the pEYFP-C1 vector-transfected cells.
DISCUSSION
Previous structure-activity study on PMAT suggested possible involvement of aromatic residues in high affinity interaction with substrates (6). In this study, we aim to identify these residues. We confined our analysis to TM1-6 as our previous studies suggested that the major substrate recognition site of PMAT is contained within this region (16). Among the three aromatic residues (Tyr, Trp, Phe), Tyr and Trp were previously shown to be involved in interaction with organic cation substrates (e.g. MPP+) by OCTs (19, 20), which share a large substrate overlap with PMAT (6). Additionally, as PMAT substrates are mostly hydrophilic cations, we suspect that Phe may be too hydrophobic to directly interact with an organic cation in an aqueous pore. Therefore, we particularly focused on Tyr and Trp in the current study.
We first analyzed the six Tyr and one Trp residues on TM1-6 by alanine substitution (Fig. 2). Our results showed that alanine replacement at Y67, Y140 and W156 did not significantly affect transporter activity, suggesting that these residues are not essential for PMAT function. It is also noted that mutant Y140A showed relatively normal transport activity towards various organic cations despite a significantly lower cell surface expression. This could imply an enhanced affinity for substrates and/or a higher intrinsic transport activity as compared to the WT PMAT, resulting in an apparently “normal” transport activity towards the tested substrates. Alanine substitution at Y71 and Y166 impaired PMAT cell surface expression, suggesting Tyr residues at these two positions are important for protein stability and membrane targeting. Importantly, alanine substitution at Y85 and Y112 resulted in normal plasma membrane expression but nearly complete loss in transport activity, suggesting that Y85 and Y112 are crucial for PMAT transport.
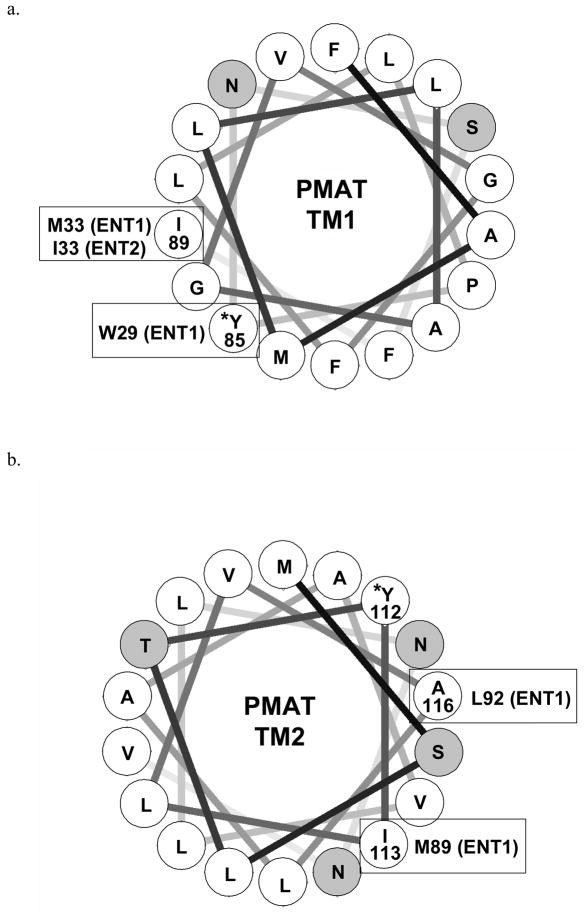
To further probe the structural requirement at Y85 and Y112, further mutational analysis was carried out. Our results showed that at Y85, aromatic substitution with Trp or Phe can fully restore PMAT activity whereas non-aromatic replacement (Ala, Ser) is not allowed, suggesting that the presence of an aromatic side chain is essential for PMAT function (Fig. 3). Detailed analysis further revealed that the effect of Trp substitution at Y85 influenced transport kinetics in a substrate-dependent manner (Fig. 5 and Table 1). Interestingly, multiple sequence alignment showed that at the equivalent position of Y85, a Trp residue is conserved among all other SLC29 members, ENT1-3 (Fig. 6a). It should be noted that nucleosides also contain at least one heterocyclic aromatic ring on the purine or pyrimidine base. In human ENT1, random mutagenesis studies have identified Trp29 as an important determinant for hENT1 nucleoside selectivity and may interact with nucleosides and nucleoside inhibitor via ring-stacking interactions (22). Trp29 lies closely to Met33, another functionally important component of the substrate/inhibitor binding site in ENT1 (23). Met33 is responsible for the high affinity binding of hENT1 to its classic inhibitor NBMPR, whereas isoleucine substitution (Ile33) in hENT2 underlies its low sensitivity to the inhibitor (23, 24). Helical wheel analysis revealed that in PMAT Y85 (corresponds to W29 in hENT1) lies closely to I89 (corresponds to M33 in hENT1 and I33 in hENT2), occupying a position that is known to form part of the substrate/inhibitor binding site in the ENTs (Fig. 7a). Together, the data suggest that Y85 and TM1 in PMAT is an important component of the substrate recognition and permeation pathway. Y85 may recognize an aromatic feature in PMAT substrates presumably through π-π interaction. Alternatively, the residue may structurally help to form the substrate recognition and translocation pathway by interacting with other nearby aromatic residues via ring-stacking interactions.
Figure 7.
Helical wheel analysis of (a) TM1 and (b) TM2 of PMAT. The transmembrane domains are assumed to be standard α-helix and each residue is plotted every 100° around the center of a circle. The figures show the projection of the positions of the residues on a plane perpendicular to the helical axis. Hydrophobic residues are shown in white, and hydrophilic residues are shown in gray. Residues in PMAT identified to be functionally important in this study are marked by asterisks*. Functionally important residues previously identified in ENT1 and/or ENT2 with positions corresponding or close to Y85 and Y112 in PMAT are indicated in bracket.
Y112 on TM2 is indispensable for organic cation transport function of PMAT. Non-aromatic replacement (Y112A, Y112S) is not allowed (Fig. 4). Even conservative substitution with Phe, which differs from Tyr only by missing a hydroxyl group on the phenyl ring, resulted in a non-functional transporter (Fig. 4). These data suggest that at position 112, both a hydroxyl group and an aromatic ring are necessary for normal transporter function. The specific role of the phenolic OH group is less clear, but it may act as an electron donor to stabilize the positive charge carried by an organic cation substrate. Interestingly, Trp substitution at Y112 partially restored transport activity towards 5-HT and dopamine, but severely impaired MPP+ transport (Fig. 4). Detailed kinetic analysis revealed that mutant Y112W showed a universal reduction in its Vmax towards all three tested substrates, suggesting that the Tyr residue at this position is essential for optimal organic cation transport function of PMAT. Our kinetic studies further uncovered that the substantial reduction in MPP+ transport observed in mutant Y112W is due to a combined impairment in both apparent binding affinity (i.e. increase in Km) and maximal transport velocity (Vmax) (Table 1). On the other hand, for 5-HT and dopamine, the reduction in Vmax was partially compensated by a simultaneous increase in binding affinity (i.e. decrease in Km), resulting in transport efficiencies that were still 60–70% of the WT PMAT. This substrate-dependent effect implies there may exist multiple substrate binding sites within the translocation pathway. Given that the substrate specificity of PMAT is broad and includes a variety of compounds with diverse structures, it is likely that PMAT protein may contain a promiscuous binding pocket, in which conservative substitution of key residues may have different impact on the binding and transport of different substrates. Multiple sequence alignment showed that at the equivalent position of Y112, ENT1 transporters possess an Ala while ENT2 and 3 contain a Ser residue (Fig. 6a). The roles of these residues in ENT function have not been analyzed. However, M89 and L92 in hENT1 TM2 have been shown to be important for nucleoside and inhibitor interactions (25, 26). M89 and L92 in hENT1 correspond to I113 and A116 in PMAT respectively (Fig. 6a). Helical wheel analysis suggested that Y112, I113 and A116 in PMAT locate in close proximity to one another on the same hemisphere of TM2 (Fig. 7b). Together these data suggest that TM2 in PMAT participates in forming the organic cation permeation pathway and Y112 specifically interacts with organic cations through both its aromatic ring and phenolic OH group.
In summary, we identified Y85 and Y112 that are crucial for the catalytic activity of PMAT function and influence substrate selectivity of the transporter. Y112 may specifically interact with organic cation substrates via its phenolic OH group. Together with our previous finding (16), our studies suggest that TM1, TM2 and TM5 are critical constituents of the substrate permeation pathway in PMAT. Although the proposed secondary structure of PMAT (Fig. 1) awaits experimental validation, results from our recent immunocytochemistry studies, using two epitope-specific polyclonal antibodies directed towards the predicted N-terminus and the last intracellular loop, are in good agreement with the proposed 11-TM model (12, 14). Such an 11-TM model has been experimentally confirmed in ENT1 (27), and TM1, TM2 and TM5 were also previously shown to be important components of the substrate recognition and translocation pathway in ENT1 (22, 23, 25, 26). Thus, our results indicate that PMAT and the ENT proteins share a similar tertiary structure, and the unique substrate selectivity of PMAT is due to evolutionary replacement of a set of key residues in the substrate recognition domains of the transporter.
ABBREVIATIONS
- PMAT
Plasma membrane monoamine transporter
- ENT
equilibrative nucleoside transporter
- 5-HT
serotonin
- MPP+
1-methyl-4-phenylpyridinium
- OCT
organic cation transporter
- WT
wildtype
- MDCK
Madin-Darby canine kidney
- YFP
yellow fluorescence protein
- TM
transmembrane
- NBMPR
nitrobenzylmercaptopurine riboside (nitrobenzylthioinosine)
Footnotes
This work is supported by the National Institutes of Health Grant GM066233.
References
- 1.Kong W, Engel K, Wang J. Mammalian nucleoside transporters. Curr Drug Metab. 2004;5:63–84. doi: 10.2174/1389200043489162. [DOI] [PubMed] [Google Scholar]
- 2.Baldwin SA, Beal PR, Yao SY, King AE, Cass CE, Young JD. The equilibrative nucleoside transporter family, SLC29. Pflugers Arch. 2004;447:735–743. doi: 10.1007/s00424-003-1103-2. [DOI] [PubMed] [Google Scholar]
- 3.Baldwin SA, Yao SY, Hyde RJ, Ng AM, Foppolo S, Barnes K, Ritzel MW, Cass CE, Young JD. Functional characterization of novel human and mouse equilibrative nucleoside transporters (hENT3 and mENT3) located in intracellular membranes. J Biol Chem. 2005;280:15880–15887. doi: 10.1074/jbc.M414337200. [DOI] [PubMed] [Google Scholar]
- 4.Govindarajan R, Leung GP, Zhou M, Tse CM, Wang J, Unadkat JD. Facilitated mitochondrial import of antiviral and anticancer nucleoside drugs by human equilibrative nucleoside transporter-3. Am J Physiol Gastrointest Liver Physiol. 2009;296:G910–922. doi: 10.1152/ajpgi.90672.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Engel K, Zhou M, Wang J. Identification and characterization of a novel monoamine transporter in the human brain. J Biol Chem. 2004;279:50042–50049. doi: 10.1074/jbc.M407913200. [DOI] [PubMed] [Google Scholar]
- 6.Engel K, Wang J. Interaction of organic cations with a newly identified plasma membrane monoamine transporter. Mol Pharmacol. 2005;68:1397–1407. doi: 10.1124/mol.105.016832. [DOI] [PubMed] [Google Scholar]
- 7.Fujita T, Urban TJ, Leabman MK, Fujita K, Giacomini KM. Transport of drugs in the kidney by the human organic cation transporter, OCT2 and its genetic variants. J Pharm Sci. 2006;95:25–36. doi: 10.1002/jps.20536. [DOI] [PubMed] [Google Scholar]
- 8.Koepsell H, Endou H. The SLC22 drug transporter family. Pflugers Arch. 2004;447:666–676. doi: 10.1007/s00424-003-1089-9. [DOI] [PubMed] [Google Scholar]
- 9.Wright SH, Dantzler WH. Molecular and cellular physiology of renal organic cation and anion transport. Physiol Rev. 2004;84:987–1049. doi: 10.1152/physrev.00040.2003. [DOI] [PubMed] [Google Scholar]
- 10.Zhang L, Brett CM, Giacomini KM. Role of organic cation transporters in drug absorption and elimination. Annu Rev Pharmacol Toxicol. 1998;38:431–460. doi: 10.1146/annurev.pharmtox.38.1.431. [DOI] [PubMed] [Google Scholar]
- 11.Zhou M, Xia L, Wang J. Metformin transport by a newly cloned proton-stimulated organic cation transporter (plasma membrane monoamine transporter) expressed in human intestine. Drug Metab Dispos. 2007;35:1956–1962. doi: 10.1124/dmd.107.015495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xia L, Zhou M, Kalhorn TF, Ho HT, Wang J. Podocyte-specific expression of organic cation transporter PMAT: implication in puromycin aminonucleoside nephrotoxicity. Am J Physiol Renal Physiol. 2009;296:F1307–1313. doi: 10.1152/ajprenal.00046.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dahlin A, Xia L, Kong W, Hevner R, Wang J. Expression and immunolocalization of the plasma membrane monoamine transporter in the brain. Neuroscience. 2007;146:1193–1211. doi: 10.1016/j.neuroscience.2007.01.072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Xia L, Engel K, Zhou M, Wang J. Membrane localization and pH-dependent transport of a newly cloned organic cation transporter (PMAT) in kidney cells. Am J Physiol Renal Physiol. 2007;292:F682–690. doi: 10.1152/ajprenal.00302.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhou M, Engel K, Wang J. Evidence for significant contribution of a newly identified monoamine transporter (PMAT) to serotonin uptake in the human brain. Biochem Pharmacol. 2007;73:147–154. doi: 10.1016/j.bcp.2006.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhou M, Xia L, Engel K, Wang J. Molecular determinants of substrate selectivity of a novel organic cation transporter (PMAT) in the SLC29 family. J Biol Chem. 2007;282:3188–3195. doi: 10.1074/jbc.M609421200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sundaram M, Yao SY, Ng AM, Griffiths M, Cass CE, Baldwin SA, Young JD. Chimeric constructs between human and rat equilibrative nucleoside transporters (hENT1 and rENT1) reveal hENT1 structural domains interacting with coronary vasoactive drugs. J Biol Chem. 1998;273:21519–21525. doi: 10.1074/jbc.273.34.21519. [DOI] [PubMed] [Google Scholar]
- 18.Yao SY, Ng AM, Vickers MF, Sundaram M, Cass CE, Baldwin SA, Young JD. Functional and molecular characterization of nucleobase transport by recombinant human and rat equilibrative nucleoside transporters 1 and 2. Chimeric constructs reveal a role for the ENT2 helix 5–6 region in nucleobase translocation. J Biol Chem. 2002;277:24938–24948. doi: 10.1074/jbc.M200966200. [DOI] [PubMed] [Google Scholar]
- 19.Popp C, Gorboulev V, Muller TD, Gorbunov D, Shatskaya N, Koepsell H. Amino acids critical for substrate affinity of rat organic cation transporter 1 line the substrate binding region in a model derived from the tertiary structure of lactose permease. Mol Pharmacol. 2005;67:1600–1611. doi: 10.1124/mol.104.008839. [DOI] [PubMed] [Google Scholar]
- 20.Gorboulev V, Shatskaya N, Volk C, Koepsell H. Subtype-specific affinity for corticosterone of rat organic cation transporters rOCT1 and rOCT2 depends on three amino acids within the substrate binding region. Mol Pharmacol. 2005;67:1612–1619. doi: 10.1124/mol.104.008821. [DOI] [PubMed] [Google Scholar]
- 21.Eisenberg D, Wilcox W, McLachlan AD. Hydrophobicity and amphiphilicity in protein structure. J Cell Biochem. 1986;31:11–17. doi: 10.1002/jcb.240310103. [DOI] [PubMed] [Google Scholar]
- 22.Paproski RJ, Visser F, Zhang J, Tackaberry T, Damaraju V, Baldwin SA, Young JD, Cass CE. Mutation of Trp29 of human equilibrative nucleoside transporter 1 alters affinity for coronary vasodilator drugs and nucleoside selectivity. Biochem J. 2008;414:291–300. doi: 10.1042/BJ20080074. [DOI] [PubMed] [Google Scholar]
- 23.Visser F, Vickers MF, Ng AM, Baldwin SA, Young JD, Cass CE. Mutation of residue 33 of human equilibrative nucleoside transporters 1 and 2 alters sensitivity to inhibition of transport by dilazep and dipyridamole. J Biol Chem. 2002;277:395–401. doi: 10.1074/jbc.M105324200. [DOI] [PubMed] [Google Scholar]
- 24.Visser F, Zhang J, Raborn RT, Baldwin SA, Young JD, Cass CE. Residue 33 of human equilibrative nucleoside transporter 2 is a functionally important component of both the dipyridamole and nucleoside binding sites. Mol Pharmacol. 2005;67:1291–1298. doi: 10.1124/mol.104.005884. [DOI] [PubMed] [Google Scholar]
- 25.Endres CJ, Unadkat JD. Residues Met89 and Ser160 in the human equilibrative nucleoside transporter 1 affect its affinity for adenosine, guanosine, S6-(4-nitrobenzyl)-mercaptopurine riboside, and dipyridamole. Mol Pharmacol. 2005;67:837–844. doi: 10.1124/mol.104.008102. [DOI] [PubMed] [Google Scholar]
- 26.Endres CJ, Sengupta DJ, Unadkat JD. Mutation of leucine-92 selectively reduces the apparent affinity of inosine, guanosine, NBMPR [S6-(4-nitrobenzyl)-mercaptopurine riboside] and dilazep for the human equilibrative nucleoside transporter, hENT1. Biochem J. 2004;380:131–137. doi: 10.1042/BJ20031880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sundaram M, Yao SY, Ingram JC, Berry ZA, Abidi F, Cass CE, Baldwin SA, Young JD. Topology of a human equilibrative, nitrobenzylthioinosine (NBMPR)-sensitive nucleoside transporter (hENT1) implicated in the cellular uptake of adenosine and anti-cancer drugs. J Biol Chem. 2001;276:45270–45275. doi: 10.1074/jbc.M107169200. [DOI] [PubMed] [Google Scholar]