Abstract
Sleeping Beauty (SB) transposase enables somatic integration of exogenous DNA in mammalian cells, but potency as a gene transfer vector especially in large mammals has been lacking. Herein, we show that hyperactive transposase system delivered by high-capacity adenoviral vectors (HC-AdVs) can result in somatic integration of a canine factor IX (cFIX) expression-cassette in canine liver, facilitating stabilized transgene expression and persistent haemostatic correction of canine hemophilia B with negligible toxicity. We observed stabilized cFIX expression levels during rapid cell cycling in mice and phenotypic correction of the bleeding diathesis in hemophilia B dogs for up to 960 days. In contrast, systemic administration of an inactive transposase system resulted in rapid loss of transgene expression and transient phenotypic correction. Notably, in dogs a higher viral dose of the active SB transposase system resulted into transient phenotypic correction accompanied by transient increase of liver enzymes. Molecular analysis of liver samples revealed SB-mediated integration and provide evidence that transgene expression was derived mainly from integrated vector forms. Demonstrating that a viral vector system can deliver clinically relevant levels of a therapeutic protein in a large animal model of human disease paves a new path toward the possible cure of genetic diseases.
Introduction
Hemophilia B is an X-linked inherited blood clotting disorder caused by mutations in the coagulation factor IX (FIX) gene. This genetic disease has a prevalence of about 1 in 30,000 Caucasian males. Current standard therapies for hemophilia B are based on protein infusion treatments utilizing recombinant proteins or plasma-derived blood components. However, limitations of this treatment option are the potential development of inhibitors, the inconvenience of repeated injections of the coagulation factor concentrates, and the expenses of replacement factor production.
As an alternative treatment strategy for hemophilia B, a plethora of liver-based gene therapy approaches have been proposed. Preclinical studies in murine and canine models for hemophilia B involving gene delivery vehicles based on retrovirus, lentivirus, adeno-associated virus (AAV), and recombinant adenovirus were performed. Moloney murine leukemia virus-based retroviral vectors for example, which integrate into the host genome, were utilized in hemophilia dogs.1 Furthermore, lentiviral vectors have been shown to be sufficient in mice but efficacy in hemophilia B dogs remains to be shown.2,3 To date the longest duration of canine FIX (cFIX) expression (8 years) in hemophilia B dogs was demonstrated after a single liver-directed injection of a recombinant AAV based on serotype 2 vector (AAV2).4 In that particular study it is claimed that extrachromosomal AAV vector genomes were responsible for long-term expression of cFIX. Unfortunately, in the context of a clinical trial in hemophilia patients, this AAV-based approach resulted in transient phenotypic correction (2 months) of the bleeding diathesis5 most likely due to an adaptive immune response directed against the incoming viral capsid.
In the present study, we utilized an integrating adenoviral hybrid-vector system for somatic integration and long-term phenotypic correction of the bleeding disorder. Various generations of recombinant adenoviral vectors were used in the past for the treatment of hemophilia B. The early generation vectors deleted for the early-transcribed adenoviral genes E1 and E3 resulted in therapeutic cFIX levels in mice and hemophilia B dogs. However, cFIX levels in mouse serum although in a therapeutic range significantly decreased over time and plasma cFIX levels of treated animals declined to undetectable levels 2 months postinjection.6 It became clear that synthesized viral antigens expressed from the early generation recombinant adenoviral genomes induced immune responses that are at least in part responsible for destruction of transduced cells. The primary immune effector cells in this process are major histocompatibility complex class I-restricted cytotoxic T-lymphocytes.7,8,9 Further deletion of the E2a, E2b, or E4 regions, while appearing safer, still resulted in some toxicity.10,11,12
The newest generation of adenoviral vectors is deleted for all viral coding sequences allowing for packaging of up to 36 kb of foreign DNA.13,14 These vectors are commonly referred to as “gutted,” gene-deleted, helper-dependent, or high-capacity adenoviral vectors (HC-AdVs). It was demonstrated that hepatic transduction utilizing HC-AdV is not accompanied by chronic toxicity due to the absence of viral gene expression.15,16 However, even HC-AdVs exhibit some dose-dependent toxicity related to the incoming viral particles (vps).17 It was shown that systemic cytokines including the early indicator of inflammation interleukin-6 as mediators of the innate immune responses were activated17,18 and an attenuated adaptive immune response was observed due to an infiltration of adenovirus-specific cytotoxic T-lymphocytes.19
Nonetheless, none of the currently available gene transfer vehicles for gene therapeutic applications is flawless and, to date, HC-AdV still represents one of the most promising gene delivery vehicles. This type of vector has been shown to result in long-term transgene expression and phenotypic correction in various species and animal models. We and others have shown that after a single injection a HC-AdV can be maintained life-long in mice and for up to 2 years in rats.16,20,21,22,23 In dogs and nonhuman primates, HC-AdV liver transduction also led to long-term transgene expression.15,16 To date, the longest period of transgene expression (up to 964 days) was observed by Brunetti-Pierri and colleagues using a HC-AdV for hepatic transduction of baboons.16,24 With respect to canine hemophilia B, there are currently two studies utilizing HC-AdV based on human adenovirus serotype 5. Our previous study demonstrated that, at a nontoxic dose, transient phenotypic correction can be achieved for several weeks after systemic administration of HC-AdV into hemophilia B dogs.25 Another study observed long-term phenotypic correction at a higher dose which in turn was associated with transient liver toxicity.26
With a very few exceptions (e.g., in embryonic stem cells)27 adenoviral vectors are believed to integrate into host chromosomes at low frequencies.28,29 To prolong the therapeutic effect after liver-based adenoviral gene transfer and to stabilize persistence of the therapeutic DNA at a lower dose, we hypothesized that somatic integration of the transgene from the episomal adenoviral vector genome into the host genome may stabilize and prolong the therapeutic effect. To test this hypothesis, we evaluated an improved version of our previously described adenovirus/Sleeping Beauty (SB) transposase hybrid-vector system30 for the first time in a canine model for hemophilia B. For high integration efficiencies into genomic DNA, the hybrid-vector technology utilized in the present study was based on a hyperactive version of the SB transposase (HSB5). This transposable element was originally generated from inactive copies of an ancestral Tc1/mariner-like transposon in fish31 and the mechanism for SB-mediated somatic integration is schematically shown in Figure 1a. By performing a mutational analysis screen the hyperactive mutant HSB5 was identified displaying tenfold higher integration efficiencies compared to wild-type SB.32 Herein, we show that this integrating viral vector system results in significantly stabilized transgene expression in rapidly dividing hepatocytes in mice. Furthermore, we demonstrate for the first time efficacy of a SB transposase system in a large animal exemplified by long-term phenotypic correction in a canine model for hemophilia B.
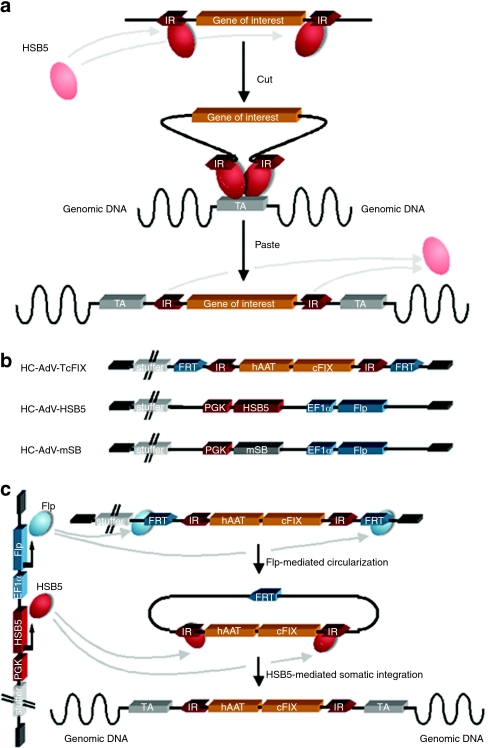
Figure 1.
Mechanism of Sleeping Beauty (SB) transposition and the high-capacity adenoviral vector (HC-AdV) system for delivery. (a) Depicted is a two-component system in which a gene of interest flanked by transposon-derived inverted repeats (IRs) is mobilized by the transposase protein provided in trans. For gene transfer experiments, the transposon is usually excised from a plasmid and integrated into a genomic target site (TA). This mechanism is commonly referred to as “cut-and-paste” mechanism. (b) HC-AdV-TcFIX represents the transposon-donor vector from which the transposon is mobilized. The transposon15 flanked by inverted repeats (IRs) and FRT sites for Flp-mediated excision. It expresses canine coagulation factor IX (cFIX) under control of the liver-specific human α-1-antitrypsin promoter (hAAT) including two liver-specific enhancers (HCR, hepatocyte control region; ApoE: apolipoprotein E). The HC-AdV-HSB5 contains a transgene expression-cassette for the hyperactive Sleeping Beauty (SB) transposase HSB5 under the control of the phosphoglycerate kinase promoter (PGK) and an expression-cassette for the Flp recombinase driven by the elongation factor-1-α promoter (EF1α). The control vector HC-AdV-mSB contains the mutated and inactive version of SB (mSB). All HC-AdVs include 22-kb stuffer DNA derived from human chromosomal DNA to optimize packaging of viral vectors. (c) For somatic integration cells are simultaneously infected with HC-AdV-TcFIX and HC-AdV-HSB5. Expressed Flp-recombinase recognizes FRT sites and mediates circularization and therefore transposon mobilization from the HC-AdV, which is essential for HSB5 functionality. Inverted repeats (IRs) of circularized intermediates are recognized by expressed hyperactive SB protein provided in trans, mediating somatic integration of the transposon into the cellular genome. HC-AdV, high-capacity adenoviral vector; HSB5, hyperactive Sleeping Beauty transposase; mSB, inactive transposase system.
Results
A two-component system for transposon mobilization from HC-AdVs
To achieve stabilized transgene expression, we used an improved version of the “two-vector-system” presented in our previous study.30 This system relies on cotransduction of one cell with two HC-AdVs. The first vector represents the transposon donor-vector (HC-AdV-TcFIX) which contains a transposon encoding the canine coagulation factor IX (cFIX)25 under the control of the liver-specific human α-1-antitrypsin promoter (hAAT) flanked by FRT recognition sites for Flp-mediated recombination (Figure 1b). The second vector (HC-AdV-HSB5) encodes Flp recombinase and the hyperactive SB transposase mutant HSB5 (Figure 1b), which displays tenfold increased integration efficiencies in comparison to the wild-type transposase.32 The mechanism of transposition from the adenoviral vector depends on two steps (Figure 1c). After both vectors cotransduce a target cell, Flp recombinase first mediates circularization of the transposon from the adenoviral vector genome. This step was necessary because adenoviral vector genomes predominantly exist as linear DNA molecule and we showed in the past that transposition mainly works from circular substrates.30 In a second step, the excised circle serves as a substrate for SB transposase-mediated somatic integration.
As a negative control for the integration machinery a derivate of the second vector (HC-AdV-mSB) containing an inactive version of SB transposase (mSB) was generated (Figure 1b). HC-AdVs were produced according to a previously described protocol for large-scale production of HC-AdV in spinner flasks.33,34 Final vector preparations were analyzed for helper virus contamination levels and the amount of transducing units per volume of the HC-AdV was determined (for details see Supplementary Materials and Methods section).
Stable transgene expression in mice after a single injection of the adenovirus/SB transposase hybrid-vector system
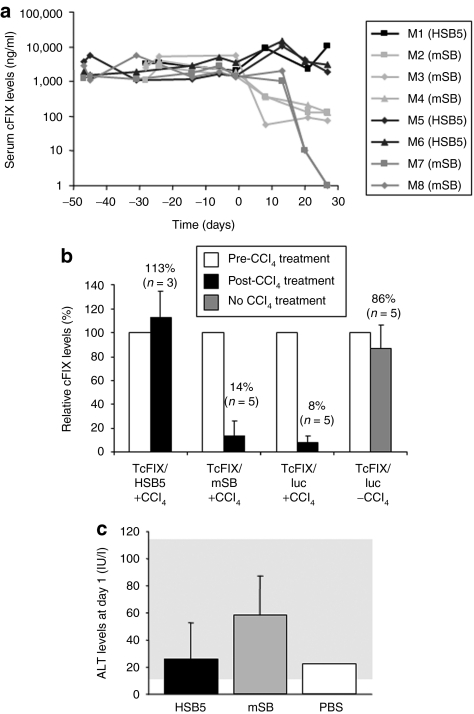
To address the question whether the new version of the adenoviral hybrid-vector system results into stable transgene expression levels in vivo, C57Bl/6 mice were coinjected at a ratio of 3:1 with the vectors HC-AdV-TcFIX and HC-AdV-HSB5, respectively. As a negative control for HSB5-mediated somatic integration, mice were treated with HC-AdV-mSB instead of HC-AdV-HSB5. In two independent experiments, animals were injected with 3 × 1010 and 1 × 1010 vps, respectively, or with a twofold higher dose. Animal injections, the viral dose, and the vector combinations, which were used, are summarized in Table 1. To demonstrate HSB5-mediated integration of the transgene expression-cassette from the adenoviral vector genome into the host chromosomal DNA, we intraperitoneally injected carbon tetrachloride (CCl4) 5–7 weeks after vector administration. CCl4 causes liver damage and therefore in treated liver tissue rapid cell cycling takes place to reconstitute normal liver size. As a consequence episomal vector forms are lost due to cell division whereas integrated transposons are maintained. To analyze stability of canine FIX expression, cFIX serum levels were measured periodically by enzyme-linked immunosorbent assay (ELISA) (Figure 2a). For mice treated with the functional SB transposase system, plasma cFIX levels maintained stable in a physiological range (cFIX levels of 2,000–10,000 ng/ml) even after injection of CCl4, whereas in mice transduced with vector encoding the inactive mSB, serum cFIX levels significantly dropped after CCl4 administration to concentrations <150 ng/ml (mice M2–M4) or undetectable levels (mice M7 and M8).
Table 1. Summary of animal injections.
Figure 2.
Stability of expression levels in mice treated with adenovirus-transposase hybrid-vectors and acute toxicity related with vectors. C57Bl/6 mice were coinjected with HC-AdV-TcFIX and HC-AdV-HSB5 at a ratio of 3:1 with a total number of 8 × 1010 viral particles (mouse M1) or 4 × 1010 viral particles (mice M5 and M6). Mice of the control groups were treated with HC-AdV-mSB instead of HC-AdV-HSB5 (8 × 1010 viral particles: mice M2–M4; 4 × 1010 viral particles: mice M7 and M8). Four to seven weeks after vector administration rapid hepatocyte proliferation was induced at day 0 by intraperitonal injection of carbon tetrachloride (CCl4) to analyze stability of transgene expression and somatic integration. (a) Canine FIX expression levels were measured periodically by ELISA. (b) Relative transgene expression levels in mice after delivery of HC-AdV-TcFIX with either HC-AdV-HSB5 (TcFIX/HSB5 + CCl4), HC-AdV-mSB (TcFIX/mSB + CCl4) or the irrelevant vector HC-AdV-luciferase (TcFIX/Luc + CCl4) as a control for persistence of the canine FIX-expressing vector without excision of the transgene by Flp recombination. To analyze persistence of transgene expression without CCl4 treatment (TcFIX/luc −CCl4), another control group solely received the vector HC-AdV-TcFIX and HC-AdV-luciferase. A total of 8 × 108 transducing units were injected at a ratio of 3:1 (transposon TcFIX vector: HSB5, mSB, or Luc vector). Mice were treated with CCl4 and serum levels of cFIX before CCl4 treatment (white bar) were defined as 100% for each group. Black bar, post-CCl4 treatment; gray bar: no CCl4 treatment. Serum cFIX levels were determined 3 weeks after CCl4 treatment for all groups. (c) Alanine transaminase (ALT) levels in serum samples of mice 1 day postinjection were measured to detect acute liver toxicity related with HC-AdV administration. Serum samples of mice injected with Dulbecco's phosphate-buffered saline (DPBS), the solvent for intravenous application of HC-AdVs were used as a negative control. cFIX, canine factor IX; ELISA, enzyme-linked immunosorbent assay; HC-AdV, high-capacity adenoviral vector; HSB5, hyperactive Sleeping Beauty transposase; mSB, inactive transposase system; PBS, phosphate-buffered saline.
As additional control, we analyzed the expression from episomal HC-AdV-TcFIX without Flp-mediated excision of the transposon. Therefore, mice were injected with the HC-AdV-TcFIX and HC-AdV-luciferase (see also Supplementary Materials and Methods section), a HC-AdV encoding for firefly luciferase replacing the vector HC-AdV-HSB5 in our experimental setup. Before injection of CCl4 similar serum, cFIX levels were detected in both groups. However, after a single injection of the liver toxin CCl4 expression levels rapidly decreased within the next 3 weeks (from serum cFIX levels of 1,300 to levels of 120 ng/ml) (Supplementary Figure S1). In sharp contrast, transgene expression levels from untreated mice remained stable.
To directly compare stability of transgene expression from episomal HC-AdV, HC-AdVs from which the transgene is excised, and HC-AdVs from which the transgene is excised and subsequently integrated into the host genome, we determined the loss rate of transgene expression for all systems with and without induction of rapid cell cycling in murine liver. We found that our integrating HC-AdV system utilizing HSB5 for somatic integration is superior in cycling cells compared to episomal HC-AdVs (Figure 2b). Moreover, we observed that the loss rate of transgene expression was independent of excision of the transposon from the adenoviral vector (Figure 2b). In conclusion, these results demonstrate that our system is clearly superior in rapidly dividing cells compared to conventionally used HC-AdVs, suggesting that this system may be useful for stable transduction of dividing cells (e.g., stem cells).
We also performed limited toxicity studies by monitoring alanine aminotransferase (ALT) levels in mouse serum samples of treated mice to detect acute toxicity associated with administration of HC-AdVs. As a negative control, mice were injected with Dulbecco's phosphate-buffered saline (PBS), the solvent for the HC-AdVs for injections. One day postinjection comparable ALT levels were measured in mice which received the HSB5 or the mSB vector system (Figure 2c). Although ALT levels in HSB5- and mSB-treated mice were slightly elevated compared to the control group treated with Dulbecco's PBS, we would like to emphasize that ALT levels remained in the normal range for all treated mice (20–80 IU/l).
Phenotypic correction after intravenous injection of the adenovirus/SB tranposase hybrid-vector system into hemophilia B dogs
After verifying that this improved version of the transposase system works efficiently in mice, we addressed the question whether transposition enables phenotypic correction in a preclinical animal model for hemophilia B. Therefore, we intravenously injected three hemophilia B dogs carrying a missense mutation in the catalytic domain of the genomic cFIX coding sequence.35 All dogs (D1, D2, and D3) were coinjected with the canine factor IX (cFIX) transposon donor vector and the transposase delivery vector at a 2:1 ratio. Virus injections, the viral dose, and the vector combinations, which were used in hemophilia B dogs, are summarized in Table 1. D1 received a total dose of 9.4 × 1011 vps/kg of the HSB5 system and D2 a 2.6-fold increased dose (2.4 × 1012 vps/kg). As a control, dog D3 was injected with the inactive version of the transposase system at a total dose of 1.2 × 1012 vps/kg, which was equivalent to the virus dose applied in dog D1.
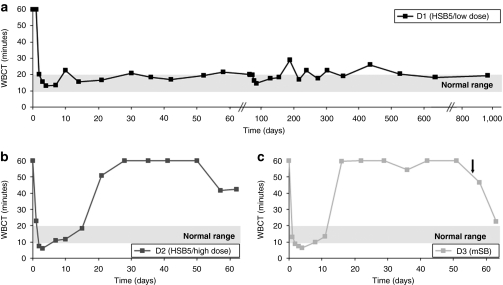
For dog D1, which received the HSB5-based hybrid-vector system, the whole blood clotting time was shortened to 18.5 minutes and stabilized for the total length of the study (960 days) (Figure 3a). This clearly demonstrated that phenotypic correction of the coagulation factor IX deficiency occurred and suggested that in contrast to our historical study utilizing a comparable dose of the cFIX encoding vector,25 stabilization of transgene expression occurred. In sharp contrast, systemic administration of the inactive transposase system (mSB) in dog D3 at a similar viral dose resulted in transient phenotypic correction for <16 days (Figure 3c). Surprisingly, dog D2, which also was treated with the HSB5-system but with an increased dose showed only transient phenotypic correction (Figure 3b). This may indicate that toxic side effects associated with the incoming viral proteins or a robust immunological response against incoming vps or against one of the transgene products may have occurred.
Figure 3.
Whole blood clotting time (WBCT) as indicator for phenotypic correction in treated hemophilia B dogs. The whole blood clotting time (WBCT) is an indicator for a functional coagulation cascade and therefore for a sufficient replacement of the inactive factor IX in hemophilia B dogs. The WBCT for untreated hemophilia B dogs is longer than 60 minutes. After injection of HC-AdVs the WBCT was measured periodically for (a) D1 (HSB5/low dose), (b) D2 (HSB5/high dose), and (c) D3 (mSB). The black arrow indicates a bleeding episode, which was treated by cFIX administration. cFIX, canine factor IX; HC-AdV, high-capacity adenoviral vector; HSB5, hyperactive Sleeping Beauty transposase; mSB, inactive transposase system.
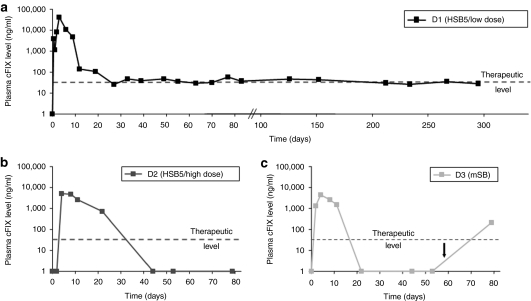
In order to monitor transgene expression, cFIX levels in canine plasma samples were monitored. We detected supraphysiological plasma cFIX levels of up to 40,000 ng/ml (normal cFIX level = 5,000 ng/ml) for dog D1 3–9 days postinjection (Figure 4a). During days 9 and 27 postinjection, we observed a rapid decline of cFIX plasma levels. However, after 4 weeks, plasma cFIX levels stabilized within a therapeutic range (60 ng/ml) for up to 295 days (latest time point analyzed by cFIX ELISA). For dogs D2 and D3 we observed physiological cFIX plasma levels 5–10 days postinjection which declined to undetectable levels 40 and 20 days postinjection for dogs D2 and D3, respectively (Figure 4b,c).
Figure 4.
Expression levels of canine factor IX in treated hemophilia B dogs. The expression level of canine factor IX (cFIX), which is encoded by the transposon, was measured by ELISA. Normal cFIX plasma levels of healthy dogs are between 500 and 12,000 ng/ml. Notably, 50 ng/ml are sufficient for effective coagulation (therapeutic level). Plasma levels of cFIX were measured in hemophilia B dog (a) D1 (HSB5/low dose), (b) D2 (HSB5/high dose), and (c) D3 (mSB). The black arrow indicates the administration of cFIX after a bleeding episode. ELISA, enzyme-linked immunosorbent assay; HSB5, hyperactive Sleeping Beauty transposase; mSB, inactive transposase system.
Laboratory measurements and toxicity profile in hemophilia B dogs
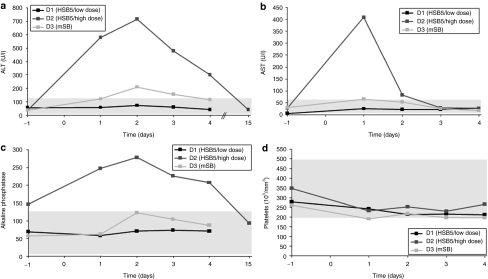
To investigate whether vector related toxicity occurred, we performed a plethora of laboratory measurements. We monitored aspartate aminotransferase (normal range, 12–118 U/l) and ALT (normal range, 15–66 U/l) levels in plasma of dogs D1, D2, and D3. We found that ALT and aspartate aminotransferase levels were elevated in dog D2, whereas transaminase levels for dogs D1 and D3 remained in a normal range (Figure 5a,b). Quantification of alkaline phosphatase levels revealed that this parameter was increased in dog D2, whereas levels stayed in a normal range for dogs D1 and D3 (Figure 5c). In concordance with these results, we also observed the most significant drop of platelets in dog D2 (Figure 5d). Furthermore, all other monitored parameters remained in a normal range for dogs D1 and D3. All laboratory measurements for treated dogs D1, D2, and D3 are summarized in Supplementary Table S1.
Figure 5.
Acute toxicity profile of treated hemophilia B dogs. Plasma samples of dogs D1 (HSB5/low dose), D2 (HSB5/high dose), and D3 (mSB) were collected at indicated time points and analyzed for markers related with acute liver toxicity. (a) Alanine aminotransferase levels (ALT; normal range 12–118 U/l) were measured as well as (b) aspartate aminotransferase levels (AST; normal range 15–66 U/l), and (c) alkaline phosphatase levels (normal range 5–131 U/l). In addition (d) the number of platelets (normal range 200,000–500,000 per mm3) were quantified for all three dogs. HSB5, hyperactive Sleeping Beauty transposase; mSB, inactive transposase system.
Lack of antibodies against cFIX and detection of low levels for neutralizing antiadenoviral antibodies
To further investigate the potential reasons for transient phenotypic correction in dogs D2 and D3 and the rapid decrease of cFIX plasma levels in dog D1 during the first weeks postinjection, we searched for anti-cFIX inhibitors and measured neutralizing antiadenoviral antibody levels. The Bethesda inhibitor assay was used to detect anti-cFIX inhibitors in dog plasma before and after injection, but no antibodies against the transgene-encoded product were detected in either dog (data not shown). Thus, the decrease in cFIX transgene expression was not caused by a humoral immune response directed against the recombinant cFIX protein.
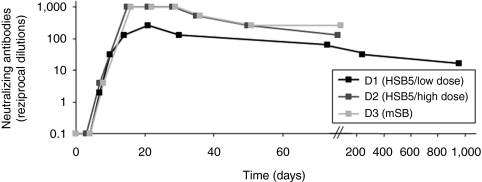
For analysis of the humoral immune response directed against the adenoviral vector, we examined dog serum at various time points for the presence of antiadenoviral neutralizing antibodies. For this purpose, we used a neutralization assay, measuring the inhibitory effect of antiadenoviral antibodies contained in canine serum samples by infection of 293 cells with an adenoviral vector encoding a reporter gene. Neutralizing antibodies for dog D1 were monitored until the end of the experiment (960 days postinjection) and for dogs D2 and D3 up to 80 days postinjection (latest time points measured). Highest levels were measured 2–3 weeks post-treatment (Figure 6). Although the biological relevance of this finding is not clear at the time, injection of HC-AdVs at a similar dose resulted in comparable or even higher levels of neutralizing antibodies in a previous study,25 indicating that these levels are normal for the injected vector dose.
Figure 6.
Neutralizing antibodies directed against adenoviral vectors in treated hemophilia B dogs. As an indicator for the adaptive immune response challenged by the HC-AdVs, neutralizing antibodies directed against the vector were measured. Therefore, the highest dilution of canine serum was determined, which is able to inhibit 50% transduction efficiencies of a reporter virus in 293 cells. The neutralizing-antibody titer was defined as the reciprocal of this dilution. HC-AdV, high-capacity adenoviral vector.
Molecular analysis of transgene persistence and detection of transposition events in canine liver
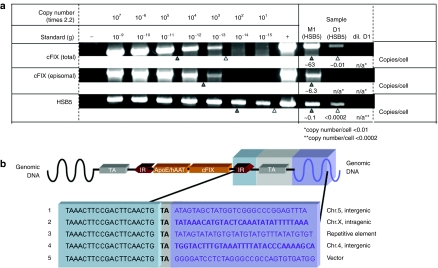
To demonstrate that the cFIX transposon underwent transposition in canine liver and to analyze the fate of transduced vector DNA, we performed detailed molecular analyses of liver-derived canine genomic DNA. First, we addressed the question whether episomal vector forms were maintained in liver cells or whether they were lost over time. Therefore, we performed semiquantitative PCR analysis of liver genomic DNA isolated from dog D1 960 days postinjection specifically detecting vector genome sequences contained in adenoviral vector genomes (amplified regions are depicted in Supplementary Figure S2). We observed that episomal vector forms either containing the cFIX transposon (HC-AdV-TcFIX and the excised circular cFIX transposase substrate) or the hyperactive transposase (HC-AdV-HSB5) were below 0.01 and 0.0002 genome copy numbers per cell, respectively (Figure 7a). These results indicated that episomal vector genome numbers were strongly reduced over time. In contrast, 0.01 copy numbers per cell of the cFIX transposon were maintained in canine liver (Figure 7a) suggesting that cFIX expression was mainly derived from integrated transposons. However, more detailed studies need to be performed and at the time we cannot exclude whether transgene expression from episomal vector genomes may also contribute to measurable cFIX expression levels and phenotypic correction in dog D1. Similar tendencies for the vector status were obtained in mice (Figure 7a). In addition canine genomic DNA was analyzed for excised and episomal circular intermediates, but no evidence for the excised transposons could be shown (Supplementary Figure S3).
Figure 7.
Molecular status of vector genomes and detection of transposition events in transduced liver cells. Hyperactive Sleeping Beauty transposase (HSB5)-transduced liver samples of mouse M1 (28 days post-CCl4 treatment) and dog D1 (960 days postinjection) were analyzed regarding episomal HC-AdV genomes and canine factor IX (cFIX) transposon integrated into the host genome. (a) Fate of vector genomes in transduced canine and murine liver was analyzed by PCR. Top, total number of coagulation factor IX transgenes; middle, detection of intact and episomal transposase donor-vector genomes (high-capacity adenoviral vector HC-AdV-TcFIX); bottom, PCR specifically detecting Sleeping Beauty transposase encoding sequences. D1, liver genomic DNA from dog D1 (500 ng); dil. D1, liver genomic DNA from dog D1 (10 ng); M1, liver genomic DNA from mouse M1 (10 ng). Serial DNA standard dilutions ranged from 1 ng (10−9 g; 2.2 × 107 genome copies) to 1 fg (10−15 g; 22 genome copies) were analyzed and spiked with 50-ng genomic DNA derived from 293 cells to simulate canine genomic DNA in samples. Genomic DNA obtained from HeLa cells, infected with the corresponding viral vectors was used as positive control (+) whereas DNA from noninfected HeLa cells served as negative control (–). Vector genome copies are presented as copy numbers per liver cell (copy number/cell). (b) Identification of sites of insertion in canine liver cell genomes after HSB5-mediated transposition. Genomic regions containing the transposon were isolated, sequenced, and blasted against the canine genome database. Five integration events are depicted and the corresponding chromosomal regions are mentioned. HC-AdV, high-capacity adenoviral vector.
To determine sites of insertion after transposition, we applied a PCR-based assay, which detects chromosomal DNA linked to the integrated transposon inverted repeats (IRs). We identified transposition events with intact IR and integration into a target site-dinucleotide in genomic DNA of canine liver cells derived from dog D1 (Figure 7b; detailed blast results are depicted in Supplementary Figure S4).
Discussion
This study evaluated for the first time efficacy of the SB transposase system in a large animal model. Herein, a hyperactive transposon-based vector system for somatic integration was delivered into hemophilia B dogs by a HC-AdV. We found that one dog (D1) showed long-term phenotypic correction (Figure 3a) of the clotting disorder (3 years) and persistent expression of cFIX (Figure 4a) which was mainly derived from integrated transgenes (Figure 7a). Furthermore, there was no measurable acute liver toxicity (Figure 5a–d).
However, application of a twofold higher viral dose (2.4 × 1012 vps/kg body weight) showed transient albeit complete correction of the hereditary genetic disease in canines (Figure 3b). Therefore, increasing the viral dose resulted in transient liver toxicity (Figure 5a–d) and may present one potential reason for the strong decrease of plasma cFIX levels observed in dog D2. A similar or even higher dose as compared to dog D2 was used in previous studies for phenotypic correction of canine hemophilia.26,36,37 In concordance with our present study, these studies also measured transient hepatotoxicity after systemic administration of the HC-AdV at a viral vector load of 1–3 × 1012 vps/kg body weight. These dose-dependent toxic effects are supported by our historical data in hemophilia B dogs showing that a lower dose of 0.9 × 1011 vps/kg resulted in undetectable liver toxicity and no thrombocytopenia.25
As speculated already in other studies performed in canines, our data emphasize that only a small therapeutic window for HC-AdV may exist. This dose threshold effect, for which a linear increase in viral vector dose does not correlate with protein expression, was also shown in mice.38 However, the minimal dose in mice for achieving long-term transgene expression seems to be lower compared to canines. This underscores the species-dependent differences after systemic administration of adenoviral vectors and that direct extrapolation across species may not be possible.
In the present study, we explored for the first time a hyperactive SB transposase for somatic integration from an adenoviral vector. We believe that utilizing HSB5 with a tenfold higher transposition activity in vitro in comparison to wild-type SB which was used in our previous study25 was probably a critical factor for achieving stabilized transgene expression in a large animal model. However, the actual integration efficiency of the HSB5-based system and the grade of improvement in comparison to our previously described two-vector system utilizing wild-type SB transposase30 needs to be investigated in more detail. To further optimize our integrating vector system it may be interesting to explore a novel SB transposase mutant with 100-fold increased integration efficiencies (SB100×).39 In fact, utilizing SB100× may allow further decreasing the viral vector dose for achieving sufficient integration and stabilized transgene expression, although a minimal dose which yet remains to be determined will still be required for cotransduction of cells with both vectors. Another approach for reducing the effective viral vector dose may be the use of hyperactive cFIX variants as shown by Brunetti-Pierri et al., who delivered a hyperactive human FIX expression-cassette using an adenoviral vector resulting in an improved therapeutic effect.40 Moreover, with respect to targeting liver cells, our approach could be optimized by utilizing more efficient routes of administration. For instance, the recently introduced pseudo-hydrodynamic balloon catheter injection method was shown to increase liver-transduction efficiency.24
Taking a closer look at the antiadenoviral neutralizing-antibody levels (Figure 6) for all treated dogs, an increase was observed 2 weeks after administration. During this time period, plasma cFIX levels decreased to low or even undetectable levels for all dogs. Because no significant anti-cFIX inhibitor levels were detected, this increase in antiadenoviral neutralizing-antibody levels could suggest that a robust adaptive immune response against the administrated viral vector capsids or cells presenting viral capsid proteins occurred. In particular the cellular immune response might be one major reason for the subsequent clearance of transduced cells and therefore the rapid decline in cFIX antigen levels. However, in the present study, we only analyzed antiadenoviral neutralizing-antibody levels in hemophilia B dogs and it is important to emphasize that not only the humoral immune response but also the innate immune system may play an important role.
Another safety issue of our hybrid-vector system may be the presence of HSB5 and Flp recombinase encoding DNA sequences in our vectors. Besides the fact, that for the SB transposase system overproduction inhibition could occur, which might potentially be the reason for transient effects in dog D2, high levels of SB transposase and/or Flp recombinase might result in genotoxicity or other adverse effects. Furthermore, at the time it is not known whether long-term expression in vivo of either SB transposase and/or Flp recombinase causes any side effects such as unwanted recombination of host genomic DNA which might even occur at low expression rates. However, our results indicated that episomal vector genomes were strongly reduced over time or even completely cleared from the organ. This could be shown for the vector encoding HSB5 as well as for cFIX (Figure 7a). These results are in concordance with previous data showing low frequencies of adenoviral vectors for homologous or heterologous integration events in transduced hepatocytes in mice.28,29
Regarding the safety for SB transposase-mediated integration, it is established that SB transposase mediates somatic integration into target site-dinucleotides.31 Up to now no insertional mutagenesis is known for gene therapy studies utilizing SB transposases and a recent study revealed that SB shows a random integration pattern with a small bias toward genes.41 Moreover, integration preferences were not significantly influenced by transcriptional activity. Although the dog-specific genomic sequence database from National Center for Biotechnology Information (NCBI) is not complete,42,43 we have begun to analyze insertion sites after SB-mediated somatic integration from the adenoviral vector into the canine genome (Figure 7b). We realize that the number of insertion sites my not be representative at this point, but our genome-wide screen for sites of transposition events from the adenoviral vector in murine liver cells revealed a random integration pattern without any preferences for integration into transcriptionally active sites (data not shown).
The combination of the SB transposase system with gutless adenovirus showed significantly stabilized transgene expression during rapid cell cycling (Figure 2a). Furthermore, we showed that in the presence of cell division our novel vector system was superior to conventionally utilized and episomally maintained HC-AdVs (Figure 2b). However, we are aware of the fact that hepatocytes are in most cases quiescent cells and therefore, stable liver-transduction for the treatment of hemophilia B may not be the perfect setup and model disease for our novel vector system. Moreover, it was also demonstrated that a single injection of an HC-AdV without integration machinery can result in long-term expression and phenotypic correction.16,20,21,22,23 However, for instance for stable transduction of stem cells, progenitor cells or any other cell type which may undergo extensive cell division, our system may be a very valuable tool.
It is of note that in a previously published study, we found that HC-AdV genomes seem to be more stable during cell division compared to nonviral DNA.44 However, within the present study we found that after injection of HC-AdV-TcFIX alone and subsequent CCl4 treatment for induction of cell cycling transgene expression levels rapidly declined. This loss in transgene expression was as extensive as in mice which received the mSB encoding vector. Although this is somewhat different to our previously published study it is important to point out that in contrast to our newly performed experiment our previous study utilized partial hepatectomy for induction of cell cycling resulting into 2–3 cell divisions of each hepatocyte. In contrast, treatment with CCl4 is harsher and results into extensive liver necrosis and subsequently also more cell divisions. This in turn may lead to a more extensive loss of episomal vector genomes. This is on concordance with our previous study in which we also observed a dramatic loss of transgene expression and vector genome copies after CCl4 treatment.45
It is of note, that adenoviral vectors were utilized in the past to deliver other than SB-based integration machineries derived from retrotransposons, retrovirus, AAV, and bacteriophage-derived integrase phiC31 (reviewed in ref. 46). At the time, none of the currently available systems for somatic integration may be perfect with respect to genotoxicity and SB-mediated integration may represent a valuable alternative, however, site-specific integration into a single and “safe” genomic locus may be the ideal option. Towards this goal, SB fusion proteins exploring zinc-fingers for site-specific DNA binding32,45 and zinc-finger nucleases in the context of viral vectors46 are currently being explored.
Last but not least, one important challenge which needs to be addressed in the future is the complexity of the design of this adenovirus/SB transposase hybrid-vector system. To increase integration efficiency the ratio between transposons and SB transposase proteins should be optimized including the ratio between vectors and the promotors driving the transcription of SB transposase and Flp recombinase. Moreover, utilizing an inducible system for expression of the integration machinery might reduce genotoxic and immunogenic side effects and may decrease an overproduction inhibition effect. Finally, we believe that exploring a “one vector-strategy” including all components for somatic integration (therapeutic DNA and the respective recombinase) in one vector genome is an option to overcome the necessity of cotransduction of one cell with two vectors.
In summary, this study provides for the first time novel insights into efficacy of adenoviral hybrid-vectors for somatic integration and the SB system in a larger animal model. For stable transduction and treatment of tissues with a high-cell turn-over rate, our system represents a valuable tool for treatment. For instance, treatment of diseases affecting the skin or ex vivo correction of hematopoietic stem cells may be feasible utilizing our vector system. We believe that with further improvements this system may even pave the way toward clinical applications.
Materials and Methods
Generation of HC-AdVs. See Supplementary Materials and Methods section for detailed description of cloning procedures for generation of adenoviral production plasmids and HC-AdV amplification, purification, and titration.
Animal studies. C57Bl/6 mice were kept and treated according to the guidelines of the Ludwig Maximilians University of Munich (Munich, Germany). All mice were tail vein injected using a total volume of 200 µl. Viral vector preparations were diluted in Dulbecco's PBS. Rapid cell cycling of murine liver cells was induced by intraperitoneal administration of 50 µl CCl4 (Sigma-Aldrich, Taufkirchen, Germany) solution (1:1 dilution in mineral oil).
Inbred hemophilia B dogs were from the University of North Carolina at Chapel Hill (Chapel Hill, NC). These hemophilia B dogs carry a missense mutation in the catalytic domain of the FIX-coding sequence resulting in undetectable FIX activity.35 Dog studies were performed under the guidelines of the University of North Carolina. For vector administration, dogs were sedated with Domitor (750 µg/m2 body surface area) and the vector dilution (~18.5 ml) followed by ~10 ml PBS for vector wash was infused by peripheral vein injection at 0.5 ml/minute into the right foreleg. During injection heart rate, blood pressure, and temperature were monitored. Routine laboratory measurements before and after adenoviral administration were performed as described earlier.25,47
Measurement of ALT serum levels in mice. For quantification of ALT levels in murine serum, we followed the instructions of the ALT detection kit (Randox, Crumlin, UK) as described earlier.45 We applied 15 µl of murine serum for each reaction.
Blood analysis after adenoviral-mediated gene transfer into hemophilia B dogs. Blood samples were obtained periodically from hemophilia B dogs and the whole blood clotting time was determined as previously described.25,47 A Bethesda inhibitor assay was used to detect anti-canine FIX inhibitors in dog serum. In brief, dog plasma samples were mixed with pooled normal canine plasma and incubated at 37 °C. The anti-canine FIX inhibitor titer was calculated from the residual FIX activity of each sample.
ELISA. The cFIX ELISA measuring cFIX levels in murine serum and canine plasma was performed.48 Briefly, to coat ELISA plates, the polyclonal rabbit anti-cFIX primary antibody (RACIX-IG; Affinity Biologicals, Ancaster, ON, Canada) was diluted 1:500 for dog and 1:200 for mouse samples. After incubation with samples obtained from treated animals the sheep horseradish peroxidase–conjugated secondary antibody (SACIX-HRP; Affinity Biologicals) was diluted 1:1,000 and 1:200 for mouse and canine samples, respectively. For generation of a standard curve, dilutions of pooled dog plasma with a normal range of 5–11.5 µg/ml cFIX antigen were used.
Detection of neutralizing antiadenoviral antibodies. The principle of this test was to analyze at which dilution the canine serum potentially containing antiadenoviral antibodies is able to inhibit 50% of the transduction of a reporter virus. Two hundred and ninety three cells were cultured in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum. One day before performing the assay, 293 cells were seeded into 96-well tissue culture plates containing 100 µl medium. To destroy heat-sensitive complement components, dog serum was incubated at 56 °C for 40 minutes. Serum samples were diluted in twofold steps in Dulbecco's modified Eagle's medium with 10% heat-inactivated fetal bovine serum in a total volume of 50 µl. We added 50 µl of the lacZ-expressing reporter virus Ad-RSV/lacZ6 diluted with Dulbecco's modified Eagle's medium containing 10% heat-inactivated fetal bovine serum (final concentration of 1 × 109 plaque-forming units/ml). After incubation of 1 hour at 37 °C, the serum virus mixture was applied to 293 cells with 80% confluency. After 24 hours, cells were fixed with 0.5% glutaraldehyde, washed with PBS, and stained in PBS-containing 3 mmol/l potassium ferricyanide, 3 mmol/l potassium ferrocyanide, 2 mmol/l MgCl2, and 1 mg/ml 5-bromo-4-chloro-3-indolyl-β-𝒹-galactopyranoside (X-gal). As a negative control serum samples derived from untreated dogs and PBS were used. The neutralizing antiadenovirus antibody titer was defined as the reciprocal of the highest dilution of serum at which the infectivity of the reporter virus was decreased by 50%.
Molecular characterization of vector genomes by PCR. To analyze whether adenoviral vector genomes were maintained in murine and canine liver and to determine the amount of the integrated cFIX encoding transposons, we isolated genomic DNA from liver tissue (~100 mg). PCR analysis was performed using GoTaq polymerase (Promega, Mannheim, Germany). For mouse samples 10 ng and for canine samples 500 and 10 ng (dil. D1) of isolated genomic DNA were subjected to PCR.
Semiquantitative PCRs for detection of total number of canine coagulation factor IX (cFIX) encoding DNA were run with 45 cycles at 95 °C for 45 seconds, 53 °C for 45 seconds, and 72 °C for 60 seconds using oligonucleotides ApoE for (5′- CCC AGA GAC TGT CTG ACT CA -3′) and cFIXshort.rev (5′-GAG ACA CAC CTC ATT ACA TA-3′). This PCR amplifies a 998-bp DNA fragment spanning the liver-specific hAAT promoter and the first part of the cFIX complementary DNA. Therefore, it detects episomal versions of the cFIX transgene (excised circles after Flp-recombinase recombination and complete adenoviral vector genomes of HC-AdV-TcFIX) as well as integrated copies of the cFIX encoding transposon.
To address the question whether episomal HC-AdV-TcFIX genomes (Figure 2b) were present, a touchdown-PCR specific for a 1,455-bp fragment of HC-AdV-TcFIX was run with 35 cycles at 95 °C for 45 seconds, 65 °C for 45 seconds, 72 °C for 90 seconds, decreasing the annealing temperature each cycle for 0.3 °C and utilizing the oligonucleotides pICeu (5′-TAA CTA TAA CGG TCC TAA GGT AGC -3′) and pApoE/hAAT-3 (5′- AGA TCA GGG GGA TCA TTC ACT GTC CCA GGT CA-3′).
For detection of HC-AdV-HSB5 a touchdown-PCR was performed with 35 cycles at 95 °C for 45 seconds, 65 °C for 45 seconds, 72 °C for 80 seconds, decreasing the annealing temperature at each cycle for 0.3 °C and using the oligonucleotides PGK-Pro_5_ BglII (5′- AGA TCT ACC GGG TAG GGG AGG CGC-3′) and HSB5'1,000 (5′-GCT CCC AAG GAT GAA CCA GAC TTG-3′) amplifying a 585 bp region.
Circular intermediates were detected by touchdown-PCR (product: 700 bp). Thirty-five cycles at 95 °C for 45 seconds, 65 °C for 45 seconds, 72 °C for 50 seconds, decreasing annealing temperature each cycle for 0.3 °C were performed and the oligonucleotides fwFRTcyclization (5′-CCA AGC TGT TTA AAG GCA CA-3′) and rev1FRTcyclization (5′-GTA GGT CAC GGT CTC GAA GC-3′) were used.
For calculation of genome copy numbers per cell we performed serial DNA standard dilutions of plasmids pHC-AdV-TcFIX and pHC-AdV-HSB5 ranging from 1 ng (10−9 g; 2.2 × 107 copies) to 1 fg (10−15 g; 22 copies). Dilutions were spiked with genomic DNA derived from HeLa cells. For calculation of genome copy numbers per cell, we took into account that one copy of a diploid canine and mouse genome contains 5 billion and 5.7 billion bp, respectively.
Analysis of sites of insertion after transposon-mediated somatic integration in canine liver cells. Genomic DNA isolated from canine liver cells was used to determine sites of insertion by the BD GenomeWalkerTM Kit from BD Biosciences Clontech (Heidelberg, Germany). Therefore four genomic DNA libraries were produced with restriction enzyme nucleases that create blunt-ended DNA fragments (StuI, EcoRV, PvuII, and SspI) and a linker was ligated to the DNA fragments. Subsequently a two-step PCR was performed in which one primer binds to the IR of the IR and the other primer binds to the linker (primer pair for first PCR: IR-specific primer 1, 5′-cct taa gac agg gaa tct tta ctc gga-3′ and adopter primer 1, 5′-gta ata cga ctc act ata ggg c-3′ primer pair for the nested PCR: IR-specific primer 2, 5′-ggc taa ggt gta tgt aaa ctt ccg act -3′ and adopter primer 2, 5′-act ata ggg cac gcg tgg t-3′). PCR products were subcloned using the Zero Blunt TOPO PCR cloning kit (Invitrogen, Karlsruhe, Germany) and analyzed by sequencing. Sequencing results were blasted against a dog-specific genomic sequence data base from NCBI (http://www.ncbi.nlm.nih.gov/projects/genome/guide/dog/).
SUPPLEMENTARY MATERIAL Figure S1. Expression levels from episomal HC-AdVs. C57Bl/6 mice were co-injected with HC-AdV-TcFIX and HC-AdV-luciferase (n=10) at a ratio of 3: 1 (transposon TcFIX vector: luciferase vector) with a total number of 8x108 transducing units. Three weeks after vector administration (on day 21 virus post-injection) one group (n=5) was injected with carbon tetrachloride (+CCl4) to induce hepatocyte proliferation. (a) Stability of transgene expression was monitored by performing an ELISA two and three weeks post CCl4 treatment (days 36 and 42). (b) The control group (n=5) was not treated with CCl4 (-CCl4). Figure S2. PCR setups for specific detection of total cFIX and episomal HC-AdVs. Primers are presented as horizontal arrows. (i) For detection of all DNA sequences encoding the cFIX expression cassette, primer binding sites in the liver-specific hAAT-promotor and the cFIX encoding sequence were chosen. Therefore, interferences with the genomic cFIX encoding sequence were avoid and only transposon encoded expression cassettes were detected independent of the molecular status of the transposon (HC-AdV, circular intermediate or integrated transposon). (ii) Primer-set for amplification of the intact episomal HC-AdV-TcFIX genome without excision of the transposon. (iii) Primer-set specifically detecting Sleeping Beauty Transposase encoding sequences. Figure S3. Detection of circular intermediates in transduced canine liver cells. Additional PCR analysis to detect specifically episomal circular intermediates after Flp-mediated recombination was performed. (a) Primer binding site are depicted. (b) Two dilutions of liver genomic DNA from D1 (D1: 500 ng; D1 dil.: 10 ng) were analyzed. As a positive control (+) genomic DNA obtained from Hela cells infected with the corresponding viral vectors was used, whereas DNA from non-infected Hela cells served as negative control (–). Figure S4. Detailed description of integration sites after SB-mediated transposition into genomic DNA of canine liver cells. Table S1. Laboratory measurements of dogs D1 (HSB5), D2 (HSB5, high dose), and D3 (mSB). Materials and Methods.
Acknowledgments
The authors thank Philip Ng for providing the producer cell line 116 and the helper virus AdNG163R-2. This work was supported in part by DFG grants SFB 455 and SPP 1230, the Wilhelm Sander-Foundation, EU Framework Programme 7 (Persistent Transgenesis), the Friedrich-Baur-Foundation (A.E.) and the National Institutes of Health NHLBI HL064274 (M.A.K). W.Z. was supported by a fellowship from the Chinese Scholarship Council in cooperation with Northwestern A&F University (Yangling, China).
Supplementary Material
Expression levels from episomal HC-AdVs. C57Bl/6 mice were co-injected with HC-AdV-TcFIX and HC-AdV-luciferase (n=10) at a ratio of 3: 1 (transposon TcFIX vector: luciferase vector) with a total number of 8x108 transducing units. Three weeks after vector administration (on day 21 virus post-injection) one group (n=5) was injected with carbon tetrachloride (+CCl4) to induce hepatocyte proliferation. (a) Stability of transgene expression was monitored by performing an ELISA two and three weeks post CCl4 treatment (days 36 and 42). (b) The control group (n=5) was not treated with CCl4 (-CCl4).
PCR setups for specific detection of total cFIX and episomal HC-AdVs. Primers are presented as horizontal arrows. (i) For detection of all DNA sequences encoding the cFIX expression cassette, primer binding sites in the liver-specific hAAT-promotor and the cFIX encoding sequence were chosen. Therefore, interferences with the genomic cFIX encoding sequence were avoid and only transposon encoded expression cassettes were detected independent of the molecular status of the transposon (HC-AdV, circular intermediate or integrated transposon). (ii) Primer-set for amplification of the intact episomal HC-AdV-TcFIX genome without excision of the transposon. (iii) Primer-set specifically detecting Sleeping Beauty Transposase encoding sequences.
Detection of circular intermediates in transduced canine liver cells. Additional PCR analysis to detect specifically episomal circular intermediates after Flp-mediated recombination was performed. (a) Primer binding site are depicted. (b) Two dilutions of liver genomic DNA from D1 (D1: 500 ng; D1 dil.: 10 ng) were analyzed. As a positive control (+) genomic DNA obtained from Hela cells infected with the corresponding viral vectors was used, whereas DNA from non-infected Hela cells served as negative control (–).
Detailed description of integration sites after SB-mediated transposition into genomic DNA of canine liver cells.
Laboratory measurements of dogs D1 (HSB5), D2 (HSB5, high dose), and D3 (mSB).
REFERENCES
- Kay MA, Rothenberg S, Landen CN, Bellinger DA, Leland F, Toman C, et al. In vivo gene therapy of hemophilia B: sustained partial correction in factor IX-deficient dogs. Science. 1993;262:117–119. doi: 10.1126/science.8211118. [DOI] [PubMed] [Google Scholar]
- Brown BD, Cantore A, Annoni A, Sergi LS, Lombardo A, Della Valle P, et al. A microRNA-regulated lentiviral vector mediates stable correction of hemophilia B mice. Blood. 2007;110:4144–4152. doi: 10.1182/blood-2007-03-078493. [DOI] [PubMed] [Google Scholar]
- Vandendriessche T, Thorrez L, Acosta-Sanchez A, Petrus I, Wang L, Ma L, et al. Efficacy and safety of adeno-associated viral vectors based on serotype 8 and 9 vs. lentiviral vectors for hemophilia B gene therapy. J Thromb Haemost. 2007;5:16–24. doi: 10.1111/j.1538-7836.2006.02220.x. [DOI] [PubMed] [Google Scholar]
- Niemeyer GP, Herzog RW, Mount J, Arruda VR, Tillson DM, Hathcock J, et al. Long-term correction of inhibitor-prone hemophilia B dogs treated with liver-directed AAV2-mediated factor IX gene therapy. Blood. 2009;113:797–806. doi: 10.1182/blood-2008-10-181479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manno CS, Pierce GF, Arruda VR, Glader B, Ragni M, Rasko JJ, et al. Successful transduction of liver in hemophilia by AAV-Factor IX and limitations imposed by the host immune response. Nat Med. 2006;12:342–347. doi: 10.1038/nm1358. [DOI] [PubMed] [Google Scholar]
- Kay MA, Landen CN, Rothenberg SR, Taylor LA, Leland F, Wiehle S, et al. In vivo hepatic gene therapy: complete albeit transient correction of factor IX deficiency in hemophilia B dogs. Proc Natl Acad Sci USA. 1994;91:2353–2357. doi: 10.1073/pnas.91.6.2353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Ertl HC., and, Wilson JM. MHC class I-restricted cytotoxic T lymphocytes to viral antigens destroy hepatocytes in mice infected with E1-deleted recombinant adenoviruses. Immunity. 1994;1:433–442. doi: 10.1016/1074-7613(94)90074-4. [DOI] [PubMed] [Google Scholar]
- Yang Y, Xiang Z, Ertl HC., and, Wilson JM. Upregulation of class I major histocompatibility complex antigens by interferon gamma is necessary for T-cell-mediated elimination of recombinant adenovirus-infected hepatocytes in vivo. Proc Natl Acad Sci USA. 1995;92:7257–7261. doi: 10.1073/pnas.92.16.7257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Li Q, Ertl HC., and, Wilson JM. Cellular and humoral immune responses to viral antigens create barriers to lung-directed gene therapy with recombinant adenoviruses. J Virol. 1995;69:2004–2015. doi: 10.1128/jvi.69.4.2004-2015.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amalfitano A, Hauser MA, Hu H, Serra D, Begy CR., and, Chamberlain JS. Production and characterization of improved adenovirus vectors with the E1, E2b, and E3 genes deleted. J Virol. 1998;72:926–933. doi: 10.1128/jvi.72.2.926-933.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrews JL, Kadan MJ, Gorziglia MI, Kaleko M., and, Connelly S. Generation and characterization of E1/E2a/E3/E4-deficient adenoviral vectors encoding human factor VIII. Mol Ther. 2001;3:329–336. doi: 10.1006/mthe.2001.0264. [DOI] [PubMed] [Google Scholar]
- Gorziglia MI, Kadan MJ, Yei S, Lim J, Lee GM, Luthra R, et al. Elimination of both E1 and E2 from adenovirus vectors further improves prospects for in vivo human gene therapy. J Virol. 1996;70:4173–4178. doi: 10.1128/jvi.70.6.4173-4178.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parks RJ, Chen L, Anton M, Sankar U, Rudnicki MA., and, Graham FL. A helper-dependent adenovirus vector system: removal of helper virus by Cre-mediated excision of the viral packaging signal. Proc Natl Acad Sci USA. 1996;93:13565–13570. doi: 10.1073/pnas.93.24.13565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parks RJ., and, Graham FL. A helper-dependent system for adenovirus vector production helps define a lower limit for efficient DNA packaging. J Virol. 1997;71:3293–3298. doi: 10.1128/jvi.71.4.3293-3298.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunetti-Pierri N, Stapleton GE, Palmer DJ, Zuo Y, Mane VP, Finegold MJ, et al. Pseudo-hydrodynamic delivery of helper-dependent adenoviral vectors into non-human primates for liver-directed gene therapy. Mol Ther. 2007;15:732–740. doi: 10.1038/sj.mt.6300102. [DOI] [PubMed] [Google Scholar]
- Morral N, Parks RJ, Zhou H, Langston C, Schiedner G, Quinones J, et al. High doses of a helper-dependent adenoviral vector yield supraphysiological levels of α1-antitrypsin with negligible toxicity. Hum Gene Ther. 1998;9:2709–2716. doi: 10.1089/hum.1998.9.18-2709. [DOI] [PubMed] [Google Scholar]
- Brunetti-Pierri N, Palmer DJ, Beaudet AL, Carey KD, Finegold M., and, Ng P. Acute toxicity after high-dose systemic injection of helper-dependent adenoviral vectors into nonhuman primates. Hum Gene Ther. 2004;15:35–46. doi: 10.1089/10430340460732445. [DOI] [PubMed] [Google Scholar]
- Mane VP, Toietta G, McCormack WM, Conde I, Clarke C, Palmer D, et al. Modulation of TNFα, a determinant of acute toxicity associated with systemic delivery of first-generation and helper-dependent adenoviral vectors. Gene Ther. 2006;13:1272–1280. doi: 10.1038/sj.gt.3302792. [DOI] [PubMed] [Google Scholar]
- Muruve DA, Cotter MJ, Zaiss AK, White LR, Liu Q, Chan T, et al. Helper-dependent adenovirus vectors elicit intact innate but attenuated adaptive host immune responses in vivo. J Virol. 2004;78:5966–5972. doi: 10.1128/JVI.78.11.5966-5972.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehrhardt A., and, Kay MA. A new adenoviral helper-dependent vector results in long-term therapeutic levels of human coagulation factor IX at low doses in vivo. Blood. 2002;99:3923–3930. doi: 10.1182/blood.v99.11.3923. [DOI] [PubMed] [Google Scholar]
- Kim IH, Józkowicz A, Piedra PA, Oka K., and, Chan L. Lifetime correction of genetic deficiency in mice with a single injection of helper-dependent adenoviral vector. Proc Natl Acad Sci USA. 2001;98:13282–13287. doi: 10.1073/pnas.241506298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiedner G, Morral N, Parks RJ, Wu Y, Koopmans SC, Langston C, et al. Genomic DNA transfer with a high-capacity adenovirus vector results in improved in vivo gene expression and decreased toxicity. Nat Genet. 1998;18:180–183. doi: 10.1038/ng0298-180. [DOI] [PubMed] [Google Scholar]
- Toietta G, Mane VP, Norona WS, Finegold MJ, Ng P, McDonagh AF, et al. Lifelong elimination of hyperbilirubinemia in the Gunn rat with a single injection of helper-dependent adenoviral vector. Proc Natl Acad Sci USA. 2005;102:3930–3935. doi: 10.1073/pnas.0500930102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunetti-Pierri N, Stapleton GE, Law M, Breinholt J, Palmer DJ, Zuo Y, et al. Efficient, long-term hepatic gene transfer using clinically relevant HDAd doses by balloon occlusion catheter delivery in nonhuman primates. Mol Ther. 2009;17:327–333. doi: 10.1038/mt.2008.257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehrhardt A, Xu H, Dillow AM, Bellinger DA, Nichols TC., and, Kay MA. A gene-deleted adenoviral vector results in phenotypic correction of canine hemophilia B without liver toxicity or thrombocytopenia. Blood. 2003;102:2403–2411. doi: 10.1182/blood-2003-01-0314. [DOI] [PubMed] [Google Scholar]
- Brunetti-Pierri N, Nichols TC, McCorquodale S, Merricks E, Palmer DJ, Beaudet AL, et al. Sustained phenotypic correction of canine hemophilia B after systemic administration of helper-dependent adenoviral vector. Hum Gene Ther. 2005;16:811–820. doi: 10.1089/hum.2005.16.811. [DOI] [PubMed] [Google Scholar]
- Ohbayashi F, Balamotis MA, Kishimoto A, Aizawa E, Diaz A, Hasty P, et al. Correction of chromosomal mutation and random integration in embryonic stem cells with helper-dependent adenoviral vectors. Proc Natl Acad Sci USA. 2005;102:13628–13633. doi: 10.1073/pnas.0506598102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harui A, Suzuki S, Kochanek S., and, Mitani K. Frequency and stability of chromosomal integration of adenovirus vectors. J Virol. 1999;73:6141–6146. doi: 10.1128/jvi.73.7.6141-6146.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephen SL, Sivanandam VG., and, Kochanek S. Homologous and heterologous recombination between adenovirus vector DNA and chromosomal DNA. J Gene Med. 2008;10:1176–1189. doi: 10.1002/jgm.1246. [DOI] [PubMed] [Google Scholar]
- Yant SR, Ehrhardt A, Mikkelsen JG, Meuse L, Pham T., and, Kay MA. Transposition from a gutless adeno-transposon vector stabilizes transgene expression in vivo. Nat Biotechnol. 2002;20:999–1005. doi: 10.1038/nbt738. [DOI] [PubMed] [Google Scholar]
- Ivics Z, Hackett PB, Plasterk RH., and, Izsvák Z. Molecular reconstruction of Sleeping Beauty, a Tc1-like transposon from fish, and its transposition in human cells. Cell. 1997;91:501–510. doi: 10.1016/s0092-8674(00)80436-5. [DOI] [PubMed] [Google Scholar]
- Yant SR, Huang Y, Akache B., and, Kay MA. Site-directed transposon integration in human cells. Nucleic Acids Res. 2007;35:e50. doi: 10.1093/nar/gkm089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jager L, Hausl MA, Rauschhuber C, Wolf NM, Kay MA., and, Ehrhardt A. A rapid protocol for construction and production of high-capacity adenoviral vectors. Nat Protoc. 2009;4:547–564. doi: 10.1038/nprot.2009.4. [DOI] [PubMed] [Google Scholar]
- Palmer D., and, Ng P. Improved system for helper-dependent adenoviral vector production. Mol Ther. 2003;8:846–852. doi: 10.1016/j.ymthe.2003.08.014. [DOI] [PubMed] [Google Scholar]
- Evans JP, Brinkhous KM, Brayer GD, Reisner HM., and, High KA. Canine hemophilia B resulting from a point mutation with unusual consequences. Proc Natl Acad Sci USA. 1989;86:10095–10099. doi: 10.1073/pnas.86.24.10095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown BD, Shi CX, Powell S, Hurlbut D, Graham FL., and, Lillicrap D. Helper-dependent adenoviral vectors mediate therapeutic factor VIII expression for several months with minimal accompanying toxicity in a canine model of severe hemophilia A. Blood. 2004;103:804–810. doi: 10.1182/blood-2003-05-1426. [DOI] [PubMed] [Google Scholar]
- McCormack WM, Jr, Seiler MP, Bertin TK, Ubhayakar K, Palmer DJ, Ng P, et al. Helper-dependent adenoviral gene therapy mediates long-term correction of the clotting defect in the canine hemophilia A model. J Thromb Haemost. 2006;4:1218–1225. doi: 10.1111/j.1538-7836.2006.01901.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bristol JA, Shirley P, Idamakanti N, Kaleko M., and, Connelly S. In vivo dose threshold effect of adenovirus-mediated factor VIII gene therapy in hemophiliac mice. Mol Ther. 2000;2:223–232. doi: 10.1006/mthe.2000.0120. [DOI] [PubMed] [Google Scholar]
- Mátés L, Chuah MK, Belay E, Jerchow B, Manoj N, Acosta-Sanchez A, et al. Molecular evolution of a novel hyperactive Sleeping Beauty transposase enables robust stable gene transfer in vertebrates. Nat Genet. 2009;41:753–761. doi: 10.1038/ng.343. [DOI] [PubMed] [Google Scholar]
- Brunetti-Pierri N, Grove NC, Zuo Y, Edwards R, Palmer D, Cerullo V, et al. Bioengineered factor IX molecules with increased catalytic activity improve the therapeutic index of gene therapy vectors for hemophilia B. Hum Gene Ther. 2009;20:479–485. doi: 10.1089/hum.2008.084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yant SR, Wu X, Huang Y, Garrison B, Burgess SM., and, Kay MA. High-resolution genome-wide mapping of transposon integration in mammals. Mol Cell Biol. 2005;25:2085–2094. doi: 10.1128/MCB.25.6.2085-2094.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirkness EF, Bafna V, Halpern AL, Levy S, Remington K, Rusch DB, et al. The dog genome: survey sequencing and comparative analysis. Science. 2003;301:1898–1903. doi: 10.1126/science.1086432. [DOI] [PubMed] [Google Scholar]
- Lindblad-Toh K, Wade CM, Mikkelsen TS, Karlsson EK, Jaffe DB, Kamal M, et al. Genome sequence, comparative analysis and haplotype structure of the domestic dog. Nature. 2005;438:803–819. doi: 10.1038/nature04338. [DOI] [PubMed] [Google Scholar]
- Ehrhardt A, Xu H., and, Kay MA. Episomal persistence of recombinant adenoviral vector genomes during the cell cycle in vivo. J Virol. 2003;77:7689–7695. doi: 10.1128/JVI.77.13.7689-7695.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jager L., and, Ehrhardt A. Persistence of high-capacity adenoviral vectors as replication-defective monomeric genomes in vitro and in murine liver. Hum Gene Ther. 2009;20:883–896. doi: 10.1089/hum.2009.020. [DOI] [PubMed] [Google Scholar]
- Jager L., and, Ehrhardt A. Emerging adenoviral vectors for stable correction of genetic disorders. Curr Gene Ther. 2007;7:272–283. doi: 10.2174/156652307781369074. [DOI] [PubMed] [Google Scholar]
- Snyder RO, Miao C, Meuse L, Tubb J, Donahue BA, Lin HF, et al. Correction of hemophilia B in canine and murine models using recombinant adeno-associated viral vectors. Nat Med. 1999;5:64–70. doi: 10.1038/4751. [DOI] [PubMed] [Google Scholar]
- Xu L, Gao C, Sands MS, Cai SR, Nichols TC, Bellinger DA, et al. Neonatal or hepatocyte growth factor-potentiated adult gene therapy with a retroviral vector results in therapeutic levels of canine factor IX for hemophilia B. Blood. 2003;101:3924–3932. doi: 10.1182/blood-2002-10-3050. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Expression levels from episomal HC-AdVs. C57Bl/6 mice were co-injected with HC-AdV-TcFIX and HC-AdV-luciferase (n=10) at a ratio of 3: 1 (transposon TcFIX vector: luciferase vector) with a total number of 8x108 transducing units. Three weeks after vector administration (on day 21 virus post-injection) one group (n=5) was injected with carbon tetrachloride (+CCl4) to induce hepatocyte proliferation. (a) Stability of transgene expression was monitored by performing an ELISA two and three weeks post CCl4 treatment (days 36 and 42). (b) The control group (n=5) was not treated with CCl4 (-CCl4).
PCR setups for specific detection of total cFIX and episomal HC-AdVs. Primers are presented as horizontal arrows. (i) For detection of all DNA sequences encoding the cFIX expression cassette, primer binding sites in the liver-specific hAAT-promotor and the cFIX encoding sequence were chosen. Therefore, interferences with the genomic cFIX encoding sequence were avoid and only transposon encoded expression cassettes were detected independent of the molecular status of the transposon (HC-AdV, circular intermediate or integrated transposon). (ii) Primer-set for amplification of the intact episomal HC-AdV-TcFIX genome without excision of the transposon. (iii) Primer-set specifically detecting Sleeping Beauty Transposase encoding sequences.
Detection of circular intermediates in transduced canine liver cells. Additional PCR analysis to detect specifically episomal circular intermediates after Flp-mediated recombination was performed. (a) Primer binding site are depicted. (b) Two dilutions of liver genomic DNA from D1 (D1: 500 ng; D1 dil.: 10 ng) were analyzed. As a positive control (+) genomic DNA obtained from Hela cells infected with the corresponding viral vectors was used, whereas DNA from non-infected Hela cells served as negative control (–).
Detailed description of integration sites after SB-mediated transposition into genomic DNA of canine liver cells.
Laboratory measurements of dogs D1 (HSB5), D2 (HSB5, high dose), and D3 (mSB).