Abstract
Loss of the q22 band of chromosome 16 is a frequent genetic event in breast cancer, and the candidate tumor suppressor gene, ATBF1, has been implicated in breast cancer by genomic deletion, transcriptional down-regulation, and association with better prognostic parameters. In addition, estrogen receptor (ER)-positive breast cancer expresses a higher level of ATBF1, suggesting a role of ATBF1 in ER-positive breast cancer. In this study, we examined whether and how ATBF1 affects the ER function in breast cancer cells. We found that ATBF1 inhibited ER-mediated gene transcription, cell growth, and proliferation in ER-positive breast cancer cells. In vitro and in vivo immunoprecipitation experiments revealed that ATBF1 interacted physically with the ER and that multiple domains in both ATBF1 and ER proteins mediated the interaction. Furthermore, we demonstrated that ATBF1 inhibited ER function by selectively competing with the steroid receptor coactivator AIB1 but not GRIP1 or SRC1 for binding to the ER. These findings not only support the concept that ATBF1 plays a tumor-suppressive role in breast cancer, they also provide a mechanism for how ATBF1 functions as a tumor suppressor in breast cancer.
Keywords: Breast Cancer, Hormone Receptors, Receptor Regulation, Tumor Promoter, Tumor Suppressor, AIB1, ATBF1, ER Function
Introduction
Breast cancer is the most common malignancy in women, with approximately one in eight women affected in their lifetime. Endogenous estrogens are thought to play an important role in the initiation and progression of breast cancer. Longer exposures to estrogens result in an increased risk for breast cancer (1–4). The biological function of estrogens is believed to be mediated largely by estrogen receptors, mainly the alpha subtype (ERα or ER2 hereafter) (5). In normal mammary epithelial cells, the level of ER fluctuates during the menstrual cycle in response to cyclical changes in estrogens. In breast cancer, however, the normal control of ER expression and function is interrupted, which contributes significantly to the initiation and progression of breast cancer (6, 7). ER regulates the expression of a variety of target genes through the recruitment of coregulators, including coactivators and corepressors that mediate local chromatin remodeling as well as communications with the basal transcriptional apparatus (8). Coregulators are often present in limiting amounts in cells, and modifications of their levels of expression and/or structure lead to alterations in the ER function (8). Coactivators such as steroid receptor coactivators (SRCs) are both necessary and sufficient to initiate the ER-mediated gene transcription (9).
On gene promoters that contain estrogen response elements (EREs), SRCs (also referred to as p160 family) including AIB1 (NCOA3/ACTR/TRAM1/SRC3), GRIP1 (TIF2/SRC2), and SRC1, form heterodimers, as either AIB1·GRIP1 or AIB1·SRC-1, to bind directly to ER to enhance its transcriptional activity in a hormone-dependent fashion. Three nuclear receptor (NR) boxes, also known as LXXLL motifs, are centrally located in all three SRC proteins to mediate their binding to a nuclear receptor (10). The significance of SRCs in breast cancer has been an area of extensive study. AIB1 is the only SRC family member that is strongly amplified and overexpressed in human breast cancer and implicated in tamoxifen resistance (11–15). AIB1 promotes tumorigenesis and is essential for HER2-driven oncogenesis in mice (16–18). In contrast, corepressors are crucial for balancing the ER-mediated transactivation by controlling the magnitude of estrogen responses, leading to inhibition of ER or repression of ER binding to DNA and conferring an active repression of ER target genes (19). Loss of ER corepressors promotes breast cancer and leads to resistance to hormone therapy (19). Therefore, studying ER and its coregulators may help develop diagnostic and therapeutic strategies for breast cancer.
AT motif-binding factor 1 (ATBF1) is a transcription factor of 3703 amino acids containing one ATPase A-motif, two DEAH box-like sequences, four homeodomains, and 23 zinc finger motifs involved in transcriptional regulations and protein-protein interactions (20). ATBF1 appears to function as a tumor suppressor gene in prostate cancer, as its inactivating somatic mutations and genomic deletion have frequently been detected (21), and inherited mutations could confer an increased risk for prostate cancer (22). In breast cancer, the ATBF1 locus is frequently deleted and ATBF1 mRNA expression is significantly down-regulated, although few mutations have been detected (23–26). ATBF1 mRNA is expressed at significantly higher levels in breast cancers without lymph node metastasis, with smaller tumor volumes (<2 cm) or that were ER-positive (26). These observations suggest a role for ATBF1 as a tumor suppressor gene in breast cancer in which ER may be involved.
In this study, we investigated whether and how ATBF1 affects the ER function in ER-positive breast cancer cells. We show that ATBF1 inhibited ER-mediated gene transcription, cell growth, and proliferation and that ATBF1 modulated the ER function by selectively competing with AIB1 but not GRIP1 or SRC1 for binding to ER.
EXPERIMENTAL PROCEDURES
Plasmid Constructs
Plasmids of estrogen-responsive reporter ERE-TK-Luc and pCMV-ER were kindly provided by Drs. Eliot Rosen and Saijun Fan of Albert Einstein College of Medicine (27). The FLAG-ER plasmid was constructed by subcloning the open reading frame (ORF) of the ER gene into the FLAG-pcDNA3 vector (Invitrogen). HA-GRIP1 plasmid in the pSG5 vector was kindly provided by Dr. Michael Stallcup of the University of Southern California (28). Plasmids of FLAG-AIB1 in the FLAG-TAG-2B vector and FLAG-SRC1 in the pSG5 vector were kindly provided by Drs. Sophia Tsai and Bert O'Malley of Baylor College of Medicine (29). Plasmids of FLAG-ATBF1 and HA-ATBF1 were obtained by subcloning the ORF of the ATBF1 gene from the pcMATBF1w plasmid, a gift from Dr. Yutaka Miura (30), into FLAG-pcDNA3 and HA-pKXUa1 vectors from Dr. J. T. Murphy of Emory University (31). Deletion mutants of ATBF1 and ER were created by PCR-based approaches with FLAG-ATBF1 and pCMV-ER plasmids as templates and HA-pcDNA3 (Invitrogen) and FLAG-pcDNA3 as vectors. Primer sequences for creating deletion mutants ATBF1A, ATBF1B, ATBF1C, ATBF1D, ATBF1E, ATBF1F, ATBF1D1, ATBF1D2, ATBF1D3, ATBF1D4, ATBF1D5, ATBF1D6, ATBF1D7, ATBF1F1, ATBF1F2, ATBF1F3, ATBF1F4, ATBF1F5, and ATBF1F6 for ATBF1 and ERN, ERC, ER1, ER2, ER3, ER4, and ER5 for ER are available on request.
Cell Lines and Reagents
For details, see the supplemental “Materials and Methods.”
Cell Growth and Proliferation Assays
Cell growth and proliferation were analyzed by conducting sulforhodamine B (SRB) staining and DNA synthesis assays. ER-positive breast cancer cell lines T-47D and MCF-7 cultured in phenol-free media supplemented with 5% charcoal dextran-stripped FBS for 3 days were seeded in 12-well plates at a density of 0.5 × 105 cells/well with [2-14C]thymidine added into the medium for DNA synthesis assay as an internal control. On the following day, after being washed three times with medium to remove unincorporated thymidine, T-47D cells were transfected with FLAG-ATBF1 or control plasmids, and MCF-7 cells were transfected with siRNA against ATBF1 or control siRNA. Eight hours after transfection, E2 or its vehicle control alcohol was added to culture media (the E2 final concentration was 1 μm). Forty-eight hours after transfection, media in culture plates for the SRB staining assay were replaced with that containing 800 μg/ml G418, and cells were cultured for another 8 days with media changed every 2 days. Media in plates for DNA synthesis assay were replaced with that containing [methyl-3H]thymidine, and cells were incubated for another 4 h for thymidine incorporation. Cells for the SRB staining assay were washed three times with PBS after selection, fixed with 10% trichloroacetic acid, and stained with SRB. Protein amounts, which indicate the cell number, were measured as optical densities as described previously (21). Cells for DNA synthesis assay were washed three times with PBS and transferred onto glass microfiber filters (VWR Scientific) to measure 3H and 14C radioactivities in ScintiSafe Econo 1 solution (Fisher Scientific) using an LS6500 multipurpose scintillation counter (Beckman Coulter). The rates of DNA synthesis were indicated by the ratio between 3H and 14C readings in each sample.
Luciferase Reporter Assay
A luciferase reporter assay in transfected cells was performed as described previously (32).
RNA Interference (RNAi)
Chemically synthesized small interfering RNAs (siRNA; Dharmacon, Chicago) were used for RNAi. Four pieces of ATBF1-specific siRNA targeting different regions of ATBF1 mRNA, including 5′-AGCGGAGAUUUGAAACCAU-3′ (siRNA 1), 5′-AGAAUAUCCUGCUAGUACA-3′ (siRNA 2), 5′-CAGGAGGAAUCGUUUAAGC-3′ (siRNA 3), and 5′-GAAGAACAAAUUGGUCAUC-3′ (siRNA 4), were chemically synthesized and tested for efficiency in the knockdown of ATBF1 mRNA by real time PCR. siRNA 2 showed the highest efficiency in ATBF1 knockdown and was used throughout the study. Two negative control siRNAs were used. One was chemically synthesized luciferase siRNA with a sequence of 5′-CUUACGCUGAGUACUUCGAUU-3′. The other was purchased from Dharmacon and had a sequence of 5′-AAUUCUCCGAACGUGUCACGU-3′, which does not match any human genomic sequences (33). SiPORT NeoFX and lipid reagents (Ambion, Austin, TX) were used for siRNA transfection. The efficiency of RNAi was monitored by either real time PCR or Western blot analysis.
Gene Expression Analysis
Gene expression was measured by real time PCR as described previously (32, 34). The primer sequences were as follows: ATBF1, 5′-TGTTCCAGATCGAGATGGGAAT-3′ and 5′-CTTTCCCAGATCCTCTGAGGTTT-3′; cathepsin D (CATD), 5′-ACTGCTGGACATCGCTTGCT-3′ and 5′-TGTCAAACGAGGTACCATTCTTC-3′; EBAG9, 5′-CCAGATGGGAGCACAGGTTTCTCTAG-3′ and 5′-TTCCTTTGTTGTTCGGCTGCTCTC-3′; and GAPDH, 5′-GTGGTCCAGGGGTCTTACTC-3′ and 5′-TTCAACAGCGACACCCACTC-3′.
Immunoprecipitation (IP) and Western Blot Analysis
Cells for different treatments grown in 100-mm dishes were washed twice with ice-cold PBS and then lysed in modified radioimmune precipitation assay buffer (150 mm NaCl, 50 mm Tris-HCl, pH 7.5, 1% CA630, 0.25% Na-deoxycholate, and protease inhibitor mixture). Cell lysates were centrifuged to collect supernatants, which were then incubated overnight with different antibodies, as indicated in the legends for Figs. 4, 6A, S1, and S2, and then for another 1 h with protein A-agarose (Millipore, Billerica, MA) with rotation at 4 °C. After washing three times with modified radioimmune precipitation assay buffer, immunoprecipitates were released by boiling for 10 min in 50 μl of 2× SDS-PAGE loading buffer, resolved in 10–15% SDS-PAGE, and then blotted with different antibodies, as indicated in the legends for Figs. 4, 6A, S1, and S2, following standard Western blot procedures.
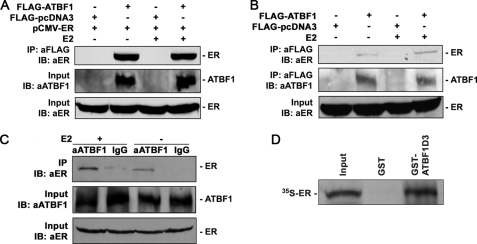
FIGURE 4.
Detection of protein-protein interactions between ATBF1 and ER by both IP and GST pulldown assays. A, ATBF1 and ER double-negative 22Rv1 cells were cotransfected with FLAG-ATBF1 and pCMV-ER plasmids, and cell lysates were precipitated with anti-FLAG affinity gel and immunoblotted (IB) with anti-ER antibody. B, T-47D cells, which express endogenous ER, were transfected with FLAG-ATBF1, and cell lysates were precipitated with anti-FLAG affinity gel and blotted with anti-ER antibody. C, cell lysates from MCF-7 cells, which express both ATBF1 and ER, were precipitated with anti-ATBF1 antibody against amino acid residues 1–160 and blotted with anti-ER antibody. Cells were treated with 1 μm E2 or its vehicle alcohol. The expression of ATBF1 and ER in cell lysate was confirmed as shown in the lower two panels in A–C. D, in vitro translated 35S-labeled ER protein was pulled down by GST-ATBF1D3 recombinant protein in a GST pulldown assay. ATBF1D3 is one of the deletion mutants of ATBF1 protein that interacts with ER protein.
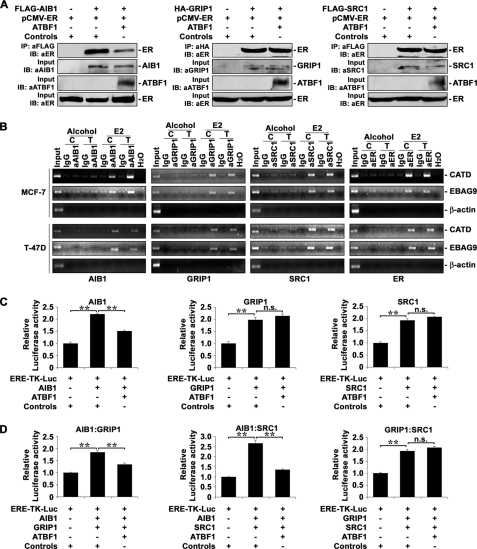
FIGURE 6.
Competition of ATBF1 with AIB1 but not GRIP1 or SRC1 for binding to ER. A, expression of ATBF1 decreased the amount of AIB1-bound ER (left panel) but not GRIP1- or SRC1-bound ER (middle and right panels). Plasmids for SRCs, ER, and ATBF1 were cotransfected into 22Rv1 cells, and co-IP and immunoblotting (IB) were performed with affinity gels as indicated. Input, indicates samples without co-IP. B, knockdown of ATBF1 in MCF-7 cells increased and expression of ATBF1 in T-47D cells decreased the binding of AIB1, but not that of GRIP1, SRC1 or ER, to promoters of two ER target genes, CATD and EBAG9, as detected by ChIP assays. Cell line names are indicated at the left, gene names at the right, antibody names and cell treatments at the top, and names of molecules examined at the bottom of the panels. Normal IgG was used as the negative control for antibodies against AIB1 (aAIB1), GRIP1 (aGRIP1), SRC1 (aSRC1), and ER (aER). Alcohol was used to dissolve E2 and as the negative control for E2. H2O was used as the negative control for PCR. Human β-actin gene was used as the negative control for ER target genes. The C indicates negative controls for transfection of siRNA against luciferase gene in MCF-7 cells or FLAG-pcDNA3 vector control plasmid in T-47D cells, and T indicates transfection of siRNA against ATBF1 in MCF-7 cells or FLAG-ATBF1 plasmid in T-47D cells. Input indicates cell lysates not subjected to ChIP. E2 treatment was at 1 μm. C, expression of ATBF1 inhibited AIB1-enhanced ER activity (left), but not that by GRIP1 (middle) or SRC1 (right), as determined by luciferase reporter assays in T-47D cells transfected with the indicated plasmids. D, ATBF1 inhibited AIB1·GRIP1 (left)- or AIB1·SRC1 (middle)- but not GRIP1·SRC1-enhanced (right) ER activity. n.s., not significant. **, p < 0.005.
GST Pulldown Assay
A glutathione S-transferase (GST) pulldown assay was performed following our published procedures (35), and GST-ATBF1D3 fusion protein was expressed and purified as described above under “Cell Lines and Reagents.” ER protein was produced by in vitro translation in the presence of [35S]methionine (GE Healthcare) using TnT Quick-coupled Transcription/Translation Systems (Promega) as described previously (36). Ten μg of GST-ATBF1D3 recombinant protein or GST alone was mixed with 10 μl of in vitro translated ER protein, and the mixture was incubated in the GST pulldown binding buffer (10 mm HEPES, pH 7.6, 3 mm MgCl2, 100 mm KCl, 5 mm EDTA, 5% glycerol, and 0.5% CA-630) with rotation at 4 °C for 2 h. Fifty μl of glutathione-Sepharose beads was then added to the mixture and incubated on a shaker at 4 °C for another 2 h. The beads were washed three times with GST pulldown binding buffer and boiled in 50 μl of 2× SDS-PAGE loading buffer. Released proteins were resolved on 10% SDS-PAGE gels that were then dried and exposed to x-ray films to detect the pulled down proteins.
Chromatin Immunoprecipitation (ChIP) Assay
MCF-7 and T-47D cells were treated as indicated in the legend of Fig. 6B, and ChIP assays were performed using the ChIP assay kit (Millipore) with anti-AIB1, anti-GRIP1, anti-SRC1, and anti-ER antibodies following our published procedures (36). Precipitated DNA was analyzed by PCR with the following promoter-specific primers for CATD and EBAG9 genes: CATD, 5′-GGTTTCTCTGGAAGCCCTGTAG-3′ and 5′-TCCTGCACCTGCTCCTCC-3′; EBAG9, 5′-ATTGTCTGCCCTTCGCCGT-3′ and 5′-TTTGGAGGCTGCGTGCTTT-3′. Human β-actin gene was used as a control with the following primers: 5′-CGGAGGGCGCCCCAACTCAG-3′ and 5′-GCGCGCGCGGCCCCAGAACA-3′.
Statistical Analysis
A Student's t test or Fisher's exact test was used to determine the p values for comparisons between different groups. A p value less than 0.05 was considered statistically significant.
RESULTS
ATBF1 Protein Expression in Breast Cancer Cell Lines
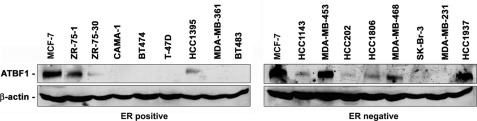
To identify proper cell models for studying ATBF1, we evaluated ATBF1 protein expression in a panel of breast cancer cell lines by Western blotting. In nine ER-positive breast cancer cell lines tested, the level of ATBF1 protein was higher in MCF-7 cells, moderate in ZR-75-1 cells, lower in HCC1395 and ZR-7-30 cells, and not detectable in the remaining cell lines including T-47D (Fig. 1). Among eight ER-negative breast cancer cell lines tested, ATBF1 protein expression was higher in MDA-MB-453 and HCC1937, moderate in MDA-MB-468, lower but detectable in HCC1143, HCC202, HCC1806, and SK-BR-3, and not detectable in MDA-MB-231 (Fig. 1). On the basis of these results, we selected ER-positive cell lines MCF-7 and T-47D and the ER-negative line MDA-MB-231 for further use. We also used the prostate cancer cell line 22Rv1 as an ER and ATBF1 double-negative cell line because it does not express ER (37) and has a frameshift mutation that truncates the majority of ATBF1 protein (21).
FIGURE 1.
Expression of ATBF1 protein in ER-positive and ER-negative breast cancer cell lines. Names of cell lines are at the top, and protein names are at the left. Expression of ATBF1 protein in MCF-7 cells served as the positive control in the panel of ER-negative cell lines and β-actin as the loading control.
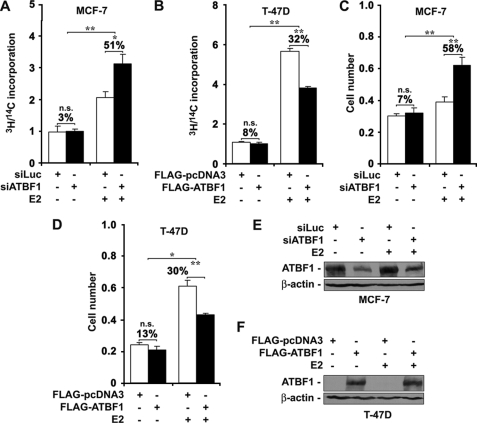
ATBF1 Inhibits ER-mediated Cell Growth and Proliferation in ER-positive Breast Cancer Cells
It is well established that ER plays a promoting role in the growth and proliferation of breast cancer cells (6, 7). We reasoned that as a transcription factor ATBF1 might interact with ER to modulate its function in regulating the cellular responses of mammary epithelial cells to estrogens in breast cancer. To test this idea, we performed cell growth and proliferation assays to examine whether ATBF1 inhibited ER-promoted cell growth and proliferation. In these experiments, cells were grown in phenol-free media supplemented with 5% charcoal dextran-stripped FBS for 3 days before use. In ER-positive MCF-7 cells, which have a higher level of ATBF1 protein (Fig. 1), knockdown of ATBF1 enhanced the rate of cell proliferation by 51% (p < 0.05) when E2 treatment was applied as measured by DNA synthesis rates, but it had no significant effect in cells without E2 treatment (Fig. 2A). In ER-positive T-47D cells in which ATBF1 protein was not detectable (Fig. 1), reconstitution of ATBF1 expression had no significant effect on cell proliferation in the absence of E2, but it inhibited cell proliferation by 32% (p < 0.01) in the presence of E2 (Fig. 2B).
FIGURE 2.
Effect of ATBF1 on ER-mediated cell growth and proliferation in ER-positive breast cancer cell lines MCF-7 and T-47D treated with or without E2. A and B, from DNA synthesis assays; C and D, from SRB staining assays. In MCF-7 cells where ATBF1 is highly expressed, ATBF1 was knocked down by RNAi, whereas in T-47D cells, which express little ATBF1, expression plasmids were transfected to express ATBF1. ATBF1 expression in transfected cells was confirmed by Western blotting in both cell lines (E and F). Transfection conditions are indicated in each panel. The final concentration of E2 in the media was 1 μm. n.s., not significant. *, p < 0.05; **, p < 0.005. SiLuc, negative control siRNA against the luciferase gene; siATBF1, ATBF1 siRNA.
An SRB staining assay further confirmed the observations from the DNA synthesis assay. Although knockdown of ATBF1 in MCF-7 cells did not significantly change the number of cells after 8 days without E2 treatment, it increased the number of cells by 58% (p < 0.01) in the presence of E2 (Fig. 2C), indicating an inhibitory role of ATBF1 in ER-mediated cell growth. Consistently, in T-47D cells, reconstitution of ATBF1 expression did not significantly inhibit cell growth in the absence of E2 but suppressed cell growth by 30% (p < 0.005) in the presence of E2 (Fig. 2D). Knockdown of ATBF1 in MCF-7 cells and restored expression of ATBF1 in T-47D cells were confirmed by Western blotting (Fig. 2, E and F). These results indicate that when ER is activated, ATBF1 inhibits ER-mediated cell growth and proliferation in ER-positive breast cancer cells.
ATBF1 Inhibits ER-mediated Gene Transcription
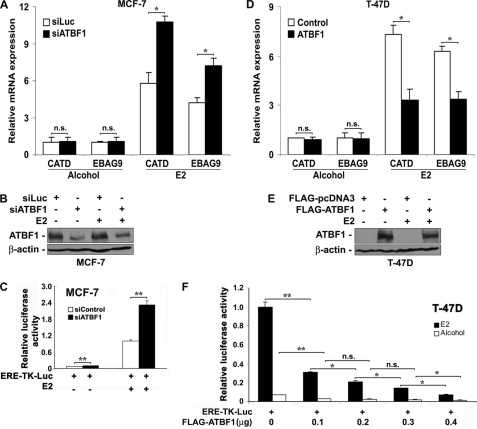
ER promotes cell growth and proliferation by regulating the expression of a variety of target genes (8). Therefore, we examined whether ATBF1 also inhibited the ER-mediated gene transcription. We first examined the effect of ATBF1 on the expression of endogenous ER target genes. We selected two well established ER target genes, CATD and EBAG9 (10), for analysis. In MCF-7 cells, whereas the expression of these genes was low and knockdown of ATBF1 had no significant effect on gene expression in the absence of E2, knockdown of ATBF1 significantly increased the expression of both CATD and EBAG9 in the presence of E2 (Fig. 3A). Consistently in T-47D cells, reconstitution of ATBF1 expression significantly inhibited the expression of both CATD and EBAG9 in the presence of E2 but not in the absence of E2 (Fig. 3D). Knockdown of ATBF1 in MCF-7 cells and restored expression of ATBF1 in T-47D cells were confirmed by Western blotting (Fig. 3, B and E). These results suggest that ATBF1 inhibits ER-mediated gene transcription in ER-positive breast cancer cells.
FIGURE 3.
Inhibitory effect of ATBF1 on ER-mediated transcriptional activity. A–C, knockdown of ATBF1 in ER-positive MCF-7 cells, which was confirmed by Western blotting (B), significantly increased the transcription of ER target genes (A) and the estrogen-responsive element-mediated reporter activity (C) caused by E2 treatment. SiLuc, siControl, and siATBF1, siRNAs against the luciferase gene, negative control siRNA (purchased from Dharmacon), and siRNAs against ATBF1, respectively. D–F, overexpression of ATBF1 in T-47D cells, as verified by Western blotting (E), significantly inhibited E2-driven transcription of ER target genes (D) and estrogen-responsive reporter activity (F). FLAG-ATBF1 plasmid was used to restore ATBF1 expression with FLAG-pcDNA3 as the control. E2 treatment was at 1 μm, and its vehicle alcohol was used as the control. n.s., not significant. *, p < 0.05; **, p < 0.005.
We further examined the effect of ATBF1 on the transactivation activity of ER using an estrogen-responsive promoter. In ER and ATBF1 double-positive MCF-7 cells, ATBF1 expression was knocked down by siRNA transfection, which was followed by transfection of the ERE-TK-Luc estrogen-responsive promoter reporter plasmid. Without E2 treatment, the reporter showed little activity, regardless of ATBF1 knockdown; but when E2 was added, the reporter showed a high level of activity, and knockdown of ATBF1 significantly enhanced the activity (Fig. 3C). The efficiency of ATBF1 knockdown was confirmed by real time PCR (data not shown). Consistently, in ER-positive but ATBF1-negative T-47D cells, cotransfection of ERE-TK-Luc reporter plasmid and different amounts of FLAG-ATBF1 plasmid showed that although the reporter had little activity without E2 treatment, it had a strong activity in the presence of E2, and ATBF1 expression significantly inhibited the reporter activity in a dose-dependent manner (Fig. 3F). Consistent results were also obtained in two other ER-negative cell lines, MDA-MB-231 and 22Rv1, in which, when the ER pathway was activated by transfecting an ER expression plasmid and adding E2 to the medium, a significant reporter activity was detected, and the transfection-mediated ATBF1 expression significantly inhibited the reporter activity in a dose-dependent manner (data not shown). Similar to effects in T-47D cells, when ER was not reconstituted the reporter had no activity in MDA-MB-231 and 22Rv1 cells, and ATBF1 had no effect on the promoter activity (data not shown). These results further indicate that ATBF1 inhibits the ER function in the gene transcription and that the inhibition is E2-dependent.
Protein-Protein Interaction between ATBF1 and ER
ER regulates the expression of a variety of genes through recruiting and interacting with coregulators including coactivators and corepressors to mediate the local chromatin remodeling and communicate with the basal transcriptional apparatus (8, 38, 39). ATBF1 is also a transcription factor bound to DNA (40). It is therefore possible that ATBF1 interacts directly with ER. To test this hypothesis, we first examined the peptide sequence of ATBF1 for the existence of an NR-box motif (-LXXLL-), which has been shown to mediate protein interaction with ER (41). One consensus NR box (LXXLL) and 31 of its variants LXXXL (leucine at the fourth residue is substituted) were identified (Fig. 5A). These NR boxes could be specifically recognized by ER, thus mediating an interaction with ER (41).
FIGURE 5.
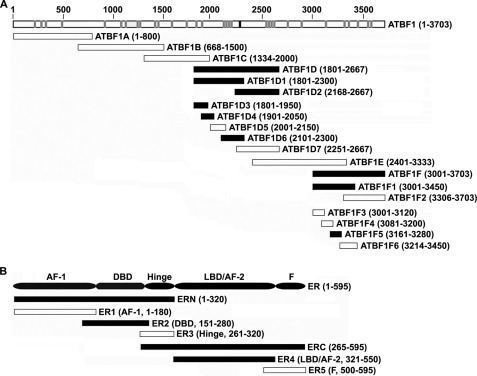
Mapping of interacting domains in ATBF1 (A) and ER (B) proteins by co-IP assays. A, summary of the mapping results for different deletion mutants of ATBF1. Full-length ATBF1 protein (residues 1–3703) is shown at the top, with the consensus LXXLL NR motif marked by a black box and 31 LXXXL variants marked by gray boxes. B, summary of mapping results for ER deletion mutants interacting with ATBF1. Full-length ER protein is shown at the top with different domains indicated. Black and white bars indicate positive and negative interactions with ER, respectively, with the name of each mutant and the residues spanned shown to the right of each bar. The results of co-IP and Western blotting for both panels are shown in supplemental Figs. S1 and S2.
We then examined whether ATBF1 and ER co-existed in the same protein complex. The ER and ATBF1 double-negative 22Rv1 cells grown in hormone-depleted media were transfected with expression constructs for FLAG-ATBF1 and ER (pCMV-ER), treated with E2, and subjected to immunoprecipitation with anti-FLAG affinity gel and Western blotting. ER protein was precipitated efficiently in ATBF1- and ER-transfected cells regardless of E2 treatment but not in control cells transfected with an empty vector and ER plasmid (Fig. 4A). These results indicate a specific interaction between ATBF1 and ER proteins in transfected cells.
To further determine whether ATBF1 interacts with ER, the FLAG-ATBF1 expression plasmid was transfected into ER-positive T-47D cells, and immunoprecipitation with anti-FLAG affinity gel was performed. Endogenous ER was detectable in the protein complexes precipitated only in FLAG-ATBF1-transfected T-47D cells but not in control cells (Fig. 4B). Finally, we performed immunoprecipitation with anti-ATBF1 antibody in MCF-7 cells, which express higher levels of ATBF1 protein and ER, and Western blotting with anti-ER antibody. Endogenous ER was clearly detected in the protein complexes precipitated with anti-ATBF1 antibody (Fig. 4C). Taken together, these results further indicate a protein interaction between ATBF1 and ER. Again, the endogenous interaction between ATBF1 and ER was detected regardless of E2, although more ER could have been precipitated when E2 was present.
We also performed a GST pulldown assay to examine whether the interaction between ATBF1 and ER occurred directly or was mediated by certain adaptor proteins. Considering that ATBF1 is a large protein (about 400 kDa) and thus would be challenging to express in full size in bacteria, we used a smaller deletion mutant, ATBF1D3, which was found to still interact with ER (see “Mapping of Domains That Mediate the Interaction between ATBF1 and ER” below). We expressed GST-fused ATBF1D3 (GST-ATBF1D3) recombinant protein in Escherichia coli, incubated it with 35S-labeled ER protein, and performed pulldown assays. The ER protein was efficiently pulled down by GST-ATBF1D3 but not by GST alone (Fig. 4D), indicating a direct interaction between ATBF1 and ER.
Mapping of Domains That Mediate the Interaction between ATBF1 and ER
To further evaluate the interaction between ATBF1 and ER and to identify specific domains of ATBF1 that mediate the interaction, we created different deletion mutants of ATBF1 and ER and analyzed them for their role in the interaction by co-IP and Western blotting. First, six overlapping deletion mutants that spanned the entire ATBF1 coding region, including ATBF1A to ATBF1F, were created in a HA-tagged form and co-transfected with FLAG-ER into 22Rv1 cells, which were then subjected to co-IP and Western blotting. ATBF1D and ATBF1F interacted with ER, but ATBF1A, ATBF1B, ATBF1C, and ATBF1E did not (Fig. 5A and supplemental Fig. S1A). To further narrow down the regions in ATBF1D and ATBF1F that mediate the interaction between ATBF1 and ER, we then created smaller deletion mutants for ATBF1D and ATBF1F and tested for their interactions with ER. After two rounds of deletion mapping, four deletion mutants spanning three independent regions of ATBF1 still showed interactions with ER, including ATBF1D3 and ATBF1D4 (residues 1800–2050), ATBF1D6 (residues 2100–2300), and ATBF1F5 (residues 3161–3280). Further deletion of these three mutants spanned by ATBF1D3, ATBF1D4, ATBF1D6, and ATBF1F5 abolished their interactions with ER (Fig. 5A and supplemental Fig. S1, B–D). The deletion mutant spanned by ATBF1D6 (residues 2100–2300) contains the consensus LXXLL NR binding motif and four of its variants. The deletion mutant between residues 1800 and 2050 spanned by ATBF1D3 and ATBF1D4 contains two LXXXL NR box variants, whereas the deletion mutant between residues 3161 and 3280 spanned by ATBF1F5 does not contain any LXXLL or LXXXL motif.
We also mapped specific ER domains that mediate the interaction between ER and ATBF1 using these same approaches (Fig. 5B and supplemental Fig. S2, A and B). Two independent domains of ER, the DNA binding domain (DBD/ER2) and ligand binding domain/activation function domain 2 (LBD/AF-2/ER4), showed positive interactions with ATBF1, whereas the AF-1, hinge, or F domain did not (Fig. 5B and supplemental Fig. S2, A and B). Both the DBD and LBD/AF2 domains are frequently involved in the interaction of ER with its coregulators (19). These mapping data for interacting domains further indicate that ATBF1 interacts with ER regardless of E2.
ATBF1 Inhibits ER Function by Selectively Competing with AIB1 for Binding to ER
We explored the mechanism by which ATBF1 inhibits the ER function. ATBF1 protein contains one consensus NR binding motif (LXXLL) and 31 of its LXXXL variants; one of the three ATBF1 deletion mutants that still interact with the ER contains the consensus LXXLL motif and four of its variants (Fig. 5A). A previous study showed that all three SRCs, including AIB1, SRC1, and GRIP1, are both necessary and sufficient to initiate ER-mediated gene transcription, and all need the consensus LXXLL motif for their binding to ER (9). We reasoned that ATBF1 might compete with SRCs for their binding to ER to thus block the ER function. In testing this hypothesis, we first performed co-IP experiments to examine whether ATBF1 competed with all three SRCs for their binding to ER (Fig. 6A). Interestingly, expression of ATBF1 decreased the amount of ER that was pulled down by the antibody against AIB1 but not by those against GRIP1 or SRC1 (Fig. 6A). These results suggest that ATBF1 can selectively compete with the ER coactivator AIB1 for binding to ER and thus inhibit the ER function. The interaction between ATBF1 and all three SRCs was not detectable under the same co-IP condition (data not shown).
We then performed ChIP assays to further evaluate whether ATBF1 completed with SRCs for their binding to ER in promoters of the ER target genes, CATD and EBAG9. Both MCF-7 and T-47D cell lines treated with or without E2 were used in ChIP experiments. Although MCF-7 cells were transfected with siRNA against ATBF1 to knock down ATBF1 expression or with siRNA against the luciferase gene as a control, T-47D cells were transfected with the ATBF1 expression plasmid to express ATBF1 or with a vector control. Consistent with co-IP results (Fig. 6A), knockdown of ATBF1 expression in MCF-7 cells increased, whereas expression of ATBF1 in T-47D cells decreased, the amount of promoter DNA bound by AIB1 but not promoter DNA bound by GRIP1 or SRC1 (Fig. 6B). As reported, no obvious promoter occupancy was detected for any of the SRCs when E2 was absent (Fig. 6B). These results further support the notion that ATBF1 selectively competes with AIB1 for inhibiting ER function.
We also examined whether ATBF1 modulated the recruitment of ER onto estrogen-responsive elements in promoters of its target genes by ChIP assays in the same cell systems. Interestingly, modulation of ATBF1 expression in either MCF-7 or T-47D cells did not affect the binding of ER to promoters of CATD and EBAG9 (Fig. 6B, far right panels). These results suggest that, although it affects the binding of AIB1 to ER and gene promoters, ATBF1 does not directly interfere with the binding of ER to gene promoters.
To functionally evaluate whether ATBF1 suppresses the SRC-mediated transactivation activity of ER, an expression plasmid for each of three SRCs was cotransfected with plasmids for the ERE-TK-Luc estrogen-responsive promoter reporter and ATBF1 into T-47D cells cultured in normal media with hormones from normal FBS. Luciferase assay was then performed. Expression of ATBF1 significantly inhibited the AIB1-enhanced ER-mediated reporter activity but not that enhanced by GRIP1 or SRC1 (Fig. 6C).
Considering that SRCs form heterodimers, either as AIB1·GRIP1 or AIB1·SRC-1, to bind to and modulate ER transcriptional activity (10), we also tested the effect of ATBF1 on the transcriptional activity of ER enhanced by a combination of two SRCs using the same assay. When a combination of any two SRCs was expressed, ATBF1 still inhibited ER-mediated reporter activity when AIB1 was involved (either AIB1·GRIP1 or AIB1·SRC1) (Fig. 6D). ATBF1 had little effect when GRIP1 and SRC1 were co-expressed (Fig. 6D). Consistent with a previous report (42), each of three SRCs or a combination of any two significantly enhanced the estrogen-responsive reporter activity (Fig. 6, C and D). These results further support the notion that ATBF1 inhibits the ER function by selectively competing with AIB1 for binding to ER.
DISCUSSION
In this study, we have demonstrated that ATBF1 significantly inhibits the ER function by selectively competing with AIB1 for binding to ER in ER-positive breast cancer cells. The inhibitory effect was detectable only when both E2 and ER were present. Because ER signaling is oncogenic to the mammary gland and other types of tissues, these results support the notion that ATBF1 is a tumor suppressor gene in human cancer, which was suggested in previous studies showing frequent genomic deletions of the ATBF1 locus in human tumors including breast cancer, frequent somatic mutations of ATBF1 in prostate cancer, the inhibition of cell proliferation by ATBF1 in prostate cancer cells, and an association of germ-line mutations of ATBF1 with an increased risk of prostate cancer (21, 22). In particular, although ATBF1 is not frequently mutated in breast cancer (23), it is significantly down-regulated in breast cancer, and the transcriptional down-regulation is significantly associated with a poorer survival in patients with breast cancer (23, 26). Therefore, although the mechanism for the transcriptional down-regulation of ATBF1 is not understood at present, such a down-regulation could lead to hyperactive ER function, thus contributing to the development and progression of breast cancer.
As a nuclear receptor, ER exerts its function by regulating the expression of target genes (9). Considering that ATBF1 is also a transcription factor that functions in the nucleus, it is likely that ATBF1 inhibits ER function by inhibiting the expression of its target genes. Our results support this hypothesis. Knockdown of ATBF1 expression in MCF-7 cells increased (Fig. 3A), but reconstitution of ATBF1 in ATBF1-negative T-47D cells decreased, E2-mediated expression of ER target genes (Fig. 3D). The same function of ATBF1 was also detected when an estrogen-responsive luciferase reporter was applied (Fig. 3, C and F).
As to how ATBF1 modulates the ER function, our results suggest that ATBF1 interacts with ER at the protein level, and such an interaction interferes with the recruitment of the ER coactivator AIB1 to ER. In ER-positive breast cancer cells, binding of E2 to ER leads to conformational changes in ER, resulting in the recruitment of coactivators such as AIB1, GRIP1, and SRC1, as well as corepressors, to ER and regulation of gene transcription (43). Our extensive IP experiments demonstrated that not only full-length ATBF1 but also its three small deletion mutants, involving residues 1801–2050, 2101–2300, and 3161–3280, interacted with ER (Figs. 4 and 5). In addition, IP experiments showed that both the DBD and LBD of ER were involved in the interaction (Fig. 5B). Furthermore, the interaction appears to be direct and independent of DNA and other transcription factors, as in vitro synthesized ER and ATBF1 deletion mutant also interact with each other in the cell-free GST pulldown assay (Fig. 4D). It is also worth noticing that the interaction between ATBF1 and ER is E2-independent, because E2 treatment was not essential for ATBF1 to form a complex with ER (Figs. 4 and 5).
The binding of ATBF1 to ER interfered with the binding of AIB1 but not GRIP1 or SRC1 to ER, resulting in a repression of the E2-induced transactivation activity for ER target genes. First, ATBF1 was not released from E2-bound ER, as the interaction between ATBF1 and ER was still detected in the presence of E2 (Fig. 4). Second, among all three SRC members that share nuclear receptor binding motifs with ATBF1, only the binding of AIB1, but not that of GRIP1 or SRC1, to both ER (Fig. 6A) and promoters of ER target genes (Fig. 6B) was decreased by ATBF1. Consistently, only the transactivation activity of AIB1, but not that of GRIP1 or SRC1, was decreased by ATBF1 (Fig. 6, C and D). We noticed that the inhibitory effect of ATBF1 on cell growth and proliferation was significant only when ER was activated. When E2 was absent from the media, ATBF1 had little effect. Such an E2-dependent inhibitory effect of ATBF1 is in accordance with the role of AIB1 in the ER-mediated gene transcription (42). These results provide a mechanism for how ATBF1 inhibits the ER function.
ATBF1 is expressed in normal breast tissues, but its expression is often reduced in breast cancer (23, 26). The ATBF1 locus is also frequently deleted in breast cancer (44), which could also compromise the function of ATBF1 by reducing its expression and/or revealing mutations. Taken together with the fact that ER signaling is oncogenic to the breast, it is likely that down-regulation of ATBF1 could lead to an enhanced ER function, thus accelerating the development and progression of breast cancer. A previous study demonstrated that ER-positive breast cancers express a higher level of ATBF1 mRNA compared with ER-negative breast cancers (26). Although the underlying mechanism and significance for this correlation is currently unknown, it is possible that cells express more ATBF1 to balance or better control the ER function when the ER function becomes higher.
AIB1 is a well established ER-interacting nuclear oncoprotein in breast cancer (16). AIB1 is the only SRC family member that is strongly amplified and overexpressed in human breast cancer and is implicated in tamoxifen resistance (11–15). Depletion of AIB1 in MCF-7 cells reduces estrogen-mediated cell growth, inhibition of apoptosis, colony formation in soft agar, and tumor growth in nude mice (45). Overexpression of AIB1 alone transforms human mammary epithelial cells (46). Transgenic overexpression of AIB1 in mice induces mammary tumors (18), whereas knock-out of AIB1 in mice delays mammary ductal growth and protects the mammary gland from carcinogen-induced tumorigenesis (47). Overexpression of AIB1 reduces the antagonist activity of tamoxifen-bound ER; breast cancers with high levels of AIB1 and HER-2/neu are resistant to tamoxifen because of an increase in estrogen agonist activity (12, 48). Therefore, our results in this study suggest that functional inactivation of ATBF1 by genomic deletion, transcriptional down-regulation, or other mechanisms could lead to increased AIB1 activity and thus cause phenotypic alterations similar to those induced by AIB1 overexpression in the development and progression of breast cancer as well as hormone resistance in the therapy of breast cancer. In this regard, we have developed a mouse model in which ATBF1 can be specifically knocked out in mammary epithelial cells, and we initiated investigations on whether deletion of ATBF1 enhances cell proliferation and causes mammary tumors as transgenic overexpression of AIB1 in mice does. Knock-out of Atbf1 at least significantly increased cell proliferation of mammary epithelial cells and enhanced mammary gland duct branching.3
In summary, we found that ATBF1 inhibited ER-mediated cell growth and proliferation as well as gene transcription by selectively competing with AIB1 for binding to ER. These findings support the notion that ATBF1 is a tumor suppressor in breast cancer and that ATBF1 is a negative regulator of ER in the development and progression of breast cancer.
Supplementary Material
This work was supported, in whole or in part, by National Institutes of Health Grant CA85560 (to J.-T. D.). This work was also supported by Department of Defense Grant W81XWH-08-1-0328 (to X.-Y. D.) and by Georgia Cancer Coalition Award GCC130042 (to X.-Y. D.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental “Materials and Methods” and Figs. S1 and S2.
M. Li, X. Y. Dong, X. Sun, and J. T. Dong, unpublished data.
- ER
- estrogen receptor
- ERE
- estrogen response element
- SRC
- steroid receptor coactivator
- NR
- nuclear receptor
- SRB
- sulforhodamine B
- CATD
- cathepsin D
- IP
- immunoprecipitation
- DBD
- DNA binding domain
- LBD
- ligand binding domain
- E2
- 17β-estradiol.
REFERENCES
- 1.Platet N., Cathiard A. M., Gleizes M., Garcia M. (2004) Crit. Rev. Oncol. Hematol. 51, 55–67 [DOI] [PubMed] [Google Scholar]
- 2.Bryant J., Fisher B., Dignam J. (2001) J. Natl. Cancer Inst. Monogr. 30, 56–61 [DOI] [PubMed] [Google Scholar]
- 3.Fisher B., Jeong J. H., Dignam J., Anderson S., Mamounas E., Wickerham D. L., Wolmark N. (2001) J. Natl. Cancer Inst. Monogr. 30, 62–66 [DOI] [PubMed] [Google Scholar]
- 4.Henderson I. C. (1993) Cancer 71, Suppl. 6, 2127–2140 [DOI] [PubMed] [Google Scholar]
- 5.Zhang H., Sun L., Liang J., Yu W., Zhang Y., Wang Y., Chen Y., Li R., Sun X., Shang Y. (2006) EMBO J. 25, 4223–4233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lemieux P., Fuqua S. (1996) J. Steroid Biochem. Mol. Biol. 56, 87–91 [DOI] [PubMed] [Google Scholar]
- 7.Ferguson A. T., Davidson N. E. (1997) Crit. Rev. Oncog. 8, 29–46 [DOI] [PubMed] [Google Scholar]
- 8.Cottone E., Orso F., Biglia N., Sismondi P., De Bortoli M. (2001) Int. J. Biol. Markers 16, 151–166 [DOI] [PubMed] [Google Scholar]
- 9.Shang Y., Hu X., DiRenzo J., Lazar M. A., Brown M. (2000) Cell 103, 843–852 [DOI] [PubMed] [Google Scholar]
- 10.Zhang H., Yi X., Sun X., Yin N., Shi B., Wu H., Wang D., Wu G., Shang Y. (2004) Genes Dev. 18, 1753–1765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Iwase H., Omoto Y., Toyama T., Yamashita H., Hara Y., Sugiura H., Zhang Z. (2003) Breast Cancer Res. Treat. 80, 339–345 [DOI] [PubMed] [Google Scholar]
- 12.Shou J., Massarweh S., Osborne C. K., Wakeling A. E., Ali S., Weiss H., Schiff R. (2004) J. Natl. Cancer Inst. 96, 926–935 [DOI] [PubMed] [Google Scholar]
- 13.Su Q., Hu S., Gao H., Ma R., Yang Q., Pan Z., Wang T., Li F. (2008) Oncology 75, 159–168 [DOI] [PubMed] [Google Scholar]
- 14.Lahusen T., Henke R. T., Kagan B. L., Wellstein A., Riegel A. T. (2009) Breast Cancer Res. Treat. 116, 225–237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhao W., Zhang Q., Kang X., Jin S., Lou C. (2009) Biochem. Biophys. Res. Commun. 380, 699–704 [DOI] [PubMed] [Google Scholar]
- 16.Anzick S. L., Kononen J., Walker R. L., Azorsa D. O., Tanner M. M., Guan X. Y., Sauter G., Kallioniemi O. P., Trent J. M., Meltzer P. S. (1997) Science 277, 965–968 [DOI] [PubMed] [Google Scholar]
- 17.Fereshteh M. P., Tilli M. T., Kim S. E., Xu J., O'Malley B. W., Wellstein A., Furth P. A., Riegel A. T. (2008) Cancer Res. 68, 3697–3706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Torres-Arzayus M. I., Font de Mora J., Yuan J., Vazquez F., Bronson R., Rue M., Sellers W. R., Brown M. (2004) Cancer Cell 6, 263–274 [DOI] [PubMed] [Google Scholar]
- 19.Dobrzycka K. M., Townson S. M., Jiang S., Oesterreich S. (2003) Endocr. Relat. Cancer 10, 517–536 [DOI] [PubMed] [Google Scholar]
- 20.Morinaga T., Yasuda H., Hashimoto T., Higashio K., Tamaoki T. (1991) Mol. Cell. Biol. 11, 6041–6049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sun X., Frierson H. F., Chen C., Li C., Ran Q., Otto K. B., Cantarel B. L., Vessella R. L., Gao A. C., Petros J., Miura Y., Simons J. W., Dong J. T. (2005) Nat. Genet. 37, 407–412 [DOI] [PubMed] [Google Scholar]
- 22.Xu J., Sauvageot J., Ewing C. M., Sun J., Liu W., Isaacs S. D., Wiley K. E., Diaz L., Zheng S. L., Walsh P. C., Isaacs W. B. (2006) Prostate 66, 1082–1085 [DOI] [PubMed] [Google Scholar]
- 23.Sun X., Zhou Y., Otto K. B., Wang M., Chen C., Zhou W., Subramanian K., Vertino P. M., Dong J. T. (2007) J. Cancer Res. Clin. Oncol. 133, 103–105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cleton-Jansen A. M., van Eijk R., Lombaerts M., Schmidt M. K., Van't Veer L. J., Philippo K., Zimmerman R. M., Peterse J. L., Smit V. T., van Wezel T., Cornelisse C. J. (2008) BMC Cancer 8, 105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kai K., Zhang Z., Yamashita H., Yamamoto Y., Miura Y., Iwase H. (2008) BMC Cancer 8, 262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang Z., Yamashita H., Toyama T., Sugiura H., Ando Y., Mita K., Hamaguchi M., Kawaguchi M., Miura Y., Iwase H. (2005) Clin. Cancer Res. 11, 193–198 [PubMed] [Google Scholar]
- 27.Fan S., Wang J., Yuan R., Ma Y., Meng Q., Erdos M. R., Pestell R. G., Yuan F., Auborn K. J., Goldberg I. D., Rosen E. M. (1999) Science 284, 1354–1356 [DOI] [PubMed] [Google Scholar]
- 28.Chen D., Ma H., Hong H., Koh S. S., Huang S. M., Schurter B. T., Aswad D. W., Stallcup M. R. (1999) Science 284, 2174–2177 [DOI] [PubMed] [Google Scholar]
- 29.Wu R. C., Qin J., Hashimoto Y., Wong J., Xu J., Tsai S. Y., Tsai M. J., O'Malley B. W. (2002) Mol. Cell. Biol. 22, 3549–3561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Miura Y., Tam T., Ido A., Morinaga T., Miki T., Hashimoto T., Tamaoki T. (1995) J. Biol. Chem. 270, 26840–26848 [DOI] [PubMed] [Google Scholar]
- 31.Murphy T. J., Pavlath G. K., Wang X., Boss V., Abbott K. L., Robida A. M., Nichols J., Xu K., Ellington M. L., Loss J. R., 2nd (2002) Methods Enzymol. 345, 539–551 [DOI] [PubMed] [Google Scholar]
- 32.Dong X. Y., Chen C., Sun X., Guo P., Vessella R. L., Wang R. X., Chung L. W., Zhou W., Dong J. T. (2006) Cancer Res. 66, 6998–7006 [DOI] [PubMed] [Google Scholar]
- 33.Liu X., Yue P., Zhou Z., Khuri F. R., Sun S. Y. (2004) J. Natl. Cancer Inst. 96, 1769–1780 [DOI] [PubMed] [Google Scholar]
- 34.Dong X. Y., Rodriguez C., Guo P., Sun X., Talbot J. T., Zhou W., Petros J., Li Q., Vessella R. L., Kibel A. S., Stevens V. L., Calle E. E., Dong J. T. (2008) Hum. Mol. Genet. 17, 1031–1042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chen C., Sun X., Guo P., Dong X. Y., Sethi P., Cheng X., Zhou J., Ling J., Simons J. W., Lingrel J. B., Dong J. T. (2005) J. Biol. Chem. 280, 41553–41561 [DOI] [PubMed] [Google Scholar]
- 36.Guo P., Dong X. Y., Zhang X., Zhao K. W., Sun X., Li Q., Dong J. T. (2009) J. Biol. Chem. 284, 6071–6078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hartel A., Didier A., Pfaffl M. W., Meyer H. H. (2003) J. Steroid Biochem. Mol. Biol. 84, 231–238 [DOI] [PubMed] [Google Scholar]
- 38.Shi B., Liang J., Yang X., Wang Y., Zhao Y., Wu H., Sun L., Zhang Y., Chen Y., Li R., Zhang Y., Hong M., Shang Y. (2007) Mol. Cell. Biol. 27, 5105–5119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.McDonnell D. P., Norris J. D. (2002) Science 296, 1642–1644 [DOI] [PubMed] [Google Scholar]
- 40.Yasuda H., Mizuno A., Tamaoki T., Morinaga T. (1994) Mol. Cell. Biol. 14, 1395–1401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Singh R. R., Kaluarachchi K., Chen M., Rayala S. K., Balasenthil S., Ma J., Kumar R. (2006) J. Biol. Chem. 281, 25612–25621 [DOI] [PubMed] [Google Scholar]
- 42.Shao W., Keeton E. K., McDonnell D. P., Brown M. (2004) Proc. Natl. Acad. Sci. U. S. A. 101, 11599–11604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Thorat M. A., Turbin D., Morimiya A., Leung S., Zhang Q., Jeng M. H., Huntsman D. G., Nakshatri H., Badve S. (2008) Histopathology 53, 634–641 [DOI] [PubMed] [Google Scholar]
- 44.Gronwald J., Jauch A., Cybulski C., Schoell B., Bohm-Steuer B., Lener M., Grabowska E., Gorski B., Jakubowska A., Domagala W., Chosia M., Scott R. J., Lubinski J. (2005) Int. J. Cancer 114, 230–236 [DOI] [PubMed] [Google Scholar]
- 45.List H. J., Lauritsen K. J., Reiter R., Powers C., Wellstein A., Riegel A. T. (2001) J. Biol. Chem. 276, 23763–23768 [DOI] [PubMed] [Google Scholar]
- 46.Louie M. C., Revenko A. S., Zou J. X., Yao J., Chen H. W. (2006) Mol. Cell. Biol. 26, 3810–3823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kuang S. Q., Liao L., Wang S., Medina D., O'Malley B. W., Xu J. (2005) Cancer Res. 65, 7993–8002 [DOI] [PubMed] [Google Scholar]
- 48.Osborne C. K., Bardou V., Hopp T. A., Chamness G. C., Hilsenbeck S. G., Fuqua S. A., Wong J., Allred D. C., Clark G. M., Schiff R. (2003) J. Natl. Cancer Inst. 95, 353–361 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.