Abstract
Mucin type O-glycosylation is a highly conserved form of post-translational modification initiated by the family of enzymes known as the polypeptide α-N-acetylgalactosaminyltransferases (ppGalNAcTs in mammals and PGANTs in Drosophila). To address the cellular functions of the many PGANT family members, RNA interference (RNAi) to each pgant gene was performed in two independent Drosophila cell culture lines. We demonstrate that RNAi to individual pgant genes results in specific reduction in gene expression without affecting the expression of other family members. Cells with reduced expression of individual pgant genes were then examined for changes in viability, morphology, adhesion, and secretion to assess the contribution of each family member to these cellular functions. Here we find that RNAi to pgant3, pgant6, or pgant7 resulted in reduced secretion, further supporting a role for O-glycosylation in proper secretion. Additionally, RNAi to pgant3 or pgant6 resulted in altered Golgi organization, suggesting a role for each in establishing or maintaining proper secretory apparatus structure. Other subcellular effects observed included multinucleated cells seen after RNAi to either pgant2 or pgant35A, suggesting a role for these genes in the completion of cytokinesis. These studies demonstrate the efficient and specific knockdown of pgant gene expression in two Drosophila cell culture systems, resulting in specific morphological and functional effects. Our work provides new information regarding the biological roles of O-glycosylation and illustrates a new platform for interrogating the cellular and subcellular effects of this form of post-translational modification.
Keywords: Adhesion, Drosophila, Glycosylation, RNA Interference (RNAi), Secretion, Drosophila Cell Culture, Mucin-type O-Glycosylation, pgant
Introduction
Mucin type O-linked glycosylation is an evolutionarily conserved post-translational modification found on secreted and membrane-bound proteins in many diverse species (1–4). Initiation of mucin type O-linked glycosylation is catalyzed by a family of glycosyltransferases known as the UDP-N-acetylgalactosamine:polypeptide N-acetylgalactosaminyltransferases (ppGaNTases or ppGalNAcTs in mammals and PGANTs in Drosophila)2 (1, 3, 5–8). It is known that there are as many as 18–20 family members in mammals,3 with many isoforms displaying overlapping tissue distribution and substrate specificity, resulting in the potential for significant functional redundancy (1–3, 6, 9, 10). Indeed, mouse models deficient in individual isoforms remain viable, although phenotypes associated with lymphocyte homing, blood coagulation (11), and hyperphosphatemia (12) are observed. Genome analysis revealed that there are fewer family members in Drosophila melanogaster (6, 7, 13), making the fly a more tractable model system in which to investigate the biological role of O-linked glycosylation. Furthermore, our laboratory has found the widespread presence of O-glycans throughout development and in many diverse organ systems, suggesting a crucial and conserved role for O-linked glycosylation in cellular and molecular processes employed during Drosophila development (13, 14). Indeed, studies from our laboratory have found that one member of this family, pgant35A, plays a role in proper epithelial tube formation during Drosophila embryogenesis (5, 15). Additionally, recent work has demonstrated that another family member (pgant3) modulates proper cell adhesion during wing formation; mutations in this transferase cause improper epithelial cell adhesion, which results in a wing blistering phenotype in adults (16). Interestingly, mutations in pgant3 appear to disrupt the secretion of an extracellular matrix protein involved in integrin-mediated cell adhesion in the wing (17).
In the past few years, RNA interference (RNAi) in Drosophila cell culture has become an increasingly popular method for rapidly investigating the function of many genes at the cellular and subcellular level (18–21). Drosophila cells will efficiently take up and process long dsRNA, resulting in specific decreases in gene expression. Given the technical challenges associated with imaging subcellular structures in Drosophila tissues in vivo, this methodology provides a facile way to investigate cellular phenotypes associated with the knockdown of specific genes. In an effort to gain more information about the functional role of O-glycosylation at the cellular level, we employed RNAi to each family member in two independent Drosophila cell lines: S2R+ cells, an adherent cell line of embryonic origin (21–23), and S2 cells, a non-adherent cell line of embryonic origin that can become adherent when plated on concanavalin A-coated surfaces (24). Our results demonstrate that RNAi in either Drosophila cell line can efficiently and specifically knock down the transcript levels of each individual pgant gene. Interestingly, we find that certain pgants are able to influence secretion in these cell culture systems, supporting our previous in vivo studies (17). Additionally, we find evidence for the role of certain pgants in the structure of the Golgi apparatus. Finally, we provide data suggesting a role for other pgant family members in cytokinesis. These studies demonstrate the utility of this technique for examining the subcellular details of phenotypes observed in vivo as well as provide a platform for discovering novel functions of other members of this multigene family.
EXPERIMENTAL PROCEDURES
dsRNA Preparation
For the generation of dsRNA, cDNAs from each pgant were amplified using primers containing T7 RNA polymerase binding sites and gene-specific sequences to produce ∼500-bp fragments containing T7 promoters at the 5′ ends. We selected regions for dsRNA generation by using the off-target sequence search tool on the Drosophila RNAi Screening Center (DRSC) website in an effort to minimize any potential off-target effects. The off-target size was set as low as 16 nucleotides for each analysis, and no off-target effects were predicted for any of the dsRNA regions used in this study. Two regions for dsRNA generation were chosen for each pgant gene to verify knockdown and phenotypes observed. All primer sequences are listed in supplemental Table 1.
PCR products from the above-mentioned primer pairs were purified and used as templates to produce RNA using the MEGASCRIPT T7 transcription kit (Ambion). RNA was LiCl-precipitated, resuspended in water, incubated at 65 °C for 30 min, and then slow-cooled to room temperature to allow annealing. dsRNA formed was then stored at −20 °C.
RNAi in Drosophila Cell Culture
Drosophila cells were grown in Schneider's medium (Invitrogen) with 10% heat-inactivated fetal bovine serum (FBS) (Invitrogen) at 25 °C in culture flasks. RNAi was performed as described on the DRSC website. Briefly, 2 × 105 cells in 250 μl of serum-free media were added to each well of a 24-well plate. dsRNA (7 μg) was added to each well, and plates were mixed back and forth. The cells were then incubated for 30 min at room temperature before adding 750 μl of medium containing 10% FBS. Cells were grown for 4 days at 25 °C. Each experiment was performed with two independent dsRNAs to each pgant to verify gene specific knockdown and observed phenotypes.
Quantitative Real-time PCR
Cells were lysed, and RNA was extracted using the RNAqueous-4 PCR kit (Ambion). cDNA synthesis was performed using the iScript cDNA synthesis kit (Bio-Rad). Real-time PCR primers for each pgant were designed using Beacon Designer software (Bio-Rad) and are shown in supplemental Table 2. Quantitative RT-PCR was performed on a MyiQ real time PCR thermocycler (Bio-Rad) using the SYBR-Green PCR Master Mix (Bio-Rad). Analyzed products were assayed in triplicate and in multiple independent experiments.
Cell Staining and Analysis
After dsRNA treatment, cells were fixed in 4% formaldehyde, phosphate-buffered saline (PBS) (Electron Microscopy Science) and then washed twice in PBS with 0.1% Triton X-100. To detect the Golgi apparatus, cells were stained with either anti-GM130 (Abcam) (dilution, 1:50) or anti-dSyx16 (25) (a kind gift of W. Trimble) (dilution 1:400) at room temperature for 1 h, then washed in PBS and incubated with FITC-conjugated anti-rabbit IgG antibody (dilution, 1:100) (Jackson ImmunoResearch Laboratories) or Cy3-conjugated anti-rabbit IgG antibody (dilution 1:100), respectively, at room temperature for 1 h. Counterstaining was then performed using 4′,6-diamidino-2-phenylindole, dihydrochloride (DAPI) (Sigma) or SYBR Green (Invitrogen) followed by washes in PBS. Actin staining was performed using TRITC-phalloidin (Sigma). To detect apoptosis, cells were stained with caspase-3 antibody (cleaved caspase-3 Asp-175 antibody) (Cell Signaling Technology) or with TUNEL staining using an in situ cell death detection kit, TMR red (Roche Applied Science). To detect lysosomes, cells were stained with the LEP-100 antibody (dilution, 1:50) (Developmental Studies Hybridoma Bank) as described previously (26). Cells were examined using a Zeiss Axiophot microscope.
Quantitative cell adhesion assays were performed on S2R+ cells after incubation with dsRNA as described previously (17). Briefly, S2R+ cells were harvested and resuspended at a density of 2 × 105 cells/ml. 100 μl of cell suspension was seeded into each well and then incubated for an additional 2 h at 25 °C. After removing the non-adherent cells, 150 μl of 0.2% crystal violet (Sigma) aqueous solution was added to stain the cells that remained attached. Wells were washed, and cells were dissolved in 150 μl of 1% SDS. The optical density at 570 nm was measured using a Microplate Reader (TECAN).
Protein Secretion Assays
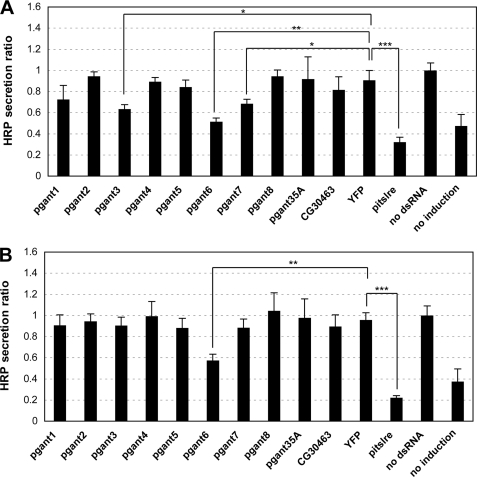
2 × 105 S2R+ or S2 cells in 250 μl of Schneider's medium with 10% FBS were added to each well of a 24-well plate. Cells were co-transfected with 0.2 μg of the pMT-ss-HRP-V5 secretion reporter construct (18) (a kind gift of F. Bard) and 1.6 μg of dsRNA using Effectene transfection reagent (Qiagen). After incubation for 3 days, HRP expression was induced by adding copper sulfate to the media at a final concentration of 500 μm. Transfected cells were incubated for 24 h, and then the supernatant was collected. 50 μl supernatant from each transfection was added to 50 μl of ECL reagent (Pierce) in a 96-well plate. The luminescence was measured with a plate reader at 450 nm. The secretion under each condition was defined as “HRP secretion ratio,” which was calculated by dividing the luminescence value of each sample by the luminescence value of the control cells not treated with dsRNA. The values shown in Fig. 3 represent the average of two independent experiments (each of which was assayed six times to obtain an average value).
FIGURE 3.
RNAi to certain pgants in S2R+ and S2 cells results in decreased secretion. S2R+ (A) and S2 (B) cells were transfected with the inducible pMT-ss-HRP-V5 secretion reporter construct (18) and treated with dsRNA to each pgant, pitslre (positive control), or YFP or untreated as described under “Experimental Procedures.” After induction, HRP secreted into the media was quantitated, normalized, and expressed as HRP secretion ratio as described under “Experimental Procedures.” Error bars = S.D. *, p < 0.05; **, p < 0.01; ***, p < 0.001.
HRP Glycosylation Assays
S2R+ cells were transfected with the pMT-V5 parental vector or pMT-ss-HRP-V5 secretion vector (kind gifts of F. Bard), and expression was induced by the addition of copper sulfate as described above. After induction for either 1 or 2 days, medium from each transfection was collected. 5 μl of cell media were electrophoresed under reducing conditions on 4–12% SDS-PAGE gradient gels. Gels were transferred to nitrocellulose; membranes were then blocked with 1× blocking buffer (Sigma) and incubated with either the V5-HRP antibody (1:2000, Invitrogen), the helix pomatia (HPA)-HRP lectin (1:1000, Sigma), or the peanut agglutinin (PNA)-HRP lectin (1:1000, EY Laboratories, Inc.). For the data in supplemental Fig. 5D, 10 μl of media from cells transfected with pMT-ss-HRP-V5 (and induced for 2 days) were either untreated or incubated with 1 μl each of peptide N-glycosidase F, sialidase A, and/or O-glycanase (Prozyme) for 3 h at 37 °C after denaturation with 2.5 μl of denaturation solution (Prozyme). Glycosidase-treated and untreated samples were electrophoresed, blotted, and probed with the V5-HRP antibody as described above.
Endocytosis Assay
After dsRNA treatment for 4 days, S2R+ or S2 cells were collected and washed 3 times with ice-cold PBS (pH 8.0) and then resuspended in PBS. Endocytosis assays were carried out as described previously (27). Briefly, 200 μl of 10 mm sulfo-NHS-biotin (Pierce) were added per ml of cell suspension. Cells were incubated for 3 min and then washed 3 times with PBS to remove the sulfo-NHS-biotin. Cells were incubated for an additional 15 min to allow internalization. Finally, cells were fixed and stained with streptavidin conjugated with Cy3 (Jackson ImmunoResearch Laboratories, 1:500) to visualize internalized sulfo-NHS-biotin. dsRNA to Chc (the gene encoding clathrin), a known effector of endocytosis (28), was used as a positive control.
RESULTS
Nine pgant Genes Are Expressed in S2R+ and S2 Cells
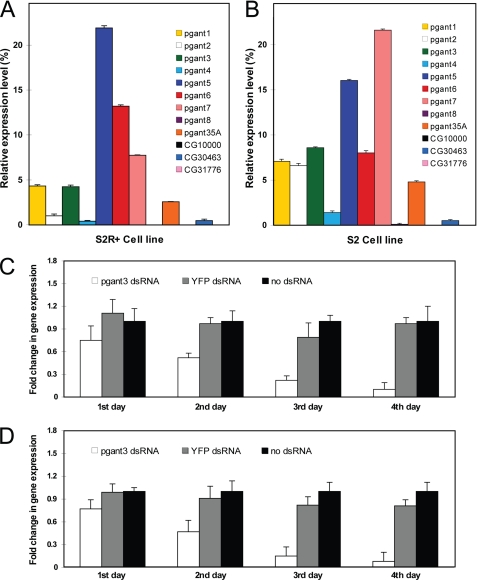
The Drosophila cell lines S2R+ and S2 were used to study the cellular effects of each pgant gene. Both cell lines have been used extensively to study cell adhesion, morphology, and subcellular architecture (21–24). S2R+ is an embryonic cell line that adheres to surfaces via an integrin-dependent mechanism (21–23). The S2 cell line is a non-adherent, embryonic line that can become adherent upon plating on concanavalin A-coated surfaces (24); however, this adhesion is distinct from that observed with S2R+ cells in that it is integrin-independent. Before embarking on the RNAi experiments, we performed quantitative PCR to determine which pgant family members are expressed in each cell line. All primer pairs were tested for optimal annealing and amplification so that relative levels of transcript could be compared among genes. In S2R+ cells (Fig. 1A), the expression of pgant5 is highest followed by pgant6, pgant7, pgant1, pgant3, pgant35A, pgant2, pgant4, and CG30463 (a putative pgant). No expression of pgant8 was detected in S2R+ cells. In S2 cells (Fig. 1B), pgant7 is expressed most abundantly, followed by pgant5, pgant3, pgant6, pgant1, pgant2, pgant35A, pgant4, and CG30463. Very low expression of pgant8 was seen in S2 cells. No expression was detected for CG31776 or CG10000 (two additional putative pgants) in either cell line. Based on these results, we performed RNAi to the pgant family members with detectable expression in these cells.
FIGURE 1.
Expression of pgants in Drosophila S2R+ and S2 cells and time course of RNAi-mediated transcript knockdown. Expression levels of pgant genes in S2R+ (A) and S2 (B) cells as determined by quantitative real time PCR. RNA levels were normalized to GAPDH. The time course shows reduction in pgant3 gene expression after RNAi treatment for 1, 2, 3, or 4 days as determined by quantitative PCR for S2R+ (C) and S2 (D) cells. Shown are data from cells treated with no dsRNA, dsRNA to YFP (negative control), or dsRNA to pgant3. RNA levels were normalized to 18 S rRNA. S.D. are shown.
RNAi to Each pgant Results in a Specific Decrease in Gene Expression
dsRNA to each pgant was directly added to the medium of both S2 and S2R+ cells in culture as described previously to induce RNAi, as these cells are known to take up nucleic acids without the need for transfection reagents (21). To determine optimal time of dsRNA treatment for efficient gene knockdown, we monitored pgant gene expression by quantitative PCR after various times of dsRNA exposure. We found maximal transcript decreases for both cell lines after 4 days of incubation with dsRNA (Fig. 1, C and D). Therefore, all dsRNA incubations were hereafter carried out for 4 days. The expression of every pgant family member was monitored by quantitative PCR after each individual dsRNA treatment to assess the specificity of each gene knockdown (supplemental Figs. 1 and 2). Our results demonstrated that only the targeted pgant gene showed a significant decrease in transcript level, indicating that the RNAi is specific to the desired gene and that there is no compensatory up or down-regulation of the transcription of other family members in either cell line (supplemental Figs. 1 and 2). Finally, gene knockdown and the phenotypes observed were independently verified by repeating all RNAi experiments using a different dsRNA for each gene (supplemental Table 1 and Figs. 3 and 4).
RNAi to Certain pgants Can Influence Cell Morphology, Cytoskeletal Organization, and Cell Adhesion
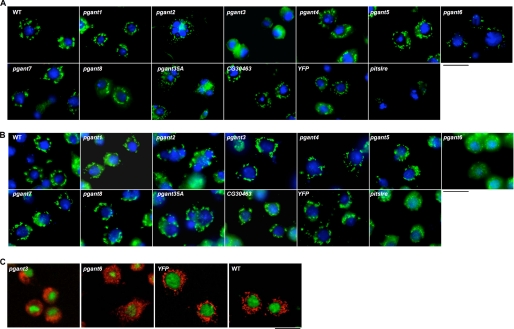
Initially, dsRNA-treated cells were monitored for changes in cell morphology, adhesion, and viability. Cells were stained with phalloidin to detect changes in actin filament organization and to investigate changes in cell shape and spreading after dsRNA treatment. Additionally, cell adhesion changes were assayed by quantitating the number of cells remaining attached, as described previously (17). We used dsRNA to mys, a gene encoding βPS integrin, as a positive control for changes in cell morphology and adhesion phenotypes, as RNAi to this gene is known to cause changes in cell shape and cell adhesion in S2R+ cells (19). As seen in Fig. 2A, untreated S2R+ cells or cells treated with YFP dsRNA had highly developed actin-based lamellae and were well spread on cell culture dishes. However, cells treated with mys dsRNA became round and non-adherent, with a dramatic actin fibril reorganization (Fig. 2A). As we reported recently, pgant3 dsRNA resulted in a similar cytoskeletal reorganization and non-adherent phenotype (17), supporting a role for pgant3 in the integrin-mediated cell adhesion that was seen previously in vivo (16, 17). Thus, this cell culture system recapitulates certain biological effects seen in the organism, validating its use to further examine the effects of O-glycosylation. Interestingly, cytoskeletal changes and cell adhesion defects were not seen upon RNAi to the other pgant family members in S2R+ cells, suggesting a unique role for pgant3 in the cell adhesive mechanisms specific to this cell line (Fig. 2A and data not shown). In S2 cells, which employ a different mechanism of adhesion on concanavalin A-coated surfaces, no disruption of cytoskeletal organization or cell adhesion was seen after RNAi to any of the pgant family members (Fig. 2B and data not shown). However, a consistent and reproducible reduction in cell spreading was seen after RNAi to pgant7 (Fig. 2B).
FIGURE 2.
Morphological analysis of S2R+ and S2 cells treated with dsRNA to each pgant. S2R+ (A) and S2 (B) cells were treated with dsRNA to the gene denoted at the top of each box and were then stained to visualize the actin cytoskeleton (phalloidin, red) and nuclei (DAPI, blue). WT = no dsRNA treatment. Black size bar = 20 μm.
RNAi to pgant3, pgant6, or pgant7 Results in Decreased Secretion
Recent studies from our group have demonstrated that pgant3 influences cell adhesion in vivo by affecting secretion of a matrix protein involved in integrin-mediated cell adhesion (17). Additionally, previous work by other groups has suggested that O-glycosylation influences secretion (29–32). To address whether O-glycosylation can influence secretion in this cell culture system, we transfected S2R+ and S2 cells with an inducible, HRP secretion construct used previously to identify genes involved in secretion (18). This construct contains a secretion signal sequence (ss) attached to the HRP reporter under the control of a Cu2+-inducible promoter. The HRP reporter is N-glycosylated in Drosophila cells, indicating that it is transiting through the secretory apparatus (supplemental Fig. 5). Cells were co-transfected with the HRP reporter construct and either no dsRNA, dsRNA to YFP, dsRNA to pitslre (a gene involved in Golgi organization and secretion, to serve as a positive control) (18), or dsRNA to individual pgant genes. After 3 days, expression of the HRP reporter construct was induced. After an additional 1–2 days of induction, HRP that had been secreted into the cell media was quantitated (Fig. 3). The amount of secreted HRP was significantly reduced relative to controls in S2R+ cells treated with RNAi to pgant3, pgant6, or pgant7 (Fig. 3A). Similarly, secretion was reduced in cells treated with RNAi to the positive control gene, pitslre (18). In S2 cells, dsRNA treatment with pgant6 resulted in decreased HRP secretion (Fig. 3B). This study demonstrates that the loss of pgant3, pgant6, or pgant7 in S2R+ cells or the loss of pgant6 in S2 cells results in decreased secretion, suggesting unique, cell type-specific functions for specific pgant family members. Because the HRP reporter is not O-glycosylated in Drosophila cells (supplemental Fig. 5), our data indicate that the effects of pgant3, pgant6, or pgant7 RNAi on secretion are not due to HRP being directly glycosylated by these enzymes. Taken together, our data suggest that certain pgants can influence secretion and lend further support to a role for pgant3 in secretion events observed in vivo (17).
The influence of the pgants on secretion prompted us to ask whether RNAi to the pgants could also affect endocytosis. S2R+ and S2 cells were treated with dsRNAs as described above. As a positive control for detecting defects in endocytosis, cells were treated with dsRNA to Chc, the gene encoding clathrin, which is known to mediate endocytosis (28). Cells were then briefly exposed to sulfo-NHS-biotin, washed, and incubated to allow endocytosis to occur. As expected, RNAi to Chc significantly reduced the amount of endocytosis occurring in both S2 and S2R+ cells, as evidenced by the lack of the internalized sulfo-NHS-biotin (supplemental Fig. 6). However, no significant change in endocytosis in either cell line was observed upon RNAi to any of the pgants (supplemental Fig. 6).
RNAi to pgant3 or pgant6 Alters Golgi Organization
Because of the effect of certain pgants on secretion, we next examined whether there was a concomitant change in the subcellular architecture of the secretory apparatus by staining dsRNA-treated cells with markers specific for the Golgi apparatus (GM130 and dSyx16). GM130 is a golgin located in the cis-Golgi, and dSyx16 is a Drosophila Q-SNARE present in the cisternal compartment adjacent to the cis-Golgi (25). As a control for detecting defects in Golgi architecture, we treated cells with dsRNA to pitslre (18). Upon examining dsRNA-treated S2R+ cells, specific alterations in Golgi appearance were seen upon RNAi to pgant3 (Fig. 4A). In pgant3 dsRNA-treated cells, the Golgi apparatus (detected by the anti-GM130 antibody) was localized to one side of the cell and had a hazy, diffuse appearance, as opposed to the evenly distributed, punctate appearance characteristic of YFP dsRNA-treated or untreated cells (Fig. 4A). Similar results were seen using the anti-dSyx16 antibody (Fig. 4C). This difference in Golgi appearance was reproducible and seen with two different pgant3 dsRNAs. Additionally, differences in GM130 and dSyx16 staining in S2R+ cells were also seen with RNAi to pgant6; in this instance, the Golgi staining was greatly reduced, as was the quantity and size of punctate structures seen (Fig. 4, A and C). This difference in Golgi appearance was also reproducible with different pgant6 dsRNAs. No differences were seen upon treatment of S2R+ cells with dsRNA to other pgant family members (Fig. 4A). Additionally, no differences in the lysosomal compartments were observed upon dsRNA treatment with any pgants in S2R+ cells (data not shown).
FIGURE 4.
Golgi staining of S2R+ and S2 cells treated with dsRNA to each pgant. S2R+ (A) and S2 (B) cells were treated with dsRNA to each gene denoted at the top of each box and were then stained to visualize the Golgi apparatus using the anti-GM130 antibody (green) and nuclei (DAPI, blue). C, S2R+ cells were also stained with the anti-dSyx16 antibody (red). WT = no dsRNA treatment. Black size bar = 20 μm.
In S2 cells treated with dsRNA to pgant6, the Golgi apparatus was also irregular, with much smaller and very diffuse punctate structures seen (Fig. 4B). This difference was reproducible and seen with two different pgant6 dsRNAs. This result is also supported by a prior genome-wide RNAi screen that identified pgant6 as one of many genes that resulted in altered Golgi organization and secretion (18). No differences in Golgi structure in S2 cells were seen upon dsRNA treatment with the other pgant family members. Additionally, no effects were seen on lysosomal structures in S2 cells (data not shown). Thus, RNAi to pgant3 or to pgant6 each affected Golgi structure in unique ways, suggesting that pgant3 and pgant6 may each be involved in unique aspects of Golgi organization and/or function in a cell type-specific manner, thus influencing secretion in different cell contexts.
RNAi to Other pgant Family Members Results in Other Subcellular Effects
By labeling DNA, we investigated cell division and cell viability after dsRNA treatment. RNAi to pgant2 or to pgant35A resulted in cells with multiple nuclei, suggesting a role for these genes in cytokinesis (Figs. 2 and 4). This phenotype was seen in both S2 and S2R+ cells and was verified using independent dsRNAs to pgant2 and pgant35A in each cell type. This phenotype was not seen upon performing RNAi to the other pgant family members. Additionally, we looked for evidence of apoptosis after dsRNA treatment by staining cells with TUNEL. However, no significant apoptosis was seen upon RNAi to individual pgant family members in either cell line (data not shown).
DISCUSSION
This study demonstrates the utility of RNAi in Drosophila cell culture to further interrogate the cellular and subcellular effects of glycosylation. Here, we demonstrate that dsRNA to the pgant-encoded transferase family can produce rapid, efficient, and specific knockdown of transcripts, allowing one to detect subcellular changes that may not be discernable in vivo. dsRNA to any given pgant specifically decreased expression of that gene and had little or no effect on the expression levels of other family members, allowing us to interrogate the specific cellular effects of individual pgants. By using cell lines with different morphological and cell adhesive properties, we are able to discern which phenotypes are cell type-specific and which are common to both cell types. Using this system, we have further verified the role of pgant3 in integrin-dependent cell adhesion events in S2R+ cells (17). In the current study we shed further light on the biological effects of pgant3 in these cells. Here we find that RNAi to pgant3 in S2R+ cells results in both reduced secretion and altered Golgi structure. In pgant3 dsRNA-treated cells, the Golgi is diffuse in appearance, lacking the normal punctate structure; secretion of a reporter construct is also significantly reduced. Studies from our laboratory indicate that the loss of pgant3 in the developing wing leads to altered secretion of extracellular matrix proteins involved in integrin-mediated cell adhesion in vivo (17). Attempts to visualize Golgi structure in the mutant wing discs have not been successful given the challenges associated with obtaining a clear image of this diffuse, subcellular structure in the elongated, irregularly shaped cells comprising the wing disc. Thus, this cell culture system may represent a more tractable way to visualize subcellular changes associated with alterations in pgant3 function. The results from this cell culture system support a role for pgant3 in proper secretion and suggest that altered Golgi structure may be associated with this secretion defect in vivo.
In addition to the Golgi and secretory effects seen for pgant3, we observed similar effects upon knockdown of another pgant family member. For example, RNAi to pgant6 in S2 and S2R+ cells resulted in changes in Golgi structure, albeit distinct from those seen with RNAi to pgant3 in S2R+ cells. These experiments are supported by a prior genome-wide screen that identified pgant6 as one gene that influenced Golgi structure (18). And like pgant3, we also discovered that the Golgi structural changes seen with RNAi to pgant6 were accompanied by reduced secretion. Because the secretion reporter protein is not normally O-glycosylated, the secretion changes seen are not due to the direct glycosylation of this protein by either PGANT3 or PGANT6. Our data suggest that PGANT3 and PGANT6 affect secretion by altering the normal structure and, therefore, function of the Golgi apparatus. Golgi architecture is known to be maintained and regulated by a number of protein families that influence vesicular trafficking and membrane dynamics as well as specific phosphorylation events that lead to Golgi dispersal and reaggregation during the mitotic cycle (33–37). PGANTs may exert their effects through the direct modification of key Golgi proteins, thereby influencing their stability, location, or ability to form proper complexes with other regulatory proteins. For example, in yeast, O-mannosylation stabilizes Sec20p, a t-SNARE protein involved in regulating vesicular traffic within the secretory apparatus (38). Alternatively, it remains possible that the PGANTs, being type II transmembrane Golgi-resident proteins, may play a structural role in Golgi dynamics independent of their enzymatic activity. However, studies from our group indicate that PGANT3 catalytic activity is specifically required for proper secretion in the developing Drosophila wing (17). Future studies will examine alterations in the levels as well as glycosylation and phosphorylation status of key proteins known to regulate Golgi structure. Additionally, Golgi dynamics will be visualized in real time to determine whether Golgi structural changes are associated with the cell cycle.
Our studies also provided the first evidence for the role of O-glycans in cytokinesis. Cells treated with dsRNA to either pgant2 or pgant35A displayed multiple nuclei, suggesting a role for these genes in some aspect of the final stages of cell division. This phenotype was observed in both S2 and S2R+ cells, suggesting the disruption of a function held in common between these two cell types. It is known that efficient transport through the Golgi and endoplasmic reticulum complexes is crucial for the increase in membrane synthesis required during cell division. Indeed, mutations in genes encoding Golgi-associated proteins that affect Golgi architecture and membrane delivery also interfere with proper cytokinesis (39–42). Also, in a screen for genes involved in cytokinesis, the second most abundant category obtained were those involved in membrane trafficking and organization (43). Finally, brefeldin A, which is known to interfere with vesicular trafficking, Golgi function, and membrane delivery, also results in cytokinesis defects (44). PGANT2 and PGANT35A may affect the stability or efficient trafficking of key components involved in new membrane formation, such that loss of either enzyme interferes with the efficient completion cytokinesis. We have previously found that mutations in pgant35A disrupt the transport of certain proteins to the apical surface of tracheal cells. Future studies will focus on identifying targets of each enzyme as well as defining when the disruption of cell division is occurring.
We are currently employing these cell lines to identify the substrates of the individual pgants responsible for specific morphological defects outlined here. We initially attempted to identify substrates for pgant3 by comparing lectin banding patterns on Western blots of dsRNA-treated versus untreated cells; however, no changes in bands were seen between the samples (data not shown). This is likely due to the fact that only a portion of the dsRNA-treated cell population actually received the dsRNA and, thus, had reduced levels of pgant3 transcripts; thus, we are examining the glycoproteins of a mixed population of cells (cells expressing wild type levels of pgant3 as well as cells expressing reduced levels of pgant3). Therefore, we suspect that we may not be able to detect changes in glycoprotein levels in this mixed population. Future work will focus on developing stable, inducible RNAi-expressing lines to obtain a homogeneous population of cells with reduced pgant gene expression to enable identification of in vivo substrates.
Altogether, we have demonstrated that this approach represents an efficient technique for interrogating the specific contribution of individual members of multigene families to cellular and subcellular morphology and function. The data presented here support and enhance our understanding of the biological role of certain pgants in vivo. Future work will continue to use a combination of the in vitro approach outlined here as well as in vivo analysis to gain a more thorough understanding of the biological role of O-glycans during eukaryotic development.
Supplementary Material
Acknowledgments
We thank our colleagues for many helpful discussions. We also thank Dr. W. Trimble, Dr. F. Bard, and the Developmental Studies Hybridoma Bank for kindly providing antibodies, plasmids, and other reagents.
This work was supported, in whole or in part, by National Institutes of Health (Intramural Research Program of the NIDCR).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Tables 1 and 2 and Figs. 1–6.
J. Raman and L. Tabak, personal communication.
- ppGaNTase or ppGalNAcT or PGANT or pgant
- UDP GalNAc:polypeptide N-acetylgalactosaminyltransferase
- TRITC
- tetramethylrhodamine isothiocyanate
- SNARE
- soluble NSF attachment protein receptor.
REFERENCES
- 1.Ten Hagen K. G., Fritz T. A., Tabak L. A. (2003) Glycobiology 13, 1R–16R [DOI] [PubMed] [Google Scholar]
- 2.Hang H. C., Bertozzi C. R. (2005) Bioorg. Med. Chem. 13, 5021–5034 [DOI] [PubMed] [Google Scholar]
- 3.Tian E., Ten Hagen K. G. (2009) Glycoconj. J. 26, 325–334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ten Hagen K. G., Zhang L., Tian E., Zhang Y. (2009) Glycobiology 19, 102–111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ten Hagen K. G., Tran D. T. (2002) J. Biol. Chem. 277, 22616–22622 [DOI] [PubMed] [Google Scholar]
- 6.Ten Hagen K. G., Tran D. T., Gerken T. A., Stein D. S., Zhang Z. (2003) J. Biol. Chem. 278, 35039–35048 [DOI] [PubMed] [Google Scholar]
- 7.Schwientek T., Bennett E. P., Flores C., Thacker J., Hollmann M., Reis C. A., Behrens J., Mandel U., Keck B., Schäfer M. A., Haselmann K., Zubarev R., Roepstorff P., Burchell J. M., Taylor-Papadimitriou J., Hollingsworth M. A., Clausen H. (2002) J. Biol. Chem. 277, 22623–22638 [DOI] [PubMed] [Google Scholar]
- 8.Hagen F. K., Nehrke K. (1998) J. Biol. Chem. 273, 8268–8277 [DOI] [PubMed] [Google Scholar]
- 9.Kingsley P. D., Ten Hagen K. G., Maltby K. M., Zara J., Tabak L. A. (2000) Glycobiology 10, 1317–1323 [DOI] [PubMed] [Google Scholar]
- 10.Young W. W., Jr., Holcomb D. R., Ten Hagen K. G., Tabak L. A. (2003) Glycobiology 13, 549–557 [DOI] [PubMed] [Google Scholar]
- 11.Tenno M., Ohtsubo K., Hagen F. K., Ditto D., Zarbock A., Schaerli P., von Andrian U. H., Ley K., Le D., Tabak L. A., Marth J. D. (2007) Mol. Cell. Biol. 27, 8783–8796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ichikawa S., Sorenson A. H., Austin A. M., Mackenzie D. S., Fritz T. A., Moh A., Hui S. L., Econs M. J. (2009) Endocrinology 150, 2543–2550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tian E., Ten Hagen K. G. (2006) Glycobiology 16, 83–95 [DOI] [PubMed] [Google Scholar]
- 14.Tian E., Ten Hagen K. G. (2007) Glycobiology 17, 820–827 [DOI] [PubMed] [Google Scholar]
- 15.Tian E., Ten Hagen K. G. (2007) J. Biol. Chem. 282, 606–614 [DOI] [PubMed] [Google Scholar]
- 16.Zhang L., Zhang Y., Ten Hagen K. G. (2008) J. Biol. Chem. 283, 34076–34086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang L., Tran D. T., Ten Hagen K. G. (2010) J. Biol. Chem. 285, 19491–19501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bard F., Casano L., Mallabiabarrena A., Wallace E., Saito K., Kitayama H., Guizzunti G., Hu Y., Wendler F., Dasgupta R., Perrimon N., Malhotra V. (2006) Nature 439, 604–607 [DOI] [PubMed] [Google Scholar]
- 19.Kiger A. A., Baum B., Jones S., Jones M. R., Coulson A., Echeverri C., Perrimon N. (2003) J. Biol. 2, 27.1–27.15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lum L., Yao S., Mozer B., Rovescalli A., Von Kessler D., Nirenberg M., Beachy P. A. (2003) Science 299, 2039–2045 [DOI] [PubMed] [Google Scholar]
- 21.Boutros M., Kiger A. A., Armknecht S., Kerr K., Hild M., Koch B., Haas S. A., Paro R., Perrimon N. (2004) Science 303, 832–835 [DOI] [PubMed] [Google Scholar]
- 22.Yanagawa S., Lee J. S., Ishimoto A. (1998) J. Biol. Chem. 273, 32353–32359 [DOI] [PubMed] [Google Scholar]
- 23.Friedman A., Perrimon N. (2006) Nature 444, 230–234 [DOI] [PubMed] [Google Scholar]
- 24.Jani K., Schöck F. (2007) J. Cell Biol. 179, 1583–1597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xu H., Boulianne G. L., Trimble W. S. (2002) J. Cell Sci. 115, 4447–4455 [DOI] [PubMed] [Google Scholar]
- 26.Mellén M. A., de la Rosa E. J., Boya P. (2008) Cell Death Differ. 15, 1279–1290 [DOI] [PubMed] [Google Scholar]
- 27.Behr M., Wingen C., Wolf C., Schuh R., Hoch M. (2007) Nat. Cell Biol. 9, 847–853 [DOI] [PubMed] [Google Scholar]
- 28.Korolchuk V. I., Schütz M. M., Gómez-Llorente C., Rocha J., Lansu N. R., Collins S. M., Wairkar Y. P., Robinson I. M., O'Kane C. J. (2007) J. Cell Sci. 120, 4367–4376 [DOI] [PubMed] [Google Scholar]
- 29.Delannoy P., Kim I., Emery N., De Bolos C., Verbert A., Degand P., Huet G. (1996) Glycoconj. J. 13, 717–726 [DOI] [PubMed] [Google Scholar]
- 30.Huet G., Hennebicq-Reig S., de Bolos C., Ulloa F., Lesuffleur T., Barbat A., Carrière V., Kim I., Real F. X., Delannoy P., Zweibaum A. (1998) J. Cell Biol. 141, 1311–1322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gouyer V., Leteurtre E., Delmotte P., Steelant W. F., Krzewinski-Recchi M. A., Zanetta J. P., Lesuffleur T., Trugnan G., Delannoy P., Huet G. (2001) J. Cell Sci. 114, 1455–1471 [DOI] [PubMed] [Google Scholar]
- 32.Potter B. A., Hughey R. P., Weisz O. A. (2006) Am. J. Physiol. Cell. Physiol. 290, C1–C10 [DOI] [PubMed] [Google Scholar]
- 33.Lowe M., Gonatas N. K., Warren G. (2000) J. Cell Biol. 149, 341–356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Derby M. C., Lieu Z. Z., Brown D., Stow J. L., Goud B., Gleeson P. A. (2007) Traffic 8, 758–773 [DOI] [PubMed] [Google Scholar]
- 35.Ramirez I. B., Lowe M. (2009) Semin. Cell Dev. Biol. 20, 770–779 [DOI] [PubMed] [Google Scholar]
- 36.Barr F. A. (2009) Semin. Cell Dev. Biol. 20, 780–783 [DOI] [PubMed] [Google Scholar]
- 37.Wei J. H., Seemann J. (2009) Semin. Cell Dev. Biol. 20, 810–816 [DOI] [PubMed] [Google Scholar]
- 38.Weber Y., Prill S. K., Ernst J. F. (2004) Eukaryot. Cell 3, 1164–1168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Farkas R. M., Giansanti M. G., Gatti M., Fuller M. T. (2003) Mol. Biol. Cell 14, 190–200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Polevoy G., Wei H. C., Wong R., Szentpetery Z., Kim Y. J., Goldbach P., Steinbach S. K., Balla T., Brill J. A. (2009) J. Cell Biol. 187, 847–858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Robinett C. C., Giansanti M. G., Gatti M., Fuller M. T. (2009) J. Cell Sci. 122, 4526–4534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Xu H., Brill J. A., Hsien J., McBride R., Boulianne G. L., Trimble W. S. (2002) Dev. Biol. 251, 294–306 [DOI] [PubMed] [Google Scholar]
- 43.Echard A., Hickson G. R., Foley E., O'Farrell P. H. (2004) Curr. Biol. 14, 1685–1693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Skop A. R., Bergmann D., Mohler W. A., White J. G. (2001) Curr. Biol. 11, 735–746 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.