Abstract
We have developed a robust high content assay to screen for novel estrogen receptor alpha (ERα) agonists and antagonists by quantitation of cytoplasmic to nuclear translocation of an estrogen receptor chimera in 384-well plates. The screen utilizes a green fluorescent protein tagged-glucocorticoid/estrogen receptor (GFP-GRER) chimera which consisted of the N-terminus of the glucocorticoid receptor fused to the human ER ligand binding domain. The GFP-GRER exhibited cytoplasmic localization in the absence of ERα ligands, and translocated to the nucleus in response to stimulation with ERα agonists or antagonists. The BD Pathway 435 imaging system was used for image acquisition, analysis of translocation dynamics, and cytotoxicity measurements. The assay was validated with known ERα agonists and antagonists, and the Library of Pharmacologically Active Compounds (LOPAC 1280). Additionally, screening of crude natural product extracts demonstrated the robustness of the assay, and the ability to quantitate the effects of toxicity on nuclear translocation dynamics. The GFP-GRER nuclear translocation assay was very robust, with z' values >0.7, CVs<5%, and has been validated with known ER ligands, and inclusion of cytotoxicity filters will facilitate screening of natural product extracts. This assay has been developed for future primary screening of synthetic, pure natural products, and natural product extracts libraries available at the National Cancer Institute at Frederick.
Keywords: estrogen receptor, high content screening, cell-based assay, nuclear translocation, cytotoxicity
INTRODUCTION
The estrogen receptor (ER) is a ligand-activated transcription factor, which is a member of the steroid/nuclear receptor super family. In women, ERα is expressed in the brain, cardiovascular system, uterus, bone and liver, and is the predominate form expressed in breast cancer. ER-mediated signal transduction is a complex pathway, which regulates cellular proliferation, differentiation and reproductive physiology. Elevated estrogen levels can lead to initiation, promotion and progression of breast tumors by several pathways in postmenopausal women. Estrogen production from the ovaries ceases following menopause, and the source of estrogen in postmenopausal women is conversion of androgens to estrogens in peripheral tissues, including the breast[1]. ER signaling through the nucleus, mitochondria, and non-genomic signaling at the plasma membrane lead to rapid cell proliferation that may lead to elevated mutation rates, altered cell-cycle control, and inhibition of apoptosis which perpetuate the growth and survival of the cancer cell [2, 3]. Additionally, metabolic conversion of estrogen to genotoxic or mutagenic metabolites by the phase I detoxification pathway may result in DNA adduct formation or oxidative DNA damage[4, 5]. These mechanisms of carcinogenesis mediated through estrogen signaling illustrate the importance of targeting ERα for therapeutic intervention.
Many of the drugs used for breast cancer therapeutics and hormone replacement therapy present adverse side effects, therefore, our goal was to identify compounds which target the estrogen receptor, which may exhibit reduced adverse side effects. These side effects are often related to the mixed agonistic/antagonist activity of a given drug, which is dependent on tissue, cell, promoter, co-activator or co-repressor expression profiles. While hormone replacement therapy (HRT) reduces menopausal symptoms, maintains bone mineral density and decreases the risk of colon cancer, these drugs also elevate the risk for the development of breast cancer, coronary heart disease, stroke, and Alzheimer's disease and blood clots[6–8]. Selective estrogen receptor modulators (SERMs) are drugs that elicit agonism or antagonism depending on tissue, cell, promoter, co-regulator expression. Tamoxifen is a SERM that is used for the treatment of hormone-responsive breast cancer, which is typically used as an adjuvant therapy after surgery and radiation. While tamoxifen exhibits ER antagonistic activity in the mammary tissue, this drug also exhibits partial agonistic activity in the uterine tissue, which increases the risk for development of endometrial cancer and uterine sarcoma[9]. Nearly half of patients do not respond to tamoxifen treatment, and patients with metastatic disease are certain to develop tamoxifen resistance, while 30–50% of patients with early stage ER-positive breast cancer that are administered tamoxifen relapse with resistant disease[10]. Tamoxifen has been demonstrated to induce non-alcoholic steatohepatitis in humans, which is a fatty acid disease that can develop into hepatocarcinoma or cirrhosis of the liver[11, 12]. Furthermore, several studies in rats have demonstrated that tamoxifen is hepatocarcinogenic[13], and this carcinogenesis is based on the ability of tamoxifen to be both a tumor initiator and tumor promoter in the liver[14, 15].
For these reasons, as well as others, there is a need for discovery of new effective drugs for breast cancer treatment, which can ameliorate the adverse side effects associated with drugs currently available. In addition, new agents are needed for the development of combination therapies that could prevent the onset of endocrine-resistant disease associated with monotherapy. This study describes a high-content nuclear translocation imaging screen, which used ERα as a therapeutic target for the discovery of new hormone replacement therapies (HRT) and breast cancer therapeutics.
MATERIALS AND METHODS
Chemicals and reagents
Dulbecco's modification of eagle's medium (DMEM), Dulbecco's phosphate-buffered saline (DPBS), and G418 were purchased from Mediatech (Manassas, VA). The penicillin/streptomycin solution, trypsin-EDTA, staurosporine, and 37% formaldehyde were purchased from Sigma-Aldrich (St. Louis, MO). Sodium butyrate was purchased from Millipore (Billerica, MA). The Nunc glass-bottom 96-well and 384-well plates and Hyclone characterized fetal bovine serum (FBS) and charcoal/dextran-treated FBS were acquired from Thermo Fisher (Pittsburgh, PA). Hoechst 33342 was obtained from Invitrogen (Carlsbad, California). 17-β estradiol (E2), propyl pyrazoletriol (PPT) diethylstilbestrol (DES), 4-hydroxy-tamoxifen (4OHT), diarylpropionitrile (DPN), genistein and the LOPAC 1280 library were acquired from Sigma-Aldrich (St. Louis, MO). The natural product extracts library was obtained from the Natural Products Branch, Developmental Therapeutics Program, at the National Cancer Institute-Frederick (Frederick, MD).
Cell culture
The 6020 cells were cultured in DMEM, supplemented with 100 U/ml penicillin, 100 μg/ml streptomycin, 1 mg/ml G418 and 10% charcoal/dextran-treated FBS, which will be referred to as complete media. The C127 cells were grown in DMEM supplemented with 100 U/ml penicillin, 100 μg/ml streptomycin, and 10% charcoal/dextran-treated FBS. Cells were cultured in a humidified incubator at a 37°C, 5% CO2, 95% air environment.
Library plate dilutions
Chemical libraries are typically screened at 10 μM final concentrations for pure synthetic compounds and 10 μg/ml for natural product extracts, and dilution plates were made in complete media containing charcoal/dextran-treated FBS. Dilutions were made using the Biomek FX liquid handling instrument (Fullerton, CA). DMSO (0.5% final concentration) served as the negative control, and estradiol (5 μM final concentration) was used for the positive control for the assay. Due to time constraints on the third day of the assay, dilution plates were made the day before and frozen overnight at −20°C.
Nuclear translocation assay
The 6020 cells were plated into 96-well or 384-well Nunc glass-bottom black plates at 8,000 cells/well or 3,500 cells/well, respectively, and cultured in complete media. The 6020 cells were seeded into 384 plates at 50 μl/well (or 100 μl/well for 96-well plates) using a Bio-Tek MicroFill microplate dispenser (Winooski, VT) and incubated at 37°C, 5% CO2, 95% air overnight. Twenty-four hours after seeding the cells into assay plates, cells were treated with 70 mM sodium butyrate (25 μl/well for 384-well plates and 50 μl/well for 96-well plates) using a Beckman FX liquid handling device (Fullerton, CA). Cells were treated with sodium butyrate for 24 hours to induce expression of the GFP-GRER chimera, which is located in a silent region of the genome. The following morning cells were treated with ER ligand dose-response plates or chemical library plates for 6 hours. The Beckman FX was used to add 10 μl of the ER ligand dose response plates or chemical dilution plates to the assay plates. After a 6 hour treatment with dose responses or chemical library plates, the assay plates were subjected to fixation with 4% formaldehyde for 45 minutes. After fixation, the assay plates were washed 5 times with 100 μl of DPBS using a Bio-Tek plate washer (Winooski, VT). The plates were stained with 0.3 μg/ml Hoechst 33342 in PBS overnight at 4°C, which was added (15 μl/well) using a Bio-Tek MicroFill microplate dispenser to the assay plates. The next day, assay plates were washed twice with 100 μl of DPBS using a Bio-Tek plate washer (Winooski, VT). Plates were sealed with aluminum sealing tape and were barcoded with a Velocity-11 VCode Bar Code Label Print and Apply Station (Menlo Park, CA).
Image acquisition and nuclear translocation analysis
Images were acquired using a BD Pathway 435 imaging system (Rockville, MD) integrated with a Thermo CRS catalyst-5 robotic arm (Waltham, MA) and a Symbol barcode reader (Schaumburg, IL) for unattended imaging. An Olympus 20× 0.75 NA objective was used for image acquisition, four sites (montages) were acquired per well, each well >100 cells, and GFP (180 ms, gain=10) and Hoechst (30ms, gain=0) filters were used. Translocation dynamics were quantitated with BD Attovision software using a ring-based (2 output) algorithm, and a GFP threshold of 300–4095 gray values was used for segmentation of the cytoplasmic area. Nuclear areas were defined by Hoechst staining, and GFP intensity was measured in that defined region of interest. The nuclear/cytoplasmic ratios were measured, and % nuclear translocation was reported. Images were written to a terabyte server during image acquisition. Attovision and Image Data Explorer software (BD Biosciences, Rockville, MD) were used for analysis of image data, and data mining. Image Data Explorer software and SigmaPlot (San Jose, California) software were used to calculate EC50 values and generate dose response curves.
Cytotoxicity assays
The 6020 cells were seeded into Nunc 96-well glass-bottom plates at 10,000 cells/well in complete media (100 μl/well) and incubated for 24 hours at 37°C, 5% CO2, 95% air. The following morning the cells were treated with 70 mM sodium butyrate (final concentration) to induce GFP-GRER expression for 24 hours before staurosporine treatment. Cells were treated with a staurosporine dose-response for 4 hours, which was followed by formaldehyde fixation and Hoechst staining as previously described. Image acquisition and translocation analysis were conducted as previously described. In addition to percent nuclear translocation, nuclear area, cytoplasmic area and regions of interest (ROIs) were measured to assess cytotoxicity.
Hit determination
The z' values were calculated as described in Zhang, et. al [16]. Compounds that elicited a nuclear translocation value of ≥72% were identified as primary hits, which is three standard deviations from the mean percent nuclear translocation with 5 μM estradiol. These hits were analyzed for average nuclear area and compounds that generated a nuclear area below 380 pixels were eliminated as false positives due to cytotoxicity. The hits were further evaluated for the number of cells or ROIs, and wells with less than 30 ROIs were eliminated. The remaining primary hits were cherry picked and re-tested in the 6020 cell line and in the parental C127 cell line. Cherry picking experiments confirm the activity in the 6020 cell line and also eliminate false positives caused by fluorescent compounds localized to the nucleus in the parental C127 cell line.
RESULTS
Nuclear translocation of GFP-GRER
The ERα undergoes nucleocytoplasmic shuttling, yet the nuclear compartment is strongly favored even in the absence of hormonal ligands. The glucocorticoid receptor (GR) is cytoplasmic in the absence of its ligands due to the presence of a nuclear localization signal (NLS) at the N-terminus, which regulates its translocation from the cytoplasm to the nucleus. Nuclear localization of the ERα in the absence of hormone presented a problem for development of a nuclear translocation screen. Consequently, we used a cell line that stably expressed the GFP-GRER chimeric receptor, which consisted of the N-terminus, DNA binding domain, hinge and partial ligand binding domain regions of the glucocorticoid receptor (GR), which was fused with the human ERα ligand binding domain and tagged with the green fluorescent protein (GFP). This chimera was stably transfected into a murine mammary adenocarcinoma cell line, and the generation and characterization of this 6020 cell line has been previously described [17]. Previous experiments have demonstrated that the GFP-GRER remains cytoplasmic in the absence of estradiol-like compounds and that binding of both ER agonists and antagonists will cause nuclear translocation of the receptor, and exposure to GR ligands does not translocate the GFP-GRER to the nuclear compartment [17]. Furthermore, previous work had demonstrated that the GFP-GRER does not undergo cytoplasmic to nuclear translocation in response to stimulation with glucocorticoid ligands[17]. The GFP-GRER chimera has the potential to identify full agonists and antagonists, mixed agonist-antagonist ligands and SERMs, therefore, it will be necessary in secondary assay to differentiate the ligand activity on ER signaling, and the effects on native ER. Additonally, previous work has demonstrated that the gene reporter assays using GFP-GRER exhibited a dose dependent increase in transcriptional activity with increasing estradiol treatment, and a 10 fold increase in luciferase activity occurred with 100nM estradiol treatment[17]. In these studies the calculated EC50 values for translocation and transactivation using GFP-GRER were 46nM, and 80nM, respectively[17].
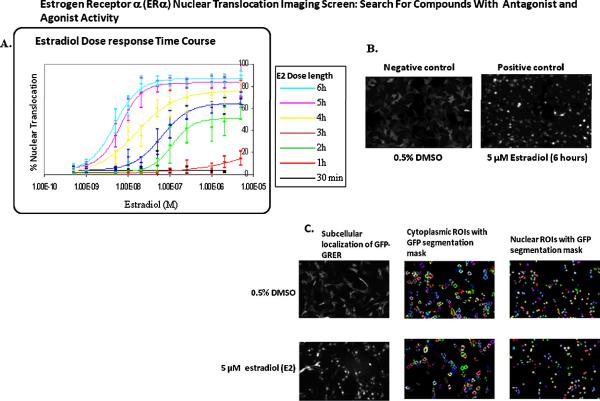
Estradiol is the natural hormone for ERα and subsequently, was selected as the positive control for the assay. The 6020 cells were plated into seven 384-well glass-bottom plates. The following morning the expression of the GFP-GRER was induced with sodium butyrate treatment (70 mM final) for approximately 24 hours. Forty eight hours post plating, the cells were treated with an estradiol dose-response for 0.5, 1, 2, 3, 4, 5, and 6 hours, followed by fixation and nuclear staining (Fig 1A). As indicated in Fig. 1A, a six hour treatment resulted in maximal translocation of GFP-GR. The cells treated with DMSO (negative control) exhibited cytoplasmic localization of the GFP-GRER, whereas nuclear localization of the receptor occurred with 5 μM estradiol treatment (Fig. 1B). Images were acquired using the BD Pathway 435 imaging system, and GFP and Hoechst dyes were captured as indicated in the materials and methods. Four images (montages) were acquired per well for each wavelength for a total of eight images per well. Hoechst 33342 dye was used to segment the nucleus, and this measurement was used to quantitate the GFP intensity in the nuclear and cytoplasmic compartments. The cytoplasmic ROI for GFP fluorescence was measured by a 20 pixel dilation from the nuclear boundary, and only thresholded GFP was quantitated in the cytoplasmic region (Fig. 1C). This eliminates measurement of background fluorescence that can occur with standard ring-based algorithms since only areas containing 6020 cells expressing GFP are quantitated, and not background fluorescence often included in the measurement caused by a circle drawn around the nuclear boundary and areas of the plate were the cells are not growing are measured. Segmented ROIs and segmentation masks are depicted overlayed on the images of the 6020 cells treated with DMSO and estradiol controls (Fig. 1C). The image acquisition for each 384-well plate is approximately 2 hours, followed by image analysis and data mining, so we expect to screen 30 of the 384 plates per week, which translates into roughly screening 10,560 samples per week.
FIG. 1.
Quantitation of nuclear translocation of GFP-GRER. (A) The 6020 cells were plated at 3,500 cells/well into seven 384-well glass-bottom plates. The plates were treated with estradiol dose responses for 30 minutes, 1, 2, 3, 4, 5, and 6 hours. Nuclear translocation was quantitated and presented graphically as indicated in the materials and methods. Results displayed as the mean ± SD, and n=32 wells for each dose of estradiol. (B) Images depict the subcellular localization of GFP-GRER upon treatment with the controls, 0.5% DMSO, and 5 μM estradiol at the 6 hour dose. (C) Images of cells treated for 6 hours with DMSO and estradiol controls with overlays of the segmentation masks for the cytoplasmic and nuclear regions of interest (ROIs).
Comparison of dilution media on estradiol dose response curves
The 6020 cells growing in 384-well plates were treated with estradiol dose responses diluted in PBS, DMEM (with phenol red), and complete media (with phenol red and charcoal/dextran-treated FBS) in triplicate plates, and prepared for image acquisition as indicated in the material and methods. The E2 dose response curves reconstituted in PBS and DMEM produced very similar curves, which are nearly indistinguishable (Fig. 2A). The dose response made in complete media generated an EC50 value of 43.4 nM and exhibited a higher potency as compared with the EC50 values for PBS, and DMEM, which were 100.6 nM and 149.2 nM, respectively. Based on these results, we decided to make all compound library dilutions in complete media.
FIG. 2.
Nuclear translocation results of estradiol dose responses diluted in various media. (A) Estradiol doses was diluted in PBS, DMEM, or complete media (containing charcoal, dextran-treated FBS), and the resulting dose response curves are depicted graphically. N=32 wells per dose on each plate and 3 plates were used per media type. (B) Estradiol dose responses were diluted in complete media containing regular FBS or charcoal, dextran-treated FBS, with corresponding dose response graphs shown. Results displayed as the mean ± SD, where N=32 wells per plate and 5 plates were used for each media type. (C) Estradiol dose responses mad e in complete media (containing charcoal, dextran-treated FBS) with or without phenol red and corresponding dose response curves are depicted. Results displayed as the mean ± SD, where n=32 well per plate and 6 plates were tested per media. (D) Estradiol dose response curves using complete media containing charcoal, dextran-treated FBS in quadruplicate 384-well plates. Results displayed as the mean ± SD, where N=32 wells per plate.
Effect of FBS on the dose response of estradiol
Additionally, growth factors and hormones in FBS may influence estrogen receptor studies by stimulating nuclear translocation of the receptor, but charcoal-stripped FBS eliminates these growth factors or steroids that can skew ER responses. Estradiol dose responses were generated in complete media containing FBS or charcoal, dextran-treated FBS, and were added to six 384-well plates containing 6020 cells for each media type. The nuclear translocation dose response curves for these data demonstrated that the hormones or growth factors in FBS do affect translocation by increasing the potency and maximal percent translocation (Fig. 2B). Consequently, charcoal/dextran-treated FBS was used in all aspects of the screening assay.
Evaluation of effects of phenol red on nuclear translocation
In previous studies, phenol red has been demonstrated to be a weak ER ligand [18], so we completed estradiol dose responses that were reconstituted in complete media containing charcoal/dextran-treated FBS with or without phenol red in 384-well plates. We plated the 6020 cells into twelve 384-well glass-bottom plates at 3,500 cells/well, induced GFP-GR expression with sodium butyrate, and treated 6 plates with estradiol dose responses diluted phenol red complete media, and 6 plates with estradiol dose responses generated in phenol red-free complete media. The dose response curves for estradiol reconstituted in complete media containing charcoal/dextran-treated FBS with or without phenol red are nearly identical (Fig. 2C), therefore we decided to use complete media containing phenol red for the assay.
Based on these studies examining the effect of media type on nuclear translocation dynamics, DMEM media containing phenol red, and charcoal/dextran-treated FBS was implemented into the screening assay. The 384-well estradiol dose response in quadruplicate plates produced an EC50 of 33.5 nM, an average z' of 0.73, and an average 4.5% CV, which demonstrated that the assay was very robust and amenable to high throughput screening in 384-well format (Fig. 2D).
Known ER ligands translocate GFP-GRER to the nucleus
Several known ER ligands were tested to validate the GFP-GRER nuclear translocation assay. Dose responses were generated in 96-well plates for estradiol, propyl pyrazole triol (PPT), 4-hydroxy-tamoxifen (4OHT), diethylstilbestrol (DES), and genistein (Fig.3). Estradiol, PPT, and DES are ERα agonists and all produced dose-response curves with EC50 values of 34 nM, 141 nM, and 129 nM, respectively. Genistein and 4OHT are ERα antagonists that generated EC50 values of 19.4 μM and 50 nM, respectively. Genistein is a low affinity isoflavone phytoestrogen, which demonstrates that the assay is robust enough to detect lower affinity ligands. Diarylpropionitrile (DPN) is a selective ERβ ligand, and therefore, dose response curves were generated with this compound to test the assay specificity for ERα ligands (Fig. 3). DPN did not induce cytoplasmic to nuclear translocation of the ERα, which demonstrates that the assay is specific for ERα ligands. This chimeric receptor is not as sensitive to ER ligands as compared to native ER, which is to be expected, but does demonstrate specificity for ER ligands. Additionally, previous studies have demonstrated that the GFP-GRER chimera is capable of inducing transcription upon estradiol treatment using gene reporter assays, with and estradiol induced translocation EC50 of 46nM and a transactivation EC50 of 80nM [17]. This chimera will be useful in identifying new ER ligands from the primary screen, but the secondary assays will explore the ligand effects on ER signaling with the native receptor to determine agonist or antagonist activity, and whether the hits are effecting protein-protein interactions, nuclear translocation, or transcriptional activity.
FIG. 3.
Dose response curves depicting nuclear translocation dynamics of known ER ligands. Dose responses were generated for estradiol, propyl pyrazole triol (PPT), 4-hydroxy-tamoxifen (4OHT), diethylstilbestrol (DES), and genistein. Diarylpropionitrile (DPN) is a selective ERβ ligand that was used to demonstrate that the assay is specific for ERα ligands. These dose responses were dosed to 6020 cells plated in to five 96-well glass-bottom plates for 6 hours. Results displayed as the mean ± SD, where N=8 wells per dose and 5 plates were tested per ligand.
Hit identification
Hits were identified with nuclear translocation values three standard deviations from the positive control, which resulted in a hit limit of 72% or greater. Cellular images of hits were examined to confirm activity, and these hits were cherry picked and tested in quadruplicate in the assay and parental cell lines to confirm activity and eliminate false positive results. Examination of compound activity in the parental non-transfected cell line, which does not express the GFP-GRER construct, facilitates elimination of false positives caused by fluorescent compounds that localize to the nuclear compartment. Fluorescence intensity images for GFP and Hoechst of the 6020 (assay) and parental cell line treated with DMSO and 5 μM estradiol are depicted in figure 4. In the estradiol control, nuclear translocation is 97% in the 6020 cell line, while in the parental cell line treated with estradiol, translocation is measured at 0%, which demonstrated that estradiol is not a fluorescent compound, and that fluorescence in the nucleus is due to presence of GFP-GRER (Fig.4). The DMSO control exhibits 8% translocation in the 6020 cell line, and 0% translocation in the parental cell line. Hoechst images are displayed for both the 6020 and parental cell lines, which demonstrated that lack of translocation or GFP fluorescence is not due to absence of cells in the parental cell line. An example of a false positive due to compound fluorescence is shown in Fig. 5A. Treatment with adriamycin exhibited 97% nuclear translocation in the 6020 cell line and 95% translocation in the parental cell line (Fig. 5A), which indicated a false positive due to fluorescence in the non-transfected parental cell line. False positive translocation results caused by fluorescent compounds exhibit nuclear localization, possibly by binding to the DNA. Adriamycin is a fluorescent anthracycline antibiotic, which intercalates DNA, and is used as an anticancer agent. Additionally, an example of a hit compound is shown in Fig. 5B, which demonstrates that treatment with raloxifene exhibited 93% translocation in the assay line, and 0% in the parental cell line. Hoechst images in both 6020 and parental cell lines are shown to demonstrate that cells are present in the images of the parental cell line. Raloxifene is a SERM that exhibits ERα antagonistic activity in the breast and uterine tissue and agonistic activity in the bone, which has been approved for the treatment of osteoporosis and for prevention of invasive breast cancer [19]. This method of testing the hit compounds in the parental cell line will eliminate false positive fluorescent compounds that commonly occur when screening natural products.
FIG. 4.
Hit confirmation. DMSO and estradiol controls were retested in the 6020 (assay) cell line, and in the non-transfected parental cell line in quadruplicate. Images of DMSO and estradiol-treated cells are depicted by showing GFP and hoechst fluorescence images in both cell lines. Analysis for nuclear translocation was performed as described in the materials and methods. Images depict fluorescence of the GFP-GRER, and its subcellular location.
FIG. 5.
Elimination of false positive translocation results caused by compound fluorescence. Hit compounds were retested in the 6020 (assay) cell line, and in the non-transfected parental cell line in quadruplicate. Analysis for nuclear translocation was performed as described in the materials and methods. Images depict fluorescence of the GFP-GRER, and its subcellular location. Compounds that exhibit nuclear fluorescence in the parental cell line are indicative of false positives caused by fluorescent compounds that localize to the nucleus.
Assessment of effects of natural product extracts on the assay
We screened a natural product extract library of 10,000 crude extracts and 1,190 primary hits were identified, which was followed by examination of cellular images of each of these hits to confirm nuclear translocation activity. Only 36 hits from the primary hits were selected for confirmation experiments, the remainder 1,154 hits were false positive results due to cytotoxicity. Cherry picking of the hits and retesting the 36 extracts in the assay and parental cell lines revealed that all the hits were false positives due to fluorescent compounds that localized to the nuclear compartment. This library did not produce any primary hits for further biological evaluation. The high level of false positives due to cytotoxicity, and the time involved in viewing images of these false positive hits demonstrated that additional filters were necessary to increase throughput and confidence in identified hits for the assay.
Effects of cytotoxicity on image analysis of translocation dynamics
Many compounds and natural product extracts elicit toxicity in cell-based screening campaigns. In order to assess the effects cytotoxicity on nuclear translocation, cell morphology and cellular image analysis we tested staurosporine, which is an inducer of apoptosis. The 6020 cells were plated into five 96-well plates and the following day they were treated with sodium butyrate for 24 hours. The cells were then treated with a staurosporine dose-response for 4 hours (Fig. 6A). These results demonstrated that staurosporine elicited false positive nuclear translocation results. The dose response curve for staurosporine is similar to the dose response curve for estradiol. These morphological changes caused by cytotoxicity were evident upon examination of the images, which consisted of rounding of cells, fragmentation or shrinkage of the nucleus, and shrinkage or loss of the cytoplasmic area, which the image analysis algorithm identifies as a compound exhibiting nuclear translocation (Fig. 6B). Loss of cytoplasmic area is apparent in the fluorescent micrographs of GFP-GRER, and the size of the nuclei stained with Hoechst is reduced with increasing dose of staurosporine (Fig. 6B). Consequently, an additional algorithm was developed to eliminate cytotoxic false positives by measuring the nuclear area, cytoplasmic area, and the number of cells or regions of interest (ROIs). The nuclear and cytoplasmic areas were quantitated for the staurosporine dose responses and these images and graphs are shown in (Figure 7). The nuclear area was quantitated for the staurosporine treatment and the dose response graphs and images demonstrate a dose-dependent decrease in nuclear area (Fig. 7A). The nuclear area dose response generated a z' factor of 0.51, which demonstrated that this measurement was suitable as a parameter for hit identification. The cytoplasmic area was also measured following a staurosporine treatment, and the images of GFP-GRER and the dose response curves exhibit a dose-dependent decrease in cytoplasmic area (Fig. 7B). This parameter generated a z' factor of 0.53, which demonstrated that it was a statistically relevant measurement to select primary hits. The nuclear area value of ≤380 pixels was chosen as the cutoff value for cytotoxicity, and only cells with nuclear areas above 380 pixels were chosen as hits. We also quantitated the cytoplasmic area from the staurosporine dose response, and have implemented a cut off of ≤800 pixels in order to eliminate cytotoxic false positives, which resulted in only cells with cytoplasmic areas above 800 pixels were identified as hits. Additionally, we measured ROIs with staurosporine treatment, and these dose response curves show a decrease in cell number with increasing staurosporine concentration (data not shown). Wells with ROIs (or cell number) below 30 cells were also eliminated at cytotoxic results.
FIG. 6.
Cytotoxic compounds elicit false positive nuclear translocation results. The 6020 cells grown in five 96-well plates were treated with staurosporine dose responses to evaluate the effect of cytotoxicity on analysis of translocation dynamics. (A) Staurosporine dose response results were plotted and curves are depicted graphically. Results displayed as the mean ± SD, where N=8 wells per dose per plate and 5 plates were tested, and graphed individually. (B) Examination of the images for GFP-GRER and Hoechst 33342 clearly show cytotoxicity.
FIG. 7.
Nuclear and cytoplasmic area measurements as a predictor of cytotoxicity. The 6020 cells grown in 96-well glass-bottom plates were treated with staurosporine dose responses for 6 hours. (A) Staurosporine dose response curves generated by quantitation of nuclear area. Results are plotted and dose response curves are shown graphically, and Hoechst 33343 fluorescent images depict the nuclear area. Results displayed as the mean ± SD, where N=8 wells per dose per plate and 5 plates were tested, and plotted individually. (B) Quantitative cytoplasmic area measurements in response to treatment with increasing doses of staurosporine are shown. Results are plotted and dose response curves are shown graphically and fluorescent images of GFP-GRER shown the morphology of the cytoplasmic area. Results displayed as the mean ± SD, where N=8 wells per dose per plate and 5 plates were tested, and graphed individually.
Since levels of cytoplasmic GFP-GRER decrease with estradiol treatment as the GFP-GRER translocated to the nucleus, it was important to quantitate the nuclear and cytoplasmic areas with maximal estradiol dosing. This analysis was necessary to ensure that enough GFP was present at maximal estradiol dosing to segment the cytoplasm, and that the nuclear area was not affected by increasing levels of estradiol. We analyzed five 384-well plates seeded with 6020 cells that were treated with an estradiol dose response and the percent nuclear translocation, nuclear area, and cytoplasmic areas are depicted graphically (Fig. 8). The nuclear translocation curves for the estradiol dose response is shown in Fig. 8A, and the nuclear areas and cytoplasmic areas are illustrated in Fig. 8B, and Fig. 8C, respectively. These dose response curves demonstrate that nuclear area remains unchanged with increasing amounts of estradiol, and that the cytoplasmic area also does not decrease with increasing estradiol dose, even though the majority of GFP-GRER migrated to the nuclear compartment. Fluorescent images of the GFP-GRER and Hoechst staining for well F23, which was treated with 5 μM estradiol, are shown in Figure 8D. Segmented ROIs and segmentation masks for GFP and Hoechst fluorescent intensities are overlayed on the images, which were used to quantitate GFP signal in the cytoplasmic and nuclear areas. The GFP thresholding used for the imaging algorithm is sensitive enough to detect low levels of the receptor in the cytoplasm (Fig.8D). The threshold for GFP was set at 300–4095 gray values, and these data demonstrate that there is enough residual GFP in the cytoplasm at high estradiol dose or ~90% nuclear translocation to allow quantitation of the cytoplasmic region, and the cytoplasmic area does not decrease with increasing estradiol treatment. The nuclear area measurements also remain constant with increasing estradiol dosing. These data illustrate that these measurements will remove cytotoxic compounds without eliminating hits that cause translocation of the GFP-GRER. These additional cellular measurements will reduce the number of cytotoxic false positives and consequently produce hits with increased ERα specificity
FIG. 8.
Quantitative measurements of nuclear translocation, nuclear area, and cytoplasmic area in response to estradiol dose response treatment. 6020 cells grown in five 384-well glass-bottom plates were treated with estradiol dose responses. Results displayed as the mean ± SD, where N=32 wells per dose per plate and 5 plates were tested. (A) Nuclear translocation was quantitated in response to estradiol treatment and dose response curves were plotted. (B) Nuclear area measurements were plotted after treatment with estradiol dose responses. (C) Quantitative measurement of the cytoplasmic area after estradiol dose response treatment. (D) Fluorescent images of well F23, which has been treated with 5 μM estradiol are shown. Micrographs of the GFP-GRER and hoechst 33342 fluorescent intensities are shown with the segmentation ROIs and segmentation mask overlays for the nuclear and cytoplasmic area measurements are shown.
Inclusion of cytotoxicity profiles for hit identification
Additional cellular measurements were incorporated into the screening process to facilitate the reduction of false positives due to cytotoxicity, which is particularly prevalent when screening natural product extracts. In addition to nuclear translocation, nuclear and cytoplasmic areas (pixels) and regions of interest (ROIs) were used to specify which samples met the criteria for hit identification. Images were analyzed and hits were defined by nuclear translocation values of 72% or greater, nuclear area above 380 pixels, cytoplasmic area above 800 pixels and ROIs of 30 or greater. These parameters were used to screen and additional set of 10,000 natural product extracts in 384-well plates. Examination of hit images treated with natural products revealed that compounds that clearly elicited toxicity were not being eliminated as hits based on cytoplasmic area alone (Fig. 9). For example, we examined images and numerical data from plates treated with natural product extracts library to demonstrate that the hit identification using nuclear translocation, ROI, nuclear area, and cytoplasmic area are confirming the image data (Fig. 9). Images and quantitative measurements of cellular parameters are shown for the DMSO and estradiol controls, and for two wells treated with natural product extracts. DMSO and estradiol controls fall within the parameters for hit selection and cytotoxicity analysis, and visual inspection of the images corroborate this result. Images of well B24 exhibit morphology consistent with cytotoxicity (Figure 9). Quantitative measurements of the cellular parameters for well B24 generated a translocation value of 100%, which would indicate a hit based on translocation alone. Although the nuclear area for well B24 is 256 pixels, and there are 8 ROIs, which is indicative of toxicity, but the cytoplasmic area fails to predict cytotoxicity since the cytoplasmic area is 1675 pixels (Fig 9). Well E2 shows cytotoxicity in the image data, and in the quantitative measurements for nuclear area, and ROI, but does not indicate cytotoxicity when looking at the cytoplasmic area value alone. For example, well E2 has a 94% translocation value, which would indicate a hit, but the images of well E2 clearly exhibit cytotoxicity (Fig. 9). The nuclear area for well E2 is 282 pixels, and there are only 16 ROIs indicating cytotoxicity, but the cytoplasmic area is 1038 pixels which is not consistent with the toxicity profiles conducted with staurosporine dose response experiments. Nuclear area and ROIs are the most sensitive cytotoxicity measurements and correlate with qualitative examination of the images, therefore, nuclear translocation, ROIs, and nuclear area were used as parameters to select hits based on nuclear translocation and elimination of cytotoxic false positives, and cytoplasmic area was eliminated as a parameter to identify hits. Based on these results, we decided to use nuclear translocation values of 72% or greater, nuclear area above 380 pixels, and ROIs of 30 or greater for hit identification when the next set of 10,000 natural product extract samples were screened. Inclusion of nuclear area and ROIs with nuclear translocation parameters in our hit identification, resulted in approximately 75% reduction in false positives as compared to nuclear translocation alone. The remaining 21 hits were cherry picked and tested in the 6020 (assay) cell line and in the parental cell line (C127), which resulted in elimination of all 21 hits either due to non-confirmation of activity or false positives due to compound fluorescence in the nucleus in the parental cell line.
FIG. 9.
Identification of false positives by using additional measurement parameters for cytotoxicity. Fluorescent images of GFP and hoechst staining depict the subcellular localization of the GFP-GRER. Quantitative parameters measured by the image analysis software are displayed next to the fluorescent images.
Screening library used for assay validation
The Library of Pharmacologically Active Compounds (LOPAC 1280) is comprised of 1,280 pharmacologically active compounds plated in 384-well format, which is commonly used for assay validation, not hit identification, and evaluates the assay specificity. The screen identified 5 hits from the LOPAC 1280 library, which were retested in the 6020 assay and parental cell line to confirm nuclear translocation. From these 5 hits, two of the hits produced translocation values below 72% and did not confirm activity. One of these hits showed nuclear fluorescence in the non-transfected parental cell line, which indicated that the compound was an inherently fluorescent compound that possible bound to the DNA. Two hits confirmed activity and did not show nuclear fluorescence in the parental cell line. One of the compounds was estrone, which is a metabolite of estradiol, and the other compound was estradiol. Both of these compounds are known estrogen receptor agonists, which further validated our screen. There were no other hits identified other than the ER ligands from the LOPAC library, which together with our data on known ER ligands, confirms the specificity of our screen.
DISCUSSION
We have developed a high throughput imaging assay to identify compounds that target the ERα. The cellular imaging screen was developed to screen synthetic compounds, pure natural products and natural product extract libraries for compounds that translocate the GFP-GRER chimera from the cytoplasm to the nucleus. The BD Pathway 435 imaging system and Attovision and BD Image Data Explorer software were used to acquire images, quantify nuclear translocation dynamics, and assess cytotoxicity.
Cellular imaging has several advantages over traditional high throughput screening methods. The ability to directly examine cellular images and quantitate cellular responses allows the investigator to further explore the effect of a particular compound on the subcellular translocation of a specific target, morphological changes, effects on compartmentalized areas of the cell, or specific organelles, and the overall physiology of the cell population. Inspection of cellular images not only permits visualization of the response, but is also useful when determining whether a hit is real or a false positive. False positive results are quite problematic, and can often be caused by fluorescent cellular debris, dust, cytotoxicity, or inherently fluorescent compounds.
High throughput screening campaigns have been reported for ER ligands using estrogen response element-driven luciferase or beta lactamase reporter systems, which screen for compounds that modulate transcriptional activity at the gene level[20, 21]. In contrast, the potential drug targets from the screen we have developed allow for simultaneous detection of agonists or antagonists, and compounds that may modulate other aspects of ER signaling, such as, nuclear transport, selective chaperone interactions, and proteosome degradation. The assay reported here has the capability to identify compounds that modulate several aspects of ER signaling as compared to a reporter assay, which would only identify compounds that affect transcriptional activity.
Native ER is not useful for a nuclear translocation assay since it is localized predominately in the nucleus, even in the absence of hormone. Subsequently, the GFP-GRER chimera was designed to enable cytoplasmic localization of the unstimulated receptor, and nuclear localization with agonist or antagonist treatment. This chimera has been validated with known ER agonists and antagonists, and with the LOPAC 1280 library. This GFP-GRER chimeric receptor is not as sensitive to ER ligands as compared to native ER, which is to be expected, but does demonstrate specificity for ER ligands. Differences between ligand specificities the GFP-GRER and native receptor should be expected due to the differences in protein structure between the native ER and the chimeric receptor hybrid, therefore, this screen will be utilized as a tool to discover ER ligands, and the ligand sensitivity and biological implications on the native receptor will be further explored in subsequent secondary assays. Consequently, comparison of ligand specificities between the GFP-GRER chimera and native ER should be qualitative not quantitative.
In addition to nuclear translocation assays, previous ERE-driven gene reporter studies have demonstrated that the GFP-GRER chimera induced transcription with ER agonist (estradiol, PPT, DES) treatment, and repressed transcription in response to cotreatment with estradiol and an ER antagonist (tamoxifen, ICI 182,780) [17], which occurs with native ER receptor. Furthermore, dose response experiments using high content assays will provide a glimpse of the toxicity profile of the hit compound, and we can determine the maximum tolerated dose in the 6020 cell line and the parental cell line. Future work will involve primary screening over 160,000 samples from our synthetic, pure natural product, and natural product extracts libraries to identify new ER ligands, and the secondary assays will explore the ligand effects on ER signaling with the native receptor to determine agonist or antagonist activity, and whether the hits are effecting protein-protein interactions, nuclear translocation, or transcriptional activity.
The nuclear translocation imaging assay we have developed for discovery of novel ER ligands is very robust, with average z' factor values above 0.7, and CV values under 5%. The assay has been developed in both 96-well and 384-well format, and it has been validated with known ER agonists and antagonists, and the LOPAC 1280 library. Additionally, we have preliminary screened a small sample library of natural product extracts to evaluate the robustness of the assay and examine the effect of cytotoxicity on nuclear translocation dynamics in preparation for the primary screening of our natural product extract libraries containing over 120,000 samples. These experiments have demonstrated that cytotoxicity does affect the ability of the analysis software to identify hits based solely on nuclear translocation, and cytotoxic compounds create false positive hits for nuclear translocation of the GFP-GRER due to changes in cellular morphology. Based on these data, we implemented additional screening parameters to filter out false positives caused by cytotoxicity. Identification of primary hits in the screening campaign will be selected with nuclear translocation values of 72% or greater, nuclear area above 380 pixels, and ROIs of 30 or greater. We have also implemented a method for elimination of false positive hits caused by fluorescent compounds by subjecting the compounds to quadruplicate testing in the 6020 assay cell line and in the non-transfected parental cell line, which will further improve the specificity of primary hits identified in the full scale screening campaign. In summary, we have developed a robust cellular imaging assay for identification of novel ER ligands through nuclear translocation. Our future work will utilize this assay to perform a primary screen of over 160,000 samples, which will exploit the synthetic, pure natural product, and natural product extracts libraries currently available at the National Cancer Institute.
ACKNOWLEDGEMENTS
This project has been funded in whole or in part with federal funds from the National Cancer Institute, National Institutes of Health, under contract HHSN26120080001E. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the U.S. Government. This Research was supported [in part] by the Intramural Research Program of the NIH, National Cancer Institute, Center for Cancer Research.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- [1].Grodin JM, Siiteri PK, MacDonald PC. Source of estrogen production in postmenopausal women. The Journal of clinical endocrinology and metabolism. 1973;36(2):207–214. doi: 10.1210/jcem-36-2-207. [DOI] [PubMed] [Google Scholar]
- [2].Russo J, Reina D, Frederick J, Russo IH. Expression of phenotypical changes by human breast epithelial cells treated with carcinogens in vitro. Cancer research. 1988;48(10):2837–2857. [PubMed] [Google Scholar]
- [3].Russo J, Calaf G, Russo IH. A critical approach to the malignant transformation of human breast epithelial cells with chemical carcinogens. Critical reviews in oncogenesis. 1993;4(4):403–417. [PubMed] [Google Scholar]
- [4].Cavalieri EL, Kumar S, Todorovic R, Higginbotham S, Badawi AF, Rogan EG. Imbalance of estrogen homeostasis in kidney and liver of hamsters treated with estradiol: implications for estrogen-induced initiation of renal tumors. Chemical research in toxicology. 2001;14(8):1041–1050. doi: 10.1021/tx010042g. [DOI] [PubMed] [Google Scholar]
- [5].Devanesan P, Todorovic R, Zhao J, Gross ML, Rogan EG, Cavalieri EL. Catechol estrogen conjugates and DNA adducts in the kidney of male Syrian golden hamsters treated with 4-hydroxyestradiol: potential biomarkers for estrogen-initiated cancer. Carcinogenesis. 2001;22(3):489–497. doi: 10.1093/carcin/22.3.489. [DOI] [PubMed] [Google Scholar]
- [6].Beral V. Breast cancer and hormone-replacement therapy in the Million Women Study. Lancet. 2003;362(9382):419–427. doi: 10.1016/s0140-6736(03)14065-2. [DOI] [PubMed] [Google Scholar]
- [7].Chlebowski RT, Hendrix SL, Langer RD, Stefanick ML, Gass M, Lane D, Rodabough RJ, Gilligan MA, Cyr MG, Thomson CA, Khandekar J, Petrovitch H, McTiernan A. Influence of estrogen plus progestin on breast cancer and mammography in healthy postmenopausal women: the Women's Health Initiative Randomized Trial. Jama. 2003;289(24):3243–3253. doi: 10.1001/jama.289.24.3243. [DOI] [PubMed] [Google Scholar]
- [8].Rossouw JE, Anderson GL, Prentice RL, LaCroix AZ, Kooperberg C, Stefanick ML, Jackson RD, Beresford SA, Howard BV, Johnson KC, Kotchen JM, Ockene J. Risks and benefits of estrogen plus progestin in healthy postmenopausal women: principal results From the Women's Health Initiative randomized controlled trial. Jama. 2002;288(3):321–333. doi: 10.1001/jama.288.3.321. [DOI] [PubMed] [Google Scholar]
- [9].Lavie O, Barnett-Griness O, Narod SA, Rennert G. The risk of developing uterine sarcoma after tamoxifen use. Int J Gynecol Cancer. 2008;18(2):352–356. doi: 10.1111/j.1525-1438.2007.01025.x. [DOI] [PubMed] [Google Scholar]
- [10].Ring A, Dowsett M. Mechanisms of tamoxifen resistance. Endocrine-related cancer. 2004;11(4):643–658. doi: 10.1677/erc.1.00776. [DOI] [PubMed] [Google Scholar]
- [11].Nemoto Y, Saibara T, Ogawa Y, Zhang T, Xu N, Ono M, Akisawa N, Iwasaki S, Maeda T, Onishi S. Tamoxifen-induced nonalcoholic steatohepatitis in breast cancer patients treated with adjuvant tamoxifen. Internal medicine (Tokyo, Japan) 2002;41(5):345–350. doi: 10.2169/internalmedicine.41.345. [DOI] [PubMed] [Google Scholar]
- [12].Bruno S, Maisonneuve P, Castellana P, Rotmensz N, Rossi S, Maggioni M, Persico M, Colombo A, Monasterolo F, Casadei-Giunchi D, Desiderio F, Stroffolini T, Sacchini V, Decensi A, Veronesi U. Incidence and risk factors for non-alcoholic steatohepatitis: prospective study of 5408 women enrolled in Italian tamoxifen chemoprevention trial. BMJ (Clinical research ed. 2005;330(7497):932. doi: 10.1136/bmj.38391.663287.E0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].White IN. The tamoxifen dilemma. Carcinogenesis. 1999;20(7):1153–1160. doi: 10.1093/carcin/20.7.1153. [DOI] [PubMed] [Google Scholar]
- [14].Williams GM, Iatropoulos MJ, Karlsson S. Initiating activity of the anti-estrogen tamoxifen, but not toremifene in rat liver. Carcinogenesis. 1997;18(11):2247–2253. doi: 10.1093/carcin/18.11.2247. [DOI] [PubMed] [Google Scholar]
- [15].Dragan YP, Fahey S, Nuwaysir E, Sattler C, Babcock K, Vaughan J, McCague R, Jordan VC, Pitot HC. The effect of tamoxifen and two of its non-isomerizable fixed-ring analogs on multistage rat hepatocarcinogenesis. Carcinogenesis. 1996;17(3):585–594. doi: 10.1093/carcin/17.3.585. [DOI] [PubMed] [Google Scholar]
- [16].Zhang JH, Chung TD, Oldenburg KR. A Simple Statistical Parameter for Use in Evaluation and Validation of High Throughput Screening Assays. J Biomol Screen. 1999;4(2):67–73. doi: 10.1177/108705719900400206. [DOI] [PubMed] [Google Scholar]
- [17].Martinez ED, Rayasam GV, Dull AB, Walker DA, Hager GL. An estrogen receptor chimera senses ligands by nuclear translocation. The Journal of steroid biochemistry and molecular biology. 2005;97(4):307–321. doi: 10.1016/j.jsbmb.2005.06.033. [DOI] [PubMed] [Google Scholar]
- [18].Berthois Y, Katzenellenbogen JA, Katzenellenbogen BS. Phenol red in tissue culture media is a weak estrogen: implications concerning the study of estrogen-responsive cells in culture. Proceedings of the National Academy of Sciences of the United States of America. 1986;83(8):2496–2500. doi: 10.1073/pnas.83.8.2496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Moen MD, Keating GM. Raloxifene: a review of its use in the prevention of invasive breast cancer. Drugs. 2008;68(14):2059–2083. doi: 10.2165/00003495-200868140-00008. [DOI] [PubMed] [Google Scholar]
- [20].Shi X, Zheng W, Schneeweis JE, Fischer PA, Strulovici B, Peekhaus NT. A short-incubation reporter-gene assay for high-throughput screening of estrogen receptor-alpha antagonists. Assay and drug development technologies. 2005;3(4):393–400. doi: 10.1089/adt.2005.3.393. [DOI] [PubMed] [Google Scholar]
- [21].Peekhaus NT, Ferrer M, Chang T, Kornienko O, Schneeweis JE, Smith TS, Hoffman I, Mitnaul LJ, Chin J, Fischer PA, Blizzard TA, Birzin ET, Chan W, Inglese J, Strulovici B, Rohrer SP, Schaeffer JM. A beta-lactamase-dependent Gal4-estrogen receptor beta transactivation assay for the ultra-high throughput screening of estrogen receptor beta agonists in a 3456-well format. Assay and drug development technologies. 2003;1(6):789–800. doi: 10.1089/154065803772613426. [DOI] [PubMed] [Google Scholar]