Abstract
Functional attributes of microbial communities are difficult to study, and most current techniques rely on DNA- and rRNA-based profiling of taxa and genes, including microarrays containing sequences of known microorganisms. To quantify gene expression in environmental samples in a culture-independent manner, we constructed an environmental functional gene microarray (E-FGA) consisting of 13,056 mRNA-enriched anonymous microbial clones from diverse microbial communities to profile microbial gene transcripts. A new normalization method using internal spot standards was devised to overcome spotting and hybridization bias, enabling direct comparisons of microarrays. To evaluate potential applications of this metatranscriptomic approach for studying microbes in environmental samples, we tested the E-FGA by profiling the microbial activity of agricultural soils with a low or high flux of N2O. A total of 109 genes displayed expression that differed significantly between soils with low and high N2O emissions. We conclude that mRNA-based approaches such as the one presented here may complement existing techniques for assessing functional attributes of microbial communities.
Culture-independent metagenomic approaches using DNA-based techniques (e.g., 16S rRNA gene sequencing [11, 16, 28]) have gained popularity in the analysis of microbial communities. While DNA-based analyses identify taxonomic groups, the processes performed by a microbial community have to be indirectly deduced from species composition. Denaturing gradient gel electrophoresis allows taxonomic fingerprinting (42) and identifies functional groups such as nitrifiers (38). Similarly, fluorescence in situ hybridization identifies phylogenetic groups and potentially also the spatial distributions and presence of certain gene families within microbial populations (10). Another tool applied to the study of environmental microbial communities (EMCs) is the use of nucleic acid microarrays. The three types of microarrays are community genome arrays (CGAs), rRNA-based oligonucleotide arrays (PhyloChips), and functional gene arrays (FGAs) (reviewed in reference 43). CGAs analyze genomic DNA from EMCs, identify known microbes to the species level (41), and are most suitable for characterizing population changes of known microbial species. PhyloChips are constructed with oligonucleotides that match rRNA sequences of specific microorganism taxa (e.g., nitrifying bacteria [22]) and reveal greater taxonomic diversity in environmental samples than does typical rRNA gene sequencing (8, 34). Thus, PhyloChips, similar to CGAs, are best suited for detecting changes in the taxonomic composition of microbial populations. However, while rRNA levels are generally representative of cell growth and activity (27), microbes can maintain high levels of rRNA in a dormant state (30, 36), resulting in overestimation of the contribution of such organisms to microbial activity. In the context of soil function, the focus on particular microbial groups may also mask the contribution of other microbes, including unknown microbe species, to overall soil processes.
FGAs are constructed with probes targeting known genes and gene products with specific functions, such as genes involved in nitrogen cycling (33, 40), detoxification (7), or sulfate reduction (37). Alternately, FGAs can be constructed from known cDNA libraries or predicted coding sequences of specific organisms (13, 17, 19) and link expression patterns with overall physiological traits. cDNA-based FGAs monitor gene expression of transcripts from many species based on cross-hybridization to homologous genes, which is useful in light of recent insights that indicate that microbial species differ geographically for similar environments, whereas biochemical functions (and corresponding genes) are generally conserved (14). While FGAs provide insight into the physiological status of EMCs, they are biased due to their construction from known microbial genes. A new method for extracting microbial mRNA from EMCs (24) was used in the present study to construct an FGA from randomly selected clones of cDNA libraries. Such libraries have previously been used to construct microarrays for rapid transcriptional profiling of single organisms (e.g., Arabidopsis thaliana [1]). The primary advantage of a microarray containing randomly selected cDNA library clones is that it does not require prior knowledge about functional genes, genomes, or species present in the environmental sample. An FGA constructed from environmental cDNA clones may provide less biased, culture-independent expression profiling of an entire microbial community. In the present study, we report the construction and application of an environmental FGA (E-FGA) using 13,056 individual transcripts randomly captured from diverse EMCs. To our knowledge, this is the first FGA constructed directly from environmental communities. Here, we use the E-FGA to demonstrate in situ gene expression profiling of soil microbes.
Many agricultural soils emit the undesirable greenhouse gas N2O, which contributes at least 10% to global warming (18). N2O emissions from agriculture are steadily rising due to increasing use of N fertilizers (4), and emissions are positively correlated with several environmental factors, including soil N content, waterlogging, temperature, and soil pH (5, 15). N2O is generated via microbial nitrification and denitrification, as well as during the less-well-understood anammox pathway, most likely by concurrent denitrification (20, 31, 39). Although knowledge of soil microbial activity is crucial for understanding drivers of N2O emission from soil, comparatively little understanding of causal soil microbial processes exists. This is primarily due to the difficulties associated with cultivating microbes ex situ (23). We currently have insufficient knowledge about how crop systems can best be managed to manipulate microbial activity to minimize N2O emission and maximize N retention in the system, yet such knowledge is needed to devise sustainable crop systems with a low N pollution footprint. For these reasons, we chose to carry out a small-scale experiment to test whether a new method such as the E-FGA can, in principle, be used to compare mRNA profiles of agricultural soils which have different N2O fluxes as determined with field-based automatic flux chambers (6).
MATERIALS AND METHODS
Soil sampling, RNA isolation, and cDNA library construction.
Environmental microbial community samples were taken from a variety of sites in South East Queensland, Australia. These included sugarcane soils (containing 160 kg N ha−1 mineral, organic, or no fertilizer; Jacobs Well, Queensland [27°44′06.93″S, 153°19′32.37″E]; sampled on 15 May 2006; see reference 1 for soil details and history; approximately 4,500 cDNA clones were used for inclusion in the microarray [see below]), common garden soils (University of Queensland, St. Lucia [27°29′52.06″S, 153°00′45.51″E]; sampled on 12 June 2007; approximately 5,300 clones), compost (private households [27°22′12.74″S, 152°59′12.11″E]; sampled on 12 June 2007; approximately 300 clones), activated sludge from a wastewater treatment plant (denitrification-rich sample from a settling tank, Oxley [27°33′18.46″S, 152°59′30.53″E]; sampled on 18 June 2007; approximately 400 clones), a eutrophic lake sample (University of Queensland, St. Lucia [27°29′56.26″S, 153°01′00.01″E]; sampled on 18 June 2007; approximately 1,400 clones), bovine rumen samples (27°36′30.46″S, 153°14′19.74″E; sampled on 18 June 2007; approximately 600 clones), human fecal samples (approximately 200 clones), and tooth, cheek, and tongue scrapings (approximately 300 clones). mRNA was enriched from RNA extractions by selective size fractionation and removal of rRNA, converted to cDNA, and cloned into the pCR-Blunt vector (Invitrogen) using the methods outlined by McGrath et al. (24). The resulting cDNA libraries were used for construction of the microarray.
The soil samples used for the microbial expression analysis were from a sugarcane site in Mackay, Australia (21°09′08.32"S, 149°07′06.96"E; sampled on 6 November 2007, 10 am to 12 pm), where a large-scale project measuring N2O and other greenhouse gases from soils was being conducted. The soil at this site consists of a noncalcic brown clay-loam that was either fertilized with 160 kg/ha ammonium nitrate 8 days prior to sampling or unfertilized. The site was sown to sugarcane (100 to 130 cm high) on a trash blanket of the previous sugarcane crop, where greenhouse gas chamber measurements were being monitored. N2O emissions were measured directly from soil using automated greenhouse gas chambers, and across the larger sugarcane site using micromet systems coupled to a tunable diode laser, and an open-path Fourier transform infrared spectroscopy system (for more details, see reference 6). Three samples from each of the two high- and low-N2O-emitting chambers from fertilized and nonfertilized soils, respectively, were taken at a depth of 5 to 20 cm. Up to 500 g of soil was sampled and mixed before 20 g of each sample was used for total RNA extraction (24). To obtain good quantity and integrity of RNA from soil, immediate extraction of microbes from the soil had to be performed directly on site within 3 min. Briefly, soil samples were fully suspended in 20 ml water for 20 to 30 s and then left to settle for 10 s to separate larger particles (i.e., sand, stones). The supernatant was decanted into individual microcentrifuge tubes and centrifuged in a car battery-powered centrifuge for 2 min (14,000 × g) to pellet the microbial contents which were snap-frozen in liquid nitrogen and stored at −80°C to preserve microbial RNA profiles.
Microarray construction.
A total of 13,056 anonymous cDNA clones (including >9,000 clones from soil samples) were randomly chosen from the cDNA libraries for representation on microarrays. Each clone was transferred into an individual well in 96-well plates containing 10 μl of sterile water. A 2× PCR master mix was added (0.1 μl iTaq DNA polymerase [10 U/μl] from Scientifix, Australia; 2 μl iTaq buffer; 1 μl deoxynucleoside triphosphates [dNTPs; 10 mM]; 1 μl of M13 forward and reverse primer mixture [10 μM]; 5.9 μl of sterile water). PCR amplification was performed under the following conditions: 95°C for 2 min; 35 cycles of 95°C for 30 s, 52°C for 30 s, and 74°C for 2 min; and 74°C for 2 min. PCR products were then precipitated by the addition of 2 μl of ammonium acetate (3.5 M) and 50 μl of 100% ethanol and centrifugation for 15 min in a microcentrifuge at maximum speed (10,000 × g). Plates were inverted and spun briefly to remove excess ethanol before being air dried and sealed for transport. The PCR products from microtiter plates were then spotted and UV cross-linked onto a glass slide by the Australian Genome Research Facility (Melbourne, Australia). Each PCR product was spotted onto the microarray twice, with each duplicate pair being separated by a distance equal to half the entire microarray (i.e., replicate patterns above and below the midline).
cDNA labeling.
Total RNA was extracted from soil samples, and mRNA was enriched by a new procedure using gel electrophoresis-based size separation (24). A total of 1 to 2 μg of mRNA was converted into cDNA by using the SuperScript Indirect cDNA labeling kit (Invitrogen, Australia) according to the manufacturer's instructions. However, the oligo(dT) nucleotides were omitted from the reaction mixture and twice the recommended amount of random hexamer was used. The cDNA from each soil type was labeled with Alexa Fluor 532 dye according to the manufacturer's instructions (Invitrogen, Australia).
Microarray normalization.
To account for variations in spot size and density across microarrays, a PCR product was produced from an empty pCR-Blunt vector using M13 primers and amino-allyl dNTPs under the conditions described in the microarray construction section of this report. This product (called nPCR for neighboring PCR) was homologous to the flanking regions that were amplified with each cDNA clone, and hence, this sequence was present on each valid spot of the microarray (Fig. 1). Because of this, the amount of hybridization of the nPCR fragment could be used to normalize variations in DNA deposition and spotting efficiencies when comparing data between arrays. Additionally, this method would account for the localized effects that could affect binding efficiencies for each spot. The nPCR fragments were labeled with Alexa Fluor 635 dye (Invitrogen, Australia) and combined with the desired cDNA sample to be profiled that was labeled with Alexa Fluor 532. This cDNA-nPCR mixture was then used for hybridization.
FIG. 1.
Schematic view of the E-FGA surface allowing simultaneous gene expression profiling and normalization. A cDNA probe with flanking vector sequences bound to the surface of the microarray is shown. The Alexa Fluor 532-labeled cDNA and the Alexa Fluor 635-labeled nPCR can bind to the same spot, allowing accurate calculation of cDNA levels for probes with variable DNA deposition or hybridization efficiencies.
Microarray hybridization and data analysis.
After labeling, the cDNA and the nPCR normalizing probe were combined and hybridized with the microarray overnight (14 to 16 h) at 42°C by the method outlined at http://ag.arizona.edu/microarray/methods.html. After washing, the microarrays were scanned using a microarray scanner (Axon Instruments) and analysis software (GenePix 6.0). The foreground fluorescence values for each channel and spot were used for the analysis. This foreground signal had to be greater than the average background plus 2 standard deviations (SD) for both channels to be considered valid (F532 > B532avg + 2 SD and F635 > B635avg + 2 SD [2, 28]). For each valid spot, the ratio of the cDNA to the nPCR probe was calculated (F532/F635 ratio), resulting in locally normalized data. Additionally, the average fluorescence level across the entire microarray for each channel was determined and used to calculate a “scan bias” ratio for each microarray (F532avg/F635avg). This value was multiplied across all individual data ratios and then log10 transformed to produce the globally normalized log-data set used for the comparison of microarrays. Student's t test was performed to compare the normalized data sets for the duplicate spots on the three high- and three low-N2O-emitting soil samples using two-tailed heteroscedastic parameters (Microsoft Excel).
Sequence analysis and bioinformatics.
One hundred fifty-three clones were identified from the clone library and sequenced (AGRF, Brisbane, Australia). The resulting sequence files were converted to FASTA format using the DNAbaser freeware program ABI 2 FASTA Converter with the following parameters: no. of good bases = 20, window length = 16, and a quality value threshold of 18. FASTA files were batched using Fasta2MultiFasta 1.9.20.18, also from DNAbaser. The multi-FASTA file was manually edited to remove vector sequences and then uploaded to NCBI BLASTN 2.2.18+ on 28 September 2008. Sequences were aligned against the nr/nt combined data set from the GenBank, EMBL, DDBJ, and PDB databases using default parameters. Sequences shorter than 50 bases were aligned against the expressed sequence tag “Others” data set.
Nucleotide sequence accession numbers.
All microarray data (including GenBank accession numbers for sequenced genes) have been submitted to NCBI's Gene Expression Omnibus at http://www.ncbi.nlm.nih.gov/geo (accession number GEO GSE23422).
RESULTS AND DISCUSSION
Construction of an E-FGA.
Previously, we reported a new method for selective mRNA isolation and cDNA library construction from environmental samples (24). Using this approach, a cDNA microarray was constructed from mRNA directly isolated from environmental microbial communities that are active in nitrogen conversion. These included mainly soil samples (sugarcane fields, garden compost soils), some aquatic samples (eutrophic lake water and sediment, activated wastewater flocculant), and commensal samples (bovine rumen, human oral and fecal samples). A total of 13,056 clones were randomly chosen from the libraries, with a bias toward the soil-derived libraries (∼8,000 clones). Libraries constructed this way are expected to consist of 90% mRNA and 10% rRNA sequences with lengths of 100 to 800 bp and an average size of ∼300 bp (24). By comparison, cDNA libraries constructed by other methods of microbial rRNA removal (e.g., by specific capture probes or subtractive hybridization) still contain at least 50% rRNA (32).
Amplified PCR products for each clone were spotted as duplicates onto glass slides to generate an E-FGA. As a result of the PCR amplification, each DNA product was flanked by two common sequences corresponding to the region of the cloning vector between the M13-F primer site and the cloning site and the region from the cloning site to the M13-R primer site (Fig. 1). Thus, every spot on the constructed microarray contained two common short regions of ∼70 bp each. Dudley et al. (9) demonstrated that by including a foreign DNA fragment with each clone before spotting the mixture onto a microarray, the foreign DNA could be used as a control to account for bias in spotting and hybridization efficiencies. Here, we found that the flanking regions of the spotted clones can be used in a similar way, by normalizing the sample cDNA fluorescence intensity to that of the flanking regions. A schematic diagram of the hybridization is shown in Fig. 1.
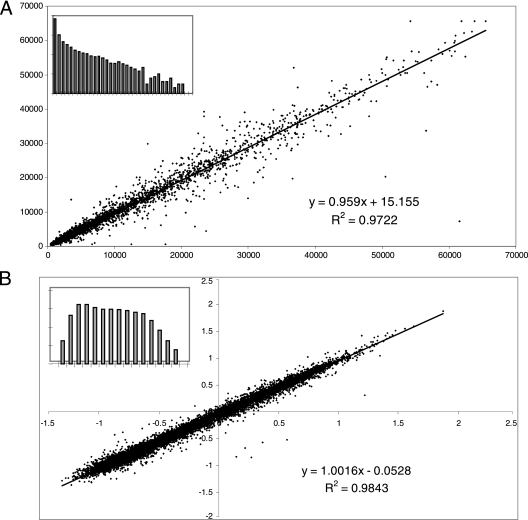
To test the normalization process, 18 microarrays were processed and analyzed. After hybridization of the cDNA-nPCR mixture, overnight incubation, and washing, the microarrays were scanned and the fluorescence signal intensities of both channels were analyzed for each spot. A comparison of the duplicate spots on the microarray was made using either the cDNA signal intensity only (raw fluorescence intensity, F-532) or the cDNA signal intensity normalized to the nPCR signal intensity (normalized intensity, F-532/F-635). For each, a linear regression analysis was performed to evaluate the congruency of duplicate spots on the array and hence the quality of the data obtained. An example of how the normalization method affected the data is shown in Fig. 2. The raw data set only takes into account signal intensities of the cDNA (Fig. 2A) and ignores localized variations in array manufacture, hybridization, or scanning. A comparison of nonnormalized data of replicate spots on the same microarray gave a correlation coefficient (r2) of 0.972. Additionally, many spots showed poor correlation (extreme outliers on the graph), and the data set was skewed to the lower end of the scale (inset, Fig. 2A). In contrast, normalization increased the internal correlation of duplicate spots to 0.984 and reduced the number of extreme outliers (Fig. 2B). Additionally, normalization removed skewness and transformed the data into a standard distribution suitable for statistical analysis (inset, Fig. 2B).
FIG. 2.
Correlations between replicate spots on a single microarray (A) before normalization (raw F532 values) and (B) after normalization using the nPCR product (F532/F635). The axes indicate (A) absolute foreground fluorescence levels and (B) normalized fluorescence values. Histograms show the relative distribution of each respective data set (inset).
A comparison of r2 values before and after normalization was performed for 18 microarrays, showing an increase in the average r2 value from 0.825 to 0.883 (Table 1). It is worth noting that the largest increases in r2 occurred when the raw data gave a particularly low level of correlation (e.g., arrays 3 and 7), where normalization resulted in an increase in r2 value of 0.25, while arrays with a high raw data correlation (e.g., arrays 12 and 18) showed a negligible increase in r2 values. Four of the microarrays showed a slight decrease in correlation values following normalization (arrays 1, 6, 13, and 17). This, however, was very minor and only the case for microarrays with relatively high prenormalization r2 values above 0.8, suggesting that the applied normalization has the greatest effect on samples with low initial correlations. Thus, when considered in combination with the associated change in the distribution of the data and the ability to account for spotting variation, the normalization procedure improved the overall validity of the analysis. Importantly, this normalization can account for variations in microarray experiments, including different efficiencies for spotting, hybridization, and scanning intensities, enabling direct comparisons of the microarrays used for different samples. To our knowledge, this is the first report of the use of flanking sequences to normalize microarray data.
TABLE 1.
Comparison of raw data (average cDNA signal intensities) and normalized data (cDNA-nPCR intensities) for 18 microarrays
| Type of data |
r2 valuea |
||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | 15 | 16 | 17 | 18 | Avg | |
| Raw | 0.886 | 0.881 | 0.632 | 0.674 | 0.797 | 0.805 | 0.661 | 0.873 | 0.722 | 0.8 | 0.71 | 0.92 | 0.912 | 0.978 | 0.967 | 0.738 | 0.916 | 0.972 | 0.825 |
| Normalized | 0.827 | 0.881 | 0.891 | 0.856 | 0.863 | 0.795 | 0.911 | 0.911 | 0.835 | 0.942 | 0.746 | 0.925 | 0.901 | 0.978 | 0.975 | 0.775 | 0.897 | 0.984 | 0.883 |
r2 values are given for each array before (raw) and after normalization (normalized).
Analysis of an E-FGA derived from soils with different N2O production rates.
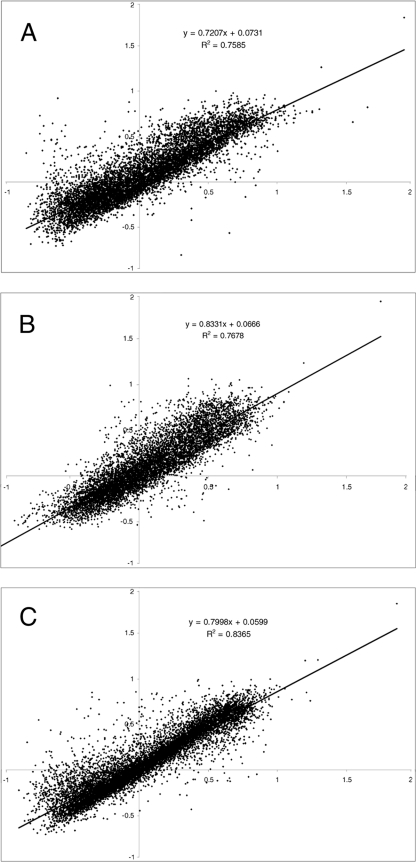
To test the potential of E-FGAs for microbial activity profiling and gene discovery, we carried out a small study of two tropical sugarcane soils which displayed different N2O emission rates. Two high- and two low-N2O-emitting sites within the same field were identified in the study by Denmead et al. (6), with average emissions of 36.97 and 2.01 ng N m−2 s−1 immediately prior to sampling, respectively. Apart from N fertilizer additions of 160 kg N ha−1 as ammonium nitrate to the sites with high N2O emissions, all of the sites had similar abiotic conditions during the sugarcane growing season (6). Soil microbial gene expression profiles were obtained using the E-FGA, and the profiles of the soils were compared by plotting the relative transcript abundances of different soil sites (Fig. 3). Relative expression levels for each gene on the microarray that passed quality control were compared between different soils by plotting the normalized gene expression levels from one soil sample to another. Soils expressing identical microbial processes would show an r2 value of 1. A comparison of gene expression levels of the two high-N2O-emitting sites (averaged from three soil samples/microarrays per site) showed some differences in the expression profile which resulted in an r2 value of 0.7585 (Fig. 3A). This indicates that while most microbial activities were similar between high-N2O-emitting sites, the expression of a range of putative genes varied. Similarly, gene expression varied between the low-N2O-emitting sites (r2 = 0.7678; Fig. 3B). The comparison of averaged data from 12 microarrays of low- versus high-N2O-emitting sites (six microarrays each) showed an r2 value of 0.8365. It is important to note that this r2 value resulted from the comparison of a total of 12 microarrays and thus cannot be directly compared with the previous r2 values based on six microarrays, which are more susceptible to deviations in gene expression in a single sample. Variability between samples from the same field may be caused by hot-spot behavior, which is described as plots of soil (some as small as a few square centimeters) that emit much more N2O than neighboring plots (35). Chambers used for N2O emission measurements were approximately 50 by 50 cm in size (6).
FIG. 3.
Microbial expression profiles comparing (A) two high-N2O-emitting sites (three soil samples and microarrays for each), (B) two low-N2O-emitting sites (three soil samples and microarrays for each), and (C) average expression levels for high- versus low-N2O-emitting sites (six soils samples/microarrays per axis). Each point represents a microarray spot. Normalized gene expression levels are shown.
Identifying microbial species, genes, and processes linked to N2O emissions.
The complete microarray data set of 12 microarrays from two low- and two high-N2O-emitting sites (three replicate soil samples per site) was analyzed to identify genes that display significantly altered expression levels between high- and low-N2O-emitting soils. Expression of 153 sequences was significantly (P < 0.05) different between high- and low-N2O-emitting soils. These genes were identified by sequencing, and 75 transcripts were matched (NCBI blast E values, <1−10) to entries in the online databases, including 66 to functional protein or rRNA coding sequences and 9 to unassigned sequences. The putative functions assigned, as with all homology-based matches, should be regarded as suggestions only until closer matches become available. The remaining 78 sequences had no significant match to GenBank entries, contained small inserts, or could not be sequenced, possibly due to high G+C content or nonviable clones.
A total of 34 genes showed a significant (E < 1−10) match to existing rRNA sequences. While the main focus of our study was to test the potential of E-FGA to identify mRNA sequences, the cDNA libraries used to construct the microarray were still expected to contain at least 10% rRNA clones (24). BLAST searches returned several entries with large portions of identical sequence, indicating that multiple rRNA clones exist on the array for some organisms (bold entries, Table 2). The signals for these spots were consistent in direction and magnitude, providing additional support for the validity of the microarray analysis. The identified rRNA sequences match to common soil bacteria, including Arthrobacter, Acidobacter, proteobacteria, Burkholderia, and Bradyrhizobium spp. Sequences similar to rRNAs of an aquatic planctomycete and the eukaryotic alga Uronema sp. were also detected, reflecting the origin of the library used to construct the E-FGA, which included lake water samples. Many of the detected gene sequences belong to uncultured microorganisms, and 15 showed homologies of less than 95%, suggesting the presence of related but as-yet-unidentified species. While further, much larger studies have to determine the roles of individual species in N2O emissions in these soils, the analysis of the differentially expressed genes that showed homology to functional microbial gene sequences can provide insight into the microbial activity in these soils.
TABLE 2.
Organisms with significantlya differential rRNA prevalence between high- and low-N2O-emitting soils which were detected as rRNA within the mRNA-dominated E-FGAb
| High/low N2O expression ratio | Matching organism (BLASTN) | E value | Identity (%) |
|---|---|---|---|
| 1.68 | Arthrobacter sp. strain SDL35 | 5E−85 | 175/175 (100) |
| 0.74 | Azoarcus sp. strain EbN1 | 7E−121 | 251/256 (98) |
| 0.81 | Bradyrhizobium sp. strain Ai4.2 | 5E−81 | 174/177 (98) |
| 1.20 | BurkholderiaphymatumSTM815 | 3E−121 | 264/275 (96) |
| 1.23 | BurkholderiaphymatumSTM815 | 7E−118 | 262/275 (95) |
| 1.21 | “Candidatus Desulforudis audaxviator” | 3E−42 | 145/174 (83) |
| 0.82 | “Candidatus Desulforudis audaxviator MP104C” | 4E−19 | 60/63 (95) |
| 0.88 | Delftia acidovorans SPH-1 | 1E−43 | 115/124 (92) |
| 0.64 | Geobacter sp. strain G02 | 3E−52 | 118/119 (99) |
| 1.20 | Geobacter uraniireducens Rf4 | 2E−102 | 113/121 (93) |
| 0.86 | Geothrix fermentans | 2E−161 | 334/345 (96) |
| 0.27 | MethylacidiphiluminfernorumV4 | 8E−103 | 257/287 (89) |
| 0.43 | MethylacidiphiluminfernorumV4 | 8E−103 | 257/287 (89) |
| 0.83 | Opitutus terrae PB90-1 | 6E−42 | 131/150 (87) |
| 0.30 | Planctomycetestrain 639 | 1E−33 | 127/156 (81) |
| 0.56 | Planctomycetestrain 639 | 5E−30 | 131/157 (83) |
| 0.79 | Rosalina sp. strain 3675 | 2E−44 | 110/114 (96) |
| 1.35 | SymbiobacteriumthermophilumIAM 14863 | 4E−17 | 88/111 (79) |
| 1.41 | SymbiobacteriumthermophilumIAM 14863 | 4E−17 | 88/111 (79) |
| 1.17 | Uncultured Acidobacteria bacterium clone 23k22 | 5E−39 | 113/124 (91) |
| 0.48 | Uncultured Acidobacteria bacterium clone 41b15 | 8E−40 | 133/158 (84) |
| 0.42 | Uncultured bacterium GRIST12 | 3E−33 | 153/198 (77) |
| 1.27 | Uncultured Bradyrhizobium sp. | 4E−71 | 152/152 (100) |
| 1.31 | Uncultured Chloroflexi bacterium isolate HS-8 | 8E−82 | 199/217 (91) |
| 0.66 | Uncultured Comamonadaceae bacterium | 0 | 391/391 (100) |
| 0.47 | Uncultured deltaproteobacterium | 2E−10 | 42/42 (100) |
| 1.27 | Uncultured Flexibacter sp. | 1E−118 | 242/243 (99) |
| 0.36 | Uncultured gammaproteobacterium | 1E−22 | 62/62 (100) |
| 0.71 | Uncultured gammaproteobacterium | 0 | 371/371 (100) |
| 0.79 | Uncultured organism clone Ast-45 | 2E−102 | 237/254 (93) |
| 0.62 | Uncultured proteobacterium | 6E−42 | 97/97 (100) |
| 0.66 | Uncultured Verrucomicrobia bacterium clone Amb_ | 4E−105 | 218/220 (99) |
| 1.76 | Unidentified bacterium UBA518602 | 4E−152 | 312/320 (97) |
| 1.30 | Uronema marinum | 1E−65 | 147/150 (98) |
P < 0.05.
Bold type indicates that multiple rRNA clones exist on the array for some organisms.
Of the identified functional genes, 24 were assignable in the Clusters of Orthologous Groups (COG) database (NCBI), which assigns metabolic functions to known genes. Of the remainder, eight genes were assigned to the unclassified group (COG category S) and three genes were absent from the COG database. In samples from high- and low-N2O-emitting soils, a total of 12 genes had mRNA expression levels that were higher in the high- than in the low-N2O-emitting soils, while the expression of 20 genes was greater in the low- than in the high-N2O-emitting soils (P < 0.05; Table 3). The genes expressed to a greater extent in the high-N2O-emitting soil may be directly involved in processes which lead to increased N2O production. Alternatively, they may indirectly promote processes which lead to conditions conducive to increased nitrification or denitrification. Further research has to determine whether this coordinated gene expression is indeed correlated with N2O emission. Nevertheless, the results show that the mRNA-based approach allows expression analysis of known and unknown microbial genes in the context of soil processes. For example, two genes with homology to nitrate reductases were expressed at higher levels in low-N2O-emitting soil than in high-N2O-emitting soils (e.g., 087g9 and 042d5, with ratios of 0.48 and 0.69, respectively) (Table 3). Dissimilatory periplasmic nitrate reductase (napA) is involved in the conversion of nitrate to nitrite during the process of denitrification (44). The product of complete denitrification is atmospheric nitrogen (N2), with one of the intermediates being N2O. The complexity of N2O production and emission has been demonstrated previously in mangrove sediment, where denitrification was attributed to the generation of most of the N2O, but N2O generated from denitrification was largely consumed, while nitrification in the surface millimeters of sediment was responsible for most of the emitted N2O (25). Thus, the greater expression of napA in low-N2O-emitting soil reported here may be a result of complete denitrification to N2. Previous studies have attempted to quantify the abundance of genes in denitrifying microbial populations using DNA-based methods (21), but Miller et al. (26) showed that N2O emission from soil was not correlated with numbers of denitrifying microbes in the soil.
TABLE 3.
Genes with significanta differential expression in high- and low-N2O-emitting soils that matched to NCBI database COG categoriesb
| Gene ID | Ratio | Homology-based match (BLAST n/x) | BLAST match organism | E value | Match (%) | COG letter(s) |
|---|---|---|---|---|---|---|
| 107e11 | 0.33 | ψ-Glu-putrescine synthetase | Escherichia coli K-12 | 4.00E−142 | 250/251 (99) | E |
| 018g2 | 0.40 | Phosphate uptake regulator, PhoU | Anaeromyxobacter dehalogenans | 3.00E−23 | 121/156 (77) | E |
| 056d7 | 1.27 | ABC transporter permease protein | Bradyrhizobium japonicum | 9.00E−36 | 149/190 (78) | E |
| 014e5 | 1.42 | Gamma-glutamyl-phosphate reductase | Escherichia coli K-12 | 0.00E+00 | 824/824 (100) | E |
| 113g5 | 1.61 | γ-Glu-putrescine synthase, γ-Glu-GABA hydrolase | Escherichia coli K-12 | 0.00E+00 | 1,087/1,131 (96) | E |
| 015c8 | 0.85 | sn-Glycerol-3-phosphate transport system permease protein | Burkholderia ambifaria AMMD | 1.00E−19 | 46/54 (85) | G |
| 116e11 | 1.19 | Conserved flagellar system protein, promoterless fragment (pseudogene) | Escherichia coli K-12 | 0.00E+00 | 1,075/1,094 (98) | N |
| 078h10 | 1.30 | Bifunctional folyl-polyglutamate synthase and dihydrofolate synthase | Escherichia coli K-12 | 0.00E+00 | 1,109/1,140 (97) | H |
| 064d2 | 0.47 | gsa1 gene for putative G alpha subunit 1 | Sordaria macrospora | 5.00E−29 | 86/92 (93) | HJ |
| 013h2 | 0.47 | Replicatory protein P | Lambda phage | 5.00E−31 | 65/65 (100) | L |
| 041h10 | 1.55 | DNA replication/recombination/repair protein | Escherichia coli W3110 | 0.00E+00 | 596/610 (97) | L |
| 101g7 | 0.46 | Nicotinoprotein alcohol dehydrogenase (aldh7-adh7 operon) | Rhodococcus ruber | 8.00E−29 | 135/175 (77) | C |
| 042d5 | 0.69 | Periplasmic nitrate reductase, large subunit | Escherichia coli K-12 | 0.00E+00 | 1,101/1,131 (97) | C |
| 095h5 | 4.08 | Molybdopterin-containing oxidoreductase catalytic subunit | Sorangium cellulosum So ce56 | 3.00E−10 | 97/133 (72) | C |
| 121c5 | 0.41 | BACc clone RP24-114C10 | Mus musculus | 2.30E+00 | 36/43 (83) | S |
| 022b2 | 0.50 | Malate dehydrogenase, FADd/NAD(P)-binding domain | Escherichia coli | 4.00E−50 | 113/113 (100) | S |
| 018a5 | 0.58 | Predicted 3-hydroxyphenylpropionic transporter | Escherichia coli W3110 | 2.00E−42 | 101/101 (100) | S |
| 017c2 | 0.60 | Predicted DNA-binding transcriptional regulator | Escherichia coli HS | 2.00E−42 | 101/101 (100) | S |
| 024c2 | 0.69 | Nucleoprotein/polynucleotide-associated enzyme | Escherichia coli K-12 substrain | 0.00E+00 | 1,055/1,078 (97) | S |
| 090h11 | 0.80 | cDNA, unknown function | Aphanomyces euteiches | 5.00E−90 | 184/184 (100) | S |
| 053h1 | 1.49 | Hypothetical protein | Escherichia coli ATCC 8739 | 6.00E−39 | 92/92 (100) | S |
| 043h4 | 1.52 | Predicted 3-hydroxyphenylpropionic transporter | Escherichia coli W3110 | 2.00E−29 | 96/107 (89) | S |
| 022e11 | 0.39 | Predicted chaperone, conserved outer membrane protein | Escherichia coli K-12 | 0.00E+00 | 684/713 (95) | R |
| 052b8 | 0.53 | Predicted lipoprotein | Escherichia coli K-12 | 0.00E+00 | 812/812 (100) | R |
| 133c8 | 0.67 | Conserved hypothetical molybdopterin oxidoreductase | Azoarcus sp. strain BH72 | 2.00E−38 | 306/435 (70) | R |
| 050e8 | 1.59 | Predicted lipoprotein | Escherichia coli K-12 | 0.00E+00 | 779/857 (90) | R |
| 096f4 | 2.45 | Acetyl coenzyme A carboxylase, biotin carboxylase | Pseudomonas mendocina ymp | 5.00E−117 | 261/277 (94) | I |
| 097e11 | 0.21 | Phosphoribosylformylglycinamidine synthase | Klebsiella pneumoniae | 1.00E−23 | 82/94 (87) | F |
| 062c11 | 1.86 | Ajudazol biosynthesis (polyketide synthase) gene cluster | Chondromyces crocatus | 2.00E−91 | 280/332 (84) | Q |
| 085e7 | 0.42 | Histidine kinase | Burk holderia phymatum STM815 | 1.00E−11 | 119/167 (71) | T |
| 085c2 | 1.22 | Cytosine deaminase and DNA-binding transcriptional dual regulator | Escherichia coli W3110 | 3.00E−96 | 201/202 (99) | NT |
| 026e11 | 0.53 | DNA-binding transcriptional dual regulator and carbonic anhydrase | Escherichia coli W3110 DNA | 0.00E+00 | 1,093/1,177 (92) | K |
This study was aimed at exploring the potential of using E-FGAs for transcriptional profiling of soil microbial communities and is limited to a very small set of genes and measurements of N2O emissions. It represents only the first attempt to correlate mRNA expression levels with N2O emission, but results are promising as they identify active microbial metabolism in the soil. E-FGAs as presented here allow linkage of microbial mRNA profiles across temporal and spatial gradients to abiotic variables and processes as influenced by management practice. Importantly, genes that have not been previously associated with certain biochemical pathways can be identified using E-FGAs. These genes may stem from unculturable or unknown microorganisms in environmental samples. Unlike DNA chips based on oligonucleotides, these cDNA microarrays also permit a high level of cross hybridization, allowing the identification and expression analysis of functional gene groups rather than individual specific genes. This is a very useful feature of E-FGAs, as distant soils had few microbial species in common (12) but shared functionally related genes to fulfill similar roles.
Functional distribution of N2O-linked genes (COG analysis).
Apart from investigating individual genes, functional profiles can be analyzed. We grouped all functionally identifiable microbial mRNA genes into broad categories in relation to their expression in N2O-emitting soils (Fig. 4). In some categories, the number of significantly differently expressed genes was biased to either a low or high N2O emission profile. For example, the functional group of signal transduction mechanisms (category T, Fig. 4) displays five genes that were elevated in low-N2O-emitting soils and one gene that was elevated in high-N2O-emitting soils. The two identified nitrate reductase-encoding genes (Table 3) belong to the COG category responsible for energy production and conversion (category C, Fig. 4). This COG category harbors five and three genes with elevated expression under conditions of low and high N2O fluxes, respectively, suggesting that these functional groups undergo general shifts under conditions that give rise to N2O genesis. While these data are informative, many more genes are generally required to perform a more complete COG analysis (e.g., see reference 14). The vast majority of genes that showed large changes in expression between the N2O states were not significant here due to the expected intrasite heterogeneity of microbial gene expression and probably the existence of N2O emission hot spots (35). Future research should use a larger sample size (soil and N2O measurement chambers) for more statistical power, greater resolution of significant differential expression, and more detailed COG analysis. The E-FGA approach developed in this study allowed cost-effective screening of a large number of genes from environmental microbial communities by using thousands of randomly selected (anonymous) cDNA clones without prior sequence knowledge, and only differentially expressed genes were sequenced. Currently less cost-effective than this approach, next-generation large-scale sequencing of microbial mRNA (e.g., see reference 14) combined with microarray construction may provide a powerful approach to rapidly analyze environmental microbial transcriptomes.
FIG. 4.
Distribution of 32 N2O-linked genes in COG functional classifications. Genes which have a significantly greater expression level in soil with high and low N2O emissions are shown above and below the horizontal line, respectively. For details of genes, see Table 3.
Conclusion.
To our knowledge, this study is the first to demonstrate the construction and use of an FGA of mRNA isolated from environmental microbial communities. This has been achieved without amplification of RNA samples, which reduces the potential for transcript bias. A new method of normalizing microarray data was devised to account for spotting and hybridization efficiencies, increasing the validity of data and enabling direct comparisons of samples analyzed across different microarrays. We tested the potential of E-FGAs for microbial gene discovery by examining differences in gene expression levels between soil microbial communities from soils with different N2O fluxes and identified candidate genes and functions that differed in soils with high or low N2O emission. The technique presented here complements existing and emerging technologies such as next-generation sequencing, as it offers certain advantages. These include (i) the fact that only differentially expressed genes require sequencing, (ii) the fact that cross hybridization may reveal transcriptional activity of gene families (rather than individual genes), and (iii) relatively low cost (approximately $150/sample, assuming the construction and use of 100 E-FGAs). It may be a valuable addition to the metatranscriptomic approach in microbial ecology to elucidate microbial function in soil and other environmental matrices.
Acknowledgments
We thank Lucy Lee and Vanessa Loo for technical assistance; Rikke Meyer, Jirko Holst, and Prakash Lakshmanan for useful discussions and comments on the manuscript; and Tom Denmead and Travis Naylor (CSIRO) for providing access to the N2O trial and emission data.
This research was funded by the Australian Department of Climate Change (formerly the Australian Greenhouse Office; project 2006000559).
Footnotes
Published ahead of print on 17 September 2010.
REFERENCES
- 1.Allen, D. E., G. Kingston, H. Rennenberg, R. C. Dalal, and S. Schmidt. 2010. Effect of nitrogen fertilizer management and waterlogging on nitrous oxide emission from subtropical sugarcane soils. Agric. Ecosyst. Environ. 136:209-217. [Google Scholar]
- 2.Campbell, E. J., P. M. Schenk, K. Kazan, I. A. Penninckx, J. P. Anderson, D. J. Maclean, B. P. Cammue, P. R. Ebert, and J. M. Manners. 2003. Pathogen-responsive expression of a putative ATP-binding cassette transporter gene conferring resistance to the diterpenoid sclareol is regulated by multiple defense signaling pathways in Arabidopsis. Plant Physiol. 133:1272-1284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chapman, S., P. M. Schenk, K. Kazan, and J. M. Manners. 2002. Using biplots to interpret gene expression patterns in plants. Bioinformatics 18:202-204. [DOI] [PubMed] [Google Scholar]
- 4.Dalal, R. C., W. Wang, G. P. Robertson, and W. J. Parton. 2003. Nitrous oxide emission from Australian agricultural lands and mitigation options: a review. Aust. J. Soil Res. 41:165-196. [Google Scholar]
- 5.Dalal, R. C., and D. E. Allen. 2008. Greenhouse gas fluxes from natural ecosystems (Turner review no. 18). Aust. J. Bot. 56:369-407. [Google Scholar]
- 6.Denmead, O. T., B. C. T. Macdonald, T. Naylor, W. Wang, B. Salter, I. White, S. Wilson, and P. Moody. 2008. Whole-of-season greenhouse gas emissions from Australian sugarcane soils. Proc. Aust. Soc. Sugar Cane Technologists 30:105-114. [Google Scholar]
- 7.Dennis, P., E. A. Edwards, S. N. Liss, and R. Fulthorpe. 2003. Monitoring gene expression in mixed microbial communities by using DNA microarrays. Appl. Environ. Microbiol. 69:769-778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.DeSantis, T. Z., E. L. Brodie, J. P. Moberg, I. X. Zubieta, Y. M. Piceno, and G. L. Andersen. 2007. High-density universal 16S rRNA microarray analysis reveals broader diversity than typical clone library when sampling the environment. Microb. Ecol. 53:371-383. [DOI] [PubMed] [Google Scholar]
- 9.Dudley, A. M., J. Aach, M. A. Steffen, and G. M. Church. 2002. Measuring absolute expression with microarrays with a calibrated reference sample and an extended signal intensity range. Proc. Natl. Acad. Sci. U. S. A. 99:7554-7559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ferrari, B. C., N. Tujula, K. Stoner, and S. Kjelleberg. 2006. Catalyzed reporter deposition-fluorescence in situ hybridization allows for enrichment-independent detection of microcolony-forming soil bacteria. Appl. Environ. Microbiol. 72:918-922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fierer, N., M. Breitbart, J. Nulton, P. Salamon, C. Lozupone, R. Jones, M. Robeson, R. A. Edwards, B. Felts, S. Rayhawk, R. Knight, F. Rohwer, and R. B. Jackson. 2007. Metagenomic and small-subunit rRNA analyses reveal the genetic diversity of bacteria, archaea, fungi, and viruses in soil. Appl. Environ. Microbiol. 73:7059-7066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fulthorpe, R. R., L. F. W. Roesch, A. Riva, and E. W. Triplett. 2008. Distantly sampled soils carry few species in common. ISME J. 2:901-910. [DOI] [PubMed] [Google Scholar]
- 13.Gao, H., Z. K. Yang, T. J. Gentry, L. Wu, C. W. Schadt, and J. Zhou. 2007. Microarray-based analysis of microbial community RNAs by whole-community RNA amplification. Appl. Environ. Microbiol. 73:563-571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gilbert, J. A., D. Field, Y. Huang, R. Edwards, W. Li, P. Gilna, and I. Joint. 2008. Detection of large numbers of novel sequences in the metatranscriptomes of complex marine microbial communities. PLoS One 3:e3042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Goodroad, L. L., and D. R. Keeney. 1984. Nitrous oxide emission from forest, marsh, and prairie ecosystems. J. Environ. Qual. 13:448-452. [Google Scholar]
- 16.Hansel, C. M., S. Fendorf, P. M. Jardine, and C. A. Francis. 2008. Changes in bacterial and archaeal community structure and functional diversity along a geochemically variable soil profile. Appl. Environ. Microbiol. 74:1620-1633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.He, Z., T. J. Gentry, C. W. Schadt, L. Wu, J. Liebich, S. C. Chong, Z. Huang, W. Wu, B. Gu, P. Jardine, C. Criddle, and J. Zhou. 2007. GeoChip: a comprehensive microarray for investigating biogeochemical, ecological and environmental processes. ISME J. 1:67-77. [DOI] [PubMed] [Google Scholar]
- 18.Houghton, J. T., Y. Ding, D. J. Griggs, M. Noguer, P. J. van der Linden, and D. Xiaosu (ed.). 2001. Climate change 2001: the scientific basis. Contribution of Working Group I to the Third Assessment Report of the Intergovernmental Panel on Climate Change. Cambridge University Press, Cambridge, United Kingdom.
- 19.Johnson, D. R., E. L. Brodie, A. E. Hubbard, G. L. Andersen, S. H. Zinder, and L. Alvarez-Cohen. 2008. Temporal transcriptomic microarray analysis of “Dehalococcoides ethenogenes” strain 195 during the transition into stationary phase. Appl. Environ. Microbiol. 74:2864-2872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kampschreur, M. J., W. R. L. Van der Star, H. A. Wielders, J. W. Mulder, M. S. M. Jetten, and M. C. M. Van Loosdrecht. 2008. Dynamics of nitric oxide and nitrous oxide emission during full-scale reject water treatment. Water Res. 42:812-826. [DOI] [PubMed] [Google Scholar]
- 21.Kandeler, E., K. Deiglmayr, D. Tscherko, D. Bru, and L. Philippot. 2006. Abundance of narG, nirK, and nosZ genes of denitrifying bacteria during primary successions of a glacier foreland. Appl. Environ. Microbiol. 72:5957-5962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kelly, J. J., S. Siripong, J. McCormack, L. R. Janus, H. Urakawa, S. El Fantroussi, P. A. Noble, L. Sappelsa, B. E. Rittmann, and D. A. Stahl. 2005. DNA microarray detection of nitrifying bacterial 16S rRNA in wastewater treatment plant samples. Water Res. 39:3229-3238. [DOI] [PubMed] [Google Scholar]
- 23.Kirk, J. L., L. A. Beaudette, M. Hart, P. Moutoglis, J. N. Klironomos, H. Lee, and J. T. Trevors. 2004. Methods of studying soil microbial diversity. J. Microbiol. Methods 58:169-188. [DOI] [PubMed] [Google Scholar]
- 24.McGrath, K. C., S. R. Thomas-Hall, C. T. Cheng, L. Leo, A. Alexa, S. Schmidt, and P. M. Schenk. 2008. Isolation and analysis of mRNA from environmental microbial communities. J. Microbiol. Methods 75:172-176. [DOI] [PubMed] [Google Scholar]
- 25.Meyer, R. L., D. E. Allen, and S. Schmidt. 2008. Nitrification and denitrification as sources of sediment nitrous oxide production: a microsensor study. Marine Chem. 110:68-76. [Google Scholar]
- 26.Miller, M. N., B. J. Zebarth, C. E. Dandie, D. L. Burton, C. Goyera, and J. T. Trevors. 2008. Crop residue influence on denitrification, N2O emissions and denitrifier community abundance in soil. Soil Biol. Biochem. 40:2553-2562. [Google Scholar]
- 27.Ramos, C., L. Mølbak, and S. Molin. 2000. Bacterial activity in the rhizosphere analyzed at the single-cell level by monitoring ribosome contents and synthesis rates. Appl. Environ. Microbiol. 66:801-809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rappé, M. S., and S. J. Giovannoni. 2003. The uncultured microbial majority. Annu. Rev. Microbiol. 57:369-394. [DOI] [PubMed] [Google Scholar]
- 29.Schenk, P. M., K. Kazan, I. Wilson, J. P. Anderson, T. Richmond, S. C. Somerville, and J. M. Manners. 2000. Coordinated plant defense responses in Arabidopsis revealed by microarray analysis. Proc. Natl. Acad. Sci. U. S. A. 97:11655-11660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schmid, M., S. Schmitz-Esser, M. Jetten, and M. Wagner. 2001. 16S-23S rDNA intergenic spacer and 23S rDNA of anaerobic ammonium-oxidizing bacteria: implications for phylogeny and in situ detection. Environ. Microbiol. 3:450-459. [DOI] [PubMed] [Google Scholar]
- 31.Schmidt, I., O. Sliekers, M. Schmid, I. Cirpus, M. Strous, E. Bock, J. G. Kuenen, and M. Jetten. 2002. Aerobic and anaerobic ammonia oxidizing bacteria—competitors or natural partners? FEMS Microb. Ecol. 39:175-181. [DOI] [PubMed] [Google Scholar]
- 32.Stewart, F. J., E. A. Ottesen, and E. F. DeLong. 2010. Development and quantitative analyses of a universal rRNA-subtraction protocol for microbial metatranscriptomics. ISME J. 4:896-907. [DOI] [PubMed] [Google Scholar]
- 33.Taroncher-Oldenburg, G., E. M. Griner, C. A. Francis, and B. B. Ward. 2003. Oligonucleotide microarray for the study of functional gene diversity in the nitrogen cycle in the environment. Appl. Environ. Microbiol. 69:1159-1171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Urich, T., A. Lanzén, J. Qi, D. H. Huson, C. Schleper, and S. C. Schuster. 2008. Simultaneous assessment of soil microbial community structure and function through analysis of the meta-transcriptome. PLoS One 3:e2527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.van den Heuvel, R. N., M. M. Hefting, N. C. G. Tan, M. S. M. Jetten, and J. T. A. Verhoeven. 2009. N2O emission hotspots at different spatial scales and governing factors for small scale hotspots. Sci. Total Environ. 407:2325-2332. [DOI] [PubMed] [Google Scholar]
- 36.Wagner, M., G. Rath, R. Amann, H. P. Koops, and K.-H. Schleifer. 1995. In situ identification of ammonia-oxidizing bacteria. Syst. Appl. Microbiol. 18:251-264. [Google Scholar]
- 37.Wagner, M., A. Loy, M. Klein, N. Lee, N. B. Ramsing, D. A. Stahl, and M. W. Friedrich. 2005. Functional marker genes for identification of sulfate-reducing prokaryotes. Methods Enzymol. 397:469-489. [DOI] [PubMed] [Google Scholar]
- 38.Wertz, S., F. Poly, X. Le Roux, and V. Degrange. 2008. Development and application of a PCR-denaturing gradient gel electrophoresis tool to study the diversity of Nitrobacter-like nxrA sequences in soil. FEMS Microb. Ecol. 63:261-271. [DOI] [PubMed] [Google Scholar]
- 39.Wrage, N., J. Lauf, A. D. Prado, M. Pinto, S. Pietrzak, S. Yamulki, O. Oenema, and G. Gebauer. 2004. Distinguishing sources of N2O in European grasslands by stable isotope analysis. Rapid Commun. Mass Spectrom. 18:1201-1207. [DOI] [PubMed] [Google Scholar]
- 40.Wu, L., D. K. Thompson, G. Li, R. A. Hurt, J. M. Tiedje, and J. Zhou. 2001. Development and evaluation of functional gene arrays for detection of selected genes in the environment. Appl. Environ. Microbiol. 67:5780-5790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wu, L., D. K. Thompson, X. Liu, M. W. Fields, C. E. Bagwell, J. M. Tiedje, and J. Zhou. 2004. Development and evaluation of microarray-based whole-genome hybridization for detection of microorganisms within the context of environmental applications. Environ. Sci. Technol. 38:6775-6782. [DOI] [PubMed] [Google Scholar]
- 42.Yu, Z., and M. Morrison. 2004. Comparisons of different hypervariable regions of rrs genes for use in fingerprinting of microbial communities by PCR-denaturing gradient gel electrophoresis. Appl. Environ. Microbiol. 70:4800-4806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhou, J. 2003. Microarrays for bacterial detection and microbial community analysis. Curr. Opin. Microbiol. 6:288-294. [DOI] [PubMed] [Google Scholar]
- 44.Zumft, W. G. 1997. Cell biology and molecular basis of denitrification. Microbiol. Mol. Biol. Rev. 61:533-616. [DOI] [PMC free article] [PubMed] [Google Scholar]