Abstract
Smoking behavior is complex, and includes multiple stages in the progression from experimentation to continued use and dependence. The experience of subjective effects, such as dizziness, euphoria, heart pounding, nausea, and high, have been associated with varying degrees of persistence and subsequent abuse/dependence of marijuana, cocaine, tobacco and alcohol (Grant et al., 2005, Wagner & Anthony, 2002). Previous studies have reported associations between neuronal nicotinic receptor (CHRN) genes and subjective effects to nicotine. We sought to replicate and expand this work by examining eight SNPs in a sample of adult smokers (n=316) who reported subjective effects following cigarette smoking in a controlled laboratory environment. Two SNPs each in the CHRNB2, CHRNB3, CHRNA6 and CHRNA4 genes were examined. A significant association was found between two SNPs and physical effects reported after smoking the first experimental cigarette. SNP rs2072658 is upstream of CHRNB2 (p value = 0.0046) and rs2229959 is a synonymous change in exon 5 of CHRNA4 (p value = 0.0051). We also examined possible functional relevance of SNP rs2072658 using an in vitro gene expression assay. These studies provided evidence that the minor allele of rs2072658 may lead to decreased gene expression, using two separate cell lines, P19 and SH-SY5Y cell lines (18% p<0.001 and 26% p<0.001 respectively). The human genetic study and functional assays suggest that variation in the promoter region of CHRNB2 gene may be important in mediating levels of expression of the β2 nicotinic receptor subunit, which may be associated with variation in subjective response to nicotine.
Keywords: Nicotinic receptors, SNP, Genetic association, Tobacco use, Subjective effects
Introduction
Tobacco use is a risk factor for six of the eight leading causes of death worldwide (Cdc, 2008). While the health risks are well known and multiple smoking cessation programs are available, the prevalence of smoking among adults in the United States remains around 21% of the adult population (Pleis et al., 2009). Multiple factors contribute to the development of nicotine dependence, and many studies have identified early factors that are important predictors for risk of dependence, such as age of initiation, early subjective effects, and co-morbidity with other drug use and psychiatric disorders (Agrawal et al., 2009, Grant et al., 2005, Wagner & Anthony, 2002).
There is strong evidence that genetic factors play an important role in smoking behavior. Twin and adoption studies suggest that roughly 50% of the variance in smoking behavior is due to genes (Heath et al., 1998, Hopfer et al., 2001, Swan & Carmelli, 1997, Swan et al., 1997, Young et al., 2006). As the primary target for nicotine, the neuronal nicotinic acetylcholine receptors (nAChRs) have been studied using pharmacological and animal genetic models for many years (Gotti et al., 2007, Gotti et al., 2006, Mineur & Picciotto, 2008). Recently, strong evidence for associations between variation in the human nAChR genes (CHRN genes) and smoking-related behaviors has emerged from genome-wide association and candidate gene studies, summarized below (Greenbaum & Lerer, 2009).
Neuronal nicotinic acetylcholine receptors are members of the neurotransmitter-gated superfamily of ion channels and are activated by acetylcholine and nicotine. They are pentameric receptors composed of α and β subunits. Six α and three β subunits are expressed in the mammalian brain. Both α and β subunits contribute to the binding sites for acetylcholine and nicotine with the exception of α5 and β3 which function as accessory subunits. Different receptor subtypes localize to different regions of the central nervous system. For example, receptors composed of α4 and β2 subunits (sometimes with α5) are the most prevalent nAChR subtype found in the brain, whereas receptors composed of α6, β2, and β3 are primarily localized to the substantia nigra, ventral tegmental area, striatum, and locus coeruleus (Gotti et al., 2006). To date, five different subtypes have been shown to be expressed on dopaminergic nerve terminals, including α4α6β2β3, α6β2β3, α6β2, α4β2 and α4α5β2 (Grady et al., 2007). Furthermore, it is clear that GABAergic neurons also express nAChRs (Lu et al., 1998), and that GABA release is modulated by α4β2 and α4α5β2 receptor subtypes (Mcclure-Begley et al., 2009).
The goal of the current study was to examine a limited number of SNPs in a subset of CHRN genes that have been previously implicated in subjective response to nicotine, using a modest sample of subjects assessed in a laboratory immediately after smoking a cigarette (Hutchison et al., 2007). The four genes included: CHRNA4, CHRNB2, CHRNB3, and CHRNA6 which encode the α4, β2, β3, and α6 nAChR subunits respectively. CHRNA4, located on chromosome 20, has been a top candidate for tobacco behavior with mixed results. A previous study examined association of subjective effects over multiple cigarettes with the CHRNA4 SNPs rs2236196 and rs6122429. The study suggested that the SNP rs2236196, located in the 3’UTR, was associated with subjective effects as well as quit success in a clinical trial. No significant associations were found with rs6122429 upstream of CHRNA4 after adjusting for multiple testing (Hutchison et al., 2007). More recently, there was suggestive evidence for association with CHRNA4 and the early subjective experience of “dizziness” when first trying cigarettes, though this association was not significant after adjustment for multiple testing (Ehringer et al., 2009). In the current study two SNPs located in the coding region of CHRNA4 were tested for association with subjective effects in regular tobacco users. These two exonic SNPs are at opposite ends of the gene and should account for a high proportion of the overall variance though too few SNPs are available in the current Hap Map data (data release #21a) to confidently characterize the linkage disequilibrium (LD) patterns in this gene.
In the CHRNB2 gene (on chromosome 1), SNP rs2072658 in the putative promoter region just upstream of exon 1 was shown to be associated with early subjective response to alcohol and tobacco in young adults (Ehringer et al., 2007b). Three other studies have recently found associations between CHRNB2 and tobacco behaviours. The first found an association between rs2072660 and initiation for smoking in young women (Greenbaum et al., 2006). The others found individuals with the minor allele of rs2072661 had substantially reduced odds of quitting smoking even with treatment, and improved effectiveness of the nicotine patch (Conti et al., 2008, Perkins et al., 2009). However, other studies of variations in the coding region of CHRNB2 have not found any association with tobacco behaviours (Ehringer et al., 2007a, Ehringer et al., 2009, Etter et al., 2009, Lueders et al., 2002, Silverman et al., 2000).
The CHRNB3 and CHRNA6 genes are contiguous on chromosome 8 and have been implicated in several studies of nicotine dependence, often being correlated with subjective response (Bierut et al., 2007, Ehringer et al., 2009, Hoft et al., 2008, Saccone et al., 2007). In two independent young adult samples, the upstream region of CHRNB3 has been associated with composite scores of early subjective response to tobacco (Zeiger et al., 2008). More recently, the specific response “dizziness” was associated with this region in a third adult sample (Ehringer et al., 2009).
Based on this literature, eight SNPs in these four genes were selected as top candidates to be examined for possible association with subjective response to smoking in adult established smokers. In order to further characterize the most significantly associated SNP, rs2072658 in CHRNB2, we assayed for its possible functional relevance in an in vitro luciferase gene expression system using both nicotine naive and chronic nicotine treated cells.
Materials and Methods
Genetic Association
Subjects
The sample included 316 daily smokers between 18 and 55 years of age (26.9 ± 9.9) recruited from the Denver/Boulder metropolitan area (for a full description of the sample (Hutchison et al., 2007)). The sample was split evenly between males and females (54.4% male) and was predominantly Causasian (79.6%), with Hispanics (8.8%), Asians (3.3%), African-Americans (2.8%), and undisclosed (5.2%) making up the remainder. Roughly half the sample consisted of unrelated individuals the other half were dizygotic twin and sib-pairs. Subject recruitment methods and protocols were approved by the Institutional Review Board at the University of Colorado.
Assessments
A demographic questionnaire was administered to collect information on sex, income, marital status, age, socioeconomic status, race at baseline, occupation, and education. A smoking history questionnaire collected information on number of years as a smoker, initial age for first cigarette smoked, number of attempts to quit, and frequency and quantity of tobacco used before this study. Nicotine dependence was assessed using the Fagerstrom test for nicotine dependence (FTND) (Heatherton et al., 1991).
The subjective responses to cigarettes were measured using the Nicotine Effects Scale (NEQ) (Hutchison et al., 2007). Participants were scheduled for an experimental session during which they smoked three experimental cigarettes (1.1 mg). Following each cigarette (spaced 25 min apart) the participants responded to a questionnaire on acutely experienced physical effects. The responses to the individual items were grouped as reported in Hutchinson et al into three composite scores (Physical effects, Cognitive effects, Rush) for each cigarette. The items which contribute to the composite Physical effects score are dizziness, palms sweating, unpleasantness, nausea, buzzing, and heart pounding. To minimize multiple testing of phenotypes, we prioritized based on previous work. Only scores from the first cigarette were used, since it was predicted to elicit the strongest responses. Based on work implicating “dizziness” as an important early subjective effect predicting nicotine dependence (Ehringer et al., 2009, Pomerleau & Pomerleau, 1984) and previous genetic associations with this item, we only analyzed the physical effects score which included “dizziness.”
Genotyping methods
Genomic DNA was isolated from buccal cell swabs and whole genome amplified using the method of Zhang et al. (Zhang et al., 1992, Zheng et al., 2001). TaqMan® assays for allelic discrimination (Applied Biosystems) were used to determine SNP genotypes, per instructions of the manufacturer under standard conditions using a Biomek 3000 Laboratory Automation Workstation (Beckman Coulter) to pipette the reactions and an ABI PRISM® 7900 instrument for PCR cycling and data collection.
Analytic Methods
Genotypes were checked for quality and for Hardy-Weinberg proportions using Haploview (Barrett et al., 2005). PLINK qfam-total option was used for genetic association analysis using the family information with 10,000 permutations (Purcell et al., 2007). QTDT was subsequently used to verify the results (Abecasis et al., 2000). Further examination of correlations and phenotypic differences was done using SPSS (SPSS Inc, Chicago, IL). The association analysis used the combined “physical” subjective effects (evenly weighted) to capture dizziness, heart pounding and other similar physical effects in the way the questionnaire and original study was designed to summarize them. The initial analysis was completed on only the eight SNPs described in Table 2, chosen to target variations previously associated with subjective effects (at the time the analysis was being conducted). A secondary analysis was done to assess genetic association between the Physical effects score and SNPs in the CHRNA5-CHRNA3-CHRNB4 cluster because of its consistent association with other nicotine behaviors and reported association with “pleasurable buzz.” (Sherva et al, 2008).
Table 2.
Description of polymorphisms examined
| SNP | Gene | Chr. | Location (NCBI) | Alleles | MAF | HWE p-value |
|---|---|---|---|---|---|---|
| rs2072658 | CHRNB2 | 1 | 151353298 | A/G | 0.027 | rare |
| rs2072660 | CHRNB2 | 1 | 151361794 | T/C | 0.23 | 0.93 |
| rs2229959 | CHRNA4 | 20 | 61451998 | A/C | 0.17 | 0.30 |
| rs2273506 | CHRNA4 | 20 | 61461383 | A/G | 0.10 | 0.04 |
| rs7004381 | CHRNB3 | 8 | 42670318 | A/G | 0.25 | 0.18 |
| rs4950 | CHRNB3 | 8 | 42671790 | A/G | 0.24 | 0.63 |
| rs2304297 | CHRNA6 | 8 | 42727356 | C/T | 0.27 | 0.35 |
| rs35389610 | CHRNA6 | 8 | 42729008 | C/T | 0.35 | 0.45 |
Functional Assays
Generation of plasmid constructs
A region of approximately 2.6 kb upstream of the CHRNB2 initiation codon was amplified from human genomic DNA (Promega Corporation), using the following forward primer 5’-TTCTTGGTGGGTATGGAAGG-3’ and the reverse primer 5’-GCTGCGAGGAGAAACCAG-3’ and Picomaxx High Fidelity PCR System (Stratagene, La Jolla, CA). PCR products were cloned into pCRII-TOPO cloning plasmids (Invitrogen, Carlsbad, CA), sequenced, and cloned fragments containing the major and minor haplotypes in this region of CHRNB2 were identified. The rare allele of rs2072658 (A replaces G) was introduced into both the major and minor haplotype constructs via site-directed mutagenesis (Quikchange II, Invitrogen, Carlsbad, CA) since it is not known whether the SNP occurs in the population within the major or minor haplotype of this region of CHRNB2. Following sequencing to confirm the success of site directed mutagenesis and verify the integrity of the sequence, the fragments were subcloned into the pGL3 basic vector upstream of the Luciferase gene using restriction enzymes SacI, SmaI and EcoRV (New England Biolabs, Ipswich, MA) and T4 ligase (Roche Applied Sciences, Indianapolis, IN). Plasmid preparations from bacterial cultures were done using FastPlasmid Mini-prep kit (Eppendorf, Westbury, NY) for small scale DNA preps or the Qiagen Plasmid Maxi Purification Kit (Qiagen, Germantown, MD) for large scale preps. Following introduction into the pGL3 basic vector the inserted DNA was re-sequenced to verify SNP alleles and check for any artificially introduced variations. Relative lengths as well as relevant polymorphisms of each construct are shown in Figure 2. Four constructs were used, two with the major haplotype background that differ only at the variant nucleotides of rs2072658 and two with the minor haplotype that also differ only at the variant nucleotides or rs2072658.
Figure 2.
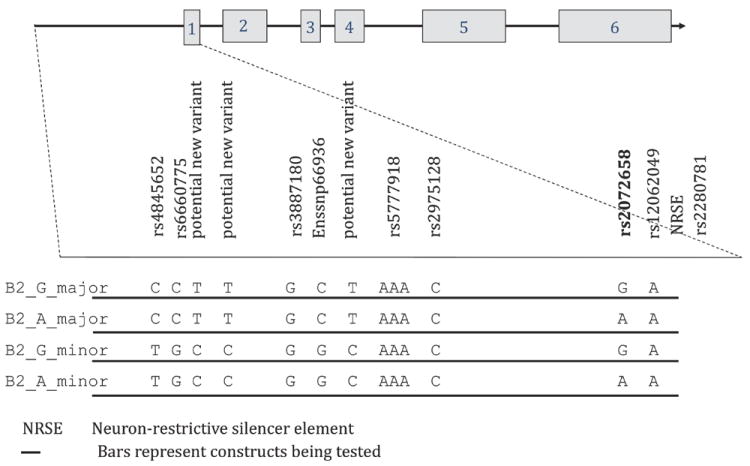
Diagram of the human CHRNB2 promoter region and known SNPs from the UCSC Genome Browser March 2006 update (http://genome.ucsc.edu/), cross-referenced with dbSNP (http://www.ncbi.nlm.nih.gov/SNP/) and Ensembl (http://www.ensembl.org/).
Cell culture and transfections
HEK293T, and SH-SH5Y cells were grown in Dulbecco’s modified Eagle’s medium supplemented with 4.5 g/liter glucose, 10% fetal bovine serum (FBS), and 1% penicillin/streptomycin. P19S18O1A1 (P19) cells were cultured in alpha MEM supplemented with fetal bovine serum (7.5%), calf serum (2.5%) and 1% penicillin/streptomycin. All three cell types were seeded at 0.3 × 106 cells per well in 24 well plates. Plasmids were transfected into HEK293T, SH-SY5Y, and P19 cells using FuGene HD transfection reagent (Roche). Each test plasmid was co-transfected with a control plasmid, and two different control plasmids were used in separate experiments. 200 ng or 400 ng of the test plasmid and 10 ng or 5 ng (for HEK293T) of the control plasmid pRL-CMV (renilla luciferase under a weak CMV promoter) were co-transfected 24 hours after seeding the cells in the plates. In separate experiments, 200 ng or 400 ng of the test plasmid and 40 ng pRL-TK (renilla luciferase under a weak TK promoter) were co-transfected 24 hours after seeding the cells in the plates. pGL3 basic vector with no insert was also used to asses background luciferase activity (empty plasmid), distinct from promoter driven expression. In some assays SH-SY5Y cells were differentiated by incubation with 75mM all-trans-retinoic acid in addition to normal growth media for 10 days prior to transfection. For the nicotine experiments, free base nicotine was added to a final concentration of 0 μM, 1 μM or 10 μM 24 hours post transfection, and the cells were maintained for another 24 hr before harvesting for luciferase activity.
Luciferase assay
Forty-eight hours after transfection, cell extracts were prepared and luciferase assays carried out in 96 well plates using the Dual-Luciferase Reporter Assay System (Promega, Madison, WI) and a PerkinElmer Victor 3V plate reader (PerkinElmer Life Sciences, Wellesley, MA) or Tecan Plate reader (Perkin Elmer, Wellesley, MA) per manufacturers’ instructions. The luciferase expression results from two separate experiments, each including four replicates, were analyzed for each condition tested. A minimum of two separate plasmid preps were tested with multiple replicates of each.
Data analysis
Luciferase activity of the test plasmid was divided by the renilla activity of the control plasmid for each sample, and values were subsequently normalized within experiments to the mean overall activity across the 96 well plate. Data were analyzed using SPSS (SPSS Inc, Chicago, IL) using one way analysis of variance [ANOVA], two-way ANOVA or Students’s t-test where appropriate. Since four different cell types were used, the threshold for experiment wide significance was p=0.0125, using Bonferroni adjustment (0.05/4).
Results
Phenotypic analysis
The “physical effects” score after the first cigarette was constructed as previously described (Hutchison et al., 2007) by an even weighting of the responses to whether the subjects felt dizziness, buzzing, nausea, heart pounding, unpleasant, and sweat on a Likert scale from 1 to 10. This composite phenotype of physical effects is significantly higher in nicotine dependent individuals vs non-dependent individuals as measured by a FTND score of 4 or more (t-test p=0.026). Interestingly the composite phenotype of physical effects is not correlated with the quantitative number of FTND symptoms in this sample, but responses to two of the items are correlated with FTND score, dizziness (r=0.146 p=0.042) and sweating (r=-0.167 p=0.019). All correlations were calculated using the subset of unrelated individuals (N=306).
Genetic association
Genotypes were tested for Hardy Weinberg equilibrium, and checked for quality by concordance between monozygotic twins and percent of genotypes successful for each individual and each SNP. All SNPs were found not to deviate significantly from expected Hardy Weinberg proportions, Table 2. Using PLINK qfam-total, a significant association was found between two of the eight SNPs tested and a sum score of the physical effects reported after smoking the first experimental cigarette, Table 3 (items listed in Table 4). The first SNP rs2072658 is just upstream of CHRNB2 (p= 0.0046, Bonferroni adjusted p = 0.037; within Caucasians p=0.0006), while the second SNP (rs2229959) is a synonymous change in exon 5 of CHRNA4 (p= 0.0051, Bonferroni adjusted p = 0.041; within Caucasians p=0.0058). QTDT indicated no evidence for population stratification with the subjective effects phenotypes for any of the SNPs, and the results within Caucasians supported the results for the entire sample. The second SNP tested in CHRNB2 was not significant, but rs2072660 is in intron 3, 8496 bp downstream of rs2072658 and the two SNPs show nearly zero r2 linkage disequilibrium. There were ten individuals who were in both this sample and the sample used in the analysis in Ehringer et al. which showed association between the SNP and early subjective effects as measured using an interview recall questionnaire (a different but related phenotype) (Ehringer et al., 2007a). Removing these individuals from the analysis resulted in a very small change in significance (rs2072658 p = 0.0046, stat=8.2, n=258, rs2229959 p = 0.0073, stat=7.3, n=248). Post Hoc analysis for association with each of the “physical” subjective effects indicated the association with rs2072658 is driven by sweating, heart pounding, and nausea (p = 0.0028, 0.0064 and 0.016 respectively). In contrast association with rs2229959 is driven by nausea, dizziness, and sweating (p = 0.007, 0.028, 0.043 respectively), Table 4.
Table 3.
Genetic association with the sum score of physical effects reported following the first experimental cigarette. All indicates analysis done on entire sample with ethnicity as a covariate and Cauc. indicates analysis within the subsample of Caucasians.
| Chr. | Gene | SNP | Risk allele | N | Empirical stat | p | Adjusted p Bonferroni | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|
|
| |||||||||||
| all | Cauc. | all | Cauc. | all | Cauc. | all | Cauc. | ||||
| 1 | CHRNB2 | rs2072658 | A | 267 | 205 | 8.2 | 17.5 | 0.0046 | 0.0006 | 0.036 | 0.005 |
| 1 | CHRNB2 | rs2072660 | - | 267 | 204 | -0.43 | -0.56 | 0.66 | 0.48 | 1 | 1 |
| 20 | CHRNA4 | rs2229959 | C | 257 | 195 | 7.3 | 9.2 | 0.0051 | 0.0058 | 0.041 | 0.046 |
| 20 | CHRNA4 | rs2273506 | A | 257 | 196 | 3.1 | 3.2 | 0.10 | 0.10 | 0.80 | 0.80 |
| 8 | CHRNB3 | rs7004381 | - | 260 | 198 | -1.1 | -0.45 | 0.25 | 0.53 | 1 | 1 |
| 8 | CHRNB3 | rs4950 | - | 265 | 201 | -0.34 | -0.97 | 0.59 | 0.36 | 1 | 1 |
| 8 | CHRNA6 | rs2304297 | - | 244 | 185 | -0.1 | 0.26 | 0.97 | 0.63 | 1 | 1 |
| 8 | CHRNA6 | rs35389610 | T | 263 | 200 | -3.9 | -6.22 | 0.042 | 0.024 | 0.336 | 0.192 |
Table 4.
Association with individual physical effects items, p<0.05 are in bold (statistic, n)
| rs2072658 | rs2229959 | |||||
|---|---|---|---|---|---|---|
|
| ||||||
| n | statistic | p-value | n | statistic | p-value | |
| Dizziness | 271 | 1.3 | 0.25 | 271 | 5.6 | 0.028 |
| Palms Sweating | 273 | 12.9 | 0.0028 | 263 | 4.7 | 0.043 |
| Unpleasantness | 270 | 0.40 | 0.52 | 260 | 7.3 | 0.007 |
| Nausea | 272 | 6.56 | 0.016 | 262 | 2.2 | 0.15 |
| Buzzing | 272 | 2.13 | 0.17 | 262 | 3.2 | 0.11 |
| Heart pounding | 272 | 10.13 | 0.0064 | 262 | 2.82 | 0.12 |
The secondary analysis for rs514743, rs680244, rs684513 and rs16969968 in CHRNA5, rs8040868 and rsrs8023462 in CHRNA3 showed no significant associations between these SNPs in the CHRNA5-CHRNA3-CHRNB4 gene cluster and the composite for physical subjective effects after controlling for multiple testing. There was suggestive evidence for association between rs16969968 and buzzing (empirical stat = 8.8 p=0.009) and rs8023462 with sweating (empirical stat = 7.1 p=0.009) (data not shown).
To further verify the association result QTDT was used, which uses a modified transmission disequilibrium test (rather than a permutation method) for analyzing family data and then combines it with non-family data (Abecasis et al., 2000). Using the –ap option in QTDT we tested for and found no evidence for stratification. For the genetic association analysis, QTDT yielded similar results as PLINK for both SNPs further supporting association between “Physical effects” reported after smoking the first experimental cigarette and rs2072658 (p = 0.0024, F=9.42, df=263) and rs2229959 (p =0.0074, F=7.3, df=250).
Functional assays
The four CHRNB2 upstream region constructs that were tested are shown in Figure 2. Experiments to assess whether 24 hr treatment with nicotine affected CHRNB2 upstream region driven-luciferase reporter gene expression revealed that nicotine treatment (1 or 10 μM) was without effect Figure 3. There also were no SNP × nicotine environment interactions observed. Therefore, data were combined across non-significant groups and the results are shown in Figure 4 and Table 5. The minor allele of rs2072658 showed decreased relative luciferase expression in SH-SY5Y, P19, and differentiated SH-SY5Y cells, and although the directional trend is the same in HEK cells, the difference was not significant.
Figure 3.
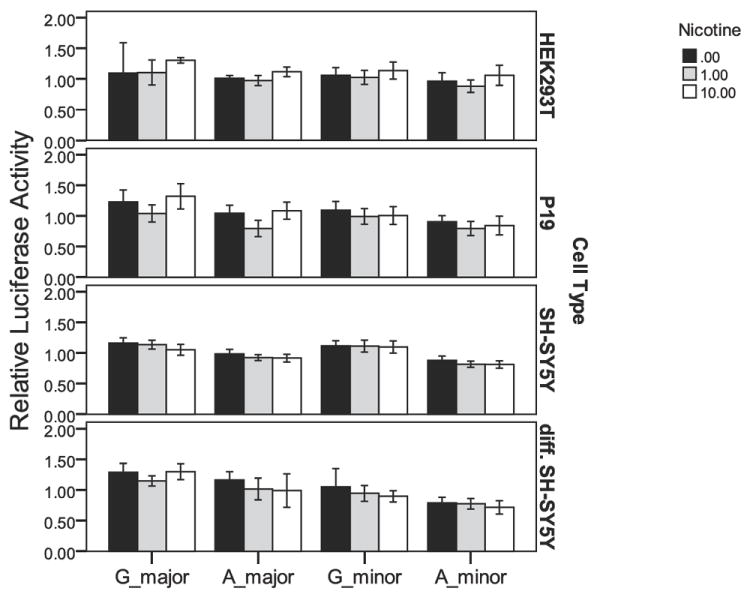
Relative luciferase activity across cell types for three concentrations of nicotine. No significant differences were observed in relative activity between 0 μM, 1 μM and 10 μM chronic nicotine exposure. Error bars represent a 95% confidence interval on the mean.
Figure 4.
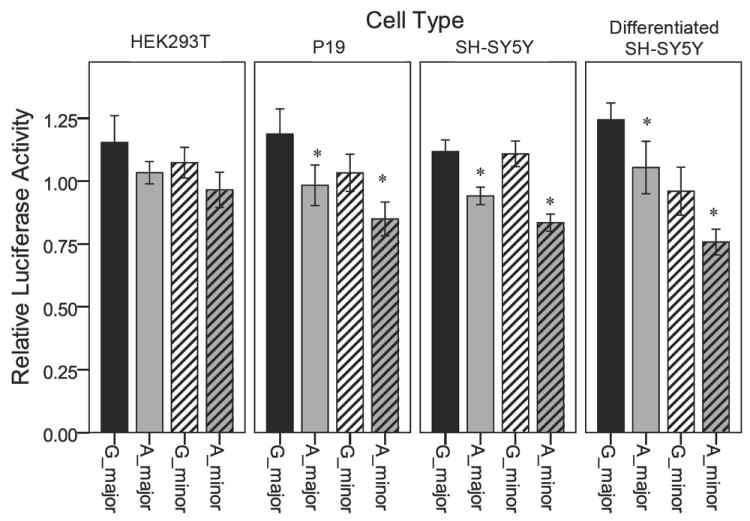
Relative luciferase activity of four constructs in four cell types. The target polymorphism rs2072658 (minor allele A shaded grey) shows a significant reduction in expression in SH-SY5Y, P19, and differentiated SH-SY5Y cells, indicated by an asterisk. This reduction in relative luciferase activity is independent of other local SNP variation (major vs minor). Error bars represent a 95% confidence interval on the mean, * indicates p<0.05.
Table 5.
Relative luciferase activity in four constructs and four cell types (also shown in Figure 4). Data were normalized. Rows 1-4 show the normalized luciferase expression compared to the control plasmid (and normalized within experiment to the overall mean signal) for each test construct. Rows 6-9 present the percent decrease for each test construct relative to the G_major plasmid.
| Construct | HEK | P19 | SH-SY5Y | SH-SY5Y differentiated | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
|
|||||||||||||
| relative luciferase intensity | G_major | 1.15±0.05 | 1.19±0.05 | 1.12±0.02 | 1.24±0.04 | ||||||||
| A_major | 1.03±0.02 | 0.98±0.04 | 0.94±0.02 | 1.05±0.05 | |||||||||
| G_minor | 1.07±0.03 | 1.03±0.04 | 1.12±0.02 | 0.96±0.05 | |||||||||
| A_minor | 0.97±0.03 | 0.85±0.03 | 0.83±0.02 | 0.76±0.03 | |||||||||
|
|
|||||||||||||
| Comparison | % | stat. (df) | p-value | % | stat. (df) | p-value | % | stat. (df) | p-value | % | stat. (df) | p-value | |
|
|
|||||||||||||
| % decrease | G_major vs A_major | 0.1 | 2.4, 10 | 0.0360 | 0.18 | 3.2, 79 | 0.0020 | 0.16 | 6.1, 116 | 1.44E-08 | 0.15 | 3.2, 59 | 0.0022 |
| G_major vs G_minor | 0.07 | 2.8, 25 | 0.010 | 0.13 | 2.6, 77 | 0.011 | 0 | 0.26, 116 | 0.795 | 0.23 | 5.0, 64 | 4.73E-06 | |
| G_major vs A_minor | 0.16 | 3.2, 25 | 0.0037 | 0.29 | 5.9, 81 | 8.09E-08 | 0.26 | 9.83, 116 | 6.96E-17 | 0.39 | 11.8, 65 | 8.21E-18 | |
| G_minor vs A_minor | 0.09 | 2.4, 36 | 0.0217 | 0.17 | 3.7, 90 | 0.0004 | 0.26 | 8.95, 116 | 6.80E-15 | 0.21 | 3.8, 65 | 0.0003 | |
|
|
|||||||||||||
The largest effect of rs2072658 was observed in SH-SY5Y (human neuronal) cells, with a 26% reduction in relative luciferase expression on the minor haplotype background (G_minor compared to A_minor) and a 16% reduction in relative luciferase expression on the major haplotype background (G_major compared to A_major) (p<0.001). No difference was observed between G_major and G_minor. Differentiated SH-SY5Y cells showed a similar pattern except that the haplotype background made a difference with a 23% reduction in relative luciferase activity observed between G_major and G_minor (p<0.001). There were no interaction effects between rs2072658 and the other SNP alleles, that is the magnitude and pattern of decrease in relative luciferase expression is roughly the same when comparing G_major vs A_major as with G_minor vs A_minor.
P19 cells also showed a small but significant decrease in relative luciferase activity due to rs2072658 in both the major and minor backgrounds (17% p<0.002, and 18% p<0.001 respectively). In HEK cells no significant differences were found between the four constructs in relative luciferase activity after adjusting for multiple testing (four cell types/conditions).
Discussion
Genetic association
Here we have examined eight SNPs in four CHRN genes selected on the basis of previous evidence for association with subjective effects to nicotine. The most significant association was between rs2072658 in CHRNB2 and smoking-induced physical effects. This result was driven by reported experiences of nausea, heart-pounding, and sweating after smoking a cigarette, and supports our previous study of adolescent early subjective effects (Ehringer et al, 2007).
The current study expands on our previous work by examining a new sample of adult subjects, using a highly detailed laboratory assessment of subjective effects. It represents the first attempt (that we know of) to replicate the association with rs2072658, so the fact that this rare SNP emerged as the most significant association provides additional evidence for its possible role in mediating these responses.
In addition, we cloned the putative promoter region with each allele of SNP rs2072658 and assayed for the effect of the SNP using a luciferase gene reporter assay. Two reporter-gene constructs were generated for each allele (G, major; A, minor) on two haplotype backgrounds. The surrounding DNA sequence of a polymorphism may influence tertiary DNA structure and/or important DNA-binding sites for key regulatory proteins, so we examined how each allele might contribute to gene expression in the context of both major and minor haplotypes. Our results indicate that in the case of rs2072858 the rare A allele decreases reporter gene expression, and this effect is independent of other local SNP variation. This does not preclude the possibility that rs2072658 interacts with SNPs in this region which were not represented in the constructs nor SNPs upstream or downstream of the cloned region. In addition, our results from multiple cell types demonstrate that other SNPs in the region are likely to affect expression in some cell types but not others. This was not unexpected, since expression of the β2 subunit is highly regulated to specific neuronal regions and cell types (Gotti et al., 2007). However, the cell type-dependent effect on expression is much smaller than the effect of the single nucleotide change at rs2072658, highlighting its possible important role in regulation.
Multiple studies have shown that upregulation of nicotinic receptors by nicotine is independent of differences in mRNA (Huang & Winzer-Serhan, 2006, Marks et al., 1992, Pauly et al., 1996). However, nicotine has been shown to increase expression of many different proteins, including transcription factors (Zhang et al., 2007). To examine whether there is support for rs2072658 making a difference in gene function we used the Transcription Element Search System (TESS; URL http://www.cbil.upenn.edu/tess). Interestingly, transcription factor LBP-1 (aka UBP-1) is predicted to bind the sequence surrounding SNP rs2072658 allele G, but not allele A. It is possible that decreased binding efficiency in the presence of the rare A allele may lead to reduced transcription of the CHRNB2 gene. Based on this in silico analysis we hypothesized that although nicotine might not be predicted to upregulate reporter gene expression from the CHRNB2 promoter, it may be possible for a particular SNP, such as rs2072658, to interrupt the binding site for a transcription factor affected by nicotine and thus lead to modulation of allele-specific differences in gene expression by nicotine. We found no effect of chronic nicotine exposure in cell culture on the relative expression. To mitigate the chance of observing or failing to observe an effect of nicotine due to changes in expression of the control plasmid a second promoter (TK) was used to check the nicotine results and no significant differences were found. This increases our confidence that there is neither an effect of nicotine on general regulation of luciferase expression in this system, nor an interaction effect between the rs2072658 genotype and nicotine, in the context of the specific CHRNB2 promoter region examined.
The only other SNP that achieved significance after accounting for multiple testing was rs2229959, which leads to a synonymous substitution in exon 5 of CHRNA4. This is consistent with the finding that rs2236196, which resides in the 3’UTR of exon 6, is associated with the subjective effects of nicotine (Hutchison et al., 2007). It is important to note that rs2236196 is in strong linkage disequilibrium with rs2229959. Ehringer et al. examined both of these SNPs and estimated linkage disequilibrium to be D’ of 0.91, but found no association with either SNP and early subjective effects to nicotine and alcohol in adolescents (Ehringer et al., 2007a). As is the case for many genetic studies, there are some SNPs and genes that are likely to replicate even in the context of different samples and phenotypes (as with rs2072658 in CHRNB2), while others SNPs are more sample and phenotype specific. In the case of rs2229959, a couple of studies have now provided evidence for association with subjective effects in adult smokers, but it remains unclear whether this gene plays a role in subjective effects in adolescents.
In a secondary analysis we looked at a few polymorphisms in the CHRNA5-CHRNA3-CHRNB4 gene cluster. There were no significant associations between these SNPs and the composite of physical effects, but rs16969968 which has been repeatedly associated with multiple smoking behaviors, showed suggestive association with “buzz”. Although not significant this is similar to the subjective effect, “pleasurable buzz” reported to be associated with the same SNP in a separate sample Sherva et al (Sherva et al., 2008). The fact that rs16969968 was not significantly associated with the composite Physical effects score, or the “dizziness” item is also consistent with our previous findings in other samples (Ehringer et al., 2009, Schlaepfer et al., 2008). Future work will be needed to further disentangle how different components of the subjective responses may be associated with various SNPs in specific CHRN genes.
Other Emerging Evidence for role of CHRNB2
Multiple recent studies have found that SNPs in CHRNB2 are associated with the effectiveness of smoking cessation aids, the failure to quit and nicotine dependence (Conti et al., 2008, Greenbaum et al., 2006, Perkins et al., 2009). The phenotypic focus of these studies was nicotine dependence and the most significant SNP (rs2072661) is located in the 3’UTR. In the present study we did not test nicotine dependence or failure to quit, but we also did not find an association with rs2072660 which shows modest LD with rs2072661. However, other studies of variations in exons of CHRNB2 also have not found any association with tobacco behaviors (Ehringer et al., 2007a, Ehringer et al., 2009, Etter et al., 2009, Lueders et al., 2002, Silverman et al., 2000). The heterogeneity of samples and slight differences in phenotypic measures contributes to this heterogeneity in results making teasing out which elements of tobacco behavior are affected by which regions of CHRNB2 a continuing challenge.
Here we measured smoking related behaviour using reported subjective effects following smoking a cigarette in the laboratory. Subjective effects to smoking are associated with nicotine dependence in this sample as well as other adult and adolescent samples (Ehringer et al., 2007a, Ehringer et al., 2009). Likewise, Pomerleau and colleagues have demonstrated the important role of subjective effects in nicotine behaviors for decades (Pomerleau & Pomerleau, 1984), more recently showing that parental history of smoking is associated with subjective response in never smokers (Pomerleau et al., 2009). Our recent genetic studies suggest that subjective effects may be a suitable nicotine-association “endophenotype” worthy of future research. It is interesting that the nicotine dependent individuals tend to endorse higher levels of what might be considered “negative” Physical response scores. This is consistent with our previous work, which includes separate samples (Zeiger et al, 2008; Ehringer et al, 2009) and one can only speculate about why this counter-intuitive phenomenon occurs. It is important to note that with only one exception, the items do not connote affective valence. Rather, the items all relate to physiological arousal, which may be related to the stimulatory effects of nicotine. On the other hand, even if the items are interpreted negatively by the participants, it is possible that individuals who decide to continue using a drug which leads to “negative” effects may represent a subgroup of people who are more severely dependent.
The β2 and α4 nAChR subunits are the most commonly expressed in the brain, and the predominant receptor subtype in brain contains both α4 and β2 subunits (Gotti et al., 2007), so it is interesting that SNPs in CHRNB2 and CHRNA4 emerged as most significant. However, we were surprised that there was no evidence for association with CHRNB3/CHRNA6, given the multiple studies that have implicated this region in smoking behavior and early subjective effects. However, as alluded to earlier, the current study may be limited by sample size, and the measure of subjective effects is different than any of the previous studies, which focused on adolescents (Zeiger et al., 2008) or specifically asked about the “first few cigarettes” (Zeiger et al., 2008; Ehringer et al., 2009). There is already evidence in the literature that age can modify the affect/association of CHRN variations with smoking traits. For example a SNP in CHRNA5 has been associated with susceptibility to nicotine dependence in individuals who begin smoking before age 16, but the association is not observed in individuals who initiate later (Weiss et al., 2008). Future studies comparing early subjective effect questionnaire measures to laboratory measures in established adult smokers may shed insight about how this endophenotype changes over time, and whether different genetic variations may contribute to different aspects of the measure.
In summary, we found that people who carry the minor (A) allele for rs2072658 report higher levels of physical subjective effects, in particular nausea, heart-pounding and sweating. This is the same direction (“risk” allele) reported in Ehringer et al. (2007a). Introduction of this SNP into a luciferase reporter vector indicates that constructs with the minor allele (A) show lower relative luciferase expression, suggesting that lower expression may lead to greater experience of physical subjective effects such as nausea, heart-pounding, and sweating. Future work examining additional variants in these genes, identifying novel rare variants through DNA resequencing in the context of specific phenotypes for human genetic studies, and expanded functional studies, will be needed to better characterize the role of CHRNB2 and CHRNA4 in mediating tobacco responses and development of dependence.
Figure 1.
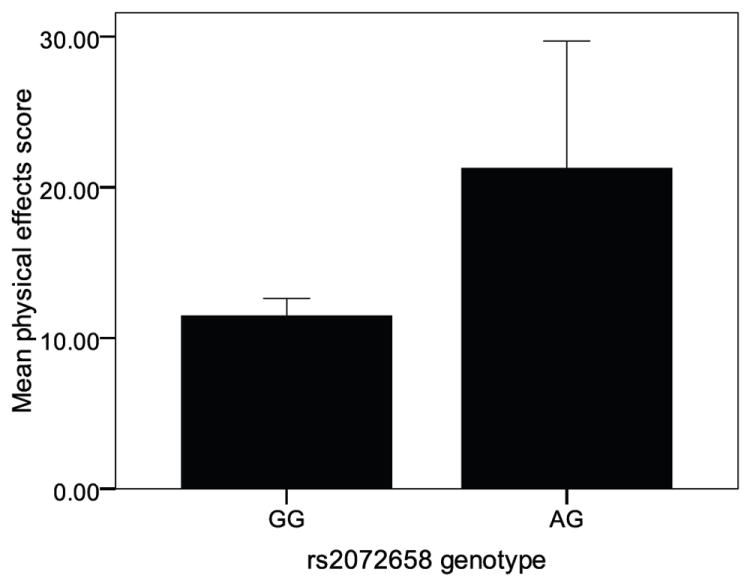
Mean physical effects score by rs2072658 genotype. Individuals with the AG genotype have significantly higher physical effects scores than individuals with the GG (common) genotype after smoking the first experimental cigarette (n=17 mean=21.3 and n=282 mean=11.6 respectively). There were no individuals in this study with the rare AA genotype. Error bars represent a 95% confidence interval on the mean, * indicates p<0.05.
Table 1.
Sample distribution of ethnicities and sexes
| N (%) | Caucasian | African-American | Asian | Hispanic | Other | Total | Age (u ± s) |
|---|---|---|---|---|---|---|---|
| Male | 147 | 6 | 9 | 19 | 15 | 197 (54.4) | 27.0 ± 10 |
| Female | 141 | 4 | 3 | 13 | 4 | 165 (45.6) | 26.7 ± 10 |
| Total | 288 (79.6) | 10 (2.8) | 12 (3.3) | 32 (8.8) | 19 (5.3) | 362 | 26.9 ± 10 |
Acknowledgments
Many thanks to Jill Miyamotto-Ditmon for help with the cell culture. This work was supported by NIH grants AA015336 (MAE), AA017889 (MAE), AA007464 (NRH), DA017637 (NRH), DA14642 (KEH) and CA81637 (KEH), CA089392 (JAS).
Footnotes
Disclosures and Potential Conflicts of Interest
Dr. Nicole R. Hoft, Dr. Jerry A. Stitzel, Dr. Kent E. Hutchison, and Dr. Marissa A. Ehringer do not have any potential conflicts of interest, financial or otherwise, relevant to the subject matter of this work.
References
- Abecasis GR, Cardon LR, Cookson WO. A general test of association for quantitative traits in nuclear families. Am J Hum Genet. 2000;66:279–292. doi: 10.1086/302698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agrawal A, Sartor CE, Lynskey MT, Grant JD, Pergadia ML, Grucza R, Bucholz KK, Nelson EC, Madden PA, Martin NG, Heath AC. Evidence for an interaction between age at first drink and genetic influences on DSM-IV alcohol dependence symptoms. Alcohol Clin Exp Res. 2009;33:2047–2056. doi: 10.1111/j.1530-0277.2009.01044.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- Bierut LJ, Madden PA, Breslau N, Johnson EO, Hatsukami D, Pomerleau OF, Swan GE, Rutter J, Bertelsen S, Fox L, Fugman D, Goate AM, Hinrichs AL, Konvicka K, Martin NG, Montgomery GW, Saccone NL, Saccone SF, Wang JC, Chase GA, Rice JP, Ballinger DG. Novel genes identified in a high-density genome wide association study for nicotine dependence. Hum Mol Genet. 2007;16:24–35. doi: 10.1093/hmg/ddl441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CDC. Morbidity and Mortality Weekly Review. 2008;57:1226–1228. [PubMed] [Google Scholar]
- Conti DV, Lee W, Li D, Liu J, Van Den Berg D, Thomas PD, Bergen AW, Swan GE, Tyndale RF, Benowitz NL, Lerman C. Nicotinic acetylcholine receptor beta2 subunit gene implicated in a systems-based candidate gene study of smoking cessation. Hum Mol Genet. 2008;17:2834–2848. doi: 10.1093/hmg/ddn181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehringer MA, Clegg HV, Collins AC, Corley RP, Crowley T, Hewitt JK, Hopfer CJ, Krauter K, Lessem J, Rhee SH, Schlaepfer I, Smolen A, Stallings MC, Young SE, Zeiger JS. Association of the neuronal nicotinic receptor beta2 subunit gene (CHRNB2) with subjective responses to alcohol and nicotine. Am J Med Genet B Neuropsychiatr Genet. 2007a;144:596–604. doi: 10.1002/ajmg.b.30464. [DOI] [PubMed] [Google Scholar]
- Ehringer MA, Clegg HV, Collins AC, Corley RP, Crowley T, Hewitt JK, Hopfer CJ, Krauter K, Lessem J, Rhee SH, Schlaepfer I, Smolen A, Stallings MC, Young SE, Zeiger JS. Association of the neuronal nicotinic receptor beta2 subunit gene (CHRNB2) with subjective responses to alcohol and nicotine. Am J Med Genet B Neuropsychiatr Genet. 2007b doi: 10.1002/ajmg.b.30464. [DOI] [PubMed] [Google Scholar]
- Ehringer MA, McQueen MB, Hoft NR, Saccone NL, Stitzel JA, Wang JC, Bierut LJ. Association of CHRN genes with “dizziness” to tobacco. Am J Med Genet B Neuropsychiatr Genet. 2009 doi: 10.1002/ajmg.b.31027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Etter JF, Hoda JC, Perroud N, Munafo M, Buresi C, Duret C, Neidhart E, Malafosse A, Bertrand D. Association of genes coding for the alpha-4, alpha-5, beta-2 and beta-3 subunits of nicotinic receptors with cigarette smoking and nicotine dependence. Addict Behav. 2009;34:772–775. doi: 10.1016/j.addbeh.2009.05.010. [DOI] [PubMed] [Google Scholar]
- Gotti C, Moretti M, Gaimarri A, Zanardi A, Clementi F, Zoli M. Heterogeneity and complexity of native brain nicotinic receptors. Biochem Pharmacol. 2007;74:1102–1111. doi: 10.1016/j.bcp.2007.05.023. [DOI] [PubMed] [Google Scholar]
- Gotti C, Zoli M, Clementi F. Brain nicotinic acetylcholine receptors: native subtypes and their relevance. Trends Pharmacol Sci. 2006;27:482–491. doi: 10.1016/j.tips.2006.07.004. [DOI] [PubMed] [Google Scholar]
- Grady SR, Salminen O, Laverty DC, Whiteaker P, McIntosh JM, Collins AC, Marks MJ. The subtypes of nicotinic acetylcholine receptors on dopaminergic terminals of mouse striatum. Biochem Pharmacol. 2007;74:1235–1246. doi: 10.1016/j.bcp.2007.07.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant JD, Scherrer JF, Lyons MJ, Tsuang M, True WR, Bucholz KK. Subjective reactions to cocaine and marijuana are associated with abuse and dependence. Addict Behav. 2005;30:1574–1586. doi: 10.1016/j.addbeh.2005.02.007. [DOI] [PubMed] [Google Scholar]
- Greenbaum L, Kanyas K, Karni O, Merbl Y, Olender T, Horowitz A, Yakir A, Lancet D, Ben-Asher E, Lerer B. Why do young women smoke? I. Direct and interactive effects of environment, psychological characteristics and nicotinic cholinergic receptor genes. Mol Psychiatry. 2006;11:312–322. 223. doi: 10.1038/sj.mp.4001774. [DOI] [PubMed] [Google Scholar]
- Greenbaum L, Lerer B. Differential contribution of genetic variation in multiple brain nicotinic cholinergic receptors to nicotine dependence: recent progress and emerging open questions. Mol Psychiatry. 2009;14:912–945. doi: 10.1038/mp.2009.59. [DOI] [PubMed] [Google Scholar]
- Heath AC, Madden PA, Martin NG. Statistical methods in genetic research on smoking. Stat Methods Med Res. 1998;7:165–186. doi: 10.1177/096228029800700205. [DOI] [PubMed] [Google Scholar]
- Heatherton TF, Kozlowski LT, Frecker RC, Fagerstrom KO. The Fagerstrom Test for Nicotine Dependence: a revision of the Fagerstrom Tolerance Questionnaire. Br J Addict. 1991;86:1119–1127. doi: 10.1111/j.1360-0443.1991.tb01879.x. [DOI] [PubMed] [Google Scholar]
- Hoft NR, Corley RP, McQueen MB, Schlaepfer IR, Huizinga D, Ehringer MA. Genetic Association of the CHRNA6 and CHRNB3 Genes with Tobacco Dependence in a Nationally Representative Sample. Neuropsychopharmacology. 2008 doi: 10.1038/npp.2008.122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hopfer CJ, Stallings MC, Hewitt JK. Common genetic and environmental vulnerability for alcohol and tobacco use in a volunteer sample of older female twins. J Stud Alcohol. 2001;62:717–723. doi: 10.15288/jsa.2001.62.717. [DOI] [PubMed] [Google Scholar]
- Huang LZ, Winzer-Serhan UH. Chronic neonatal nicotine upregulates heteromeric nicotinic acetylcholine receptor binding without change in subunit mRNA expression. Brain Res. 2006;1113:94–109. doi: 10.1016/j.brainres.2006.06.084. [DOI] [PubMed] [Google Scholar]
- Hutchison KE, Allen DL, Filbey FM, Jepson C, Lerman C, Benowitz NL, Stitzel J, Bryan A, McGeary J, Haughey HM. CHRNA4 and tobacco dependence: from gene regulation to treatment outcome. Arch Gen Psychiatry. 2007;64:1078–1086. doi: 10.1001/archpsyc.64.9.1078. [DOI] [PubMed] [Google Scholar]
- Lu Y, Grady S, Marks MJ, Picciotto M, Changeux JP, Collins AC. Pharmacological characterization of nicotinic receptor-stimulated GABA release from mouse brain synaptosomes. J Pharmacol Exp Ther. 1998;287:648–657. [PubMed] [Google Scholar]
- Lueders KK, Hu S, McHugh L, Myakishev MV, Sirota LA, Hamer DH. Genetic and functional analysis of single nucleotide polymorphisms in the beta2-neuronal nicotinic acetylcholine receptor gene (CHRNB2) Nicotine Tob Res. 2002;4:115–125. doi: 10.1080/14622200110098419. [DOI] [PubMed] [Google Scholar]
- Marks MJ, Pauly JR, Gross SD, Deneris ES, Hermans-Borgmeyer I, Heinemann SF, Collins AC. Nicotine binding and nicotinic receptor subunit RNA after chronic nicotine treatment. J Neurosci. 1992;12:2765–2784. doi: 10.1523/JNEUROSCI.12-07-02765.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClure-Begley TD, King NM, Collins AC, Stitzel JA, Wehner JM, Butt CM. Acetylcholine-stimulated [3H]GABA release from mouse brain synaptosomes is modulated by alpha4beta2 and alpha4alpha5beta2 nicotinic receptor subtypes. Mol Pharmacol. 2009;75:918–926. doi: 10.1124/mol.108.052274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mineur YS, Picciotto MR. Genetics of nicotinic acetylcholine receptors: Relevance to nicotine addiction. Biochem Pharmacol. 2008;75:323–333. doi: 10.1016/j.bcp.2007.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pauly JR, Marks MJ, Robinson SF, van de Kamp JL, Collins AC. Chronic nicotine and mecamylamine treatment increase brain nicotinic receptor binding without changing alpha 4 or beta 2 mRNA levels. The Journal of pharmacology and experimental therapeutics. 1996;278:361–369. [PubMed] [Google Scholar]
- Perkins KA, Lerman C, Mercincavage M, Fonte CA, Briski JL. Nicotinic acetylcholine receptor beta2 subunit (CHRNB2) gene and short-term ability to quit smoking in response to nicotine patch. Cancer Epidemiol Biomarkers Prev. 2009;18:2608–2612. doi: 10.1158/1055-9965.EPI-09-0166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pleis JR, Lucas JW, Ward BW. Summary health statistics for U.S. adults: National Health Interview Survey, 2008. National Center for Health Statistics. Vital Health Stat. 2009;10 [PubMed] [Google Scholar]
- Pomerleau OF, Pomerleau CS. Neuroregulators and the reinforcement of smoking: towards a biobehavioral explanation. Neurosci Biobehav Rev. 1984;8:503–513. doi: 10.1016/0149-7634(84)90007-1. [DOI] [PubMed] [Google Scholar]
- Pomerleau OF, Pomerleau CS, Snedecor SM, Finkenauer R, Mehringer AM, Langenecker SA, Sirevaag EJ. Substance use, trait measures, and subjective response to nicotine in never-smokers stratified on parental smoking history and sex. Nicotine Tob Res. 2009;11:1055–1066. doi: 10.1093/ntr/ntp099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, Maller J, Sklar P, de Bakker PI, Daly MJ, Sham PC. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saccone SF, Hinrichs AL, Saccone NL, Chase GA, Konvicka K, Madden PA, Breslau N, Johnson EO, Hatsukami D, Pomerleau O, Swan GE, Goate AM, Rutter J, Bertelsen S, Fox L, Fugman D, Martin NG, Montgomery GW, Wang JC, Ballinger DG, Rice JP, Bierut LJ. Cholinergic nicotinic receptor genes implicated in a nicotine dependence association study targeting 348 candidate genes with 3713 SNPs. Hum Mol Genet. 2007;16:36–49. doi: 10.1093/hmg/ddl438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlaepfer IR, Hoft NR, Collins AC, Corley RP, Hewitt JK, Hopfer CJ, Lessem JM, McQueen MB, Rhee SH, Ehringer MA. The CHRNA5/A3/B4 gene cluster variability as an important determinant of early alcohol and tobacco initiation in young adults. Biol Psychiatry. 2008;63:1039–1046. doi: 10.1016/j.biopsych.2007.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherva R, Wilhelmsen K, Pomerleau CS, Chasse SA, Rice JP, Snedecor SM, Bierut LJ, Neuman RJ, Pomerleau OF. Association of a single nucleotide polymorphism in neuronal acetylcholine receptor subunit alpha 5 (CHRNA5) with smoking status and with ‘pleasurable buzz’ during early experimentation with smoking. Addiction. 2008;103:1544–1552. doi: 10.1111/j.1360-0443.2008.02279.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silverman MA, Neale MC, Sullivan PF, Harris-Kerr C, Wormley B, Sadek H, Ma Y, Kendler KS, Straub RE. Haplotypes of four novel single nucleotide polymorphisms in the nicotinic acetylcholine receptor beta2-subunit (CHRNB2) gene show no association with smoking initiation or nicotine dependence. Am J Med Genet. 2000;96:646–653. [PubMed] [Google Scholar]
- Swan GE, Carmelli D. Behavior Genetic Investigations of Cigarette Smoking and Related Issues in Twins. In: Blum K, Noble EP, editors. Handbook of Psychiatric Genetics CRC. Press Inc.; Boca Raton, New York, London, Tokyo: 1997. pp. 387–406. [Google Scholar]
- Swan GE, Carmelli D, Cardon LR. Heavy consumption of cigarettes, alcohol and coffee in male twins. J Stud Alcohol. 1997;58:182–190. doi: 10.15288/jsa.1997.58.182. [DOI] [PubMed] [Google Scholar]
- Wagner FA, Anthony JC. From first drug use to drug dependence; developmental periods of risk for dependence upon marijuana, cocaine, and alcohol. Neuropsychopharmacology. 2002;26:479–488. doi: 10.1016/S0893-133X(01)00367-0. [DOI] [PubMed] [Google Scholar]
- Weiss RB, Baker TB, Cannon DS, von Niederhausern A, Dunn DM, Matsunami N, Singh NA, Baird L, Coon H, McMahon WM, Piper ME, Fiore MC, Scholand MB, Connett JE, Kanner RE, Gahring LC, Rogers SW, Hoidal JR, Leppert MF. A candidate gene approach identifies the CHRNA5-A3-B4 region as a risk factor for age-dependent nicotine addiction. PLoS Genet. 2008;4:e1000125. doi: 10.1371/journal.pgen.1000125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young SE, Rhee SH, Stallings MC, Corley RP, Hewitt JK. Genetic and environmental vulnerabilities underlying adolescent substance use and problem use: general or specific? Behav Genet. 2006;36:603–615. doi: 10.1007/s10519-006-9066-7. [DOI] [PubMed] [Google Scholar]
- Zeiger JS, Haberstick BC, Schlaepfer I, Collins AC, Corley RP, Crowley TJ, Hewitt JK, Hopfer CJ, Lessem J, McQueen MB, Rhee SH, Ehringer MA. The neuronal nicotinic receptor subunit genes (CHRNA6 and CHRNB3) are associated with subjective responses to tobacco. Hum Mol Genet. 2008;17:724–734. doi: 10.1093/hmg/ddm344. [DOI] [PubMed] [Google Scholar]
- Zhang L, Cui X, Schmitt K, Hubert R, Navidi W, Arnheim N. Whole genome amplification from a single cell: implications for genetic analysis. Proc Natl Acad Sci U S A. 1992;89:5847–5851. doi: 10.1073/pnas.89.13.5847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q, Tang X, Zhang ZF, Velikina R, Shi S, Le AD. Nicotine induces hypoxia-inducible factor-1alpha expression in human lung cancer cells via nicotinic acetylcholine receptor-mediated signaling pathways. Clin Cancer Res. 2007;13:4686–4694. doi: 10.1158/1078-0432.CCR-06-2898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng S, Ma X, Buffler PA, Smith MT, Wiencke JK. Whole genome amplification increases the efficiency and validity of buccal cell genotyping in pediatric populations. Cancer Epidemiol Biomarkers Prev. 2001;10:697–700. [PubMed] [Google Scholar]


