Abstract
Heart formation requires a highly balanced network of transcriptional activation of genes. The homeodomain transcription factor, Shox2, is essential for the formation of the sinoatrial valves and for the development of the pacemaking system. The elucidation of molecular mechanisms underlying the development of pacemaker tissue has gained clinical interest as defects in its patterning can be related to atrial arrhythmias. We have analyzed putative targets of Shox2 and identified the Bmp4 gene as a direct target. Shox2 interacts directly with the Bmp4 promoter in chromatin immunoprecipitation assays and activates transcription in luciferase-reporter assays. In addition, ectopic expression of Shox2 in Xenopus embryos stimulates transcription of the Bmp4 gene, and silencing of Shox2 in cardiomyocytes leads to a reduction in the expression of Bmp4. In Tbx5del/+ mice, a model for Holt-Oram syndrome, and Shox2−/− mice, we show that the T-box transcription factor Tbx5 is a regulator of Shox2 expression in the inflow tract and that Bmp4 is regulated by Shox2 in this compartment of the embryonic heart. In addition, we could show that Tbx5 acts cooperatively with Nkx2.5 to regulate the expression of Shox2 and Bmp4. This work establishes a link between Tbx5, Shox2 and Bmp4 in the pacemaker region of the developing heart and thus contributes to the unraveling of the intricate interplay between the heart-specific transcriptional machinery and developmental signaling pathways.
INTRODUCTION
Heart formation requires the coordinated recruitment of various transcription factors including members of the T-box and homeobox-containing gene families (1). Different family members have been implicated in vertebrate heart tissue patterning and differentiation. In early stages of development, the heart is a slow conducting linear tube. During embryogenesis, the primitive tube develops into a synchronous and regular beating heart structure initiated by a small group of specialized cells forming the pacemaker region or sinoatrial node (SAN). Cells of the SAN locate at the junction of the right atrial wall and the superior caval vein and are spontaneously active (2,3). Yet, the exact molecular mechanisms underlying pacemaker potentials and its defects leading to arrhythmias have not been fully elucidated.
One key regulator of pacemaker differentiation is the homeobox transcription factor, Shox2. This gene has a highly restricted expression pattern in the sinus venosus myocardium comprising the SAN region (4,5). Knock-down and knock-out mouse models of Shox2 have demonstrated a crucial role of this gene in the development of heart and limb (4–7). Loss of Shox2 leads to embryonic lethality owing to heart defects occurring between E11.5 and E17.5 (4,7). Furthermore, Shox2 deficiency has been shown to lead to impaired pacemaking function in embryonic Zebrafish hearts and isolated hearts of Shox2 mutant mice have shown a slower heart beat rate (4,5). Very recently, Shox2 has been shown to be a direct target of Pitx2c, which is known to prevent susceptibility to atrial arrhythmias by inhibiting left-sided pacemaker specification (8). These findings have revealed a critical role for Shox2 in heart development and pacemaker function, but the detailed molecular mechanism of Shox2 function remains unclear. To investigate the transcriptional regulation of Shox2 and to identify putative target genes, we have used Xenopus and mouse as model systems. We provide genetic evidence for an epistatic relationship between Tbx5, Shox2 and Bmp4 in a very distinct region of the developing heart.
RESULTS
Shox2 regulates the expression of Bmp4
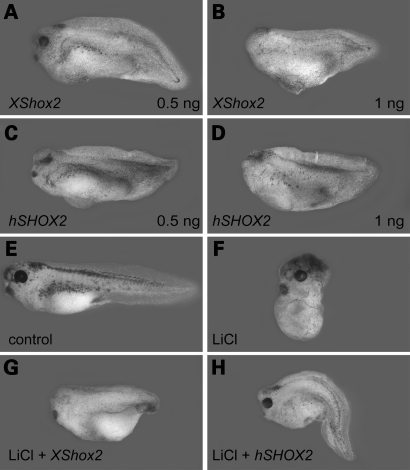
As a first step towards a functional analysis of Shox2, we cloned the full-length coding region of the Xenopus tropicalis orthologous gene. Sequence comparison between human and Xenopus Shox2 revealed an overall homology of 80% at the DNA and 85% at the amino acid level. Crucial functional features are conserved between the human and Xenopus Shox2 proteins including an identical OAR- (orthopedia, aristaless and rx homeoproteins) and homeodomain (Supplementary Material, Fig. S1). Shox2-specific transcripts can be first detected in stage 23 embryos and the expression level increases until stage 45 (Supplementary Material, Fig. S2A). Whole-mount in situ hybridization and following transverse sections of the heart anlage showed that Shox2 expression is restricted to the posterior domain of the myocardium (Supplementary Material, Fig. S2B–H), which continues to cover the dorsolateral aspect of the endocardium, anterior to its bifurcation in the sinus venosus. Endogenous Shox2 transcripts are not found in gastrula and early neurula Xenopus embryos, but when Shox2 mRNA was provided experimentally before gastrulation, defects were observed in embryonic patterning. Synthetic Shox2 mRNA was injected into each blastomere of 4-cell stage embryos. Subsequently, they were cultured until stage 36 and the dorso-anterior index (DAI) was determined. All embryos injected either with human or Xenopus Shox2 mRNA were ventro-posteriorized with DAI scores ranging from 2 to 4 (Fig. 1A–D, Supplementary Material, Table S1). The ventralized phenotype obtained by Shox2 injection was dose-dependent. Embryos injected with 0.5 ng of Shox2 mRNA exhibit reduced eyes and foreheads (Fig. 1A and C), whereas those that received higher doses (1 ng) were acephalic (Fig. 1B and D). These results indicate that ectopic Shox2 suppresses dorso-anterior structures and promotes ventro-posterior development. The ventralized phenotype observed in Xenopus embryos after injection of Shox2 RNA could be owing to the inhibition of early Wnt signaling, which also results in the loss of dorsal structures. Therefore, we analyzed the expression of the Wnt targets siamois (sia) and nodal-related-3 (Xnr3) at early gastrula stage after Shox2 injection. Whole-mount in situ hybridization for Xnr3 and quantitative reverse transcriptase–polymerase chain reaction (RT–PCR) for sia and Xnr3 demonstrated that expression of these Wnt targets was unaffected in Shox2-injected embryos (data not shown). This shows that ectopic Shox2 does not inhibit early Wnt-signaling.
Figure 1.
Ectopic Shox2 expression induces a ventralizing effect during early Xenopus development and rescues embryos partially dorsalized by LiCl. Lateral view of stage 36 embryos, radially injected at 4-cell stage with 0.5 ng (A, C) and 1 ng (B, D) Xenopus tropicalis Shox2 RNA (A, B) and human SHOX2a RNA (C, D). Shox2-injected embryos show a dose-dependent ventralizing effect. Uninjected stage 36 control embryo (E) and control embryo treated with 120 mM LiCl (F). Ventral injection of 1 ng Shox2 RNA rescues embryos partially dorsalized by LiCl (G, H).
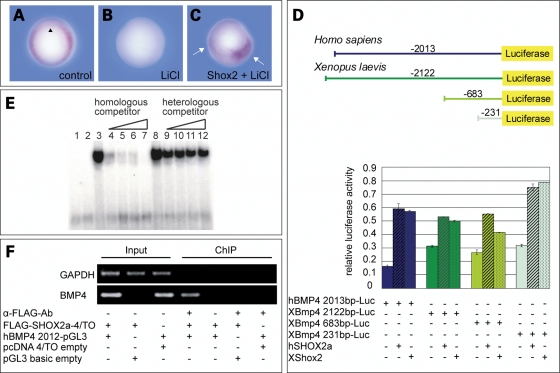
Bmp4 mediates dorsal–ventral patterning in Xenopus embryos by inhibiting differentiation of dorsal mesoderm and neural tissue. Since the phenotype in Shox2-injected embryos strongly resembled the effect of early ectopic Bmp4 expression in Xenopus, we speculated that Shox2 might induce Bmp4 transcription. We therefore inhibited zygotic transcription of Bmp4 by lithium chloride (LiCl) treatment. LiCl acts through inhibition of the glycogen synthase kinase-3β (GSK-3β), which allows activation of the Wnt/β-catenin signaling pathway and leads to dorsalization of the mesoderm (9). Induction of early Wnt signaling dorsalizes the mesoderm and the entire mesoderm acquires properties of the Spemann Organizer (dorsal mesoderm). In this situation, no Bmp4 transcripts are detected in the mesoderm. Thus, LiCl treatment inhibits ventro-posterior structures, resulting in a dorso-anteriorized phenotype with DAI scores >5 (Fig. 1F, Supplementary Material, Table S1). Ectopic expression of Shox2 antagonizes the dorsalized LiCl-phenotype and partially rescues posterior structures, suggesting an upregulation of Bmp4 by Shox2 (Fig. 1G and H, Supplementary Material, Table S1). To validate this hypothesis, we performed whole-mount in situ hybridization on Shox2-injected, LiCl-treated embryos. Bmp4-specific mRNA was almost undetectable in LiCl-treated embryos (Fig. 2B). In contrast, Shox2-injected, LiCl-treated embryos clearly showed an upregulation of Bmp4 expression in those cells derived from the Shox2-injected blastomeres (Fig. 2C). Since Shox2 acts as a transcription factor, a direct regulation of the Bmp4 promoter was possible. Comparative analysis of genomic Bmp4 sequences has indicated that exons 3, 4 and 5 of the human BMP4 gene correspond to exons 1, 2 and 3 of the orthologous Xenopus gene (Supplementary Material, Fig. S3). To identify specific DNA regulatory elements within the Bmp4 gene that are essential for the transactivation by Shox2, we used human and the corresponding Xenopus Bmp4 promoter-constructs and tested their activities by transient transfections of Cos-7 and HEK-293 cells in dual-luciferase-reporter assays. Co-transfections of the different Bmp4 reporter-constructs together with the expression plasmids for either human or X. tropicalis Shox2 led to an approximately 3-fold upregulation in luciferase activity, compared to cells transfected only with the corresponding Bmp4 reporter-constructs (Fig. 2D). By transfection of serial gene deletion-constructs, we have narrowed down the Shox2-responsive element of the Bmp4 promoter to an interval of 231 bp upstream of the mRNA start site. Within this 231 bp fragment, a 34 bp sequence motif, highly conserved between human, mouse and Xenopus was identified (Supplementary Material, Fig. S4). Electrophoretic mobility shift assay (EMSA) competitive experiments showed that a SHOX2–GST fusion protein could bind to this 34 bp BMP4-sequence motif with high affinity and was competed away by an excess of the unlabeled BMP4 oligonucleotide (Fig. 2E). A non-specific random oligonucleotide sequence did not affect the binding of the SHOX2–GST to the BMP4 oligonucleotide.
Figure 2.
Shox2 regulates the expression of Bmp4. (A) Bmp4 expression in uninjected embryos at stage 10.5 (n=35). Black arrowhead indicates the dorsal lip. (B) Bmp4 expression is reduced or absent in 87% of LiCl-treated embryos (n=31). (C) Spots of Bmp4 expression in 77% of previously Shox2-injected, LiCl-treated embryos (n=30). Shox2 RNA was injected diagonally into two blastomers of a 4-cell stage embryo. Bmp4 staining in two areas of the embryo (marked by white arrows) indicates an upregulation of Bmp4 expression corresponding to the Shox2 injection. (D) Shox2 increases the activity of the human and Xenopus laevis Bmp4 promoter upon co-transfection of Shox2 expression plasmids (1 µg) and the indicated Bmp4 reporter-constructs (1 µg) into Cos-7 cells. Data using HEK-293 cells are not shown. (E) Electrophoretic mobility shift assay of the GST–SHOX2 fusion protein on a conserved promoter oligonucleotide sequence (BMP4). An excess of non-radioactively labelled oligonucleotides (homologous competitor) reduces the DNA-binding ability of SHOX2, whereas excess of a random oligonucleotide sequence (heterologous competitor) does not affect DNA-binding. 1, free oligonucleotide; 2, GST alone; 3, GST–SHOX2; 4–7, 10-fold, 50-fold, 75-fold and 150-fold molar excess of homologous competitor; 8, GST–SHOX2; 9–12, 10-fold, 50-fold, 75-fold and 150-fold molar excess of heterologous competitor. (F) Chromatin immunoprecipitation (ChIP) assay. HEK-293 cells were co-transfected with the FLAG-SHOX2a expression or empty control vector and with either the hBMP4 2013-Luc reporter or empty control vector as indicated. Formaldehyde-crosslinked DNA was immunoprecipitated using an anti-FLAG antibody (α-FLAG-Ab) or no antibody, as a negative control. Precipitated DNA fragments (ChIP) and DNA from lysate before immunoprecipitation (Input) were subjected to PCR using primer sets amplifying the putative SHOX2-binding element in the BMP4 gene (lower panel) or GAPDH (upper panel) as a control.
To further examine the SHOX2 binding to the BMP4 promoter in vivo, we performed a Chromatin immunoprecipitation (ChIP) assay (10). An anti-FLAG antibody was used to immunoprecipitate chromatin from HEK-293 cells transiently co-transfected with the FLAG-tagged SHOX2a expression plasmid and the hBMP4 2013-Luc reporter-construct containing the putative SHOX2-binding site of the BMP4 promoter. The binding of SHOX2 to the BMP4 promoter was detected by PCR analysis using specific primers spanning the putative SHOX2-binding site within the BMP4 promoter. We obtained PCR products in the sample that was immunoprecipitated with the anti-FLAG antibody as well as in the input DNA, whereas the negative controls (no antibody control, transfections with empty vectors) failed to yield a product (Fig. 2F). Identical results were also obtained using Xenopus FLAG-tagged Shox2 and XBmp4 231 bp-Luc-constructs (data not shown). Taken together, these results indicate that SHOX2 binds specifically to a 34 bp conserved DNA interval in the Bmp4 promoter in vitro and in vivo.
Tbx5 acts upstream of Shox2 and Bmp4 in the inflow tract of the developing heart
Mutations in the T-box transcription factor gene TBX5 cause heart and limb malformations in the Holt-Oram syndrome (11,12). Because of a common heart and limb defect pattern between Tbx5 (13) and Shox2, we speculated on a possible epistatic relationship between Tbx5, Shox2 and Bmp4 in the developing heart. We have carried out two independent sets of experiments to assess this hypothesis.
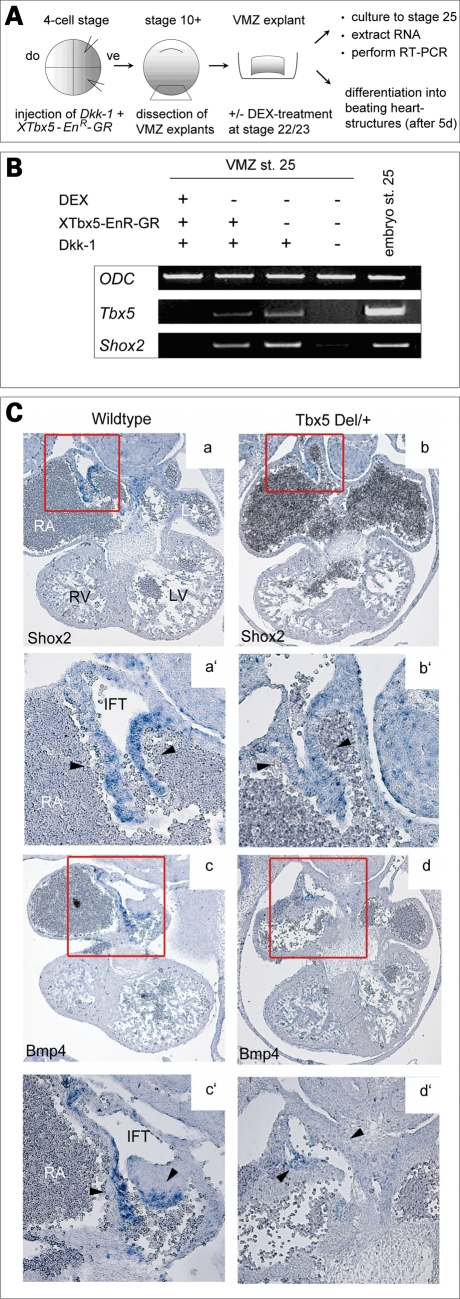
To study Tbx5-dependent Shox2 expression, we used experimentally induced heart structures in Xenopus. Injection of the Wnt antagonist Dickkopf-1 (Dkk-1) into Xenopus embryos can induce heart differentiation in explants of ventral marginal zone (VMZ) mesoderm (14). ‘In vitro hearts’ only form when Wnt is inhibited. Thus, we injected Dkk-1 mRNA together with an inducible Tbx5 Engrailed-repressor glucocorticoid-receptor fusion protein (XTbx5-EnR-GR), which blocks endogenous Tbx5 activity, into ventral blastomeres of early Xenopus embryos. XTbx5-EnR-GR was activated by dexamethasone (DEX) treatment in Dkk-1-injected VMZ explants at later embryonic stage 22/23 to avoid complete loss of heart tissue. In such samples, the heart marker Troponin is expressed, indicating that heart tissue is present under these experimental conditions (data not shown). Remarkably, RT–PCR analysis of the explants at stage 25 revealed that XTbx5-EnR-GR completely blocks Shox2 expression in Dkk-1-injected VMZ explants (Fig. 3A and B). These results indicate that Tbx5 function is critical for the expression of Shox2 in cardiac tissues.
Figure 3.
Shox2 mediates Tbx5 expression to Bmp4 signaling in the developing heart. (A) Scheme of the ventral marginal zone (VMZ) experiment. Xenopus embryos were injected (vegetal dorsal) with 1 ng Dkk-1 mRNA and 0.2 ng of XTbx5 EnR-GR DNA into two blastomeres at the 4-cell stage and VMZ explants were dissected at early gastrula stage. Explants were cultured until stage 22/23 and then treated with 0.5 µM dexamethasone (DEX)-solution to activate the injected repressor-construct. RNA was extracted at stage 25 and analysed by RT–RCR for the presence of XTbx5 and XShox2. (B) RT–PCR shows that Dkk-1 induces Tbx5 and Shox2 expression in VMZ explants. XTbx5 EnR-GR abolishes Dkk-1 induced Tbx5 and Shox2 expression after DEX-treatment. Ornithine decarboxylase was used as a loading control. (C) Section in situ hybridization on E11.5 wild-type and Tbx5del/+ mouse hearts. Murine Shox2 is strongly expressed in the inflow tract of the developing heart in wild-type embryos (a), which is markedly decreased in Tbx5del/+ embryos (b). Murine Bmp4 expression overlaps with the Shox2 expression domain in wild-type hearts (c) and is almost absent in Tbx5del/+ embryos (d). RA, right atrium; LA, left atrium; RV, right ventricle; LV, left ventricle.
Using mouse as a model, we found that expression of Shox2 and Bmp4 overlaps in the inflow tract (IFT) of the developing heart (Fig. 3Ca and c). This is consistent with the data published by Norden et al. (15). To assess the relationship between Tbx5, Shox2 and Bmp4, we have analyzed mice with a heterozygous deletion of the Tbx5 gene (Tbx5del/+). Homozygous Tbx5del/del mice do not survive past E10.5 (13). We investigated whether expression of Shox2 or Bmp4 in the IFT of the right atrium relies on Tbx5 function in a dose-dependent manner. In situ hybridization on histological sections of embryonic hearts demonstrated reduced Shox2 expression in the IFT of the Tbx5del/+ mutant mice at E11.5 (Fig. 3Ca and b). Similarly, Bmp4 expression, which overlaps with the Shox2 expression domain, is strongly reduced and almost absent in this distinct heart region of Tbx5del/+ embryos (Fig. 3Cc and d). This is consistent with data from the microarray experiments, confirmed by quantitative RT–PCR, which suggested Shox2 as one of the roughly 100 putatively downregulated genes in Tbx5-deficient mouse hearts (16). Other genes important for normal development and function of the conduction system, like α1D L-type calcium channel, Cx43 and Tbx3, were either normally expressed, or in the case of Tbx3, downregulated but still expressed in Tbx5del/+ hearts (13,16). Hence, we can state that the strongly reduced expression of Shox2 and Bmp4 in Tbx5del/+ hearts is not owing to incorrect formation of the IFT.
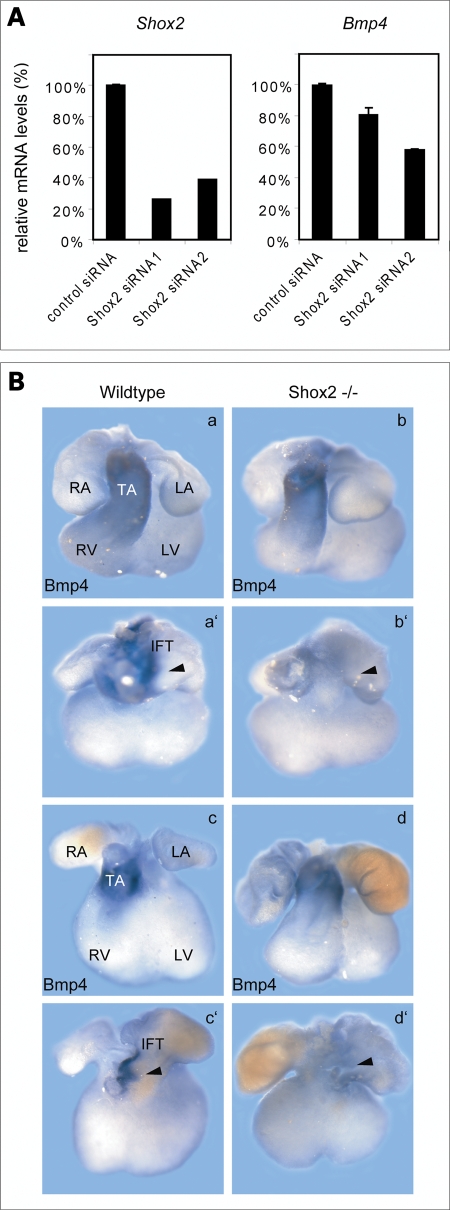
We next performed gene-specific silencing of Shox2 expression in neonatal rat cardiomyocytes by RNAi. H10 cells were transiently transfected with two different siRNAs directed against the Shox2 cDNA with a knock-down efficiency of 74% and 61% (Fig. 4A). We could also show that Shox2 silencing leads to a reduction in Bmp4 expression (Fig. 4A).
Figure 4.
Shox2 deficiency impairs Bmp4 expression. (A) H10 cells were transfected with two different Shox2 siRNAs (Shox2 siRNA1 and Shox2 siRNA2) in parallel with a control siRNA. Expression levels of Shox2 (left panel) and Bmp4 (right panel) were assessed 24 h after transfection by qRT–PCR analysis. siRNA-mediated knock-down of Shox2 results in 19–42% reduction of Bmp4 mRNA levels. All results were normalized to Hprt1 (hypoxanthine phosphoribosyltransferase 1) mRNA values. (B) Whole-mount in situ hybridization on E11.5 (a, b) and E12.5 (c, d) wild-type and Shox2−/− mouse hearts using a Bmp4 RNA probe. Both, ventral (a–d) and dorsal (a′–d′) views are shown. Murine Bmp4 is strongly expressed in the truncus arteriosus (TA) (a, c) and in the inflow tract (a′, c′) of the developing heart in wild-type embryos. In the Shox2−/− mouse hearts, Bmp4 expression is still present in the truncus arteriosus (b, d) but completely absent in the IFT (b′, d′), where Shox2 and Bmp4 expression domains overlap in the wild-type heart. RA, right atrium; LA, left atrium; RV, right ventricle; LV, left ventricle.
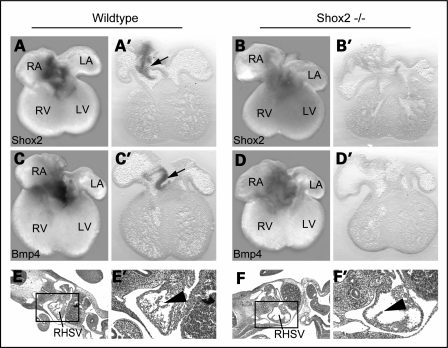
We also carried out whole-mount in situ hybridization on Shox2−/− embryonic mouse hearts (4) to confirm that Bmp4 expression depends on Shox2 function in the IFT of the developing heart. These stainings revealed that Bmp4 is completely absent in the IFT of Shox2-deficient mice, whereas a strong expression in the truncus arteriosus is still detectable (Fig. 4B). Shox2 and Bmp4 stained Shox2−/− hearts, as well as histological sections of Shox2−/− embryos, clearly showed that the IFT in Shox2-deficient mice develops properly, even if the venous valve formation is abnormal (Fig. 5). We also carried out in situ hybridization using Tbx3 and Hcn4 as markers for the pacemaker region and can show that Tbx3 and Hcn4 are downregulated but still present in Shox2−/− hearts (data not shown). These data are consistent with a recent study investigating the role of Shox2 by expression analysis of different key regulators in SAN development (5). Thus, we demonstrate that the absence of Bmp4 expression in the IFT of Shox2−/− hearts is not owing to incorrect formation of this tissue. Taken together, these data strongly suggest that Tbx5 acts upstream of Shox2, which, in turn, activates Bmp4 in the IFT of the heart.
Figure 5.
Bmp4 expression is absent in the inflow tract (IFT) of Shox2−/− embryos. Whole-mount in situ hybridization with corresponding sections (50 µm) on E12.5 wild-type and Shox2−/− mouse hearts using Shox2 (A, B) and Bmp4 (C, D) RNA probes. Murine Shox2 (A, A′) and Bmp4 (C, C′) are expressed in the IFT of wild-type hearts (indicated by black arrows), but are completely absent in the IFT of Shox2−/− hearts (B, B′, D, D′). Note that IFT tissue is still present in Shox2−/− hearts (B′, D′). Histological sections of wild-type (E, E′) and Shox2−/− (F, F′) embryos at E10.5. Squares in E and F show the right horn of the sinus venosus that is magnified in E′ and F′. Shox2-deficient embryos develop IFT tissue (F, F′), even if the venous valve formation is abnormal (black arrowhead in F′). RA, right atrium; LA, left atrium; RV, right ventricle; LV, left ventricle; RHSV, right horn of the sinus venosus.
DISCUSSION
Shox2 encodes a homeodomain transcription factor that among other features plays a critical role in heart development (4,5,17). The aim of our study was to understand the molecular mechanisms underlying Shox2 expression in the developing heart. Our data show that the transformation growth factor-β, Bmp4, is a direct target of Shox2. In contrast to the repressive effect of Shox2 on Bmp4 expression during limb development (7), we could show that in the heart, Shox2 activates Bmp4. Several lines of evidence support this conclusion. We demonstrate that ectopic expression of Shox2 in Xenopus embryos stimulates transcription of the Bmp4 gene and silencing of Shox2 in cardiomyocytes leads to a reduction in Bmp4 expression. In addition, Shox2 activates Bmp4 transcription in luciferase-reporter assays. We also show by EMSA and ChIP assays that Shox2 binds specifically to a 34 bp AT-rich sequence within the Bmp4 promoter region. Bmp4 thus represents the first gene identified as a direct Shox2 target. In loss-of-function experiments using Xenopus embryos, as well as Tbx5del/+ and Shox2−/− mice, we show that the T-box transcription factor Tbx5 is required for Shox2 expression in the IFT and that Bmp4 is regulated by Shox2 in this compartment of the embryonic heart.
Bmp4 is known to exert numerous developmental functions including different roles in cardiogenesis. Homozygosity for a null mutation, for example, leads to severe looping defects and embryonic death at E9.5 or earlier (18,19). The analysis of a hypomorphic allele has demonstrated additional later functions in atrioventricular septation, valvulogenesis and formation of the outflow tract (20,21). Both Shox2 and Bmp4 are expressed already strongly at E9.5, prior to the disturbed development in the Shox2 mutant. At this time, incorporation of the sinus venosus into the right atrium takes place and at the venous pole new myocardium is still added to the sinus venosus (22). Most probably, both processes are disturbed and contribute to the abnormalities seen in Shox2−/− embryos. Abnormal Bmp4 expression has also been reported to result in abnormal myofibrillogenesis of the precardiac mesoderm (23), which also might contribute to embryonic heart failure in Shox2-deficient embryos. The essential cardiogenic functions previously attributed to Bmp4 pertain to the atrioventricular and semilunar valves that, in contrast to the venous valves of the IFT, develop from endocardial cushions. Our analyses suggest a link between Bmp4 expression and early sinoatrial development and venous valve formation.
It was also reported that expression of Bmp4 is dysregulated in the developing heart of the zebrafish heartstrings (hst) mutant with Tbx5 deficiency (24). Although this hst mutation does not perturb atrioventricular patterning as described for other species with Tbx5 mutations, it impairs pacemaking functions, similar to that observed in Shox2 knock-down experiments with morpholino-modified antisense oligonucleotides (4,24). These data suggest that the hst mutation affecting the cardiac conduction system may also regulate the expression of downstream genes such as, for example, Shox2 and Bmp4. Overall, our data indicate that in specific structures and during different time points, Bmp4 also can act downstream of Tbx5.
Tbx5 overexpression experiments in Xenopus animal cap (AC) and dorsal marginal zone (DMZ) explants do not provide support for the regulation of Shox2 by Tbx5 alone (data not shown), suggesting that Tbx5 affects Shox2 expression also through other factors not present in the AC and DMZ explants. Previous studies have demonstrated that Tbx5 directly associates with co-factors, such as the homeodomain transcription factor Nkx2.5 and the zinc finger factor Gata4 to induce cardiac-specific genes like Nppa and Gja5 (13,25,26). Moreover, it has been shown that co-activators such as Baf60c or TAZ and YAP are important for the control of Tbx5-dependent transcription in cardiac development (27,28). Very recent work has also demonstrated that Baf60c and Gata4 cooperate with Tbx5 to set-up a cardiac gene expression program, including Nkx2.5, which transforms non-cardiac embryonic mesoderm into beating heart cells (29). It cannot be excluded that Shox2 may play a role in these processes too.
An involvement of further co-factors in cardiac development in the described epistatic relationship between Tbx5, Shox2 and Bmp4 can be assumed. In contrast to the highly restricted expression pattern of Shox2 in the sinus venosus myocardium, Tbx5 and Bmp4 are widely expressed during embryonic heart development (4,17,30,31). Owing to the fact that the overlapping expression domains of these three genes are confined to a restricted region of the IFT of the heart, it is likely that additional as yet unknown factors may contribute to control the transcriptional readout in other regions of the developing heart. To find out if heart-specific co-factors of Tbx5, such as Nkx2.5 contribute to the regulation of Shox2, we analyzed the SHOX2 promoter for TBX5 and NKX2.5 binding sites, and three putative TBX5 and three NKX2.5 binding sites upstream of the SHOX2 start codon could be identified. In luciferase-reporter assays using a SHOX2 reporter-construct containing all putative binding sites, we could demonstrate that TBX5 acts cooperatively with NKX2.5 to activate SHOX2 expression. In contrast, TBX5 alone is not able to activate the SHOX2 promoter. The same effect could be shown by using the BMP4 reporter-construct (Supplementary Material, Fig. S5). Our data indicate that Nkx2.5 acts upstream of Shox2. The findings of Espinoza-Lewis et al. (5) have shown that Shox2 can also inhibit the expression of Nkx2.5, arguing for a regulatory feedback-loop between Shox2 and Nkx2.5. These novel findings support the idea that a cardiac-specific molecular environment is necessary for Tbx5 to regulate Shox2.
In summary, we have demonstrated a previously undescribed connection of Tbx5, Shox2 and Bmp4 in a distinct subregion of the developing heart. Our results suggest that Tbx5 signaling in the IFT of the heart directly or indirectly controls the expression of the transcription factor Shox2, which regulates Bmp4 in a direct manner. Taken into account the crucial role of Shox2 in pacemaking function, an investigation in patients with atrial arrhythmia of unknown molecular cause may be of interest. Indeed, the recent genetic linkage of Tbx5 and Nkx2.5 to atrial fibrillation suggests that this pathway may be of relevance in the general population (32,33).
MATERIALS AND METHODS
Generation of plasmid-constructs
Cloning of the Xenopus Shox2 cDNA, generation of Xenopus Shox2, human SHOX2 and NKX2.5 expression-constructs, and BMP4 and SHOX2 promoter-constructs are described in the Supplementary Material.
Egg and embryo manipulations
Ovulation, in vitro fertilization, embryo culture and manipulation were performed as previously described (34). LiCl treatment was carried out as described (35).
mRNA synthesis
Capped mRNAs were synthesized from linearized plasmids using mMessage mMachine Kit (Ambion). pCS2+-SHOX2a was linearized with ApaI, pCS2+-Xtropicalis Shox2 and pCS2+-Dkk-1 were linearized with NotI. mRNAs were synthesized using SP6 RNA polymerase.
Semi-quantitative and quantitative RT–PCR
For embryos, total RNA was prepared with Trizol® reagent (Invitrogen) and for transfected H10 cells with the RNeasy®Mini Kit (Qiagen) with subsequent DNase I treatment. cDNA was synthesized using the SuperScriptTM First-Strand Synthesis System for RT–PCR (Invitrogen). Semi-quantitative PCR was performed using standard conditions. Quantitative RT–PCR (qRT–PCR) was performed on a 7500 Fast Real-Time PCR System (Applied Biosystems) using SYBR Green ROX dye (Thermo Scientific). Primer sets used are presented in Supplementary Material, Table S2.
Xenopus in situ hybridization
Whole-mount in situ hybridization and antisense probe preparation was carried out as described (36). Digoxigenin-labelled antisense RNA was synthesized from the plasmids pTOPO-X.trop-Shox2 (linearized with HindIII) and pBSK/B12-Bmp4 (35) (linearized with EcoRI) using T7 RNA polymerase.
Cell culture, transfection and luciferase assay
Cos-7 and HEK-293 cells were cultured at 37°C in Dulbecco's Modified Eagle Medium (DMEM) medium containing high glucose, supplemented with 10% fetal calf serum and antibiotics. Neonatal rat heart myocytes, immortalized with a temperature-sensitive SV40T antigen (H10 cells), were cultured at 33°C in DMEM medium (see above). For luciferase assay analysis, the cells were co-transfected in triplicates with different constructs using polyethylenimine (Sigma-Aldrich). Forty-eight hours after transfection, luciferase activity was determined and normalized to Renilla luciferase activity with a dual-luciferase assay kit (Promega). Experiments were repeated at least three times in triplicates with consistent results and representative data were shown. Knock-down experiments were performed using Stealth RNAi Duplex (Invitrogen), directed against the Shox2 cDNA (Shox2 siRNA1: 5′-GGACCAATTTCACCCTGGAACAACT-3′, Shox2 siRNA2: 5′-GCAAGGACTCCAGCATCGCCGATCT-3′). H10 cells were transiently transfected with 100 pmol siRNA in 12-well plates using Lipofectamine 2000 (Invitrogen) and the relative knock-down efficiency was determined by qRT–PCR 24 h after transfection. Stealth RNAi negative control GC high was used as a control siRNA. Representative data from three independent experiments with consistent results are shown.
Electrophoretic mobility shift assay
EMSA was performed using standard protocols. For the binding reaction, 32P-labeled, double-stranded BMP4 oligonucleotides (Supplementary Material, Table S2) were used together with purified, bacterially expressed recombinant GST–SHOX2 protein.
Chromatin Immunoprecipitation assay
HEK-293 cells were transiently co-transfected with hBMP4 2013-Luc reporter or empty control vector and with either the FLAG-SHOX2a expression vector or empty control vector using Lipofectamine 2000 transfection reagent (Invitrogen). Cells were cultured for 48 h and the ChIP assay carried out as described by Abcam protocols using 10 µg of anti-FLAG antibody (M2, Sigma-Aldrich). PCR analysis of immunoprecipitated DNA and input DNA was performed under standard conditions using primer sets amplifying the putative SHOX2-binding element in the BMP4 gene (−1241 to −1082 bp region) or GAPDH as control (Supplementary Material, Table S2).
Mouse whole-mount and section in situ hybridization
Shox2−/− and Tbx5del/+ mice were described previously (4,13). Whole-mount in situ hybridization was performed as previously reported. Digoxigenin-labelled antisense RNA was synthesized from the plasmids pCR-S-Og12 (Shox2) (17) (linearized with SacI) and pSP72-Bmp4 (linearized with AccI) using T7 RNA polymerase. Section in situ hybridization was performed on 10 µm paraffin sections using standard protocols. The murine Shox2 antisense probe was made from a cDNA clone ordered from Open Biosystems (Huntsville, USA; Cat no. EMM1032-595060) using T7 polymerase after linearizing with EcoRI. The Bmp4 antisense probe was synthesized from pSP72-Bmp4 (see above).
SUPPLEMENTARY MATERIAL
FUNDING
This work was supported by the Deutsche Forschungsgemeinschaft grant no. BL 372/4-2. Funding to pay the Open Access Charge was provided by the Medical Faculty Heidelberg.
Supplementary Material
ACKNOWLEDGEMENTS
This study is dedicated to Rüdiger Blaschke. We thank A. Gittenberger-de Groot for reading the manuscript, A. Schweikert for assistance on sectioning, G. Thomsen for providing the X. laevis Tbx5-construct, W. Knöchel for the X. laevis Bmp4-promoter plasmids, C. Niehrs for the Dkk-1 plasmid, H.-J. Hippe for the H10 cell line and B. Hogan for the murine Bmp4 in situ probe.
Conflict of Interest statement. None declared.
References
- 1.Bruneau B.G. The developmental genetics of congenital heart disease. Nature. 2008;451:943–948. doi: 10.1038/nature06801. [DOI] [PubMed] [Google Scholar]
- 2.Boyett M.R., Honjo H., Kodama I. The sinoatrial node, a heterogeneous pacemaker structure. Cardiovasc. Res. 2000;47:658–687. doi: 10.1016/s0008-6363(00)00135-8. [DOI] [PubMed] [Google Scholar]
- 3.Christoffels V.M., Smits G.J., Kispert A., Moorman A.F. Development of the pacemaker tissues of the heart. Circ. Res. 2010;106:240–254. doi: 10.1161/CIRCRESAHA.109.205419. [DOI] [PubMed] [Google Scholar]
- 4.Blaschke R.J., Hahurij N.D., Kuijper S., Just S., Wisse L.J., Deissler K., Maxelon T., Anastassiadis K., Spitzer J., Hardt S.E., et al. Targeted mutation reveals essential functions of the homeodomain transcription factor Shox2 in sinoatrial and pacemaking development. Circulation. 2007;115:1830–1838. doi: 10.1161/CIRCULATIONAHA.106.637819. [DOI] [PubMed] [Google Scholar]
- 5.Espinoza-Lewis R.A., Yu L., He F., Liu H., Tang R., Shi J., Sun X., Martin J.F., Wang D., Yang J., et al. Shox2 is essential for the differentiation of cardiac pacemaker cells by repressing Nkx2–5. Dev. Biol. 2009;327:376–385. doi: 10.1016/j.ydbio.2008.12.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cobb J., Dierich A., Huss-Garcia Y., Duboule D. A mouse model for human short-stature syndromes identifies Shox2 as an upstream regulator of Runx2 during long-bone development. Proc. Natl Acad. Sci. USA. 2006;103:4511–4515. doi: 10.1073/pnas.0510544103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yu L., Liu H., Yan M., Yang J., Long F., Muneoka K., Chen Y. Shox2 is required for chondrocyte proliferation and maturation in proximal limb skeleton. Dev. Biol. 2007;306:549–559. doi: 10.1016/j.ydbio.2007.03.518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang J., Klysik E., Sood S., Johnson R.L., Wehrens X.H., Martin J.F. Pitx2 prevents susceptibility to atrial arrhythmias by inhibiting left-sided pacemaker specification. Proc. Natl Acad. Sci. USA. 2010;107:9753–9758. doi: 10.1073/pnas.0912585107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Klein P.S., Melton D.A. A molecular mechanism for the effect of lithium on development. Proc. Natl Acad. Sci. USA. 1996;93:8455–8459. doi: 10.1073/pnas.93.16.8455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wells J., Farnham P.J. Characterizing transcription factor binding sites using formaldehyde crosslinking and immunoprecipitation. Methods. 2002;26:48–56. doi: 10.1016/S1046-2023(02)00007-5. [DOI] [PubMed] [Google Scholar]
- 11.Basson C.T., Bachinsky D.R., Lin R.C., Levi T., Elkins J.A., Soults J., Grayzel D., Kroumpouzou E., Traill T.A., Leblanc-Straceski J., et al. Mutations in human TBX5 [corrected] cause limb and cardiac malformation in Holt-Oram syndrome. Nat. Genet. 1997;15:30–35. doi: 10.1038/ng0197-30. [DOI] [PubMed] [Google Scholar]
- 12.Li Q.Y., Newbury-Ecob R.A., Terrett J.A., Wilson D.I., Curtis A.R., Yi C.H., Gebuhr T., Bullen P.J., Robson S.C., Strachan T., et al. Holt-Oram syndrome is caused by mutations in TBX5, a member of the Brachyury (T) gene family. Nat. Genet. 1997;15:21–29. doi: 10.1038/ng0197-21. [DOI] [PubMed] [Google Scholar]
- 13.Bruneau B.G., Nemer G., Schmitt J.P., Charron F., Robitaille L., Caron S., Conner D.A., Gessler M., Nemer M., Seidman C.E., et al. A murine model of Holt-Oram syndrome defines roles of the T-box transcription factor Tbx5 in cardiogenesis and disease. Cell. 2001;106:709–721. doi: 10.1016/s0092-8674(01)00493-7. [DOI] [PubMed] [Google Scholar]
- 14.Schneider V.A., Mercola M. Wnt antagonism initiates cardiogenesis in Xenopus laevis. Genes Dev. 2001;15:304–315. doi: 10.1101/gad.855601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Norden J., Grieskamp T., Lausch E., van Wijk B., van den Hoff M.J., Englert C., Petry M., Mommersteeg M.T., Christoffels V.M.,, Niederreither K., et al. Wt1 and retinoic acid signaling in the subcoelomic mesenchyme control the development of the pleuropericardial membranes and the sinus horns. Circ. Res. 2010;106:1212–1220. doi: 10.1161/CIRCRESAHA.110.217455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mori A.D., Zhu Y., Vahora I., Nieman B., Koshiba-Takeuchi K., Davidson L., Pizard A., Seidman J.G., Seidman C.E., Chen X.J., et al. Tbx5-dependent rheostatic control of cardiac gene expression and morphogenesis. Dev. Biol. 2006;297:566–586. doi: 10.1016/j.ydbio.2006.05.023. [DOI] [PubMed] [Google Scholar]
- 17.Blaschke R.J., Monaghan A.P., Schiller S., Schechinger B., Rao E., Padilla-Nash H., Ried T., Rappold G.A. SHOT, a SHOX-related homeobox gene, is implicated in craniofacial, brain, heart, and limb development. Proc. Natl Acad. Sci. USA. 1998;95:2406–2411. doi: 10.1073/pnas.95.5.2406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lawson K.A., Dunn N.R., Roelen B.A., Zeinstra L.M., Davis A.M., Wright C.V., Korving J.P., Hogan B.L. Bmp4 is required for the generation of primordial germ cells in the mouse embryo. Genes Dev. 1999;13:424–436. doi: 10.1101/gad.13.4.424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fujiwara T., Dehart D.B., Sulik K.K., Hogan B.L. Distinct requirements for extra-embryonic and embryonic bone morphogenetic protein 4 in the formation of the node and primitive streak and coordination of left-right asymmetry in the mouse. Development. 2002;129:4685–4696. doi: 10.1242/dev.129.20.4685. [DOI] [PubMed] [Google Scholar]
- 20.Jiao K., Kulessa H., Tompkins K., Zhou Y., Batts L., Baldwin H.S., Hogan B.L. An essential role of Bmp4 in the atrioventricular septation of the mouse heart. Genes Dev. 2003;17:2362–2367. doi: 10.1101/gad.1124803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu W., Selever J., Wang D., Lu M.F., Moses K.A., Schwartz R.J., Martin J.F. Bmp4 signaling is required for outflow-tract septation and branchial-arch artery remodeling. Proc. Natl Acad. Sci. USA. 2004;101:4489–4494. doi: 10.1073/pnas.0308466101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.De la Cruz M.V., Sánchez-Gómez G., Palomino M.A. The primitive cardiac regions in the straight tube heart (Stage 9) and their anatomical expression in the mature heart: an experimental study in the chick embryo. J. Anat. 1989;165:121–131. [PMC free article] [PubMed] [Google Scholar]
- 23.Nakajima Y., Yamagishi T., Ando K., Nakamura H. Significance of bone morphogenetic protein-4 function in the initial myofibrillogenesis of chick cardiogenesis. Dev. Biol. 2002;245:291–303. doi: 10.1006/dbio.2002.0637. [DOI] [PubMed] [Google Scholar]
- 24.Garrity D.M., Childs S., Fishman M.C. The heartstrings mutation in zebrafish causes heart/fin Tbx5 deficiency syndrome. Development. 2002;129:4635–4645. doi: 10.1242/dev.129.19.4635. [DOI] [PubMed] [Google Scholar]
- 25.Hiroi Y., Kudoh S., Monzen K., Ikeda Y., Yazaki Y., Nagai R., Komuro I. Tbx5 associates with Nkx2–5 and synergistically promotes cardiomyocyte differentiation. Nat. Genet. 2001;28:276–280. doi: 10.1038/90123. [DOI] [PubMed] [Google Scholar]
- 26.Garg V., Kathiriya I.S., Barnes R., Schluterman M.K., King I.N., Butler C.A., Rothrock C.R., Eapen R.S., Hirayama-Yamada K., Joo K., et al. GATA4 mutations cause human congenital heart defects and reveal an interaction with TBX5. Nature. 2003;424:443–447. doi: 10.1038/nature01827. [DOI] [PubMed] [Google Scholar]
- 27.Murakami M., Nakagawa M., Olson E.N., Nakagawa O. A WW domain protein TAZ is a critical coactivator for TBX5, a transcription factor implicated in Holt-Oram syndrome. Proc. Natl Acad. Sci. USA. 2005;102:18034–18039. doi: 10.1073/pnas.0509109102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lickert H., Takeuchi J.K., Von Both I., Walls J.R., McAuliffe F., Adamson S.L., Henkelman R.M., Wrana J.L., Rossant J., Bruneau B.G. Baf60c is essential for function of BAF chromatin remodelling complexes in heart development. Nature. 2004;432:107–112. doi: 10.1038/nature03071. [DOI] [PubMed] [Google Scholar]
- 29.Takeuchi J.K., Bruneau B.G. Directed transdifferentiation of mouse mesoderm to heart tissue by defined factors. Nature. 2009;459:708–711. doi: 10.1038/nature08039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Abdelwahid E., Rice D., Pelliniemi L.J., Jokinen E. Overlapping and differential localization of Bmp-2, Bmp-4, Msx-2 and apoptosis in the endocardial cushion and adjacent tissues of the developing mouse heart. Cell Tissue Res. 2001;305:67–78. doi: 10.1007/s004410100399. [DOI] [PubMed] [Google Scholar]
- 31.Chapman D.L., Garvey N., Hancock S., Alexiou M., Agulnik S.I., Gibson-Brown J.J., Cebra-Thomas J., Bollag R.J., Silver L.M., Papaioannou V.E. Expression of the T-box family genes, Tbx1–Tbx5, during early mouse development. Dev. Dyn. 1996;206:379–390. doi: 10.1002/(SICI)1097-0177(199608)206:4<379::AID-AJA4>3.0.CO;2-F. [DOI] [PubMed] [Google Scholar]
- 32.Holm H., Gudbjartsson D.F., Arnar D.O., Thorleifsson G., Thorgeirsson G., Stefansdottir H., Gudjonsson S.A., Jonasdottir A., Mathiesen E.B., Njolstad I., et al. Several common variants modulate heart rate, PR interval and QRS duration. Nat. Genet. 2010;42:117–122. doi: 10.1038/ng.511. [DOI] [PubMed] [Google Scholar]
- 33.Pfeufer A., van Noord C., Marciante K.D., Arking D.E., Larson M.G., Smith A.V., Tarasov K.V., Muller M., Sotoodehnia N., Sinner M.F., et al. Genome-wide association study of PR interval. Nat. Genet. 2010;42:153–159. doi: 10.1038/ng.517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Swain R.K., Katoh M., Medina A., Steinbeisser H. Xenopus frizzled-4S, a splicing variant of Xfz4 is a context-dependent activator and inhibitor of Wnt/beta-catenin signaling. Cell Commun. Signal. 2005;3:12. doi: 10.1186/1478-811X-3-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fainsod A., Steinbeisser H., De Robertis E.M. On the function of BMP-4 in patterning the marginal zone of the Xenopus embryo. EMBO J. 1994;13:5015–5025. doi: 10.1002/j.1460-2075.1994.tb06830.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Harland R.M. In situ hybridization: an improved whole-mount method for Xenopus embryos. Methods Cell Biol. 1991;36:685–695. doi: 10.1016/s0091-679x(08)60307-6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.