Abstract
One-hundred sixty-eight group B streptococcal (GBS) isolates from a Brazilian hospital were phenotypically and genotypically characterized. Isolates were recovered from human sources from April 2006 to May 2008 and classified as either invasive, noninvasive, or colonizing isolates. Classical methods for serotyping and antibiotic resistance profiling were employed. Clonal groups were also defined by pulsed-field gel electrophoresis (PFGE). Results showed that susceptibility to beta-lactam antimicrobials was predominant among the isolates. Only 4.7% were resistant to erythromycin and clindamycin. The erm(B) gene was widely detected in our GBS isolates, according to our phenotypic results (constitutive macrolide-lincosamide-streptogramin B [cMLSB] resistance phenotype), and the erm(A) gene was also detected in some isolates. MLSB resistance was restricted to strains isolated from patients with noninvasive infections and carriers. Serotype Ia was predominant (38.1%), serotype IV isolates were found at a high frequency (13.1%), and few isolates of serotype III were identified (3%). Pulsed-field gel electrophoresis results revealed a variety of types, reflecting the substantial genetic diversity among GBS strains, although a great number of isolates could be clustered into two major groups with a high degree of genetic relatedness. Three main PFGE clonal groups were found, and isolates sharing the same PFGE type were grouped into different serotypes. Furthermore, in a few cases, isolates from the same patients and possessing the same PFGE type were of different serotypes. These findings could be related to the occurrence of capsular switching by horizontal transfer of capsular genes.
In the last 40 years, Streptococcus agalactiae (group B streptococcus [GBS]) has been described as an important pathogen in neonates and pregnant women. Vaginal colonization with GBS during pregnancy is significantly associated with infections in newborns and requires investigation (40, 46, 48). Despite the reduction in the incidence of early-onset neonatal disease by using antimicrobial intrapartum prophylaxis, mortality and permanent disability rates caused by GBS continue to be significant. However, GBS has also emerged as an important pathogen in other patient groups, such as children, young adults with underlying medical conditions, and elderly individuals (40). Penicillin is the drug of choice for prevention and treatment of GBS infections, which remain universally susceptible. Erythromycin and clindamycin are recommended when risks of anaphylaxis or therapeutic failure are present. However, resistance to erythromycin and clindamycin has increased in many countries in North America (2, 6, 16, 40), Europe (21, 22, 42, 45), and Asia (24) but not in Brazil (18, 50) and other Latin American countries (23, 35, 36). Such a resistance profile is mainly due to two mechanisms: a methylase-mediated target site modification and an active efflux pump (53). In GBS, coresistance to macrolide, lincosamide, and streptogramin B (MLSB) is due to methylases encoded by erm genes that modify a ribosomal target (57). The MLSB resistance phenotype can be mediated by two classes of erm genes: (i) the widely predominant erm(B) determinant, which can be expressed either constitutively or inducibly and which is usually associated with high-level resistance (21), and (ii) erm(TR), a member of the erm(A) subclass (44), which is normally inducible and for which the level of resistance that it appears to confer depends on the contribution of drug efflux pumps (57). In GBS, the presence of a drug efflux pump [a membrane-bound protein encoded mainly by the mef(A) gene] (32) is associated with a low-level resistance pattern and confers resistance only to 14- and 15-membered ring macrolides (M phenotype) (9). Recently, mobile genetic elements have been identified in streptococcal species, including GBS carrying genes conferring macrolide resistance, suggesting that erythromycin and clindamycin resistance could be widely spread (43, 57).
GBS can be subclassified into serotypes according to the immunogenic type of their polysaccharide capsule, and serotyping methods have been used to investigate GBS epidemiology in humans. There are nine well-known serotypes described (serotypes Ia, Ib, II, III, IV, V, VI, VII and VIII). The existence of a new serotype (serotype IX) has recently been proposed (52). The predominant serotypes in the Western hemisphere are Ia, Ib, II, III, and V (39, 40); serotypes VI and VIII have been more frequently found in Japan (28); and serotypes IV and VII are rarely isolated (38).
Pulsed-field gel electrophoresis (PFGE), a molecular typing method, has been carried out for GBS strains to evaluate possible clonal relatedness and genetic diversity (1, 5, 34) in different clinical settings and for different diseases.
The aim of this study was to phenotypically and genotypically characterize GBS strains recently isolated from community and hospitalized patients in southern Brazil.
MATERIALS AND METHODS
Bacterial isolates.
A total of 168 GBS strains were isolated from outpatient and inpatient populations at the Clinical Hospital of the Federal University of Parana (HC-UFPR), a 635-bed tertiary-care teaching hospital located in Curitiba, Parana, Brazil, between April 2006 and May 2008. Strains were assigned to three different groups with regard to clinical outcome and anatomical site of isolation: (i) invasive (I) strains (n = 20) for isolates recovered from otherwise sterile body sites or samples, such as blood, cerebrospinal fluid, joint and bone biopsy specimens, and peritoneal fluid; (ii) noninvasive (NI) strains (n = 37) for isolates recovered from wound and abscess specimens, urine samples, a surgical wound, and an intravenous catheter; and (iii) colonizing (C) strains (n = 111) for isolates recovered from anogenital specimens from pregnant women and urine samples from healthy patients. Urinary tract infection samples were classified according to the results of urine culture (GBS present at >100,000 CFU/ml and the sole bacterial species isolated) and of urinalysis (pyuria and low epithelial cell count). Essentially, only one bacterial isolate from each patient was included, except for nine patients, from whom two bacterial samples were isolated.
Isolates were identified using standard biochemical tests (25). Serological grouping was also performed using a commercial latex agglutination test (Avipath-Strep; Omega Diagnostics, United Kingdom). All GBS isolates were stored at −80°C in Trypticase soy broth (HiMedia, Mumbai, India) containing 15% glycerol and 5% sheep blood for phenotypic and genotypic analysis.
Serotyping.
All isolates were serotyped by the immunodiffusion method with specific rabbit anticapsular antibodies against nine capsular polysaccharides (Ia, Ib, II, III, IV, V, VI, VII, and VIII) as previously described (29, 51). Serotype IX was not investigated. Nonserotypeable isolates were designated NT.
Antimicrobial susceptibility testing.
All GBS strains were tested for penicillin, ampicillin, erythromycin, clindamycin, levofloxacin, and vancomycin susceptibility by disk diffusion (10) and agar dilution (11) methods, as recommended by Clinical and Laboratory Standards Institute (CLSI) guidelines. The double-disk diffusion method with erythromycin and clindamycin disks was performed to determine the GBS resistance phenotypes, as described by CLSI (12). Tests were interpreted according to CLSI standards (12). Streptococcus pneumoniae ATCC 49619 and Enterococcus faecalis ATCC 29212 were used as controls.
Detection of macrolide resistance genes.
All resistant isolates, as defined by phenotypic methods, were tested for the presence of the erm(A), erm(B), and mef(A) genes by PCR amplification using primer pairs erm(A)-F (5′-GCATGACATAAACCTTCA-3′) and erm(A)-R (5′-AGGTTATAATGAAACAGA-3′), erm(B)-F (5′-GAAAAGGTACTCAACCAAATA-3′) and erm(B)-R (5′-AGTAACGGTACTTAAATTGTTTAC-3′), and mef(A)-F (5′-AGTATCATTAATCACTAGTGC-3′) and mef(A)-R (5′-TTCTTCTGGTACTAAAAGTGG-3′) under previously described conditions (14, 15, 49, 53). Streptococcus pyogenes 53157 erm(A), S. pyogenes 015195 erm(B), and S. pyogenes 06196 mef(A) were used as positive control in all PCR experiments.
Molecular typing by PFGE and dendrogram analysis.
Chromosomal DNA of all GBS isolates was prepared in agarose plugs as described previously (26, 54) and treated using 30 U of SmaI (Invitrogen, San Diego, CA) for 8 h at 30°C. The fragments were separated by PFGE in 1.2% agarose gels in a CHEF-DRIII system (Bio-Rad Laboratories, Hercules, CA) with pulse times of 3.5 to 45 s for 12 h and 1 to 5 s for 8 h at 14°C and 6 V/cm. The SmaI restriction profiles were interpreted according to the criteria of Tenover et al. (55). The Dice similarity coefficient was used to determine the similarity between each banding pattern, and a dendrogram was constructed using the unweighted-pair group method with arithmetic averages, along with the aid of Gel Pro-Analyzer and NTSYS computer software. PFGE samples were numbered 1 to 168. Isolates with similarities of >80% were clustered as highly genetically related (possibly or closely related, according to the criteria of Tenover et al. [55]) and were placed into dendrogram branches numbered 1 to 14. Genetically indistinguishable isolates (clones) were assigned with a capital letter (A, B, or C) to clusters with a minimum of 14 isolates. Isolates with similarities of <80% were considered genetically unrelated.
RESULTS
Clinical data and patient profile.
Patient profiles and their clinical data are summarized in Table 1. Among nonpregnant patients, GBS strains were frequently isolated from urine (80.7%). Skin, bone, and joint tissue samples yielded few isolates (6%), and 9.6% of the cases were identified as bacteremia. In pregnant women, most of the GBS strains were isolated from urine (58.4%), but some isolates were also obtained from vaginal and rectal specimens (38.9%). The median age of the adult patients was 34 years.
TABLE 1.
Distribution of 168 GBS isolates among patient groups and patient clinical data
| Patient group | No. (%) of patients | No. of isolates |
|||
|---|---|---|---|---|---|
| Invasive population | Noninvasive population | Colonizing population | Total | ||
| Pregnanta,b | 72 (45.3) | 10 | 67 | 77 | |
| Nonpregnanta,c | 80 (50.3) | 12 | 27 | 44 | 83 |
| Female | 51 | 2 | 19 | 32 | 53 |
| Male | 29 | 10 | 8 | 12 | 30 |
| Neonatesa | 7 (4.4) | 8 | 8 | ||
| EOSd | 4 | 4 | 4 | ||
| LOSe | 3 | 4 | 4 | ||
| Total | 159 (100) | 20 | 37 | 111 | 168 |
Five pregnant patients, three nonpregnant adults, and a neonate yielded two isolates each.
Fourteen patients ages 13 to 19 years harbored 3 NI and 12 C isolates (1 patient included 2 C isolates); 58 patients ages 20 to 64 years harbored 7 NI and 55 C isolates (3 patients harbored 2 C isolates, and 1 patient harbored both an NI and a C isolate).
Four patients ages 13 to 19 years harbored only colonizing isolates; 60 patients ages 20 to 64 years harbored 8 I, 19 NI, and 36 C isolates (2 patients harbored NI/C isolates, and 1 harbored only a noninvasive isolate); 16 patients >64 years of age harbored 4 I, 8 NI, and 4 C isolates.
EOS, early-onset syndrome. All newborns with early-onset syndrome developed bacteremia.
LOS, late-onset syndrome. Among the newborns with late-onset syndrome, one developed meningitis, one developed bacteremia, and one developed both meningitis and bacteremia.
Serotype analysis.
Table 2 shows the number of isolates for each serotype group and their clinical manifestations. Colonizing isolates were present in higher numbers and were distributed in all serotypes. Serotype IV was commonly found in infections. Serotype Ia was found in several types of infections (bacteremia, meningitis, osteomyelitis, peritonitis, skin and urinary tract infections). A higher diversity of serotypes was observed in genital and rectal isolates, as well as urine and blood samples.
TABLE 2.
Clinical manifestations and serotype distributions of 168 GBS isolates
| Isolate type and clinical manifestation or site | No. of isolates of serotypea: |
|||||||
|---|---|---|---|---|---|---|---|---|
| Ia | Ib | II | III | IV | V | NT | Total | |
| Invasive isolates | ||||||||
| Bacteremia | 4 | 2 | 3 | 2 | 2 | 1 | 14 | |
| Meningitis | 1 | 1 | 2 | |||||
| Arthritis | 1 | 1 | ||||||
| Osteomyelitis | 1 | 1 | ||||||
| Peritonitis | 2 | 2 | ||||||
| Subtotal | 8 | 2 | 4 | 3 | 2 | 1 | 20 | |
| Noninvasive isolates | ||||||||
| Puerperal infection | 1 | 1 | ||||||
| Skin infection | 2 | 1 | 1 | 1 | 5 | |||
| Urinary tract infection | 10 | 1 | 5 | 2 | 8 | 3 | 2 | 31 |
| Subtotal | 12 | 1 | 5 | 2 | 9 | 4 | 4 | 37 |
| Colonizing isolates | ||||||||
| Genitals/rectum | 12 | 5 | 6 | 1 | 1 | 2 | 3 | 30 |
| Urinary tract | 32 | 9 | 12 | 2 | 9 | 13 | 4 | 81 |
| Subtotal | 44 | 14 | 18 | 3 | 10 | 15 | 7 | 111 |
| Total | 64 | 17 | 27 | 5 | 22 | 21 | 12 | 168 |
Serotypes VI, VII, and VIII were not found in this study.
Table 3 shows that in most age groups, serotype Ia was predominant. Different serotypes were mainly found in teenagers, nonpregnant, and pregnant adults. Eight GBS strains were obtained from newborns: four strains of three different serotypes were obtained both from newborns with early-onset infections and from newborns with late-onset infections.
TABLE 3.
Serotype distribution of 168 GBS isolates in various age groups
| Patient group (age [yr])a | No. of isolates of serotype: |
|||||||
|---|---|---|---|---|---|---|---|---|
| Ia | Ib | II | III | IV | V | NT | Total | |
| Early newbornsb | 2 | 1 | 1 | 4 | ||||
| Late newbornsc | 1 | 1 | 2 | 4 | ||||
| Teenagers (13-19) | 9 | 1 | 3 | 2 | 2 | 2 | 19 | |
| Nonpregnant adults (20-64) | 23 | 6 | 10 | 3 | 9 | 10 | 2 | 63 |
| Pregnant adults (20-64) | 19 | 8 | 13 | 7 | 6 | 9 | 62 | |
| Elderly (>64) | 10 | 1 | 2 | 3 | 16 | |||
| Total | 64 | 17 | 27 | 5 | 22 | 21 | 12 | 168 |
No GBS isolate was recovered from children (1 to 12 years old).
Age 1 to 7 days.
Age 8 to 90 days.
Antimicrobial susceptibility and MLSB resistance profile.
All GBS isolates were susceptible to penicillin, ampicillin, levofloxacin, and vancomycin. Resistance to erythromycin and clindamycin was found in 4.7% of the samples (8 of 168) (Table 4). All resistant GBS strains expressed the constitutive MLSB phenotype, and these possessed the erm(B) gene, as detected by PCR. The erm(A) and erm(B) resistance genes were found concomitantly in five of these isolates, and no mef(A) gene was found. Only serotypes II (n = 3), IV (n = 2), and V (n = 3) were identified among resistant isolates.
TABLE 4.
Antibiotic susceptibilities of 168 GBS isolates
| Antibiotic | MIC (μg/ml) |
% Sb | ||
|---|---|---|---|---|
| Rangea | 50% | 90% | ||
| Penicillin G | 0.007-0.96 | 0.03 | 0.06 | 100 |
| Ampicillin | 0.015-0.5 | 0.06 | 0.06 | 100 |
| Erythromycinc | 0.03-8 | 0.03 | 0.03 | 95.3 |
| Clindamycinc | 0.03-8 | 0.03 | 0.03 | 95.3 |
| Levofloxacin | 0.25-16 | 1.0 | 2.0 | 100 |
| Vancomycin | 0.06-2 | 0.5 | 0.5 | 100 |
Range, range of concentrations tested.
S, sensitive.
Resistant isolates presented MICs of >8 μg/ml for both antibiotics.
Molecular typing by PFGE.
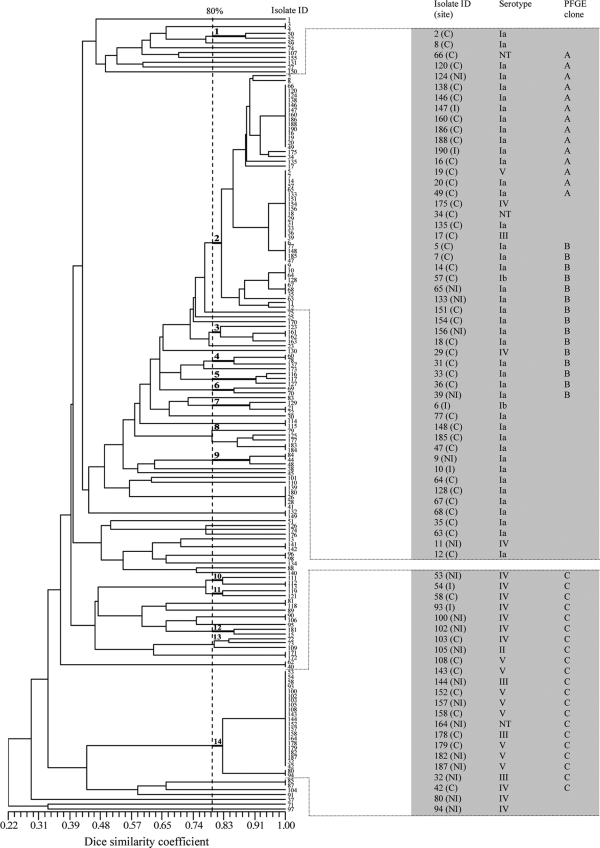
Ninety-one distinct PFGE types were identified on the basis of dendrogram analysis. According to the data shown in Fig. 1, isolates in 14 PFGE groups possessed above 80% similarity, and each group clustered at least two isolates. Of these, groups 2 and 14 clustered the highest number of GBS isolates. Also, among those PFGE groups, five included only GBS strains of the same serotype, and the remaining PFGE groups included GBS isolates of different serotypes. The other PFGE types showed less than 80% similarity and represented unrelated isolates.
FIG. 1.
Dendrogram of 168 GBS isolates constructed on the basis of PFGE patterns. A Dice coefficient similarity within at least 80% included 14 PFGE groups numbered from 1 to 14. Isolate identifications (isolate ID) are indicated in the dendrogram. An expanded view of groups 2 and 14, which contain multiple numbers of strains with degrees of similarity higher than 80%, is presented on the right in gray. The site of isolation, serotype, and PFGE grouping of those isolates are shown. Isolates 155, 184, 132, 149, 152, 179, 80, and 94 are MLSB resistant.
For PFGE groups 2 and 14, clones (100% identity on the basis of PFGE analysis) with a minimum of 14 isolates are shown in greater detail in Fig. 1, and those were designated PFGE clones A, B, and C, which accounted for 50 (29.8%) of the GBS isolates. Clone A comprised 14 isolates, 12 of serotype Ia, 1 of serotype V, and an NT isolate. Of those, 2 were invasive, 1 was noninvasive, and 11 were colonizing. Clone B contained 15 isolates, 13 of serotype Ia, 1 of serotype Ib, and 1 of serotype IV. Four isolates were noninvasive, and 11 were colonizing. Clone C consisted of 21 isolates, 8 of serotype IV and V, 3 of serotype III, 1 of serotype II, and 1 that was NT. Two strains were isolated from patients with invasive disease, 10 were collected from patients with noninvasive disease, and 9 originated from carriers. Except for serotypes Ia, IV, and V, for which clusters with 43, 10, and 8 isolates, respectively, could be seen in the dendrogram (data not shown), the other serotypes presented high degrees of genetic diversity.
PFGE analysis of the eight MLSB-resistant isolates resulted in their distribution into five PFGE types (data not shown). Four strains were clustered (>80% similarity), consisting of two clonal isolates of serotype IV and two of serotype V. Also, two strains of serotype II were clonal. The remaining two strains were not related. No invasive strains displayed MLSB resistance.
Nine patients yielded two clonal isolates each, as determined by PFGE analysis. In some cases, the pair of isolates from a patient belonged to different serotypes. From three patients, four isolates were recovered from the urinary tract at distinct times (one of serotype III and one of serotype IV from the first patient and one of serotype IV and one of V from the second one). Meanwhile, two isolates (one of serotype IV and one of serotype V) were recovered from rectal specimen culture and the urinary tract of the third patient at the same time. For the remaining six patients, a single serotype was present in each patient. In two cases, GBS samples recovered from newborns and their respective mothers were of the same serotype (serotype Ia) and also presented identical PFGE types.
DISCUSSION
Data collected from different geographic areas revealed considerable variation in the phenotypic and genotypic characteristics of GBS isolates (15, 18, 21, 22, 23, 24, 30, 31, 39, 42, 45). In Brazil, data on the distribution of serotypes, as well as the molecular epidemiology of GBS isolates, are still scarce. In the present study, we investigated the phenotypic and genotypic properties for a collection of GBS strains isolated from community and hospitalized patients in southern Brazil.
A few cases of neonatal infections with no associated mortality were attributed to GBS infection in our study. However, GBS contributed to death when some underlying disease was present in both neonates and nonpregnant adults (data not shown). Previous studies have reported that GBS have emerged as important pathogens in children, young adults with underlying medical conditions, and elderly patients (40, 47). During the study period, no child was included. A higher number of colonizing isolates was found in nonpregnant adults. However, although a small number of elderly patients (n = 16) and newborns (n = 7) has been included, most of the elderly patients and all of the neonates presented with infection. Furthermore, prematurity and low birth weight were frequently observed in neonates; and comorbidities such as cancer, diabetes mellitus, and renal disorders were found in elderly patients. Usually, newborns and patients with serious underlying diseases are at higher risk of bacterial colonization (19, 47), and this would be consistent with our observations for those patient groups.
The distribution of GBS capsular polysaccharide types has previously been reported to vary geographically. Studies performed in European, North American, and Latin American countries have demonstrated that serotype Ia, III, or V is more frequently found (2, 18, 21, 23, 28, 30, 31, 39, 52). In our study, all capsular serotypes were found except VI, VII, and VIII, which are often considered rare (2, 21, 23, 31, 39). Serotype Ia was predominant, in accordance with the findings of previous studies (18, 23, 30, 31, 35, 40), and it occurred in infection and colonizing isolates. Likewise, serotype II was also frequently seen in colonizing and infection isolates. In another Brazilian study, serotype Ib was often found in pregnant women (50). Conversely, in our study a low number of isolates were of serotype Ib; however, some of those were causing early-onset and late-onset disease. This different finding is still unclear but confirms the higher degree of variability of serotypes in distinct regions even within the same country. Overall, serotype V was described elsewhere as an important serotype causing invasive infections in adults (21, 39, 40, 48). In this study, serotype V was also identified among adult patients, but most of them consisted of colonizing isolates (15 of 21).
Our finding that showed that serotype III is of particular importance differed from the findings of previous studies, as it was frequently isolated in other countries (21, 30, 31, 35, 39, 40) and causes the majority of infections in neonates (47). Only five GBS isolates from this study were of serotype III, and none of them were from neonates. A previous Brazilian study also isolated a lower number of GBS strains of serotype III from pregnant women (50). Another study found a considerable number of strains of serotype III, but they were recovered from the milk of dairy cows (17).
The rate of occurrence of serotype IV in different parts of the world is low (21, 38, 40); however, we isolated GBS expressing this capsular polysaccharide at a relatively high frequency (n = 22; 13.1%). Furthermore, serotype IV was associated with late-onset disease and was the only serotype displaying a larger number of infectious isolates (Table 2). In contrast, serotype IV has not been described in previous Brazilian studies (18, 50).
Previous studies reported occurrence rates of NT strains ranging from 1 to 18% (3, 21, 27, 50). Our numbers (7.1%) included colonizing, noninvasive, and invasive isolates. Molecular serotyping methods would probably increase their typeability. However, a decreased capacity for capsular polysaccharide synthesis, noncapsulated phase variation, or cps gene cluster modification could prevent proper serotype identification (8, 27).
Our study confirms the high level of beta-lactam susceptibility of GBS (6, 40). Neither penicillin-resistant nor penicillin-intermediate GBS strains were found. Although full penicillin resistance has not been confirmed in GBS, some studies have reported a reduced susceptibility to penicillin (7, 13, 37), but the overall penicillin susceptibility pattern remained unchanged (6, 36, 40, 45, 50).
Erythromycin and clindamycin resistance was detected in 4.7% of the GBS isolates, and these strains were recovered from patients with noninvasive infections as well as carriers. This can be regarded as a low rate of incidence compared to incidence data from Asia, Europe, the United States, and Canada (6, 16, 24, 40, 42); but similar results were also found in other Latin American and Nordic countries (18, 23, 35, 36, 37). This could be attributed to the more intense clinical use of these antibiotics in Asia, Europe, the United States, and Canada. The presence of methylases encoded by erm genes is the most common MLSB resistance mechanism found (15, 58). The erm(B) gene was detected in all resistant GBS isolates by PCR, and the erm(A) gene was also detected in a number of these isolates. The co-occurrence of these genes has been reported in previous studies. However, this combination does not change the MIC results and the presence of erm(B) alone has been reported to lead to higher MICs (4, 18, 20). No strains were positive for mef(A). This gene has not been found in Brazilian GBS strains (18).
The high degree of genetic diversity of the GBS isolates identified in this study is reflected in the results of the PFGE profiling analysis. Out of 91 distinct PFGE types, 14 PFGE groups with at least 80% similarity were identified. Of those, two PFGE groups (groups 2 and 14) represented 43.4% of all GBS isolates. From a study with 91 GBS strains, Gherardi et al. (21) reported that 52.7% of isolates belonged to four major clonal groups.
Molecular studies have reported that GBS strains of different serotypes would share more sequence similarities than isolates of the same serotype (56). Our study found isolates of different serotypes belonging to the same clonal groups (A, B, and C) (Fig. 1). This finding may be attributed to the horizontal transfer of capsular genes among GBS isolates of different serotypes, which could be driven by the evasion of the host-mediated immune response promoted by the genetic selection and maintenance of conserved structural motifs in the polysaccharide repeat unit of all capsular serotypes (8, 56). We also observed similar PFGE types in serotypes Ia, IV, and V, while serotypes Ib, II, and III showed higher levels of heterogeneity. It has been found that the genetic variety is consistent with that found in previous studies in both large and small GBS populations (18, 21, 41).
This study contained a GBS population recovered from a great diversity of patients who did not present epidemiological relationships. Also, no relevant associations among clinical data and phenotypic or genotypic characteristics were found.
Our GBS population included a considerable number of serotype IV isolates, in contrast to what is observed in most other countries. This fact highlights the need for further studies with larger numbers of isolates in different geographic areas. Variations in the relative occurrence of serotypes in different areas or countries could potentially compromise the effectiveness of more universal vaccines. Although we have shown a high degree of genetic diversity in isolates of the same serotype, many GBS isolates had high degrees of genetic similarity. An environmental origin could be the explanation for the spread of such isolates, since our patients were epidemiologically unrelated. We also observed that one GBS clone can express distinct serotypes in the same patient. Neither serotyping nor PFGE profiling alone would be enough to determine the relatedness of GBS isolates, since either typing method can fail to discriminate unrelated isolates or to group together related isolates. In this context, Tettelin et al. (56) reported that even other commonly used strain classification methods, such as multilocus sequence typing, do not reveal the real GBS genetic diversity evidenced for them by whole-genome analysis. Genomic sequencing of higher numbers of GBS strains representing the major disease-causing serotypes has been suggested to be an adequate tool for rational vaccine design (33), and such approaches may also contribute to the understanding of the epidemiology of GBS.
Acknowledgments
We thank the staff of the Laboratório de Bacteriologia Médica/IMPPG/UFRJ for the GBS capsular serotype analyses.
We have no competing interests to declare.
Ethical approval for the project has been provided by the Research Ethics Committee of the Hospital de Clínicas, UFPR, under protocol 1772.189/2008-09.
Footnotes
Published ahead of print on 29 September 2010.
REFERENCES
- 1.Amundson, N. R., A. E. Flores, S. L. Hillier, C. J. Baker, and P. Ferrieri. 2005. DNA macrorestriction analysis of nontypeable group B streptococcal isolates: clonal evolution of nontypeable and type V isolates. J. Clin. Microbiol. 43:572-576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Andrews, J. I., D. J. Diekema, S. K. Hunter, P. R. Rhomberg, M. A. Pfaller, R. N. Jones, and G. V. Doern. 2000. Group B streptococci causing neonatal bloodstream infection: antimicrobial susceptibility and serotyping results from SENTRY centers in the Western Hemisphere. Am. J. Obstet. Gynecol. 183:859-862. [DOI] [PubMed] [Google Scholar]
- 3.Benson, J. A., A. E. Flores, C. J. Baker, S. L. Hillier, and P. Ferrieri. 2002. Improved methods for typing nontypeable isolates of group B streptococci. Int. J. Med. Microbiol. 292:37-42. [DOI] [PubMed] [Google Scholar]
- 4.Betriu, C., E. Culebras, M. Gomez, I. Rodriguez-Avial, B. A. Sanchez, M. C. Agreda, and J. J. Picazo. 2003. Erythromycin and clindamycin resistance and telithromycin susceptibility in Streptococcus agalactiae. Antimicrob. Agents Chemother. 47:1112-1114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bliss, S. J., S. D. Manning, P. Tallman, C. J. Baker, M. D. Pearlman, C. F. Marrs, and B. Foxman. 2002. Group B Streptococcus colonization in male and nonpregnant female university students: a cross-sectional prevalence study. Clin. Infect. Dis. 34:184-190. [DOI] [PubMed] [Google Scholar]
- 6.Borchardt, S. M., J. H. DeBusscher, P. A. Tallman, S. D. Manning, C. F. Marrs, T. A. Kurzynski, and B. Foxman. 2006. Frequency of antimicrobial resistance among invasive and colonizing Group B streptococcal isolates. BMC Infect. Dis. 6:57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chu, Y. W., C. Tse, G. K. Tsang, D. K. So, J. T. Fung, and J. Y. Lo. 2007. Invasive group B Streptococcus isolates showing reduced susceptibility to penicillin in Hong Kong. J. Antimicrob. Chemother. 60:1407-1409. [DOI] [PubMed] [Google Scholar]
- 8.Cieslewicz, M. J., D. Chaffin, G. Glusman, D. Kasper, A. Madan, S. Rodrigues, J. Fahey, M. R. Wessels, and C. E. Rubens. 2005. Structural and genetic diversity of group B Streptococcus capsular polysaccharides. Infect. Immun. 73:3096-3103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Clancy, J., J. Petitpas, F. Dib-Hajj, W. Yuan, M. Cronan, A. V. Kamath, J. Bergeron, and J. A. Retsema. 1996. Molecular cloning and functional analysis of a novel macrolide-resistance determinant, mefA, from Streptococcus pyogenes. Mol. Microbiol. 22:867-879. [DOI] [PubMed] [Google Scholar]
- 10.Clinical and Laboratory Standards Institute. 2009. Performance standards for antimicrobial disk susceptibility tests; approved standard, 10th ed. CLSI document M2-A10. Clinical and Laboratory Standards Institute, Wayne, PA.
- 11.Clinical and Laboratory Standards Institute. 2009. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically; approved standard, 8th ed. CLSI document M7-A8. Clinical and Laboratory Standards Institute, Wayne, PA.
- 12.Clinical and Laboratory Standards Institute. 2009. Performance standards for antimicrobial susceptibility testing; 19th informational supplement. CLSI document M100-S19. Clinical and Laboratory Standards Institute, Wayne, PA.
- 13.Dahesh, S., M. E. Hensler, N. M. Van Sorge, R. E. Gertz, Jr., S. Schrag, V. Nizet, and B. W. Beall. 2008. Point mutation in the group B streptococcal pbp2x gene conferring decreased susceptibility to beta-lactam antibiotics. Antimicrob. Agents Chemother. 52:2915-2918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.de Azavedo, J. C., R. H. Yeung, D. J. Bast, C. L. Duncan, S. B. Borgia, and D. E. Low. 1999. Prevalence and mechanisms of macrolide resistance in clinical isolates of group A streptococci from Ontario, Canada. Antimicrob. Agents Chemother. 43:2144-2147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.de Azavedo, J. C., M. McGavin, C. Duncan, D. E. Low, and A. McGeer. 2001. Prevalence and mechanisms of macrolide resistance in invasive and noninvasive group B Streptococcus isolates from Ontario, Canada. Antimicrob. Agents Chemother. 45:3504-3508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Desjardins, M., K. L. Delgaty, K. Ramotar, C. Seetaram, and B. Toye. 2004. Prevalence and mechanisms of erythromycin resistance in group A and group B Streptococcus: implications for reporting susceptibility results. J. Clin. Microbiol. 42:5620-5623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Duarte, R. S., O. P. Miranda, B. C. Bellei, M. A. Brito, and L. M. Teixeira. 2004. Phenotypic and molecular characteristics of Streptococcus agalactiae isolates recovered from milk of dairy cows in Brazil. J. Clin. Microbiol. 42:4214-4222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Duarte, R. S., B. C. Bellei, O. P. Miranda, M. A. Brito, and L. M. Teixeira. 2005. Distribution of antimicrobial resistance and virulence-related genes among Brazilian group B streptococci recovered from bovine and human sources. Antimicrob. Agents Chemother. 49:97-103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Farley, M. M., R. C. Harvey, T. Stull, J. D. Smith, A. Schuchat, J. D. Wenger, and D. S. Stephens. 1993. A population-based assessment of invasive disease due to group B Streptococcus in nonpregnant adults. N. Engl. J. Med. 328:1807-1811. [DOI] [PubMed] [Google Scholar]
- 20.Fluegge, K., S. Supper, A. Siedler, and R. Berner. 2004. Antibiotic susceptibility in neonatal invasive isolates of Streptococcus agalactiae in a 2-year nationwide surveillance study in Germany. Antimicrob. Agents Chemother. 48:4444-4446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gherardi, G., M. Imperi, L. Baldassarri, M. Pataracchia, G. Alfarone, S. Recchia, G. Orefici, G. Dicuonzo, and R. Creti. 2007. Molecular epidemiology and distribution of serotypes, surface proteins, and antibiotic resistance among group B streptococci in Italy. J. Clin. Microbiol. 45:2909-2916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gonzalez, J. J., and A. Andreu. 2005. Multicenter study of the mechanisms of resistance and clonal relationships of Streptococcus agalactiae isolates resistant to macrolides, lincosamides, and ketolides in Spain. Antimicrob. Agents Chemother. 49:2525-2527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gonzalez Pedraza Aviles, A., M. C. Ortiz Zaragoza, and R. Mota Vazquez. 2002. Serotypes and antimicrobial susceptibility of group B Streptococcus from pregnant women in Mexico. Rev. Latinoam. Microbiol. 44:133-136. [PubMed] [Google Scholar]
- 24.Hsueh, P. R., L. J. Teng, L. N. Lee, S. W. Ho, P. C. Yang, and K. T. Luh. 2001. High incidence of erythromycin resistance among clinical isolates of Streptococcus agalactiae in Taiwan. Antimicrob. Agents Chemother. 45:3205-3208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Isenberg, H. D. 2004. Clinical microbiology procedures handbook, vol. 1, 2nd ed. ASM Press, Washington, DC.
- 26.Kaufmann, M. E. 1998. Pulsed-field gel electrophoresis, p. 33-51. In N. Woodford and A. P. Johnson (ed.), Molecular bacteriology: protocols and clinical applications. Humana Press Inc., Totowa, NJ.
- 27.Kong, F., S. Gowan, D. Martin, G. James, and G. L. Gilbert. 2002. Molecular profiles of group B streptococcal surface protein antigen genes: relationship to molecular serotypes. J. Clin. Microbiol. 40:620-626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lachenauer, C. S., D. L. Kasper, J. Shimada, Y. Ichiman, H. Ohtsuka, M. Kaku, L. C. Paoletti, P. Ferrieri, and L. C. Madoff. 1999. Serotypes VI and VIII predominate among group B streptococci isolated from pregnant Japanese women. J. Infect. Dis. 179:1030-1033. [DOI] [PubMed] [Google Scholar]
- 29.Lancefield, R. C., and E. H. Freimer. 1966. Type-specific polysaccharide antigens of group B streptococci. J. Hyg. (Lond.) 64:191-203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lee, K., J. W. Shin, Y. Chong, and H. Mikamo. 2000. Trends in serotypes and antimicrobial susceptibility of group B streptococci isolated in Korea. J. Infect. Chemother. 6:93-97. [DOI] [PubMed] [Google Scholar]
- 31.Lopardo, H. A., P. Vidal, P. Jeric, D. Centron, H. Paganini, R. R. Facklam, and J. Elliott. 2003. Six-month multicenter study on invasive infections due to group B streptococci in Argentina. J. Clin. Microbiol. 41:4688-4694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Luna, V. A., P. Coates, E. A. Eady, J. H. Cove, T. T. Nguyen, and M. C. Roberts. 1999. A variety of gram-positive bacteria carry mobile mef genes. J. Antimicrob. Chemother. 44:19-25. [DOI] [PubMed] [Google Scholar]
- 33.Maione, D., I. Margarit, C. D. Rinaudo, V. Masignani, M. Mora, M. Scarselli, H. Tettelin, C. Brettoni, E. T. Iacobini, R. Rosini, N. D'Agostino, L. Miorin, S. Buccato, M. Mariani, G. Galli, R. Nogarotto, V. Nardi Dei, F. Vegni, C. Fraser, G. Mancuso, G. Teti, L. C. Madoff, L. C. Paoletti, R. Rappuoli, D. L. Kasper, J. L. Telford, and G. Grandi. 2005. Identification of a universal group B Streptococcus vaccine by multiple genome screen. Science 309:148-150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Manning, S. D. 2003. Molecular epidemiology of Streptococcus agalactiae (group B Streptococcus). Front. Biosci. 8:s1-s18. [DOI] [PubMed] [Google Scholar]
- 35.Martinez, M. A., A. Ovalle, C. Duran, I. Reid, G. Urriola, B. Garay, and M. Cifuentes. 2004. Serotypes and antimicrobial susceptibility of Streptococcus agalactiae. Rev. Med. Chil. 132:549-555. [DOI] [PubMed] [Google Scholar]
- 36.Mollerach, A., E. Mendez, R. Massa, and J. Di Conza. 2007. Streptococcus agalactiae isolated in Santa Fe, Argentina: antibiotic susceptibility and erythromycin-clindamycin resistance mechanisms. Enferm. Infecc. Microbiol. Clin. 25:67-68. [DOI] [PubMed] [Google Scholar]
- 37.Nagano, N., Y. Nagano, K. Kimura, K. Tamai, H. Yanagisawa, and Y. Arakawa. 2008. Genetic heterogeneity in pbp genes among clinically isolated group B streptococci with reduced penicillin susceptibility. Antimicrob. Agents Chemother. 52:4258-4267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Paoletti, L. C., and D. L. Kasper. 2002. Conjugate vaccines against group B Streptococcus types IV and VII. J. Infect. Dis. 186:123-126. [DOI] [PubMed] [Google Scholar]
- 39.Persson, E., S. Berg, B. Trollfors, P. Larsson, E. Ek, E. Backhaus, B. E. Claesson, L. Jonsson, G. Radberg, T. Ripa, and S. Johansson. 2004. Serotypes and clinical manifestations of invasive group B streptococcal infections in western Sweden 1998-2001. Clin. Microbiol. Infect. 10:791-796. [DOI] [PubMed] [Google Scholar]
- 40.Phares, C. R., R. Lynfield, M. M. Farley, J. Mohle-Boetani, L. H. Harrison, S. Petit, A. S. Craig, W. Schaffner, S. M. Zansky, K. Gershman, K. R. Stefonek, B. A. Albanese, E. R. Zell, A. Schuchat, and S. J. Schrag. 2008. Epidemiology of invasive group B streptococcal disease in the United States, 1999-2005. JAMA 299:2056-2065. [DOI] [PubMed] [Google Scholar]
- 41.Pillai, P., U. Srinivasan, L. Zhang, S. M. Borchardt, J. Debusscher, C. F. Marrs, and B. Foxman. 2009. Streptococcus agalactiae pulsed-field gel electrophoresis patterns cross capsular types. Epidemiol. Infect. 137:1420-1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Poyart, C., L. Jardy, G. Quesne, P. Berche, and P. Trieu-Cuot. 2003. Genetic basis of antibiotic resistance in Streptococcus agalactiae strains isolated in a French hospital. Antimicrob. Agents Chemother. 47:794-797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Puopolo, K. M., D. C. Klinzing, M. P. Lin, D. L. Yesucevitz, and M. J. Cieslewicz. 2007. A composite transposon associated with erythromycin and clindamycin resistance in group B Streptococcus. J. Med. Microbiol. 56:947-955. [DOI] [PubMed] [Google Scholar]
- 44.Roberts, M. C., J. Sutcliffe, P. Courvalin, L. B. Jensen, J. Rood, and H. Seppala. 1999. Nomenclature for macrolide and macrolide-lincosamide-streptogramin B resistance determinants. Antimicrob. Agents Chemother. 43:2823-2830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schoening, T. E., J. Wagner, and M. Arvand. 2005. Prevalence of erythromycin and clindamycin resistance among Streptococcus agalactiae isolates in Germany. Clin. Microbiol. Infect. 11:579-582. [DOI] [PubMed] [Google Scholar]
- 46.Schrag, S., R. Gorwitz, K. Fultz-Butts, and A. Schuchat. 2002. Prevention of perinatal group B streptococcal disease. Revised guidelines from CDC. MMWR Recommend. Rep. 51(RR-11):1-22. [PubMed] [Google Scholar]
- 47.Schuchat, A. 1998. Epidemiology of group B streptococcal disease in the United States: shifting paradigms. Clin. Microbiol. Rev. 11:497-513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schuchat, A. 1999. Group B Streptococcus. Lancet 353:51-56. [DOI] [PubMed] [Google Scholar]
- 49.Seppälä, H., M. Skurnik, H. Soini, M. C. Roberts, and P. Huovinen. 1998. A novel erythromycin resistance methylase gene (ermTR) in Streptococcus pyogenes. Antimicrob. Agents Chemother. 42:257-262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Simões, J. A., V. M. Alves, S. E. Fracalanzza, R. P. de Camargo, L. Mathias, H. M. Milanez, and E. M. Brolazo. 2007. Phenotypical characteristics of group B Streptococcus in parturients. Braz. J. Infect. Dis. 11:261-266. [DOI] [PubMed] [Google Scholar]
- 51.Slotved, H. C., S. Sauer, and H. B. Konradsen. 2002. False-negative results in typing of group B streptococci by the standard Lancefield antigen extraction method. J. Clin. Microbiol. 40:1882-1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Slotved, H. C., F. Kong, L. Lambertsen, S. Sauer, and G. L. Gilbert. 2007. Serotype IX, a proposed new Streptococcus agalactiae serotype. J. Clin. Microbiol. 45:2929-2936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sutcliffe, J., T. Grebe, A. Tait-Kamradt, and L. Wondrack. 1996. Detection of erythromycin-resistant determinants by PCR. Antimicrob. Agents Chemother. 40:2562-2566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Teixeira, L. M., M. G. Carvalho, V. L. Merquior, A. G. Steigerwalt, D. J. Brenner, and R. R. Facklam. 1997. Phenotypic and genotypic characterization of Vagococcus fluvialis, including strains isolated from human sources. J. Clin. Microbiol. 35:2778-2781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tenover, F. C., R. D. Arbeit, R. V. Goering, P. A. Mickelsen, B. E. Murray, D. H. Persing, and B. Swaminathan. 1995. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J. Clin. Microbiol. 33:2233-2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tettelin, H., V. Masignani, M. J. Cieslewicz, C. Donati, D. Medini, N. L. Ward, S. V. Angiuoli, J. Crabtree, A. L. Jones, A. S. Durkin, R. T. Deboy, T. M. Davidsen, M. Mora, M. Scarselli, I. Margarit y Ros, J. D. Peterson, C. R. Hauser, J. P. Sundaram, W. C. Nelson, R. Madupu, L. M. Brinkac, R. J. Dodson, M. J. Rosovitz, S. A. Sullivan, S. C. Daugherty, D. H. Haft, J. Selengut, M. L. Gwinn, L. Zhou, N. Zafar, H. Khouri, D. Radune, G. Dimitrov, K. Watkins, K. J. O'Connor, S. Smith, T. R. Utterback, O. White, C. E. Rubens, G. Grandi, L. C. Madoff, D. L. Kasper, J. L. Telford, M. R. Wessels, R. Rappuoli, and C. M. Fraser. 2005. Genome analysis of multiple pathogenic isolates of Streptococcus agalactiae: implications for the microbial “pan-genome.” Proc. Natl. Acad. Sci. U. S. A. 102:13950-13955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Varaldo, P. E., M. P. Montanari, and E. Giovanetti. 2009. Genetic elements responsible for erythromycin resistance in streptococci. Antimicrob. Agents Chemother. 53:343-353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Weisblum, B. 1985. Inducible resistance to macrolides, lincosamides and streptogramin type B antibiotics: the resistance phenotype, its biological diversity, and structural elements that regulate expression—a review. J. Antimicrob. Chemother. 16(Suppl. A):63-90. [DOI] [PubMed] [Google Scholar]