Abstract
Alternative splicing is a crucial step in the generation of protein diversity and its misregulation is observed in many human cancer types. By analyzing 143 colorectal samples using exon arrays, SLC39A14, a divalent cation transporter, was identified as being aberrantly spliced in tumor samples. SLC39A14 contains two mutually exclusive exons 4A and 4B and the exon 4A/4B ratio was significantly altered in adenomas (p = 3.6 × 10−10) and cancers (p = 9.4 × 10−11), independent of microsatellite stability status. The findings were validated in independent exon array data sets and by quantitative real-time reverse-transcription PCR (qRT-PCR). Aberrant Wnt signaling is a hallmark of colorectal tumorigenesis and is characterized by nuclear β-catenin. Experimental inactivation of Wnt signaling in DLD1 and Ls174T cells by knockdown of β-catenin or overexpression of dominant negative TCFs (TCF1 and TCF4) altered the 4A/4B ratio, indicating that SLC39A14 splicing is regulated by the Wnt pathway. An altered 4A/4B ratio was also observed in gastric and lung cancer where Wnt signaling is also known to be aberrantly activated. The splicing factor SRSF1 and its regulator, the kinase SRPK1, were found to be deregulated upon Wnt inactivation in colorectal carcinoma cells. SRPK1 was also found up-regulated in both adenoma samples (p = 1.5 × 10−5) and cancer samples (p = 5 × 10−4). In silico splicing factor binding analysis predicted SRSF1 to bind predominantly to the cancer associated exon 4B, hence, it was hypothesized that SRPK1 activates SRSF1 through phosphorylation, followed by SRSF1 binding to exon 4B and regulation of SLC39A14 splicing. Indeed, siRNA-mediated knockdown of SRPK1 and SRSF1 in DLD1 and SW480 colorectal cancer cells led to a change in the 4A/4B isoform ratio, supporting a role of these factors in the regulation of SLC39A14 splicing. In conclusion, alternative splicing of SLC39A14 was identified in colorectal tumors and found to be regulated by the Wnt pathway, most likely through regulation of SRPK1 and SRSF1.
Most colorectal cancers (CRC)1 are believed to arise from adenomas, and a hallmark of colorectal tumor initiation is the constitutive activation of the Wnt pathway, which is observed in ∼85% of colorectal tumors (1). An active Wnt pathway is characterized by nuclear β-catenin and increased expression of key cell-cycle regulators such as v-myc myelocytomatosis viral oncogene homolog (MYC). Other key mediators of the Wnt pathway are TCF4 (also called transcription factor 7-like 2 (TCF7L2)) and TCF1 (also called transcription factor 7 (TCF7)) both of which form a complex with β-catenin and regulate the expression of many downstream Wnt-responsive genes (2). The Wnt pathway also plays a significant role in other cancer types, such as lung, gastric, thyroid, and prostate cancer (3–5), and it has been shown to be involved in the regulation of RNA splicing as well (6).
Alternative splicing (AS) enhances proteome diversity, and more than 90% of human multi-exon genes undergo AS (7, 8). Splicing is a tightly regulated process, and several families of splicing factors have been described, including the SR proteins, a conserved family of RNA-binding proteins (9). AS plays a critical role during development, and defects in AS have been implicated in numerous human diseases, including cancer (10–12). Several SR proteins are activated by phosphorylation of their serines by SRSF protein kinase 1 (SRPK1), which is up-regulated in colon, breast, and pancreatic cancer (13). The best described SRPK1 target is serine/arginine-rich splicing factor 1 (SRSF1, formerly known as SF2/ASF) (14, 15), an essential splicing factor that participates in both constitutive and AS, and which was recently shown to be a proto-oncogene (12) supporting the involvement of AS in cancer development.
The aim of the study was to discover AS in colorectal adenoma and cancer samples, and we identified tumor-specific splicing of solute carrier family 39 (zinc transporter), member 14 (SLC39A14), a metal ion transporter with the capacity to transport divalent cations, such as zinc, iron, and cadmium. SLC39A14 is a glycosylated protein located on the plasma membrane, which exists in at least two isoforms that differ from each other in the cation-binding domain. The difference is caused by AS, which involves two mutually exclusive exons 4A and 4B of similar size. The two protein isoforms have different capacities to bind cadmium (Cd2+) (16–18). We found that the 4A/4B ratio in tumor samples is shifted toward exon 4B, which has an eightfold higher Cd2+ affinity. Furthermore, we showed that splicing of SLC39A14 is Wnt-regulated and that the mechanism behind this regulation may involve Wnt-specific up-regulation of SRPK1. Finally, we show that knockdown of SRSF1 also affects the splicing of SLC39A14.
EXPERIMENTAL PROCEDURES
Clinical Samples
Patient samples were obtained immediately following surgery or colonoscopy and embedded in Tissue-Tek O.C.T. Compound (Sakura Finetek, Torrance, CA), snap-frozen in liquid nitrogen, and stored at −80 °C. In total, 143 colorectal tissues were included, 33 adjacent normal mucosa samples, 56 adenomas (all microsatellite stable (MSS)), and 54 carcinomas (43 MSS and 11 with microsatellite instability (MSI)). The samples were divided in two independent sets, one for discovery of AS events and one for validation. The discovery set consisted of 24 normal mucosa samples, 49 MSS adenomas (16 tubular, 12 tubulo-villous, and 21 villous), 30 MSS carcinomas (1 Stage I, 16 stage II, and 13 stage III), and 5 MSI carcinomas (4 stage II and 1 stage III). The independent validation set was previously described (19), and consists of 9 normal mucosa samples, 7 MSS adenomas (all tubular), 13 MSS carcinomas (8 stage II and 5 stage III), and 6 MSI carcinomas (1 Stage I, 4 stage II and 1 stage III). Informed written consent was obtained from all patients, and the study was approved by the Central Denmark Region Committees on Biomedical Research Ethics.
RNA Extraction
Sample tissue composition was evaluated by hematoxylin and eosin staining, and macroscopic trimming was used when necessary to enrich the fraction of tumor cells. A minimum of 60% neoplastic cells was required before total RNA was purified from serial cryo-sections. Total RNA was purified using RNAeasy Mini elute kit (Qiagen, Venlo, Netherlands). The RNA quality was analyzed on the 2100 Bioanalyzer (Agilent, Santa Clara, CA), and samples with a 28S/18S ratio <1.0 and RNA integrity number (RIN) <6 were excluded.
Human Exon 1.0 ST Array Analysis
For the exon array discovery set, 100 ng of total RNA was labeled according to the GeneChip Whole Transcript (WT) Sense Target Labeling Assay (Affymetrix, Inc., Santa Clara, CA) and hybridized to Human Exon 1.0 ST Arrays (Affymetrix) overnight. For the exon array validation set, 1 μg of total RNA was depleted of ribosomal RNA using the RiboMinus kit (Invitrogen) and labeled as described above. Scanning was performed in an Affymetrix GCS 3000 7G scanner. To avoid batch effects, all samples were labeled and scanned in a randomized order within the two datasets.
Data Analysis
Exon array data were quantile normalized using ExonRMA16 with core probe sets (228,940 probe sets) in GeneSpring GX 10 software package (Agilent). The data were filtered to remove genes not expressed in at least half of the samples within both the normal and tumor group. A total of 9606 genes were found to be expressed in both sample groups, and these genes formed the basis for further analysis. Candidate genes for AS were found by applying a splicing ANOVA approach. A stringent p value cut-off (<10−40) was applied to retain the 100 candidates with the lowest splicing ANOVA p values. Further filtration removing genes with no exons that had a log 2 delta Splice Index (exon expression value/transcript expression value) above 1 or below −1, yielded 40 candidate genes. Splice index analysis is, however, susceptible to large differences in gene expression values, and, therefore, genes that were differentially expressed (Benjamini-Hochberg corrected p value <0.05) were removed from the analysis. This left only 11 candidate genes for further analysis; these were manually inspected in a genomic context, and SLC39A14 was selected as the most promising candidate for further study. The Student's t test was used for all other statistical comparisons.
Quantitative Real-Time Reverse-Transcription PCR
Reverse transcription of 1 μg total RNA was done using Superscript II Reverse Transcriptase (Invitrogen) and a combination of oligo(dT) and random nonamer primers. Quantitative real-time reverse-transcription PCR (qRT-PCR) was performed in triplicates on a 7500 Fast or 7900HT Real-Time PCR System (Applied Biosystems, Foster City, CA). The following predesigned TaqMan assays were used: SLC39A14 exon 4A (Hs00939122_m1), SLC39A14 exon 4B (Hs00939901_m1), SRPK1 (Hs00177298_m1), and SRSF1 (Hs00199471_m1). β-catenin and total SLC39A14 mRNA levels were measured using SYBR green with the following primers: SLC39A14-F (ggacgagaaggtcattgtgg), SLC39A14-R (gtgatcatccaggccagagt), β-cat-F (gcgtggacaatggctactca), and β-cat-R (ccgcttttctgtctggttcc). Ubiquitin C was used for normalization as previously described (20).
Public Human Exon 1.0 ST Data Sets
A colon cancer Exon 1.0 ST data set (21) was downloaded from www.affymetrix.com. This set consisted of 18 paired samples of adenocarcinoma and adjacent normal tissue. Two additional human Exon 1.0 ST data sets covering gastric and lung cancer were downloaded from the Gene Expression Omnibus: GSE13195 and GSE12236, respectively. The gastric cancer data set consisted of 48 paired samples of adenocarcinoma and adjacent normal tissue. The lung cancer data set consisted of 40 paired samples from 20 patients with lung adenocarcinomas (22). The Exon 1.0 ST bladder cancer data set consisted of 11 normal, 12 Ta, 12 T1, and 12 T2–4 samples and was previously described (19). All data sets were analyzed as described above.
Immunohistochemistry
Standard indirect staining procedures were used for IHC as previously described (23), using anti-SLC39A14 (Sigma) in a 1:1800 dilution.
Splicing Factor Binding Site Predictions
To predict splicing factor binding sites, we used the SFmap software with default settings (24). Briefly, SFmap implements the COS(WR) algorithm (25), which computes similarity scores for a given regulatory motif on the basis of information derived from its sequence environment and its evolutionary conservation. SFmap is available online at http://sfmap.technion.ac.il.
Wnt-Pathway Model System
The LS174T and DLD1 derived cell lines stably expressing inducible dominant-negative (dn)TCF1 or dnTCF4 were a kind gift from Dr. Hans Clevers (The Hubrecht Laboratory, The Netherlands), and were previously described (2). For knockdown of β-catenin, DLD1 cells were transfected with 20 nm siRNA targeting β-catenin (Dharmacon, Lafayette, CO) or 20 nm scrambled nontargeting siRNA. Transfections were carried out using Lipofectamine 2000 (Invitrogen). RNA extraction was performed using RNeasy (Qiagen). cDNA synthesis was done with random nonamer primers, and qRT-PCR was performed as described above.
SRPK1 and SRSF1 Knockdown in DLD1 and SW480 Cells
DLD1 cells were cultured in RPMI 1640 medium (GIBCO, Invitrogen) supplemented with 5% fetal calf serum and 1% penicillin-streptomycin at 37 °C and 5% CO2. SW480 (ATCC) was cultured in DMEM medium (GIBCO, Invitrogen) supplemented with 10% fetal calf serum (Invitrogen) and 1% penicillin-streptomycin (Invitrogen) at 37 °C and 5% CO2. Cells were transfected with siRNA targeting SRSF1, SRPK1, or both transcripts, (using SRSF1siRNA Dharmacon # L018672–01), and silencer select siRNA SRPK1(s13451, Ambion). As a control, we used scrambled nontargeting siRNA (Dharmacon # d-001206–14-20). All transfections were carried out using Lipofectamine 2000 (Invitrogen) according to the manufacturer's instructions. Final concentrations of 50 and 100 nm siRNA were used.
RESULTS
Global exon expression was measured in a total of 143 colorectal tissue samples covering 33 normal mucosa samples, 56 adenomas, and 54 cancer samples by using the Affymetrix Exon 1.0 ST Array. The majority of the samples (24 normal mucosa and 84 tumor samples) were used to identify genes with tumor specific AS, whereas the remaining samples (9 normal mucosa and 26 tumor samples) were used as an independent validation set. Candidate genes for tumor specific AS were found by applying a splicing ANOVA approach along with a set of stringent filtration criteria (see Experimental Procedures). SLC39A14 was identified as the most prominent candidate on the basis of both the low splicing ANOVA p value (<10−40) and the Splice Index value (exon expression value/transcript expression value).
SLC39A14 Shows Alternative Splicing in Colorectal Neoplasias
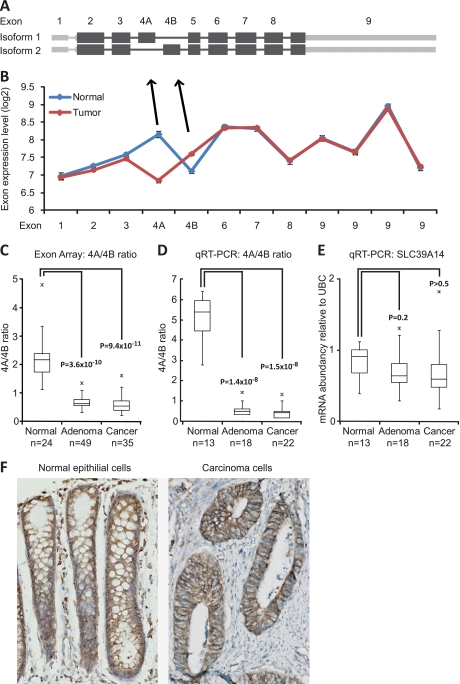
Several transcript variants of SLC39A14 are known, and the gene contains two mutually exclusive exons 4A and 4B (Fig. 1A). We found a striking difference in the expression of exons 4A and 4B between normal and tumor samples (Fig. 1B). Our data show that exon 4A is expressed at a >twofold higher level in normal mucosa samples compared with tumor samples, whereas exon 4B levels were slightly elevated in tumor samples relative to normal mucosa samples. The shift in ratio between exons 4A and 4B was consistent in both adenomas (p = 3.6 × 10−10) and carcinomas (p = 9.4 × 10−11) (independent of MSI-status) in virtually all samples (Fig. 1C). The gene expression level of SLC39A14 was at the same level in the normal mucosa and tumor samples. We made a technical validation of our exon array findings by qRT-PCR on 53 randomly selected samples used for exon array analysis. Again, we saw a dramatic shift in the inclusion rate of exons 4A and 4B between normal mucosa and tumor samples (Fig. 1D). The 4A/4B ratio changed significantly between normal mucosa samples and adenomas (p = 1.4 × 10−8) and between normal mucosa and cancer samples (p = 1.5 × 10−8) with a lower ratio in all adenoma and cancer samples compared with normal mucosa samples. The total mRNA level of SLC39A14 (both isoforms) did not change significantly (Fig. 1E). Finally, we validated our findings in two independent colorectal exon array sample sets (Supplemental Fig. 1). Total SLC39A14 protein level was evaluated by immunohistochemistry, and SLC39A14 was found to be expressed at equivalent levels in normal epithelial cells and carcinoma cells with a cellular localization corresponding to the plasma membrane as expected (Fig. 1F). Antibodies specific for either SLC39A14 isoform 1 or 2 were not available.
Fig. 1.
SLC39A14 alternative splicing is dysregulated in colorectal neoplasia. A, Exon structure of SLC39A14 isoforms 1 and 2. The isoforms differ in exon 4 usage. Exons 4A and 4B are mutually exclusive. B, Expression levels of the individual exons of SLC39A14 measured in normal mucosa (n = 24) and tumor (n = 84) samples using exon arrays. Error bars represent S.E. C, SLC39A14 exon 4A/4B ratio in normal mucosa samples, adenomas, and carcinomas, same data as in B. D, Technical validation of SLC39A14 exon 4A/4B ratio by qRT-PCR. E, The total transcript level of SLC39A14 measured by qRT-PCR on the same samples as in D. F, Total protein level of SLC39A14 detected by immunohistochemistry in normal epithelial cells and carcinoma cells, respectively.
The Wnt Pathway Regulates SLC39A14 Splicing
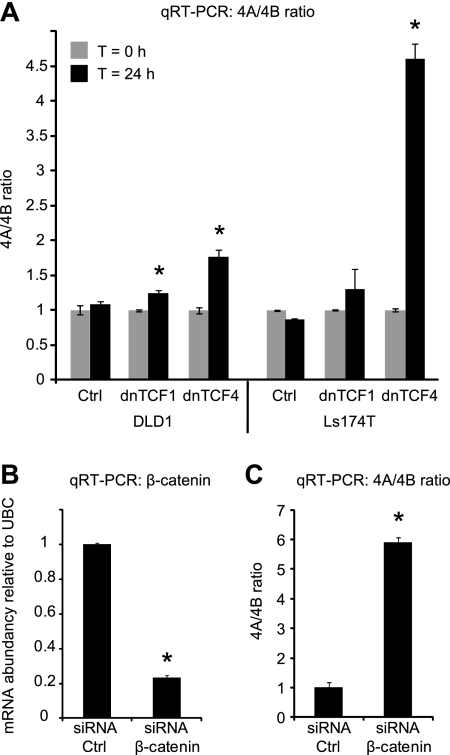
To investigate whether SLC39A14 AS is regulated by the Wnt pathway, we used two Wnt-model systems that utilize overexpression of a dominant negative form of TCF4 (dnTCF4) or TCF1 (dnTCF1) to interrupt the TCF4 or TCF1 mediated part of the Wnt pathway, respectively. Both dominant negative TCFs were overexpressed in the two colorectal cancer cell lines DLD1 and Ls174T (26–28). qRT-PCR analysis showed that the 4A/4B ratio was increased when TCF mediated Wnt signaling was turned off (Fig. 2A). The shift was most obvious in the two dnTCF4 models, but a small increase was also seen when dnTCF1 was overexpressed. This indicates that overexpression of dnTCFs lead to altered SLC39A14 splicing, and, hence, that the splicing of this gene could be regulated by Wnt signaling. To confirm that the altered splice pattern was truly caused by altered Wnt signaling, and not a mere side-effect of the dominant negative TCFs, a second model system was developed in which Wnt signaling was abrogated upstream of the TCFs by knocking down the key regulator, β-catenin, in DLD1 cells using siRNA. The knockdown of β-catenin was successful (Fig. 2B), and the effect on the splicing pattern of SLC39A14 was an even more dramatic change (∼sixfold) in the ratio of exon 4A/4B (Fig. 2C) confirming the regulatory role of the Wnt pathway on SLC39A14 splicing. The Wnt pathway has also been implicated in the development of several other cancer types such as gastric (4) and lung cancer (5). Consistent with the findings in CRC, our analysis of publicly available exon array data from these cancer types also indicated that the splicing of SLC39A14 in tumors, in contrast to the normal counterpart, was pushed toward the exon 4B containing isoform (Supplemental Fig. 2). This was not the case in bladder tumors, where Wnt signaling has not been found to play any role in tumor development (Supplemental Fig. 2D).
Fig. 2.
SLC39A14 exon 4A/4B ratio is regulated by Wnt signaling in colorectal carcinoma cells. A, dnTCF1 or dnTCF4 expression was induced in stably transfected DLD1 or Ls174T cells. The 4A/4B ratio in dnTCF1 and dnTCF4 Wnt-model systems was measured by qRT-PCR. B, qRT-PCR of β-catenin mRNA from DLD1 cells transfected with 20 nm control or β-catenin siRNA. C, 4A/4B ratio in the siRNA transfected cells described in B. Error bars represent standard deviation of three (A) or two (B and C) replicas. * = p < 0.05.
SRPK1 and SRSF1 Expression is Regulated by the Wnt Pathway
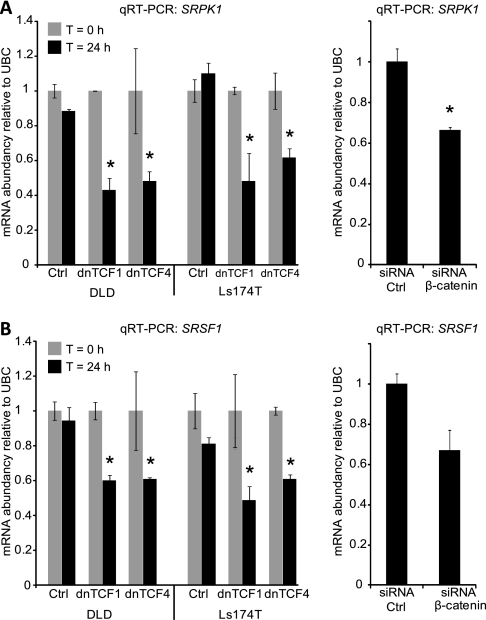
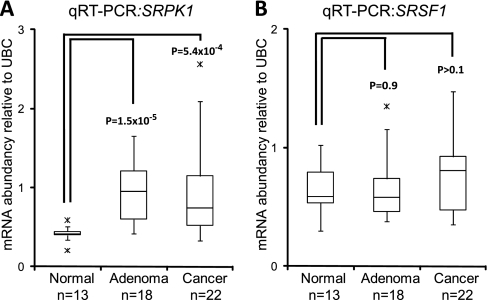
We used the Gene Ontology biological process term “RNA splicing” (GO:0008380) to identify 302 human genes involved in splicing regulation. Analysis of the expression of these splicing regulatory genes in our exon array dataset, revealed that four genes (SRPK1, SF3B3, PRPF4, and PPARGC1A) were significantly differentially expressed (p < 0.05 Benjamini-Hochberg corrected) between normal and tumor samples with a fold change of at least 1.5. The expression of the four genes was examined in an already published expression dataset of the DLD1 and Ls174T dnTCF Wnt-models described above (27). Only SRPK1 showed a twofold change in both cell lines indicating a regulatory role of the Wnt pathway on SRPK1 expression. Furthermore, a publicly available ChIP on chip dataset identifying TCF4 binding sites in the Ls174T CRC cell line (29) found a TCF4 binding site just upstream of the SRPK1 start exon (Supplemental Fig. 3A). The ChIP on chip study also revealed TCF4 binding just upstream of SRSF1 (Supplemental Fig. 3B). We measured the expression of both SRPK1 (Fig. 3A) and SRSF1 (Fig. 3B) by qRT-PCR in all Wnt models, and both SRPK1 and SRSF1 expression were consistently found to be lower when the Wnt pathway was inactivated. We then measured the expression of SRPK1 and SRSF1 in tissue samples and confirmed that SRPK1 is up-regulated in both adenoma samples (p = 1.5 × 10−5) and cancer samples (p = 5 × 10−4) (Fig. 4A), whereas SRSF1 expression was not altered significantly between normal mucosa and adenoma samples or normal mucosa and cancer samples (Fig. 4B). Similarly, a highly significant up-regulation of SRPK1 was seen in cancer samples from both gastric (Supplemental Fig. 4A) and lung cancer (Supplemental Fig. 4B). In contrast to CRC, SRSF1 expression was found to be slightly up-regulated (less than 2-fold) in both gastric (Supplemental Fig. 4C) and lung cancer (Supplemental Fig. 4D). No significant deregulation of either SRPK1 or SRSF1 was seen in bladder cancer (data not shown).
Fig. 3.
Expression of the splicing regulators SRPK1 and SRSF1 is regulated by Wnt signaling. SRPK1 and SRSF1 expression was quantified by qRT-PCR in the Wnt-model systems described in Fig. 2. A, Expression of SRPK1 in the dnTCF1, dnTCF4 (left panel) and β-catenin knockdown models (right panel). B, Expression of SRSF1 in the same models as in A. * = p < 0.05.
Fig. 4.
SRPK1 and SRSF1 expression in normal mucosa and tumor samples. SRPK1 and SRSF1 expression was quantified by qRT-PCR in colorectal tissue samples. A, Expression of SRPK1 and B, Expression of SRSF1.
Prediction of SRSF1 Binding to Exons 4A and 4B
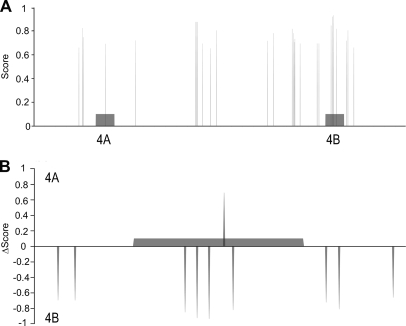
In silico predictions of splicing factor binding to exons 4A and 4B of SLC39A14 reported hits for a few splicing factors, where the most prominent predictions were for the exonic splicing enhancer SRSF1, which was predicted to bind predominantly to exon 4B, however, several binding motifs were found in the flanking introns as well (Fig. 5A). Because exons 4A and 4B are highly homologous, we aligned them and compared their SRSF1 candidate binding sites to find hits that were present in one, but not the other exon (Fig. 5B). This comparison revealed that despite the high sequence similarity between both exons (68%), several SRSF1 binding sites were present in exon 4B, but not in exon 4A. This is consistent with the higher inclusion rate of exon 4B that is seen when SRSF1 is expressed at a higher level (active Wnt pathway). However, in the colon tissue samples there was no significant difference in the expression of SRSF1 between normal and cancer samples. SRPK1 was, however, up-regulated in both adenoma and cancer samples, and because SRPK1 is capable of activating SRSF1 through phosphorylation, SRSF1 may be the splicing factor responsible for the changed SLC39A14 splicing pattern in both tissue samples and cell lines.
Fig. 5.
Prediction of SRSF1 binding sites in exon 4A and 4B and surrounding intronic sequences. A, Combined binding site scores for SRSF1 binding motifs crsmsgw and ugrwgvh. The scores summarize the clustering potential and evolutionary conservation of the candidate binding sites. B, Exon 4A and 4B specific hits. Δscores represent the difference between 4A and 4B scores in every sequence position of a pairwise alignment between both exons. Highly positive Δscore values indicates 4A specific motifs. Conversely, highly negative Δscore account for 4B specific motifs.
Knockdown of SRPK1 and SRSF1 in CRC Cell Lines Changes the 4A/4B Ratio
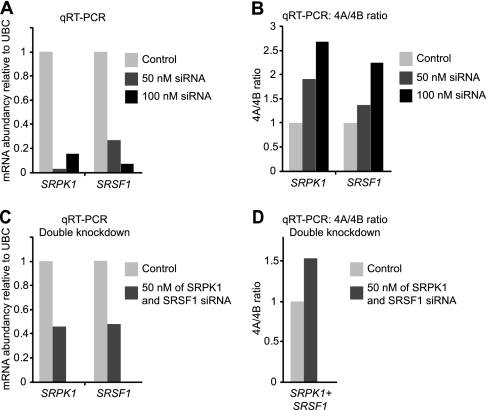
To further investigate the role of SRPK1 and SRSF1 in the AS of SLC39A14, we used siRNA to knock down SRPK1 and SRSF1 in the DLD1 and SW480 colon cancer cell lines. Knockdown of either SRPK1 or SRSF1 was successful in both cell lines (Fig. 6A and Supplemental Fig. 5A), and the expected increase in the exon 4A/4B ratio was also observed in both cell lines (Fig. 6B and Supplemental Fig. 5B) showing that individual knockdown of both SRPK1 and SRSF1 is able to change the ratio of exon 4A/4B. These results clearly support the role of both SRPK1 and SRSF1 in the regulation of SLC39A14 AS. To explore a potential cumulative effect of SRPK1 and SRSF1 on AS, we made double siRNA knockdown experiments of SRPK1 and SRSF1 in DLD1 and SW480 cells (Fig. 6C and Supplemental Fig. 5C). No cumulative effect was observed on the 4A/4B ratio (Fig. 6D, Supplemental Fig. 5D) indicating that double knockdown of SRPK1 and SRSF1 has no synergistic effect on SLC39A14 splicing.
Fig. 6.
Knockdown of SRPK1 and SRSF1 in DLD1 cells and the effect on 4A/4B ratio. A, SRPK1 and SRSF1 qRT-PCR expression in DLD1 cells treated with either control, SRPK1, or SRSF1 siRNA (50 nm or 100 nm). B, 4A/4B ratio following knockdown of SRPK1 or SRSF1 as in A. C, SRPK1 and SRSF1 qRT-PCR expression following their simultaneous knockdown in DLD1 cells treated with 100 nm siRNA in total. D, 4A/4B ratio following simultaneous knockdown of SRPK1 and SRSF1 as in C.
DISCUSSION
In the present study, we identified tumor specific AS of SLC39A14. The ratio (4A/4B) between the two mutually exclusive exons 4A and 4B was shown to be significantly higher in normal mucosa samples than in adenoma and cancer samples, regardless of adenoma type, microsatellite status, and cancer stage. In three different Wnt-pathway model systems and in two independent colorectal cancer cell lines, we have shown that the 4A/4B ratio is altered when Wnt signaling is abrogated. In addition, the same tumor specific AS pattern can be seen in gastric and lung cancer, where the Wnt pathway has been reported to be activated (4, 5). We have shown that a likely mechanism for regulating the AS of SLC39A14 is Wnt-signaling mediated up-regulation of SRPK1 in tumor samples and that knockdown of SRPK1 leads to an altered 4A/4B ratio. When the SRPK1 substrate SFSR1 is knocked down, the splicing pattern of SLC39A14 is also altered indicating that SRSF1 binds to SLC39A14 and regulate the AS of 4A/4B.
Previously, SLC39A14 has been found to be widely expressed across both human and mouse tissues, but no information about isoforms was provided (16, 17). The two isoforms of SLC39A14 that contain either exon 4A or 4B are both capable of binding zinc, iron, and cadmium, but interestingly, the isoform containing exon 4B has an eightfold higher affinity for Cd2+ than the isoform containing exon 4A (17). Hence, the data indicate that colorectal tumors, because of activated Wnt-signaling, have an increased Cd2+ uptake. In that respect, it is interesting to note that Cd2+ is a potent carcinogen, widely found in our environment, and whose uptake in the body can cause damage to multiple tissue types. Cd2+ has been found to influence apoptosis, differentiation, and cell growth, along with several other cellular processes (30). Although the direct mutagenic effect of Cd2+ is weak (31), there is strong evidence for several indirect mechanisms such as Cd2+ mediated disruption of DNA mismatch repair (30, 32, 33) and inhibition of base excision repair and nucleotide excision repair (34). Especially the inhibition of DNA mismatch repair has been linked to the development of CRC, and mismatch repair gene defects are found in the vast majority of hereditary nonpolyposis colorectal cancer (HNPCC) cases, and also in ∼20% of sporadic CRC (35). Finally, Cd2+ also seems to induce conformational changes of p53, thereby, impairing the p53 response to DNA damage (36). Altogether, this underlines the ability of Cd2+ to act indirectly as a strong carcinogen. Recently, Cd2+ has been found to induce Wnt signaling in kidney cells by disrupting the E-cadherin/β-catenin complex (31), and in mice chronic Cd2+ exposure has been found to up-regulate both Wnt ligands, Wnt receptors and Wnt activated genes in the kidney (37). These data, in addition to our findings, indicate a possible feedback mechanism for Wnt-pathway activity and cellular Cd2+ uptake.
The Wnt pathway plays a pivotal role in the development of CRC and nearly all colorectal tumors have an active Wnt pathway (1). The fact that a lower 4A/4B ratio was found in virtually all tumor samples investigated (including adenomas) is in concordance with a Wnt-regulated splicing event, and, furthermore, indicates that this is an early event in tumorigenesis. We interrupted the Wnt pathway either at the TCF1 or TCF4 as well as the β-catenin level of the pathway and confirmed the involvement of the Wnt pathway in the regulation of SLC39A14 AS. The existence of experimentally validated TCF4 binding sites both in the promoter regions of SRPK1 and SRSF1 makes these two genes obvious Wnt-targets, and both of them were found to be down-regulated when Wnt signaling was abrogated. In the two other Wnt-associated cancer types we investigated (gastric and lung) both SRPK1 and SRSF1 were significantly up-regulated in the cancer samples, although the fold change was greater for SRPK1. The observations of AS of SLC39A14 in gastric and lung cancer further support the role of the Wnt pathway in the splicing of SLC39A14. The Wnt pathway is not known to be involved in bladder cancer, and for this cancer type, we did not observe any AS of SLC39A14 or deregulation of SRPK1 or SRSF1, further indicating that AS of SLC39A14 is Wnt regulated and not a general effect of cancer development. The Wnt pathway has been reported to be involved in AS events in CRC cell lines, for example through deregulation of Srp20 (6, 38) and a few other splicing factors (39, 40). However, Wnt-regulated splicing caused by up-regulation of SRPK1 has to our knowledge not been reported before.
Splicing is a tightly regulated process that involves many factors, and SRSF1 is only one of many SR proteins that participate in splicing. SRPK1 is known to activate SR proteins by phosphorylation enabling them to migrate to the nucleus and participate in the splicing process (14). SRPK1 has been reported to be up-regulated in several human cancers including colon and breast where a knockdown of SRPK1 was shown to increase the number of cells undergoing apoptosis (13). Splicing factor binding prediction analysis showed that SRSF1 binding motifs were found primarily in exon 4B of SLC39A14, which could explain the increased presence of 4B when either SRPK1 or SRSF1 is up-regulated. We performed knockdown of both SRPK1 and SRSF1 in two colorectal cancer cell lines and found an increased 4A/4B ratio in concordance with the proposed binding of SRSF1 to exon 4B.
Cancer specific alternatively spliced isoforms have the potential to serve as cancer biomarkers and treatment targets. Several studies have identified isoforms that are potential biomarkers (41), either as general cancer markers or as potential diagnostic or prognostic markers (42). The striking difference in the 4A/4B ratio observed between normal mucosa and colorectal tumors in this study indicates that the ratio may be used as a tumor marker for identification of not only cancer, but precancerous lesions as well. Furthermore, our analyses suggest that this also applies to other tumor types that rely on Wnt activation.
In summary, we have shown AS of SLC39A14 in colorectal tumors, and our data strongly indicate that this AS is regulated by the Wnt pathway. We found that both SRPK1 and SRSF1 were regulated by the Wnt pathway and that knockdown of either SRPK1 or SRSF1 in colorectal cell lines led to a change in the 4A/4B isoform ratio supporting a role of these factors in the regulation of SLC39A14 splicing.
Acknowledgments
The excellent technical assistance of Inge-Lis Thorsen, Hanne Steen, Pamela Celis, Lisbet Kjeldsen, and Gitte Høj is gratefully appreciated. We thank Dr. Hans Clevers (The Hubrecht Laboratory, The Netherlands) for the inducible LS174T-derived and DLD1- derived cell lines. Previously described dnTCF4 gene expression profiles were kindly provided by Laurens G. van der Flier (The Hubrecht Laboratory, The Netherlands).
Footnotes
* This work was supported by The John and Birthe Meyer Foundation, the EU project GENICA Grant number 201630, The Danish Council for Independent Research Medical Sciences, the Lundbeck Foundation, and The Danish Ministry of the Interior and Health.
 This article contains supplemental Figs. 1–5.
This article contains supplemental Figs. 1–5.
1 The abbreviations used are:
- CRC
- Colorectal cancer
- AS
- alternative splicing
- dn
- dominant negative
- MSI
- Microsatellite instability
- MSS
- Microsatellite stable
- qRT-PCR
- Quantitative real-time reverse-transcription PCR.
REFERENCES
- 1. Bienz M., Clevers H. (2000) Linking colorectal cancer to Wnt signaling. Cell 103, 311–320 [DOI] [PubMed] [Google Scholar]
- 2. van de Wetering M., Sancho E., Verweij C., de Lau W., Oving I., Hurlstone A., van der Horn K., Batlle E., Coudreuse D., Haramis A. P., Tjon-Pon-Fong M., Moerer P., van den Born M., Soete G., Pals S., Eilers M., Medema R., Clevers H. (2002) The beta-catenin/TCF-4 complex imposes a crypt progenitor phenotype on colorectal cancer cells. Cell 111, 241–250 [DOI] [PubMed] [Google Scholar]
- 3. Van Scoyk M., Randall J., Sergew A., Williams L. M., Tennis M., Winn R. A. (2008) Wnt signaling pathway and lung disease. Transl Res. 151, 175–180 [DOI] [PubMed] [Google Scholar]
- 4. Clements W. M., Wang J., Sarnaik A., Kim O. J., MacDonald J., Fenoglio-Preiser C., Groden J., Lowy A. M. (2002) beta-Catenin mutation is a frequent cause of Wnt pathway activation in gastric cancer. Cancer Res. 62, 3503–3506 [PubMed] [Google Scholar]
- 5. Yue W., Sun Q., Dacic S., Landreneau R. J., Siegfried J. M., Yu J., Zhang L. (2008) Downregulation of Dkk3 activates beta-catenin/TCF-4 signaling in lung cancer. Carcinogenesis 29, 84–92 [DOI] [PubMed] [Google Scholar]
- 6. Gonçalves V., Matos P., Jordan P. (2008) The beta-catenin/TCF4 pathway modifies alternative splicing through modulation of SRp20 expression. RNA. 14, 2538–2549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Pan Q., Shai O., Lee L. J., Frey B. J., Blencowe B. J. (2008) Deep surveying of alternative splicing complexity in the human transcriptome by high-throughput sequencing. Nat. Genet. 40, 1413–1415 [DOI] [PubMed] [Google Scholar]
- 8. Wang E. T., Sandberg R., Luo S., Khrebtukova I., Zhang L., Mayr C., Kingsmore S. F., Schroth G. P., Burge C. B. (2008) Alternative isoform regulation in human tissue transcriptomes. Nature 456, 470–476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Long J. C., Caceres J. F. (2009) The SR protein family of splicing factors: master regulators of gene expression. Biochem. J. 417, 15–27 [DOI] [PubMed] [Google Scholar]
- 10. Skotheim R. I., Nees M. (2007) Alternative splicing in cancer: noise, functional, or systematic? Int J Biochem. Cell Biol. 39, 1432–1449 [DOI] [PubMed] [Google Scholar]
- 11. Faustino N. A., Cooper T. A. (2003) Pre-mRNA splicing and human disease. Genes. Dev. 17, 419–437 [DOI] [PubMed] [Google Scholar]
- 12. Karni R., de Stanchina E., Lowe S. W., Sinha R., Mu D., Krainer A. R. (2007) The gene encoding the splicing factor SF2/ASF is a proto-oncogene. Nat. Struct. Mol. Biol. 14, 185–193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Hayes G. M., Carrigan P. E., Miller L. J. (2007) Serine-arginine protein kinase 1 overexpression is associated with tumorigenic imbalance in mitogen-activated protein kinase pathways in breast, colonic, and pancreatic carcinomas. Cancer Res. 67, 2072–2080 [DOI] [PubMed] [Google Scholar]
- 14. Ma C. T., Hagopian J. C., Ghosh G., Fu X. D., Adams J. A. (2009) Regiospecific phosphorylation control of the SR protein ASF/SF2 by SRPK1. J. Mol. Biol. 390, 618–634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Ngo J. C., Giang K., Chakrabarti S., Ma C. T., Huynh N., Hagopian J. C., Dorrestein P. C., Fu X. D., Adams J. A., Ghosh G. (2008) A sliding docking interaction is essential for sequential and processive phosphorylation of an SR protein by SRPK1. Mol. Cell 29, 563–576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Taylor K. M., Morgan H. E., Johnson A., Nicholson R. I. (2005) Structure-function analysis of a novel member of the LIV-1 subfamily of zinc transporters, ZIP14. FEBS Lett. 579, 427–432 [DOI] [PubMed] [Google Scholar]
- 17. Girijashanker K., He L., Soleimani M., Reed J. M., Li H., Liu Z., Wang B., Dalton T. P., Nebert D. W. (2008) Slc39a14 gene encodes ZIP14, a metal/bicarbonate symporter: similarities to the ZIP8 transporter. Mol. Pharmacol. 73, 1413–1423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Liuzzi J. P., Aydemir F., Nam H., Knutson M. D., Cousins R. J. (2006) Zip14 (Slc39a14) mediates non-transferrin-bound iron uptake into cells. Proc. Natl. Acad. Sci. U.S.A. 103, 13612–13617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Thorsen K., Sørensen K. D., Brems-Eskildsen A. S., Modin C., Gaustadnes M., Hein A. M., Kruhøffer M., Laurberg S., Borre M., Wang K., Brunak S., Krainer A. R., Tørring N., Dyrskjøt L., Andersen C. L., Orntoft T. F. (2008) Alternative splicing in colon, bladder, and prostate cancer identified by exon array analysis. Mol. Cell Proteomics 7, 1214–1224 [DOI] [PubMed] [Google Scholar]
- 20. Andersen C. L., Jensen J. L., Ørntoft T. F. (2004) Normalization of real-time quantitative reverse transcription-PCR data: a model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets. Cancer Res. 64, 5245–5250 [DOI] [PubMed] [Google Scholar]
- 21. Gardina P. J., Clark T. A., Shimada B., Staples M. K., Yang Q., Veitch J., Schweitzer A., Awad T., Sugnet C., Dee S., Davies C., Williams A., Turpaz Y. (2006) Alternative splicing and differential gene expression in colon cancer detected by a whole genome exon array. BMC. Genomics. 7, 325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Xi L., Feber A., Gupta V., Wu M., Bergemann A. D., Landreneau R. J., Litle V. R., Pennathur A., Luketich J. D., Godfrey T. E. (2008) Whole genome exon arrays identify differential expression of alternatively spliced, cancer-related genes in lung cancer. Nucleic. Acids. Res. 36, 6535–6547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Andersen C. L., Christensen L. L., Thorsen K., Schepeler T., Sørensen F. B., Verspaget H. W., Simon R., Kruhøffer M., Aaltonen L. A., Laurberg S., Ørntoft T. F. (2009) Dysregulation of the transcription factors SOX4, CBFB and SMARCC1 correlates with outcome of colorectal cancer. Br. J. Cancer 100, 511–523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Paz I., Akerman M., Dror I., Kosti I., Mandel-Gutfreund Y. (2010) SFmap: a web server for motif analysis and prediction of splicing factor binding sites. Nucleic Acids Res. 38 Suppl, W281–285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Akerman M., David-Eden H., Pinter R. Y., Mandel-Gutfreund Y. (2009) A computational approach for genome-wide mapping of splicing factor binding sites. Genome. Biol. 10, R30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Schepeler T., Mansilla F., Christensen L. L., Orntoft T. F., Andersen C. L. (2007) Clusterin expression can be modulated by changes in TCF1-mediated Wnt signaling. J Mol Signal 2, 6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Van der Flier L. G., Sabates-Bellver J., Oving I., Haegebarth A., De Palo M., Anti M., Van Gijn M. E., Suijkerbuijk S., Van de Wetering M., Marra G., Clevers H. (2007) The Intestinal Wnt/TCF Signature. Gastroenterology 132, 628–632 [DOI] [PubMed] [Google Scholar]
- 28. van de Wetering M., de Lau W., Clevers H. (2002) WNT signaling and lymphocyte development. Cell 109 Suppl, S13–19 [DOI] [PubMed] [Google Scholar]
- 29. Hatzis P., van der Flier L. G., van Driel M. A., Guryev V., Nielsen F., Denissov S., Nijman I. J., Koster J., Santo E. E., Welboren W., Versteeg R., Cuppen E., van de Wetering M., Clevers H., Stunnenberg H. G. (2008) Genome-wide pattern of TCF7L2/TCF4 chromatin occupancy in colorectal cancer cells. Mol. Cell. Biol. 28, 2732–2744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Wieland M., Levin M. K., Hingorani K. S., Biro F. N., Hingorani M. M. (2009) Mechanism of cadmium-mediated inhibition of Msh2-Msh6 function in DNA mismatch repair. Biochemistry 48, 9492–9502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Chakraborty P. K., Lee W. K., Molitor M., Wolff N. A., Thévenod F. (2010) Cadmium induces Wnt signaling to upregulate proliferation and survival genes in sub-confluent kidney proximal tubule cells. Mol. Cancer 9, 102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Jin Y. H., Clark A. B., Slebos R. J., Al-Refai H., Taylor J. A., Kunkel T. A., Resnick M. A., Gordenin D. A. (2003) Cadmium is a mutagen that acts by inhibiting mismatch repair. Nat. Genet. 34, 326–329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Banerjee S., Flores-Rozas H. Cadmium inhibits mismatch repair by blocking the ATPase activity of the MSH2-MSH6 complex. Nucleic. Acids Res. 33, 1410–1419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Schwerdtle T., Ebert F., Thuy C., Richter C., Mullenders L. H., Hartwig A. (2010) Genotoxicity of soluble and particulate cadmium compounds: impact on oxidative DNA damage and nucleotide excision repair. Chem. Res. Toxicol. 23, 432–442 [DOI] [PubMed] [Google Scholar]
- 35. Peltomäki P. (2003) Role of DNA mismatch repair defects in the pathogenesis of human cancer. J Clin. Oncol. 21, 1174–1179 [DOI] [PubMed] [Google Scholar]
- 36. Méplan C., Mann K., Hainaut P. (1999) Cadmium induces conformational modifications of wild-type p53 and suppresses p53 response to DNA damage in cultured cells. J. Biol. Chem. 274, 31663–31670 [DOI] [PubMed] [Google Scholar]
- 37. Chakraborty P. K., Scharner B., Jurasovic J., Messner B., Bernhard D., Thevenod F. (2010) Chronic cadmium exposure induces transcriptional activation of the Wnt pathway and upregulation of epithelial-to-mesenchymal transition markers in mouse kidney. Toxicol Lett. [DOI] [PubMed] [Google Scholar]
- 38. Gonçalves V., Matos P., Jordan P. (2009) Antagonistic SR proteins regulate alternative splicing of tumor-related Rac1b downstream of the PI3-kinase and Wnt pathways. Hum. Mol. Genet. 18, 3696–3707 [DOI] [PubMed] [Google Scholar]
- 39. Shitashige M., Naishiro Y., Idogawa M., Honda K., Ono M., Hirohashi S., Yamada T. (2007) Involvement of splicing factor-1 in beta-catenin/T-cell factor-4-mediated gene transactivation and pre-mRNA splicing. Gastroenterology 132, 1039–1054 [DOI] [PubMed] [Google Scholar]
- 40. Sato S., Idogawa M., Honda K., Fujii G., Kawashima H., Takekuma K., Hoshika A., Hirohashi S., Yamada T. (2005) beta-catenin interacts with the FUS proto-oncogene product and regulates pre-mRNA splicing. Gastroenterology 129, 1225–1236 [DOI] [PubMed] [Google Scholar]
- 41. Brinkman B. M. (2004) Splice variants as cancer biomarkers. Clin. Biochem. 37, 584–594 [DOI] [PubMed] [Google Scholar]
- 42. Ghigna C., Valacca C., Biamonti G. (2008) Alternative splicing and tumor progression. Curr. Genomics. 9, 556–570 [DOI] [PMC free article] [PubMed] [Google Scholar]