Abstract
Recent data from several laboratories have provided evidence that the newly fertilized oocyte inherits epigenetic signals from the sperm chromatin that are required for proper embryonic development. For the purposes of this review, the term epigenetic is used to describe all types of molecular information that are transmitted from the sperm cell to the embryo. There are at least six different forms of epigenetic information that have already been established as being required for proper embryogenesis in mammals or for which there is evidence that it may do so. These are (i) DNA methylation; (ii) sperm-specific histones, (iii) other chromatin-associated proteins; (iv) the perinuclear theca proteins; (v) sperm-born RNAs and, the focus of this review; and (vi) the DNA loop domain organization by the sperm nuclear matrix. These epigenetic signals should be considered when designing protocols for the manipulation and cryopreservation of spermatozoa for assisted reproductive technology as necessary components for effective fertilization and subsequent embryo development.
Keywords: embryogenesis, epigenetics, sperm DNA, sperm nuclear matrix, DNA loop domain organization
Introduction
Basic and clinical fertility research has benefited from the interest of many other fields in the unique biological systems involved in reproduction. The discoveries from laboratories focused on other interests but using fertility as a model have provided important insights into the molecular and biological mechanisms of fertility, often with direct consequences to clinical research and practice. The role of the sperm nucleus in fertility is an important example. Several years ago, a group of researchers interested in the basic principles governing nuclear function developed a model in which Xenopus oocyte extracts could be induced to form nuclei around purified DNA added to the system.1 These synthetic nuclei had double plasma membranes characteristic of normal nuclei and condensed the DNA into histone-bound chromatin. They were also functionally competent in that they could replicate, although inefficiently, the foreign DNA that was used to induce nuclear formation2, 3 and transcribe the exogenous DNA into RNA.4 These studies established that the oocyte cytoplasm contains all the factors that are required to fold naked DNA into functional chromatin and form a nucleus, de novo. They supported the idea that the spermatozoon's sole function was to deliver the genetic information into the oocyte in pristine condition. In this model, the reorganization of the paternal chromatin after fertilization was absolute, and no aspects of DNA packaging in the sperm cell were maintained in the newly fertilized zygote. The only contribution of the father to the progeny was the paternal DNA sequence.
More recent data, however, suggest that this model was incomplete, and the zygote also inherits certain elements from sperm chromatin that are necessary signals for proper development of the embryo. Our laboratory focused on one of these, the organization of DNA by the sperm nuclear matrix,5 but this is not the only aspect of sperm chromatin that is transferred to the newly fertilized egg. Evidence suggests that some sperm histones may also be inherited by the paternal pronuclear chromatin from the sperm cell,6, 7 and several studies have demonstrated that sperm-born RNA is also delivered to the egg.8, 9, 10 The clinical implications of these new discoveries about the variety of information inherited by the zygote from the sperm cell for assisted reproductive technologies (ARTs) are that we need to take care to protect these important molecular signals when storing and manipulating spermatozoa in the clinic. Here, we will discuss the current models for the contribution of sperm cell to the developing embryo and the specific implications for ARTs.
Sperm chromatin structure compartmentalizes active sites
Spermatozoa have the most unique chromatin structure among all known cell types. It is the most condensed DNA known in eukaryotic cells, and its structure is impervious to electron microscopy.11, 12 This condensation is accomplished during spermiogenesis when protamines replace histones as the major DNA-binding protein in sperm chromatin.13, 14 Protamines coil the sperm DNA into tightly packed toroids that approach a crystalline-like state of condensation.15, 16 Many excellent reviews have been written about this important family of proteins and how they function to package sperm DNA;17 hence a review of toriod structure will not be necessary here. However, one consequence of this packaging will be discussed in the light of a recently proposed model for sperm DNA packaging.18
It has long been known that a small portion of mammalian sperm DNA remains associated with histones in mature mammalian spermatozoa.19, 20, 21, 22 Recently, two separate laboratories have demonstrated that these remnant histones are associated with specific DNA sequences.23, 24 Similarly, it has been shown that sperm DNA is also organized into loops by the sperm nuclear matrix.25, 26 These data prompted a recently proposed model for sperm chromatin structure in which most of the DNA is coiled into inaccessible protamine toroids, and the DNA between each toroid is attached to the nuclear matrix18 (Figure 1, condensed chromatin, lower left). Sometimes the entire loops remain bound to histones, but these are supported structurally by neighboring protamine-bound loops.
Figure 1.
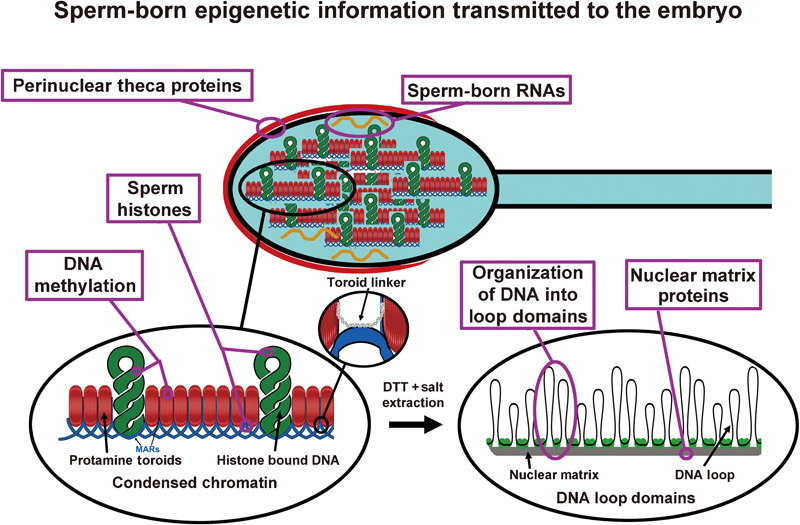
Sperm-born epigenetic information. This diagram illustrates some of the molecular information that the sperm nucleus transmits to the oocytes, much of which may have direct influences on development. DNA methylation is the best-known example of non-DNA sequence information that is required for embryogenesis, but other potentially important elements have been described. Sperm DNA is tightly condensed by protamines into toroids (lower left inset), but some histones remain bound to the chromatin. The DNA is organized into loop domains that are required for DNA replication in the oocyte. Proteins of the nuclear matrix and perinuclear theca are also delivered to the oocyte. DTT, dithiothreitol; MARs, matrix attachment regions.
The aspect of protamine binding that will be discussed here is that this unique chromatin structure naturally fractionates the non-protamine-bound chromatin. Exogenous nucleases can digest histone-bound chromatin, but cannot penetrate the protamine toroids to digest most of the sperm chromatin.27, 28, 29, 30 As discussed below, evidence is accumulating that these nuclease-sensitive, toroid linker, nuclear matrix-associated chromatin sites are active centers of sperm chromatin that confer molecular instructions to the zygote. Thus, the protamines, which most likely serve to protect the paternal genome during the transit that it must endure before fertilization, also serve to concentrate these active centers in the sperm nucleus into the only accessible chromatin to the oocyte. That is, the protamines sequester 90–99% of the sperm DNA (depending on the species) into an inaccessible crystalline lattice.22, 28, 30, 31, 32, 33 The remaining minor portion of the paternal DNA, which is sensitive to nucleases and other DNA-binding proteins, may also represent the seeds of function in the paternal pronucleus. Thus, the condensation of sperm DNA by protamines leaves only a small fraction of the genome in the spermatozoon that remains accessible to the DNA-binding proteins that are required to activate DNA replication and gene transcription. These may, in fact, be the most important sites for the initiation of paternal genome function in the early embryo.
Inherited epigenetic information
These active sites of sperm chromatin at the protamine toroids may contain important information for the developing embryo in addition to the DNA sequence. Any such information will be referred to as epigenetic in this review. Here, we will briefly mention several possible epigenetic moieties that may contain important signals for embryogenesis. The first, methylation, is the best-known example, and is not expected to be sequestered into these active sites, but distributed throughout the paternal genome.
DNA methylation
In 1984, two laboratories clearly demonstrated that for mammals the paternal and maternal genomes were not equal.34, 35 By transferring pronuclei using micromanipulation techniques, this group demonstrated that fertilized mouse oocytes that had two male pronuclei or two female pronuclei could not develop. The molecular mechanism for this difference turned out to be differential methylation of imprinted genes in the oocyte and sperm DNA.36, 37 More recent work using cloning techniques helped narrow the timing of the establishment of these differential methylation patterns to early embryonic development.38 In this study, the point at which the nuclei of primordial germ cells, the cells that differentiate into oogonia or spermatogonia, can no longer support development of a mouse by cloning, corresponded to the establishment of sex-specific methylation patterns. Many excellent reviews have been written about methylation and imprinting, and this will not be discussed here. The important point for this review is that methylation is the most well-characterized example of the epigenetic contribution of the sperm nucleus to the developing embryo. Without proper paternal methylation, the embryo cannot develop. More importantly, several studies have shown that this particular type of methylation can be altered in ART.39, 40, 41 DNA methylation is a covalent modification of DNA. If this form of epigenetic modification of the chromatin is susceptible to techniques used in ART, it is likely that the non-covalent chromatin modifications, discussed below, are also susceptible.
Sperm histones: inheritance of higher order chromatin structure
The continuing presence of histones in fully mature spermatozoa raised the question of whether these were left as residual chromatin representing incomplete chromatin remodeling during spermiogenesis, or whether the relatively small histone-bound fraction of sperm DNA had a functional significance. An early attempt to address this question demonstrated that some individual sequences could be attributed to the protamine versus histone-bound fraction of sperm chromatin.29 Recently, two groups focused on the genome-wide distribution of histones in human sperm nuclei and found evidence for small and large segments of sperm chromatin that were specifically associated with histones.24, 30 These data supported the idea that during chromatin condensation, some histones remain associated with specific sequences of the sperm DNA, suggesting a programmed distribution rather than residual deposition. Furthermore, there now exists evidence to support a functional role for these residual sperm histones in the newly fertilized oocyte. In the mouse, histone variants H4 acetylated on K8 or K12 (H4K8ac or H4K12ac)42 and in humans, histone H3.1 and H3.26 were inherited by the newly fertilized oocyte from the sperm nucleus. Histone covalent modifications are associated with a variety of nuclear function including transcriptional control, chromatin packaging and DNA methylation. Thus, it seems probable that the sperm cell contributes epigenetic signals for the function of the paternal genome in the form of histone modifications. The same is most likely true for the oocyte, although this is not surprising because the maternal chromosomes remain bound to histones throughout fertilization.
If this proves to be correct, it would have important implications for ARTs. Histones are much more easily extracted from DNA than protamines, and histone-bound DNA is much more susceptible to virtually all types of DNA-damaging agents than is protamine-bound DNA. Once again, this is a component of sperm structure that needs to be taken into account when analyzing techniques for sperm manipulation and cryopreservation.
Non-histone, sperm nuclear proteins
Several laboratories have published proteomic analyses of sperm proteins using mass spectrometry.43, 44 For comprehensive reviews of this subject, see Aitken and Baker45 and Castillo and Oliva.46 Analyses of the different areas of the spermatozoon are sure to follow, and at least one group has published a proteomic analysis of the rat sperm nuclear matrix.47 We have reported evidence that the DNA loop domain organization in the sperm nucleus is required for embryogenesis (see below). This infers that at least a portion of the proteins of the nuclear matrix may also be inherited by the newly fertilized embryo. The sperm cell also enters the oocyte with the perinuclear theca still attached. This organelle also contains a host of proteins, one of which is an extranuclear located histone H2B.48 Many of these proteins are likely to be incorporated into the functioning paternal pronucleus, and may also be counted as part of the epigenetic inheritance of the embryo from the spermatozoon.
Sperm nuclear RNA
Recently, several independent laboratories have demonstrated that the fully mature spermatozoon contains several types of RNAs.8, 9, 10 These RNAs are also thought to be carried into the oocyte with the spermatozoa. Because many of these are microRNAs with known functions in transcriptional regulation, it is possible that these RNAs contribute to the regulation of the paternal genome in the one-cell embryo. If so, sperm-born RNAs can be considered as another form of epigenetic information that is passed from the father to the embryo.
DNA loop domain organization by the sperm nuclear matrix: transmission of a function scaffold for the paternal genome
It has long been known that sperm DNA, similar to that of somatic cells, is organized into loop domains of about 20–50 kb that are attached at their bases to the a proteinaceous structure termed the nuclear matrix.49, 50, 51, 52 The work from our laboratory has focused on the hypothesis that sperm DNA loop domain organization is inherited by the newly fertilized embryo and that this organization is required for proper embryonic development.53, 54, 55 This idea was a logical extension of the work that had been done in somatic cells on the function of the nuclear matrix. Several different laboratories had revealed that DNA was replicated at the base of the loops, with the nuclear matrix serving as the scaffold on which the replication machinery was assembled.56, 57, 58, 59 The DNA is reeled through the fixed replication ‘factory' on the nuclear matrix. Origins of DNA replication are initially located on the nuclear matrix.60, 61 Many groups have also suggested that RNA transcription takes place on the nuclear matrix by similarly fixing the transcription machinery to one site on the nuclear matrix.49, 52, 62, 63, 64 The nuclear matrix clearly has a role in the function of somatic cell DNA.
A similar role is almost certainly played by the nuclear matrix of the paternal nucleus in the one-cell embryo. The question is whether the DNA loop domain organization was constructed de novo in the embryo during sperm decondensation and the subsequent nuclear formation, or is the paternal loop attachment structure inherited by the embryo from the sperm cell? This latter possibility is plausible, as nuclear matrices in somatic cells have been shown to expand when histones are extracted. It is therefore possible that the nuclear matrix in the round spermatid contracts during spermiogenesis when the chromatin is condensed by protamine deposition and then expands again in the embryo. But is this what happens?
Recent evidence from different laboratories suggests that this might, indeed, be the case. The first piece of supporting evidence comes from the cloning field. The fact that transferring adult, somatic cell nuclei (nuclear transfer or cloning) into enucleated oocytes results in live births demonstrates that the entire DNA required for embryogenesis remains intact throughout life.65, 66 However, the efficiency of cloning remains extremely low—only 2–5% of nuclear transfer embryos develop to term. Most researchers in the field believe that this is due to chromatin reprogramming, but the exact nature of the chromatin elements that need to be altered is unknown. One group has recently demonstrated that adult erythrocyte nuclei preincubated in mitotic egg extracts could replicate much more efficiently than nuclei that were not preincubated.67 The most obvious chromatin structure that was altered in the preincubated nuclei was the DNA loop domain organization, which was markedly shortened. This suggests that the organization of DNA by the nuclear matrix is crucial for DNA replication in the zygote. In support of this, we have recently demonstrated that replication of the mouse paternal genome in the one-cell embryo requires the sperm nuclear matrix and the proper DNA attachment sites.25 When original attachment configuration of DNA to the nuclear matrix is disrupted, the DNA is no longer replicated.25, 68
These studies support the model that the embryo inherits the DNA loop domain organization of the paternal genome from the sperm cell. The sperm nuclear matrix may serve as a ‘function scaffold' on which the DNA is replicated. Thus, it is the three-dimensional organization of sperm DNA, in addition to the DNA sequence, that is required for one of the first steps of embryogenesis—the replication of the paternal genome. The sites of DNA attachment to the nuclear matrix are called matrix attachment regions and correspond to the ‘seeds of chromatin function', located at the protamine toroid linker regions, mentioned above. This is one additional level of chromatin structure that must be preserved in sperm micromanipulation and cryopreservation techniques used in ART.
The sperm nuclear matrix as a possible checkpoint for chromatin stability
The sperm nuclear matrix, which is required for proper embryogenesis, also has a function in live, mature spermatozoa before fertilization. Again, this function was tested in spermatozoa because of its role in somatic cells. When cells undergo apoptosis, the first step of DNA degradation is the reversible cleavage of DNA by topoisomerase II located at the bases of the DNA loop domains on the nuclear matrix.69, 70, 71 This cleavage results in the degradation of the entire chromatin into loop-sized fragments of 60–100 kb. In most cases, these double-stranded DNA breaks can actually be reversed by inducing the topoisomerase II to religate the broken strands of DNA. In the second step, one or more nucleases initiate a more complete digestion of the DNA and this step cannot be reversed.72, 73, 74 Although the exact mechanism of this degradation is unknown, some evidence points to a direct interaction between topoisomerase II and nucleases. This model suggests that the DNA attachment site to the nuclear matrix and a closely associated topoisomerase II serves as a checkpoint or simply as the initiation point for the DNA degradation in apoptosis. Topoisomerase II is one of the components of the sperm nuclear matrix proteins (Oliva R, pers. commun., 2010), and may also be inherited by the embryo.
We have demonstrated that mouse spermatozoa can be induced to digest the entire paternal genome into 20–50 kb fragments and that this digestion can be reversed with EDTA, a typical reversal reagent for topoisomerase II cleavage.75 Evidence suggests that the point of cleavage is the matrix attachment regions, the same sites that we have described as harboring the origins of replication for the paternal pronucleus. These are the only accessible parts of the sperm chromatin to most types of DNA-damaging agents, including enzymes. These active chromatin regions in the sperm cell may also function before fertilization as checkpoint regions for the integrity of sperm chromatin structure.
Conclusion
There are at least six components of the sperm nucleus other than the DNA that have either already been shown to be inherited by the paternal nucleus or for which there is evidence that suggests that they are. These are DNA methylation, sperm-specific histones, other chromatin-associated proteins such as topoisomerase II, the perinuclear theca proteins, sperm-born RNAs and the focus of this review, the DNA loop domain organization by the sperm nuclear matrix. Other chemical signals in the form of lipids or carbohydrates may also be discovered in the future—the list of epigenetic components presented here is almost certainly not complete. The molecularly diverse groups of epigenetic signals that are transferred to the oocyte by the spermatozoon also speak of the complex nature of inheritance.
As with many other aspects of reproductive biology, this conclusion has two important implications—one for cell and molecular biology and one for clinical reproduction. The first has just been mentioned, that inheritance is much more complex than the transmission of the information embedded in the DNA sequence of the parents. The parental chromatin also provides a complex series of instructions for the proper execution of the genetic program encoded in the DNA in the form of epigenetic signals. The second implication is another caution for the clinical infertility. Evidence has already been reported that one of the six types of epigenetic signals that may be transmitted from the sperm to the embryo, methylation, may be disrupted by ART.39, 40, 41 This raises the possibility that other, less stable, epigenetic signals may also be disrupted by the gamete manipulation used in ART procedures. Fortunately, the vast majority of children born from ARTs are normal, and any potential hazards will be minor, if they exist at all. Still, a better understanding of the epigenetic contributions of the sperm to the embryo may increase embryo survival in vitro before transplantation to the mother and/or increase the stability of pregnancies.
The study of the still enigmatic mammalian sperm chromatin continues to provide new insights into reproductive biology. However, because of its unique function, and unique divestment of most of the normal chromatin attributes during spermiogenesis, it also provides important foundations for chromatin structure in somatic cells.
Acknowledgments
This work was supported by an NIH/NICHD Grant R01HD060722 to W. Steven Ward.
The authors declare no competing financial interests.
References
- Harland RM, Laskey RA. Regulated replication of DNA microinjected into eggs of Xenopus laevis. Cell. 1980;21:761–71. doi: 10.1016/0092-8674(80)90439-0. [DOI] [PubMed] [Google Scholar]
- Blow JJ, Laskey RA. Initiation of DNA replication in nuclei and purified DNA by a cell-free extract of Xenopus eggs. Cell. 1986;47:577–87. doi: 10.1016/0092-8674(86)90622-7. [DOI] [PubMed] [Google Scholar]
- Newport J. Nuclear reconstitution in vitro: stages of assembly around protein-free DNA. Cell. 1987;48:205–17. doi: 10.1016/0092-8674(87)90424-7. [DOI] [PubMed] [Google Scholar]
- Ullman KS, Forbes DJ. RNA polymerase III transcription in synthetic nuclei assembled in vitro from defined DNA templates. Mol Cell Biol. 1995;15:4873–83. doi: 10.1128/mcb.15.9.4873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaman JA, Yamauchi Y, Ward WS. Function of the sperm nuclear matrix. Arch Androl. 2007;53:135–40. doi: 10.1080/01485010701329378. [DOI] [PubMed] [Google Scholar]
- van der Heijden GW, Ramos L, Baart EB, van den Berg IM, Derijck AA, et al. Sperm-derived histones contribute to zygotic chromatin in humans. BMC Dev Biol. 2008;8:34. doi: 10.1186/1471-213X-8-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Heijden GW, van den Berg IM, Baart EB, Derijck AA, Martini E, et al. Parental origin of chromatin in human monopronuclear zygotes revealed by asymmetric histone methylation patterns, differs between IVF and ICSI. Mol Reprod Dev. 2009;76:101–8. doi: 10.1002/mrd.20933. [DOI] [PubMed] [Google Scholar]
- Lalancette C, Miller D, Li Y, Krawetz SA. Paternal contributions: new functional insights for spermatozoal RNA. J Cell Biochem. 2008;104:1570–9. doi: 10.1002/jcb.21756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan W, Morozumi K, Zhang J, Ro S, Park C, et al. Birth of mice after intracytoplasmic injection of single purified sperm nuclei and detection of messenger RNAs and MicroRNAs in the sperm nuclei. Biol Reprod. 2008;78:896–902. doi: 10.1095/biolreprod.107.067033. [DOI] [PubMed] [Google Scholar]
- Carrell DT. Contributions of spermatozoa to embryogenesis: assays to evaluate their genetic and epigenetic fitness. Reprod Biomed Online. 2008;16:474–84. doi: 10.1016/s1472-6483(10)60454-3. [DOI] [PubMed] [Google Scholar]
- Fawcett DW, Anderson WA, Phillips DM. Morphogenetic factors influencing the shape of the sperm head. Dev Biol. 1971;26:220–51. doi: 10.1016/0012-1606(71)90124-2. [DOI] [PubMed] [Google Scholar]
- Yanagimachi R, Noda YD. Electron microscope studies of sperm incorporation into the golden hamster egg. Am J Anat. 1970;128:429–62. doi: 10.1002/aja.1001280404. [DOI] [PubMed] [Google Scholar]
- Marushige K, Marushige Y, Wong TK. Complete displacement of somatic histones during transformation of spermatid chromatin: a model experiment. Biochemistry. 1976;15:2047–53. doi: 10.1021/bi00655a004. [DOI] [PubMed] [Google Scholar]
- Brewer LR, Corzett M, Balhorn R. Protamine-induced condensation and decondensation of the same DNA molecule. Science. 1999;286:120–3. doi: 10.1126/science.286.5437.120. [DOI] [PubMed] [Google Scholar]
- Hud NV, Downing KH, Balhorn R. A constant radius of curvature model for the organization of DNA in toroidal condensates. Proc Natl Acad Sci USA. 1995;92:3581–5. doi: 10.1073/pnas.92.8.3581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hud NV, Vilfan ID. Toroidal DNA condensates: unraveling the fine structure and the role of nucleation in determining size. Annu Rev Biophys Biomol Struct. 2005;34:295–318. doi: 10.1146/annurev.biophys.34.040204.144500. [DOI] [PubMed] [Google Scholar]
- Balhorn R. The protamine family of sperm nuclear proteins. Genome Biol. 2007;8:227. doi: 10.1186/gb-2007-8-9-227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward WS. Function of sperm chromatin structural elements in fertilization and development. Mol Hum Reprod. 2010;16:30–6. doi: 10.1093/molehr/gap080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manfredi Romanini MG, Biggiogera M, Formenti D, Fraschini A, Garagna S, et al. Sperm-chromatin maturation in the mouse. A cytochemical approach. Histochemistry. 1986;84:484–91. doi: 10.1007/BF00482981. [DOI] [PubMed] [Google Scholar]
- Tanphaichitr N, Sobhon P, Taluppeth N, Chalermisarachai P. Basic nuclear proteins in testicular cells and ejaculated spermatozoa in man. Exp Cell Res. 1978;117:347–56. doi: 10.1016/0014-4827(78)90148-9. [DOI] [PubMed] [Google Scholar]
- Puwaravutipanich T, Panyim S. The nuclear basic proteins of human testes and ejaculated spermatozoa. Exp Cell Res. 1975;90:153–8. doi: 10.1016/0014-4827(75)90368-7. [DOI] [PubMed] [Google Scholar]
- Bench GS, Friz AM, Corzett MH, Morse DH, Balhorn R. DNA and total protamine masses in individual sperm from fertile mammalian subjects. Cytometry. 1996;23:263–71. doi: 10.1002/(SICI)1097-0320(19960401)23:4<263::AID-CYTO1>3.0.CO;2-I. [DOI] [PubMed] [Google Scholar]
- Hammoud S, Liu L, Carrell DT. Protamine ratio and the level of histone retention in sperm selected from a density gradient preparation. Andrologia. 2009;41:88–94. doi: 10.1111/j.1439-0272.2008.00890.x. [DOI] [PubMed] [Google Scholar]
- Linnemann AK, Platts AE, Krawetz SA. Differential nuclear scaffold/matrix attachment marks expressed genes. Hum Mol Genet. 2009;18:645–54. doi: 10.1093/hmg/ddn394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaman JA, Yamauchi Y, Ward WS. The sperm nuclear matrix is required for paternal DNA replication. J Cell Biochem. 2007;102:680–8. doi: 10.1002/jcb.21321. [DOI] [PubMed] [Google Scholar]
- Martins RP, Ostermeier GC, Krawetz SA. Nuclear matrix interactions at the human protamine domain: a working model of potentiation. J Biol Chem. 2004;279:51862–8. doi: 10.1074/jbc.M409415200. [DOI] [PubMed] [Google Scholar]
- Sotolongo B, Lino E, Ward WS. Ability of hamster spermatozoa to digest their own DNA. Biol Reprod. 2003;69:2029–35. doi: 10.1095/biolreprod.103.020594. [DOI] [PubMed] [Google Scholar]
- Pittoggi C, Renzi L, Zaccagnini G, Cimini D, Degrassi F, et al. A fraction of mouse sperm chromatin is organized in nucleosomal hypersensitive domains enriched in retroposon DNA. J Cell Sci. 1999;112:3537–48. doi: 10.1242/jcs.112.20.3537. [DOI] [PubMed] [Google Scholar]
- Gatewood JM, Cook GR, Balhorn R, Bradbury EM, Schmid CW. Sequence-specific packaging of DNA in human sperm chromatin. Science. 1987;236:962–4. doi: 10.1126/science.3576213. [DOI] [PubMed] [Google Scholar]
- Hammoud SS, Nix DA, Zhang H, Purwar J, Carrell DT, et al. Distinctive chromatin in human sperm packages genes for embryo development. Nature. 2009;460:473–8. doi: 10.1038/nature08162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adenot PG, Mercier Y, Renard JP, Thompson EM. Differential H4 acetylation of paternal and maternal chromatin precedes DNA replication and differential transcriptional activity in pronuclei of 1-cell mouse embryos. Development. 1997;124:4615–25. doi: 10.1242/dev.124.22.4615. [DOI] [PubMed] [Google Scholar]
- Churikov D, Siino J, Svetlova M, Zhang K, Gineitis A, et al. Novel human testis-specific histone H2B encoded by the interrupted gene on the X chromosome. Genomics. 2004;84:745–56. doi: 10.1016/j.ygeno.2004.06.001. [DOI] [PubMed] [Google Scholar]
- Gineitis AA, Zalenskaya IA, Yau PM, Bradbury EM, Zalensky AO. Human sperm telomere-binding complex involves histone H2B and secures telomere membrane attachment. J Cell Biol. 2000;151:1591–8. doi: 10.1083/jcb.151.7.1591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGrath J, Solter D. Completion of mouse embryogenesis requires both the maternal and paternal genomes. Cell. 1984;37:179–83. doi: 10.1016/0092-8674(84)90313-1. [DOI] [PubMed] [Google Scholar]
- Surani MA, Barton SC, Norris ML. Development of reconstituted mouse eggs suggests imprinting of the genome during gametogenesis. Nature. 1984;308:548–50. doi: 10.1038/308548a0. [DOI] [PubMed] [Google Scholar]
- Monk M. Genomic imprinting. Memories of mother and father. Nature. 1987;328:203–4. doi: 10.1038/328203a0. [DOI] [PubMed] [Google Scholar]
- Reik W, Collick A, Norris ML, Barton SC, Surani MA. Genomic imprinting determines methylation of parental alleles in transgenic mice. Nature. 1987;328:248–51. doi: 10.1038/328248a0. [DOI] [PubMed] [Google Scholar]
- Yamazaki Y, Low EW, Marikawa Y, Iwahashi K, Bartolomei MS, et al. Adult mice cloned from migrating primordial germ cells. Proc Natl Acad Sci USA. 2005;102:11361–6. doi: 10.1073/pnas.0504943102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Bouc Y, Rossignol S, Azzi S, Steunou V, Netchine I, et al. Epigenetics, genomic imprinting and assisted reproductive technology. Ann Endocrinol (Paris) 2010;71:237–8. doi: 10.1016/j.ando.2010.02.004. [DOI] [PubMed] [Google Scholar]
- Gomes MV, Huber J, Ferriani RA, Amaral Neto AM, Ramos ES. Abnormal methylation at the KvDMR1 imprinting control region in clinically normal children conceived by assisted reproductive technologies. Mol Hum Reprod. 2009;15:471–7. doi: 10.1093/molehr/gap038. [DOI] [PubMed] [Google Scholar]
- Manipalviratn S, DeCherney A, Segars J. Imprinting disorders and assisted reproductive technology. Fertil Steril. 2009;91:305–15. doi: 10.1016/j.fertnstert.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Heijden GW, Derijck AA, Ramos L, Giele M, van der Vlag J, et al. Transmission of modified nucleosomes from the mouse male germline to the zygote and subsequent remodeling of paternal chromatin. Dev Biol. 2006;298:458–69. doi: 10.1016/j.ydbio.2006.06.051. [DOI] [PubMed] [Google Scholar]
- de Mateo S, Martinez-Heredia J, Estanyol JM, Dominguez-Fandos D, Vidal-Taboada JM, et al. Marked correlations in protein expression identified by proteomic analysis of human spermatozoa. Proteomics. 2007;7:4264–77. doi: 10.1002/pmic.200700521. [DOI] [PubMed] [Google Scholar]
- Baker MA, Reeves G, Hetherington L, Aitken RJ. Analysis of proteomic changes associated with sperm capacitation through the combined use of IPG-strip pre-fractionation followed by RP chromatography LC-MS/MS analysis. Proteomics. 2010;10:482–95. doi: 10.1002/pmic.200900574. [DOI] [PubMed] [Google Scholar]
- Aitken RJ, Baker MA. The role of proteomics in understanding sperm cell biology. Int J Androl. 2008;31:295–302. doi: 10.1111/j.1365-2605.2007.00851.x. [DOI] [PubMed] [Google Scholar]
- Castillo J, Oliva R.Proteomics and the genetics of sperm chromatin condensation Asian J Androle-pub ahead of print 2010, doi: aja201065. [DOI] [PMC free article] [PubMed]
- Codrington AM, Hales BF, Robaire B. Chronic cyclophosphamide exposure alters the profile of rat sperm nuclear matrix proteins. Biol Reprod. 2007;77:303–11. doi: 10.1095/biolreprod.107.060244. [DOI] [PubMed] [Google Scholar]
- Aul RB, Oko RJ. The major subacrosomal occupant of bull spermatozoa is a novel histone H2B. Dev Biol. 2002;242:376–87. [PubMed] [Google Scholar]
- Kaplan JG, Brown DL, Chaly N, Greer WL, Prasad KV, et al. Structural and evolutionary implications of the packaging of DNA for differentiation and proliferation in the lymphocyte. J Mol Evol. 1987;26:173–9. doi: 10.1007/BF02099849. [DOI] [PubMed] [Google Scholar]
- Ward WS, Partin AW, Coffey DS. DNA loop domains in mammalian spermatozoa. Chromosoma. 1989;98:153–9. doi: 10.1007/BF00329678. [DOI] [PubMed] [Google Scholar]
- Kalandadze AG, Bushara SA, Vassetzky YS, Jr, Razin SV. Characterization of DNA pattern in the site of permanent attachment to the nuclear matrix located in the vicinity of replication origin. Biochem Biophys Res Commun. 1990;168:9–15. doi: 10.1016/0006-291x(90)91667-h. [DOI] [PubMed] [Google Scholar]
- Choudhary SK, Wykes SM, Kramer JA, Mohamed AN, Koppitch F, et al. A haploid expressed gene cluster exists as a single chromatin domain in human sperm. J Biol Chem. 1995;270:8755–62. doi: 10.1074/jbc.270.15.8755. [DOI] [PubMed] [Google Scholar]
- Ward WS, Coffey DS. DNA packaging and organization in mammalian spermatozoa: comparison with somatic cells. Biol Reprod. 1991;44:569–74. doi: 10.1095/biolreprod44.4.569. [DOI] [PubMed] [Google Scholar]
- Ward WS. The structure of the sleeping genome: implications of sperm DNA organization for somatic cells. J Cell Biochem. 1994;55:77–82. doi: 10.1002/jcb.240550109. [DOI] [PubMed] [Google Scholar]
- Sotolongo B, Ward WS. DNA loop domain organization: the three dimensional genomic code. J Cell Biochem. 2000;35:23–6. doi: 10.1002/1097-4644(2000)79:35+<23::aid-jcb1122>3.0.co;2-n. [DOI] [PubMed] [Google Scholar]
- Pardoll DM, Vogelstein B, Coffey DS. A fixed site of DNA replication in eucaryotic cells. Cell. 1980;19:527–36. doi: 10.1016/0092-8674(80)90527-9. [DOI] [PubMed] [Google Scholar]
- Berezney R, Coffey DS. Nuclear protein matrix: association with newly synthesized DNA. Science. 1975;189:291–3. doi: 10.1126/science.1145202. [DOI] [PubMed] [Google Scholar]
- Berezney R. Visualizing DNA replication sites in the cell nucleus. Semin Cell Biol. 1991;2:103–15. [PubMed] [Google Scholar]
- Gerdes MG, Carter KC, Moen PT, Jr, Lawrence JB. Dynamic changes in the higher-level chromatin organization of specific sequences revealed by in situ hybridization to nuclear halos. J Cell Biol. 1994;126:289–304. doi: 10.1083/jcb.126.2.289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson DA, Cook PR. Replication occurs at a nucleoskeleton. EMBO J. 1986;5:1403–10. doi: 10.1002/j.1460-2075.1986.tb04374.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dijkwel PA, Hamlin JL. Origins of replication and the nuclear matrix: the DHFR domain as a paradigm. Int Rev Cytol. 1995;162A:455–84. doi: 10.1016/s0074-7696(08)61236-x. [DOI] [PubMed] [Google Scholar]
- Cockerill PN, Garrard WT. Chromosomal loop anchorage of the kappa immunoglobulin gene occurs next to the enhancer in a region containing topoisomerase II sites. Cell. 1986;44:273–82. doi: 10.1016/0092-8674(86)90761-0. [DOI] [PubMed] [Google Scholar]
- Getzenberg RH. Nuclear matrix and the regulation of gene expression: tissue specificity. J Cell Biochem. 1994;55:22–31. doi: 10.1002/jcb.240550105. [DOI] [PubMed] [Google Scholar]
- Krawetz SA, Draghici S, Goodrich R, Liu Z, Ostermeier GC. In silico and wet-bench identification of nuclear matrix attachment regions. Methods Mol Med. 2005;108:439–58. doi: 10.1385/1-59259-850-1:439. [DOI] [PubMed] [Google Scholar]
- Wakayama T, Perry AC, Zuccotti M, Johnson KR, Yanagimachi R. Full-term development of mice from enucleated oocytes injected with cumulus cell nuclei. Nature. 1998;394:369–74. doi: 10.1038/28615. [DOI] [PubMed] [Google Scholar]
- Wilmut I, Schnieke AE, McWhir J, Kind AJ, Campbell KH. Viable offspring derived from fetal and adult mammalian cells. Nature. 1997;385:810–3. doi: 10.1038/385810a0. [DOI] [PubMed] [Google Scholar]
- Lemaitre JM, Danis E, Pasero P, Vassetzky Y, Mechali M. Mitotic remodeling of the replicon and chromosome structure. Cell. 2005;123:787–801. doi: 10.1016/j.cell.2005.08.045. [DOI] [PubMed] [Google Scholar]
- Yamauchi Y, Shaman JA, Boaz SM, Ward WS. Paternal pronuclear DNA degradation is functionally linked to DNA replication in mouse oocytes. Biol Reprod. 2007;77:407–15. doi: 10.1095/biolreprod.107.061473. [DOI] [PubMed] [Google Scholar]
- Lagarkova MA, Iarovaia OV, Razin SV. Large-scale fragmentation of mammalian DNA in the course of apoptosis proceeds via excision of chromosomal DNA loops and their oligomers. J Biol Chem. 1995;270:20239–41. doi: 10.1074/jbc.270.35.20239. [DOI] [PubMed] [Google Scholar]
- Li TK, Chen AY, Yu C, Mao Y, Wang H, et al. Activation of topoisomerase II-mediated excision of chromosomal DNA loops during oxidative stress. Genes Dev. 1999;13:1553–60. doi: 10.1101/gad.13.12.1553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solovyan VT, Bezvenyuk ZA, Salminen A, Austin CA, Courtney MJ. The role of topoisomerase II in the excision of DNA loop domains during apoptosis. J Biol Chem. 2002;277:21458–67. doi: 10.1074/jbc.M110621200. [DOI] [PubMed] [Google Scholar]
- Durrieu F, Samejima K, Fortune JM, Kandels-Lewis S, Osheroff N, et al. DNA topoisomerase IIalpha interacts with CAD nuclease and is involved in chromatin condensation during apoptotic execution. Curr Biol. 2000;10:923–6. doi: 10.1016/s0960-9822(00)00620-5. [DOI] [PubMed] [Google Scholar]
- Widlak P, Garrard WT. Discovery, regulation, and action of the major apoptotic nucleases DFF40/CAD and endonuclease G. J Cell Biochem. 2005;94:1078–87. doi: 10.1002/jcb.20409. [DOI] [PubMed] [Google Scholar]
- Hars ES, Lyu YL, Lin CP, Liu LF. Role of apoptotic nuclease caspase-activated DNase in etoposide-induced treatment-related acute myelogenous leukemia. Cancer Res. 2006;66:8975–9. doi: 10.1158/0008-5472.CAN-06-1724. [DOI] [PubMed] [Google Scholar]
- Shaman JA, Prisztoka R, Ward WS. Topoisomerase IIB and an extracellular nuclease interact to digest sperm DNA in an apoptotic-like manner. Biol Reprod. 2006;75:741–8. doi: 10.1095/biolreprod.106.055178. [DOI] [PubMed] [Google Scholar]


