Abstract
Transgenic mice have had a tremendous impact on biomedical research. Most researchers are familiar with transgenic mice that carry Cre recombinase (Cre) and how they are used to create conditional knockouts. However, some researchers are less familiar with many of the other types of transgenic mice and their applications. For example, transgenic mice can be used to study biochemical and molecular pathways in primary cultures and cell suspensions derived from transgenic mice, cell-cell interactions using multiple fluorescent proteins in the same mouse, and the cell cycle in real time and in the whole animal, and they can be used to perform deep tissue imaging in the whole animal, follow cell lineage during development and disease, and isolate large quantities of a pure cell type directly from organs. These novel transgenic mice and their applications provide the means for studying of molecular and biochemical events in the whole animal that was previously limited to cell cultures. In conclusion, transgenic mice are not just for generating knockouts.
Keywords: Cre, enhanced green fluorescent protein, high-throughput, renal-specific, transgenics
transgenic mice have revolutionized biomedical research. They permit the manipulation of genetic, molecular, biochemical, and physiological processes in the whole animal with nearly the same specificity seen in cell culture models. Most researchers are familiar with the Cre/loxP system and how it is used to create conditional knockouts. However, most are unfamiliar with the many other transgenic mice and their applications, novel and well-established alike. These novel mice go beyond the knockout and provide the means to study of cellular processes in the whole animal that was traditionally limited to culture models. This review highlights a few of these transgenic mice and their applications.
Transgenic Fundamentals
Working with transgenic mice can be maddening; the nomenclature alone can be confusing. For example, transgenic, knockout, knockin, homologous and nonhomologous recombination, pronuclear injection, concatemer, copy number, Cre/loxP, and gene targeting are but a few of the many terms associated with transgenic and knockout mice. Similarly, working with transgenic mice can be frustrating; e.g., mice are expensive, it takes months to generate the desired genotype/phenotype, and nonspecific expression, no expression, and loss of expression are but a few of the multiple problems one encounters when working with transgenic mice. Therefore, it is helpful to understand transgenic fundamentals prior to working with transgenic or gene-targeted mice.
Transgenic mice are created by insertion of foreign DNA or removal of native DNA. Insertion of foreign DNA occurs by nonhomologous or homologous recombination. The fundamental differences are highlighted in Figs. 1 and 2. In addition, the cost of generating of mice by nonhomologous and homologous recombination is about $2,000 and $10,000, respectively.
Fig. 1.
Schematic of nonhomologous recombination. A: a basic transgene. B: pronuclear injection of a fertilized mouse oocyte. Typical transgene consists of a cell-specific promoter placed upstream of the cDNA followed by a 3′-untranslated region [3′-UTR (polyadenylation signal)]. The promoter drives expression of the cDNA in a cell-specific manner, while the 3′-UTR promotes efficient translation of the cDNA. Unique restriction enzymes (linearized) are used to remove the transgene from the plasmid vector, and the transgene is placed in a micropipette and injected into the pronucleus of a fertilized mouse oocyte.
Fig. 2.
Schematic of homologous recombination: target locus and targeting construct (A), embryonic stem (ES) cells and feeder cells (B), and ES cell injection into mouse blastocyte (C). Targeting construct consists of arms of homology (ARM-H), loxP sites flanking exon 2 and neomycin (Neo) resistance gene, and the thymidine kinase (tk). Arms of homology consist of ∼2,000- to 5,000-bp sequences that flank the target locus; this is the critical step for homologous recombination. The loxP sites flank exon 2 and the Neo resistance gene, which will be removed later following Cre-loxP-mediated excision. The Neo resistance gene is removed by Cre-loxP excision in the ES cells, because it has been shown that Neo can cause adverse effects in the whole animal. The thymidine kinase gene is placed outside the arms of homology for selection against nonhomologous recombination. The targeting construct is electroporated into mouse ES cells, plated onto a “lawn” of feeder cells, and grown in the presence of gentamicin. Only those cells where recombination has occurred will grow. Cells are also grown in the presence of ganciclovir to select against nonhomologous recombination. If nonhomologous recombination occurs, the thymidine kinase-selectable marker is expressed and produces a toxin in the presence of ganciclovir, thereby killing all those cells. ES cells that are positive for homologous recombination are then expanded and injected into the mouse blastocyte.
Technical Considerations
Working with transgenic and knockout mice can be challenging and is often hampered by limitations and technical problems. The most common limitations involved construction of the transgene, generation and characterization of founder lines, and maintenance of a transgenic colony. Examples of the most common problems and limitations, along with recommended solutions, are highlighted below.
The typical transgene has a promoter (5′-flanking region) fused to the cDNA of interest [Cre recombinase (Cre) or enhanced green fluorescent protein (EGFP)] followed by a polyadenylation signal (3′-untranslated region). The 5′-flanking region or promoter determines the cell-specific expression of the cDNA, which is why selection of the correct promoter is so important. When a promoter is selected, the level and pattern of expression of the endogenous gene/protein in the embryo and adult should be taken into account. Gene and protein expression patterns within the developing and adult kidney can be examined using the Genitourinary Database Molecular Anatomy Project (GUDMAP: www.gudmap.org) and the renal-specific transcriptomic and proteomic databases (https://dirweb.nhlbi.nih.gov/CENTERS/CBPC/LKEM_G/LKEM/Pages/–TranscriptomicandProteomicDatabases.aspx), respectively. Once a promoter has been chosen, determining the size (number of base pairs) of the promoter is the next step.
The size of the promoter influences expression; however, the size of the promoter appears to be gene-specific, so no one size works for every transgene. For example, the Hoxb7-Cre transgene uses 1.3 kb of the Hoxb7 promoter (60), the AQP2-Cre uses ∼14 kb of the human aquaporin 2 (AQP2) promoter (42), and the ATP6V1B1-EGFP uses ∼7 kb of the human ATP6V1B1 promoter (38). The availability of unique restriction sites within the promoter and the degree of conservation among different species will influence the size of a promoter. If traditional cloning approaches fail to yield a viable transgenic mouse, bacterial artificial chromosome (BAC) transgenics or homologous recombination should be considered.
BAC transgenics use BAC clones to drive expression of the transgene. BAC clones are specialized plasmids that contain a large (150- to 350-kb) piece of chromosomal DNA. The transgene (Cre or EGFP) is cloned into the BAC clone by homologous recombination, replacing exon 1; this requires specialized Escherichia coli and cloning vectors. Once the BAC transgene is generated, it is linearized and injected into the pronucleus of the oocyte, similar to traditional transgenes. Generation of BAC transgenes is technically challenging; however, they recapitulate endogenous gene expression more faithfully, because they are likely to contain all or most of the transcriptional regulatory elements. However, similar to traditional transgenes, BAC transgenes are susceptible to gene silencing, because integration into the host genome is random. If gene silencing persists or faithful transgene expression is required, generation of a transgenic mouse by homologous recombination should be considered.
Homologous recombination is traditionally used to create knockout mice, but it can also be used to knock in a transgene. It provides faithful transgene expression by replacing the first exon with the transgene (Cre or EGFP); however, in doing so, it creates a heterozygous knockout of that allele. Therefore, this will work only if one allele is sufficient for normal function. Once a transgene has been constructed and the mouse has been generated, it is critical to confirm transgene integration and determine copy number and cell-specific expression.
PCR genotyping is the most common method used to determine transgene integration and copy number, while cell-specific expression is best examined using immunohistochemistry and fluorescence microscopy. Copy number is estimated by comparing the densities of the PCR products generated by amplification of tail DNA from transgenic and wild-type mice that had been spiked with the plasmid vector containing the transgene. Cell-specific expression is examined in tissue samples that have been labeled with an antibody against the endogenous protein and then visualized using fluorescence microscopy. However, when cell-specific expression of Cre is examined, the Cre transgenic mouse must first be crossed with a reporter mouse, such as the Rosa26-EYFP (53). This approach examines cell-specific Cre expression and activity. Finally, once a transgenic line has been generated and characterized, it is critical to maintain only those mice that continue to express the transgene faithfully.
Maintaining a transgenic mouse colony is costly, labor-intensive, and time-consuming. Routine maintenance includes PCR genotyping of all offspring and examination of transgene levels and cell-specific expression. It is recommended that the latter be performed every fourth generation to avoid breeding mice where transgene expression is diminishing, which is the most frequent problem, primarily due to epigenetic phenomena and inbreeding. Transgene silencing is typically not reversible; therefore, maintenance of separate breeding pairs of any given transgenic line is highly recommended.
Fluorescent Proteins
Fluorescent proteins represent a powerful tool to study a number of important physiological events and identify cellular structures and a particular cell type within a heterogeneous population of cells. The application of fluorescent proteins has evolved from their use in cell cultures, and fluorescent proteins are now used in transgenic mice. The following examples highlight a few of these novel applications.
Simultaneous use.
EGFP was the first fluorescent protein expressed in transgenic mice and remains the most widely used today (43). However, there is a trend toward using fluorescent proteins other than EGFP, e.g., enhanced yellow, blue, and cyan fluorescent protein (EYFP, EBFP, and ECFP) (27, 46, 59) or the monomeric red fluorescent proteins (mRFPs) (5, 45). Excitation and emission frequencies of mRFP-derived fluorescent proteins are significantly higher than those of EGFP-derived fluorescent proteins and pH- and Ca2+-sensitive dyes. Figure 3 illustrates the range of peak emission frequencies for the EGFP-derived (green) and pH- and Ca2+-sensitive (blue) dyes and the mRFP-derived (red) fluorescent proteins. Most mRFPs can be simultaneously used with EGFP-derived and pH- and Ca2+-sensitive dyes because of their higher emission frequencies.
Fig. 3.
Summary of fluorescent protein peak excitation and emissions. A: range of peak emission frequencies for enhanced green fluorescent protein (EGFP)-derived (green) and pH- and Ca2+-sensitive (blue) dyes and mouse red fluorescent protein (mRFP)-derived (red) fluorescent proteins. B: peak excitation and emission frequencies for select fluorescent proteins and pH- and Ca2+-sensitive dyes. EBFP, ECFP, and EYFP, enhanced blue, cyan, and yellow fluorescent proteins.
Simultaneous use of fluorescent proteins in transgenic mice has many valuable applications (56). For example, the role of Ca2+ signaling in liver regeneration was examined in Discosoma red fluorescent protein (DsRed)-expressing hepatocytes with use of the Ca2+-sensitive dye fura 2-AM (30). Also, mRFP-expressing tumor cells were transplanted into EGFP-expressing transgenic mice for examination of tumor-induced angiogenesis (2, 22). Similarly, the simultaneous expression of three or more fluorescent proteins in transgenic mice improved the examination of neural circuitries; i.e., adjacent neurons were easily distinguishable from each other, as were their complex cellular interactions (34). Multicolored transgenic mice are very valuable for observing multiple events, cell types, or cell-cell interactions. Because of the complex architecture and cellular heterogeneity of the kidney, transgenic mice expressing more than one fluorescent protein are ideal for such a study.
Whole animal imaging.
The use of fluorescent proteins for in vivo imaging has proved difficult, because hemoglobin absorbs most <600-nm wavelengths, which makes tissue appear opaque. In 2009, Lin et al. (33) overcame this problem by developing a fluorescent protein (Neptune) with 610- to 630-nm excitation and 660- to 700-nm emission frequencies. Neptune is an ideal fluorescent protein for in vivo imaging, because it can be visualized using standard confocal microscopy. Figure 4 illustrates the principles involved in in vivo imaging using standard fluorescent proteins and Neptune. In vivo imaging has many practical applications, including tracking cell movement and proliferation during development and monitoring physiological events.
Fig. 4.
Deep tissue imaging. Excitation (ex) and emission (em) wavelengths are required to penetrate the skin, red blood cells (RBC), and cells expressing the fluorescent protein. A: excitation and emission wavelengths at or near 600 nm are likely to be absorbed by hemoglobin (heme) within RBC and, thus, do not reach cells that express the fluorescent protein. B: excitation and emission wavelengths >600 nm are not absorbed by RBCs and are able to reach cells expressing the fluorescent protein Neptune.
Biosensors.
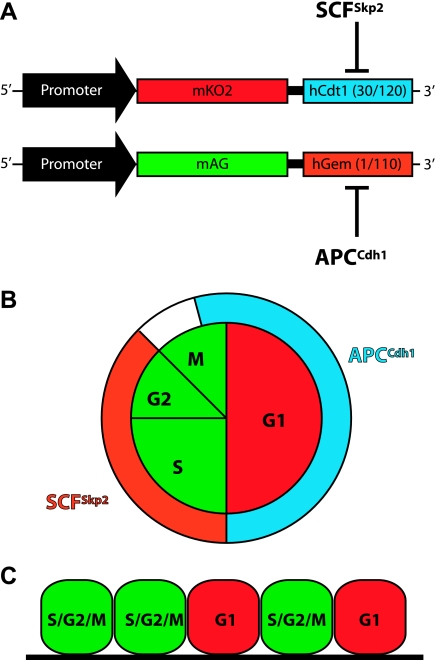
Most fluorescent proteins have a half-life of 3–5 days, which limits their utility in monitoring fast-occurring events, e.g., cell cycle progression, protein turnover, and cyclic gene expression. To overcome this limitation, DNA sequences from ubiquitin-independent or -dependent targeted proteins were added to fluorescent proteins. For example, addition of the PEST motif from the mouse ornithine decarboxylase reduced the half-life of EGFP from 5 days to <3 h (32). The PEST sequence targets proteins for ubiquitin-independent degradation (24, 35) and has been used to study cyclical gene expression in retinal cells of transgenic mice (36). Similarly, addition of antiphase oscillating proteins to fluorescent proteins permits in vivo visualization of the transition from the G1 to the S phase during the cell cycle (50, 51); this approach is referred to as fluorescent ubiquitination-based cell cycle indicator. Figure 5 provides an illustrated overview of fluorescent ubiquitination-based cell cycle indicator. Fluorescent proteins with reduced half-life should prove extremely valuable for monitoring protein turnover, transcription, and translation.
Fig. 5.
Monitoring cell cycle progression in real time. A: transgenes used to monitor cell cycle progression. The mKO2 fluorescent protein (red) was fused to a mutated human chromatin licensing and DNA replication factor 1 (hCdt1), which is expressed during the G1 phase of the cell cycle. The mAG fluorescent protein (green) was fused to a mutated human Germin (hGem), which is expressed during the S/G2/M phase of the cell cycle. SCFSkp2 and APCCdh1 are E3 ligases that are expressed during the S/G2 phase and late M and G1 phases, respectively. SCFSkp2 and APCCdh1 target hCdt1 and hGem for degradation, respectively. B: schematic representation of the cell cycle. Inner circle represents cell cycle phases, and corresponding colors show expression of the fluorescent proteins mKO2 (red) and mAG (green). Outer circle shows expression of SCFSkp2 and APCCdh1. C: cells expressing these two transgenes; i.e., green cells are in the S/G2/M phase and red cells are in the G1 phase of the cell cycle.
Biomarkers.
Native or unmodified EGFP is expressed throughout the cell; however, it can be made to localize to nearly any part of the cell by fusion to other proteins or by posttranslational modifications. Naturally, this requires that EGFP be modified before generation of the transgenic mice. Such modifications include the addition of glycosylphosphatidylinositol or myristoyl, which targets EGFP to lipid membranes (49), or fusion to histones, which localize EGFP to the nucleus (17, 25). Labeling of lipid membranes with fluorescent proteins provides an excellent high-contrast method for study of cell morphology and changes in cell volume and shape in real time without the need of fixation or staining. In 2009, Chi et al. (11) used the Hoxb7 promoter to drive expression of a myristoylated Venus fluorescent protein, which labels the lipid membranes within the cells of the ureteric bud and collecting duct of the embryonic and adult kidneys, respectively. This transgenic mouse should be very helpful in studies of cell volume regulation. Glycosylphosphatidylinositol-EGFP transgenic mice also target EGFP to the plasma membrane and have been in use for more than 10 years (29, 55). Similarly, fusion of EGFP to proteins is very helpful for study of protein trafficking and labeling of other cellular compartments. For example, fusion of EGFP to histone 2b (H2b) restricts EGFP to the nucleus (17, 25), which would be extremely helpful for cell-specific nuclear isolation from whole animals by flow cytometry. The purified nuclei could be used for nuclear proteomics and/or chromatin immunoprecipitation (ChIP-quantitative PCR or ChIP sequencing). Figure 6 illustrates the basic concepts involved in targeted localization of fluorescent proteins to the cytosol, plasma membrane, or nucleus.
Fig. 6.
Green fluorescent protein (GFP) labeling of subcellular structures. A–C: schematic overview of the transgenes glycosylphosphatidylinositol (GPI)-EGFP, myristoyl (Myr)-EGFP, and histone 2b (H2b)-EGFP transgene. For the GPI-EGFP construct, the NH2-terminal membrane translocation signal sequence from acrosin (MT) was added to the GFP NH2 terminus, and the COOH-terminal sequence from the Thy-1 NH2-terminal GPI-linked signal sequence was added to the GFP COOH terminus. For the Myr-EGFP construct, the NH2-terminal myristoylation tag from Src was added to the GFP NH2 terminus. The H2b-GFP construct was generated by fusion of the pEGFP sequence to the H2b sequence. D: cells expressing non-GFP, GFP, Myr-GFP or GPI-GFP, and H2b-GFP and corresponding localization of GFP. GFP is expressed throughout the cytoplasm in GFP-expressing cells, in the lipid membranes (plasma and nuclear) in the Myr-GFP- and GPI-GFP-expressing cells, and in the nucleus in the H2b-GFP-expressing cells.
Isolation of single cells by flow cytometry.
Because of its complex architecture and cellular heterogeneity, it is difficult to obtain cell-specific molecular and biochemical descriptions within the kidney. The lack of viable methods for selective purification of a defined cell population has hampered efforts. However, renal-specific EGFP transgenic mice make possible the direct purification of cells from the adult and embryonic kidney with use of fluorescence-activated cell sorting (FACS) (6, 7, 18, 31). FACS was used to isolate principal cells from inner medullary collecting duct primary cultures derived from the AQP2-EGFP transgenic mouse (62). Similarly, amiloride-sensitive Na+ absorption was examined in FACS-isolated collecting duct cells from transgenic mice that were heterozygous for Hoxb7-EGFP and homozygous for the BALB/c polycystic kidney (BPK) mutation (57). More recently, a proteomic profile of renal collecting duct intercalated cells was generated from EGFP-positive cells (intercalated cells) obtained directly by FACS isolation from whole kidneys following mechanical and enzymatic digestion (14). FACS-based isolation of a defined cell population is crucial for understanding cell-specific protein-DNA interactions and profiling renal cell-specific transcriptomes, proteomes, and phosphoproteomes.
Isolation of nephron fragments by large-particle flow cytometry.
Single epithelial cells of the nephron do not adequately model the basolateral-epithelial or epithelial-epithelial interactions seen in the intact nephron. In 1962, using a mild mechanical and enzymatic digestion, Burg and Orloff (9) developed a method for isolating suspensions of functionally intact nephron segments from the rabbit renal cortex; this protocol is still widely used. However, it cannot be used to isolate pure or enriched distal convoluted tubule, connecting segment (CNT), cortical collecting duct (CCD), or outer medullary collecting duct without the assistance of manual microdissection. Manual microdissection is time-consuming and laborious, requires training and skill, and does not yield enough material for routine analysis of molecular and biochemical function in the intact tubule.
To overcome this problem, in 2006, Miller et al. (39) used large-particle flow cytometry to develop a method to isolate large quantities of pure or enriched EGFP-positive collecting ducts from a single B1-EGFP transgenic mouse. The large-particle flow cytometry is referred to as complex object parameter analyzer sorting (COPAS), which permits the isolation of 12–15 cm of collecting duct in ∼1 h; the amount of tissue that can be isolated has been significantly improved using an enzymatic digestion protocol developed by Alain Doucet, Lydie Cheval, and Dominique Eladari. As a proof-of-principle, Miller and Nelson (unpublished observations) demonstrated that COPAS-isolated CNT/CCD grown on permeable supports reached confluence within 2 wk (24-mm-diameter 6-well Transwell plate), were vasopressin-responsive, and demonstrated a robust transepithelial resistance and amiloride-sensitive current (Fig. 7). Similarly, they showed that tubular suspensions of COPAS-isolated CNT/CCD produced cAMP in response to vasopressin (Fig. 7). COPAS is also being used to enrich other difficult-to-isolate nephron segments, such as the distal convoluted tubule (unpublished observations). One could envision being able to use COPAS to isolate many different nephron segments from a single kidney using a transgenic mouse that expresses multiple fluorescent proteins. This would be useful for generating segment-specific transcriptomes and proteomes and profiling global protein-DNA interactions using ChIP sequencing.
Fig. 7.
Primary cultures and tubular suspensions from complex object parameter analyzer sorter (COPAS)-isolated connecting segment (CNT)/cortical collecting duct (CCD). A: typical fluorescence confocal micrograph of primary cultures from COPAS-isolated CNT/CCD following exposure to 10−8 DDAVP for 72 h; cells demonstrated strong expression of aquaporin 2 [AQP2 (red)]. B: typical open-circuit current recording from primary cultures from COPAS-isolated CNT/CCD following exposure to 1 μM aldosterone for 4 h and then 10 μM amiloride. C: vasopressin-dependent cAMP production in COPAS-isolated CNT/CCD (200 tubules/sample). CNT/CCD were isolated and incubated at 37°C for 2 h, at which time cAMP levels were measured following 30 min of exposure to 10−8 dDAVP and 400 μM IBMX. (From L. Miller, D. Kohan, K. Strait, and R. Nelson, unpublished data).
Spatiotemporal Identification of Actively Translated mRNA
Protein abundance plays an important role in many cellular functions and is determined by the balance between protein synthesis and degradation. Proteins are synthesized by translation of mRNA, which occurs in four phases: activation, initiation, elongation, and termination. Initiation begins when the small and large ribosomal subunits (S40 and S60, respectively) bind the mRNA to form the large ribosomal complex (S80), which binds only actively translated mRNA. With use of this basic principle, actively translated mRNA can be distinguished from nontranslated mRNA by gradient centrifugation, spectroscopy, and quantitative RT-PCR or microarray analysis (28, 37), which is referred to as polysomal profiling. Traditionally, polysomal profiling is performed using cell cultures, because it requires large quantities of a homogeneous population of cells, which has prevented its wide use on tissue samples or whole animals.
To overcome this limitation, in 2008, using transgenic mice, Doyle et al. (16) and Heiman et al. (23) created a new method for isolation of mRNA-polysomal complexes from whole tissues and in a cell-specific manner; they termed this approach translating ribosome affinity purification (TRAP). TRAP involves expression of an EGFP-L10a ribosomal fusion protein in a cell-specific manner using BAC transgenics. Briefly, the tissue of interest, in this case the brain, is isolated and lysed, the cytoplasm is exposed to cycloheximide (reversibly blocking elongation), and mRNA bound to EGFP-L10a is selectively enriched by immunoprecipitation using magnetic beads labeled with anti-EGFP antibodies (Fig. 8). TRAP represents a major advancement toward the identification of actively translated proteins in the whole animal and in a spatiotemporal-specific manner.
Fig. 8.
Spatiotemporal profiling of actively translated mRNA. A: schematic of EGFP-L10a ribosome fusion transgene. B: EGFP-L10a ribosomal-expressing and -nonexpressing cells. C: EGFP-L10a ribosome-mRNA complex and native ribosome-mRNA complex. D: selective isolation of EGFP-L10a ribosome-mRNA complex using magnetic beads labeled with anti-EGFP antibodies.
Expression of Genetically Engineered Membrane Receptors
G protein-coupled receptors (GPCRs) regulate a wide variety of cellular functions by converting extracellular signals (light, toxins, nutrients, ions, and hormones) into intracellular signals via the production of second messengers, i.e., cAMP, phosphatidylinositol 4,5-bisphosphate, and Ca2+. GPCRs are notoriously difficult to study, because they are expressed in nearly every cell type; many respond to the same ligand, and some activate the same G protein, while others stimulate multiple G proteins. To overcome these limitations, a few investigators have developed genetically modified GPCRs that are activated by a synthetic ligand, and not by their endogenous ligand. These modified GPCRs are referred to as receptors activated solely by synthetic ligands (RASSLs) (12, 52, 54).
First-generation RASSLs were developed by introduction of mutations that abolished activation of the human κ-opioid receptor, a Gi-coupled receptor, by its native ligand (opioid) while retaining responsiveness to the agonist spiradoline (13). The role of the human κ-opioid receptor was then examined in taste buds (41, 61), astrocytes (54), heart (47, 48), and bone (44) by expression of this Gi-RASSL in transgenic mice followed by acute and chronic administration of spiradoline; however, spiradoline produced off-target effects. To overcome this limitation, second-generation RASSLs were developed through iterative mutations until the GPCR responded only to the inert ligand clozapine-N-oxide (4). These next-generation RASSLs were named DREADDs (designer receptors exclusively activated by designer drugs). DREADDs for Gq/11, Gs, and Gi have been expressed in transgenic mice. For example, clozapine-N-oxide-dependent activation of the hM3Dq-DREADD receptor in forebrain principal cells increased locomotion, stereotypy, and limbic seizures in transgenic mice (1). Furthermore, conditional and selective activation of Gq/11- and Gs-DREADDs in pancreatic beta cells of transgenic mice (19) improved glucose tolerance in insulin-resistant and obese mice. The myriad of GPCRs and nephron heterogeneity make the kidney an ideal organ to study GPCR signaling using DREADD transgenics.
Cre/lox Reporter System: Analysis of Cell Lineage and Fate
During development, epithelial cells appear in a process referred to as mesenchyme-epithelial transition (MET); conversely, epithelial cells are lost and replaced with fibroblasts in a process known as epithelial-mesenchyme transition (EMT). Identification of the progenitor cells that give rise to adult epithelial cells or fibroblasts is essential for understanding MET and EMT. However, it can be challenging, because results from standard approaches for cellular identification, such as immunohistochemistry, in situ hybridization, and cellular morphology, are often misleading. For example, an adult epithelial cell may express a particular gene commonly expressed in a progenitor cell; however, it does not prove that cell was derived from that progenitor cell. Genetic labeling of progenitor cells using the Cre/lox reporter system overcomes these limitations by permanently labeling the cell, thereby eliminating the need for labeling and guessing-by-association.
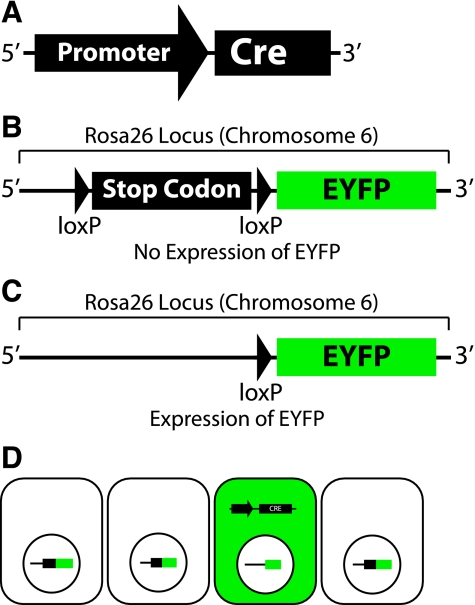
The most common Cre/lox reporter system involves crossing a Cre-expressing transgenic mouse with one carrying the Rosa26-lox-EYFP (Fig. 9). Briefly, the loxP-stop codon-loxP-EYFP allele was targeted to the Rosa26 locus, which is located on chromosome 6 in mice (53); the Rosa26 locus demonstrates ubiquitous expression (52). In the absence of Cre, the upstream stop codon prevents transcription/translation of EYFP. However, EYFP is expressed following Cre-mediated excision of the stop codon; this is nonreversible and causes all Cre-expressing cells to become EYFP-expressing cells. This is not the case if EYFP expression is driven by a promoter, which is reversible. The Cre/lox reporter system is widely used by developmental (10, 38, 58) and stem cell (15) biologists and would be helpful for studying EMT following acute kidney injury.
Fig. 9.
Cre/loxP system for cell lineage and fate. A–C: schematic of generic Cre transgene, Rosa26-loxP-stop codon-loxP-EYFP targeted allele with an intact stop codon, and Rosa26-lox-EYFP targeted allele following Cre-mediated excision of the stop codon (C). D: schematic of progenitor cells. All cells possess the Rosa26-lox-EYFP targeted allele, but only those cells that express Cre are green and will remain green over the course of development, differentiation, or dedifferentiation.
Deleter Mouse: Spatiotemporal Cell Ablation
Most researchers are familiar with conditional gene knockout, but most are unfamiliar with conditional cell knockout. Conditional cell knockout or cell ablation involves killing a cell by expression of a toxin, an inhibitor of DNA elongation, or a chemical inducer of apoptosis in a spatiotemporal-specific manner; expression can be controlled using a cell-specific promoter or the Cre/lox-Rosa26 deleter system. The most commonly expressed toxin is the A-chain of the diphtheria toxin (DTA), which is extremely toxic; a single DTA is sufficient to kill a single cell by inhibiting protein synthesis (3). Similarly, thymidine kinase inhibits DNA elongation in the presence of ganciclovir and is typically expressed using a cell-specific promoter; thymidine kinase was one of the first cDNAs to be expressed in transgenic mice (8); ganciclovir is given to the mouse. Finally, the capase-3 chemical inducer of dimerization activates capase-3-dependent apoptosis. Figure 10 illustrates the fundamentals of these three methods for causing cell death or ablation.
Fig. 10.
Conditional cell death/ablation. A and B: schematic of generic Cre transgene and Rosa26-loxP-stop codon-loxP-diphtheria toxin A-chain (DTA) targeted allele. C: schematic of cells. All cells possess the Rosa26-lox-DTA targeted allele (red), but only those cells that express Cre (red) will express DTA and are targeted for cell death/ablation. Rosa26-lox-DTA allele is represented in red for effect; it is not and should not be considered a fluorescent protein. Thymidine kinase and chemical inducer of dimerization transgenes are not shown. However, they are expressed, and their function is similar to that of DTA; i.e., only those cells expressing these “deleter” genes in conjunction with Cre are targeted for cell death. Similarly, DTA, thymidine kinase, and chemical inducer of dimerization transgenes can be expressed by targeting them to the Rosa26 locus (as shown here) or using a cell-specific promoter.
Conditional cell ablation has many applications. For example, the role of brown adipose tissue (BAT) in energy balance was investigated following the conditional ablation of BAT cells using the promoter of gene specifically expressed in BAT cells to drive expression of DTA. Results showed that BAT cells protect mice from diet-induced obesity (21). Also, the podocyte-specific Cre transgenic mouse (40) was used in conjunction with the Rosa26-DTA-176 knockin mouse (26) to ablate podocytes during nephrogenesis. This study showed that the loss of podocytes during development significantly affected the development and maintenance of the intra- and extraglomerular vasculature (26). In addition, in 2008, Haldar et al. (20) used the Cre/Rosa-LacZ reporter system to identify two types of progenitor cells involved in mouse skeletal development and then used the Cre/Rosa26-DTA ablation system to determine the role of these two progenitor cells. They showed that one of the progenitor cells contributed to the development of most skeletal muscles, while the other progenitor cell contributed to the development of a specific skeletal muscle type. The conditional cell ablation system represents a powerful tool to study the role of progenitor cells in embryogenesis, the role of a given cell type in adult physiology, and the origin of stem cells involved in tissue regeneration.
Summary
Transgenic mice have changed biomedical research by giving researchers the ability to control the expression of genes and proteins in a temporal and cell-specific manner in the whole animal. While most transgenic mice are used to knock out genes, transgenic mice have many other important and useful applications. Simultaneous use of fluorescent proteins allows labeling of multiple cells and/or observation of physiological events in a given cell type and labeling of subcellular compartments, such as lipid membranes and the nucleus, as well as observation of rapidly occurring events, such as cell cycle and cyclic gene expression, and whole animal imaging. Similarly, fluorescently labeled cells from transgenic mice can be isolated as single cells or cell clusters using FACS or COPAS. Transgenic mice can also be used to conditionally express genetically engineered proteins, such as those used to study GPCR signaling in the whole animal, and isolate actively translated mRNA from a specific cell type. Also, the Cre/lox reporter and conditional cell ablation systems are very useful for following cell lineage, fate mapping, and determining the importance of a particular cell type. This review surveyed only some of the many different transgenic mice and applications. Future transgenic mice might be engineered to monitor blood pressure, volume, ionic composition, and acid-base balance in the whole animal and in real time. Transgenic mice represent a powerful tool for physiologists to study genetic, molecular, biochemical, and physiological events in the whole animal, organ, tissue, or cell in vivo, as well as in real time, with resolution and specificity similar to that obtained in cell cultures.
DISCLOSURES
No conflicts of interest, financial or otherwise, are declared by the author.
REFERENCES
- 1. Alexander GM, Rogan SC, Abbas AI, Armbruster BN, Pei Y, Allen JA, Nonneman RJ, Hartmann J, Moy SS, Nicolelis MA, McNamara JO, Roth BL. Remote control of neuronal activity in transgenic mice expressing evolved G protein-coupled receptors. Neuron 63: 27–39, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Amoh Y, Katsuoka K, Hoffman RM. Color-coded fluorescent protein imaging of angiogenesis: the AngioMouse models. Curr Pharm Des 14: 3810–3819, 2008 [DOI] [PubMed] [Google Scholar]
- 3. Apostolov K, Barker W. Reversible increase in the saturation of C18 fatty acids induced by diphtheria toxin in tissue culture cells. Infect Immun 38: 843–847, 1982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Armbruster BN, Li X, Pausch MH, Herlitze S, Roth BL. Evolving the lock to fit the key to create a family of G protein-coupled receptors potently activated by an inert ligand. Proc Natl Acad Sci USA 104: 5163–5168, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Armstrong JJ, Larina IV, Dickinson ME, Zimmer WE, Hirschi KK. Characterization of bacterial artificial chromosome transgenic mice expressing mCherry fluorescent protein substituted for the murine smooth muscle α-actin gene. Genesis 48: 457–463, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Barrow J, Bernardo AS, Hay CW, Blaylock M, Duncan L, Mackenzie A, McCreath K, Kind AJ, Schnieke AE, Colman A, Hart AW, Docherty K. Purification and characterization of a population of EGFP-expressing cells from the developing pancreas of a neurogenin3/EGFP transgenic mouse. Organogenesis 2: 22–27, 2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Belmadani A, Jung H, Ren D, Miller RJ. The chemokine SDF-1/CXCL12 regulates the migration of melanocyte progenitors in mouse hair follicles. Differentiation 77: 395–411, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Brinster RL, Chen HY, Trumbauer M, Senear AW, Warren R, Palmiter RD. Somatic expression of herpes thymidine kinase in mice following injection of a fusion gene into eggs. Cell 27: 223–231, 1981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Burg MB, Orloff J. Oxygen consumption and active transport in separated renal tubules. Am J Physiol 203: 327–330, 1962 [DOI] [PubMed] [Google Scholar]
- 10. Chen J, Boyle S, Zhao M, Su W, Takahashi K, Davis L, Decaestecker M, Takahashi T, Breyer MD, Hao CM. Differential expression of the intermediate filament protein nestin during renal development and its localization in adult podocytes. J Am Soc Nephrol 17: 1283–1291, 2006 [DOI] [PubMed] [Google Scholar]
- 11. Chi X, Hadjantonakis AK, Wu Z, Hyink D, Costantini F. A transgenic mouse that reveals cell shape and arrangement during ureteric bud branching. Genesis 47: 61–66, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Conklin BR, Hsiao EC, Claeysen S, Dumuis A, Srinivasan S, Forsayeth JR, Guettier JM, Chang WC, Pei Y, McCarthy KD, Nissenson RA, Wess J, Bockaert J, Roth BL. Engineering GPCR signaling pathways with RASSLs. Nat Methods 5: 673–678, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Coward P, Wada HG, Falk MS, Chan SD, Meng F, Akil H, Conklin BR. Controlling signaling with a specifically designed Gi-coupled receptor. Proc Natl Acad Sci USA 95: 352–357, 1998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. DaSilva N, Pisitkun T, Belleannee C, Miller LR, Nelson R, Knepper MA, Brown D, Breton S. Proteomic analysis of V-ATPase-rich cells harvested from the kidney and epididymis by fluorescence-activated cell sorting. Am J Physiol Cell Physiol 298: C1326–C1342, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Delaunay D, Heydon K, Miguez A, Schwab M, Nave KA, Thomas JL, Spassky N, Martinez S, Zalc B. Genetic tracing of subpopulation neurons in the prethalamus of mice (Mus musculus). J Comp Neurol 512: 74–83, 2009 [DOI] [PubMed] [Google Scholar]
- 16. Doyle JP, Dougherty JD, Heiman M, Schmidt EF, Stevens TR, Ma G, Bupp S, Shrestha P, Shah RD, Doughty ML, Gong S, Greengard P, Heintz N. Application of a translational profiling approach for the comparative analysis of CNS cell types. Cell 135: 749–762, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Eakin GS, Hadjantonakis AK, Papaioannou VE, Behringer RR. Developmental potential and behavior of tetraploid cells in the mouse embryo. Dev Biol 288: 150–159, 2005 [DOI] [PubMed] [Google Scholar]
- 18. Gao X, Enikolopov G, Chen J. Direct isolation of neural stem cells in the adult hippocampus after traumatic brain injury. J Neurotrauma 25: 985–995, 2008 [DOI] [PubMed] [Google Scholar]
- 19. Guettier JM, Gautam D, Scarselli M, de Azua IR, Li JH, Rosemond E, Ma X, Gonzalez FJ, Armbruster BN, Lu H, Roth BL, Wess J. A chemical-genetic approach to study G protein regulation of beta cell function in vivo. Proc Natl Acad Sci USA 106: 19197–19202, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Haldar M, Karan G, Tvrdik P, Capecchi MR. Two cell lineages, myf5 and myf5-independent, participate in mouse skeletal myogenesis. Dev Cell 14: 437–445, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Hamann A, Flier JS, Lowell BB. Obesity after genetic ablation of brown adipose tissue. Z Ernahrungswiss 37 Suppl 1: 1–7, 1998 [PubMed] [Google Scholar]
- 22. Hayashi K, Yamauchi K, Yamamoto N, Tsuchiya H, Tomita K, Amoh Y, Hoffman RM, Bouvet M. Dual-color imaging of angiogenesis and its inhibition in bone and soft tissue sarcoma. J Surg Res 140: 165–170, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Heiman M, Schaefer A, Gong S, Peterson JD, Day M, Ramsey KE, Suarez-Farinas M, Schwarz C, Stephan DA, Surmeier DJ, Greengard P, Heintz N. A translational profiling approach for the molecular characterization of CNS cell types. Cell 135: 738–748, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Hoyt MA, Zhang M, Coffino P. Ubiquitin-independent mechanisms of mouse ornithine decarboxylase degradation are conserved between mammalian and fungal cells. J Biol Chem 278: 12135–12143, 2003 [DOI] [PubMed] [Google Scholar]
- 25. Isern J, Fraser ST, He Z, Baron MH. Developmental niches for embryonic erythroid cells. Blood Cells Mol Dis 44: 207–208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Jia Q, McDill BW, Sankarapandian B, Wu S, Liapis H, Holzman LB, Capecchi MR, Miner JH, Chen F. Ablation of developing podocytes disrupts cellular interactions and nephrogenesis both inside and outside the glomerulus. Am J Physiol Renal Physiol 295: F1790–F1798, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Katoh A, Fujihara H, Ohbuchi T, Onaka T, Young WS, 3rd, Dayanithi G, Yamasaki Y, Kawata M, Suzuki H, Otsubo H, Murphy D, Ueta Y. Specific expression of an oxytocin-enhanced cyan fluorescent protein fusion transgene in the rat hypothalamus and posterior pituitary. J Endocrinol 204: 275–285, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kitamura H, Ito M, Yuasa T, Kikuguchi C, Hijikata A, Takayama M, Kimura Y, Yokoyama R, Kaji T, Ohara O. Genome-wide identification and characterization of transcripts translationally regulated by bacterial lipopolysaccharide in macrophage-like J774.1 cells. Physiol Genomics 33: 121–132, 2008 [DOI] [PubMed] [Google Scholar]
- 29. Kondoh G, Gao XH, Nakano Y, Koike H, Yamada S, Okabe M, Takeda J. Tissue-inherent fate of GPI revealed by GPI-anchored GFP transgenesis. FEBS Lett 458: 299–303, 1999 [DOI] [PubMed] [Google Scholar]
- 30. Lagoudakis L, Garcin I, Julien B, Nahum K, Gomes DA, Combettes L, Nathanson MH, Tordjmann T. Cytosolic calcium regulates liver regeneration in the rat. Hepatology 52: 602–611, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Li L, Lili R, Hui Q, Min W, Xue W, Xin S, Jing L, Yan L, Yeqiang L, Fenrong H, Furong L, Guanxin S. Combination of GLP-1 and sodium butyrate promote differentiation of pancreatic progenitor cells into insulin-producing cells. Tissue Cell 40: 437–445, 2008 [DOI] [PubMed] [Google Scholar]
- 32. Li X, Zhao X, Fang Y, Jiang X, Duong T, Fan C, Huang CC, Kain SR. Generation of destabilized green fluorescent protein as a transcription reporter. J Biol Chem 273: 34970–34975, 1998 [DOI] [PubMed] [Google Scholar]
- 33. Lin MZ, McKeown MR, Ng HL, Aguilera TA, Shaner NC, Campbell RE, Adams SR, Gross LA, Ma W, Alber T, Tsien RY. Autofluorescent proteins with excitation in the optical window for intravital imaging in mammals. Chem Biol 16: 1169–1179, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Livet J, Weissman TA, Kang H, Draft RW, Lu J, Bennis RA, Sanes JR, Lichtman JW. Transgenic strategies for combinatorial expression of fluorescent proteins in the nervous system. Nature 450: 56–62, 2007 [DOI] [PubMed] [Google Scholar]
- 35. Loetscher P, Pratt G, Rechsteiner M. The C terminus of mouse ornithine decarboxylase confers rapid degradation on dihydrofolate reductase. Support for the pest hypothesis. J Biol Chem 266: 11213–11220, 1991 [PubMed] [Google Scholar]
- 36. Man PS, Evans T, Carter DA. Rhythmic expression of an egr-1 transgene in rats distinguishes two populations of photoreceptor cells in the retinal outer nuclear layer. Mol Vis 14: 1176–1186, 2008 [PMC free article] [PubMed] [Google Scholar]
- 37. Melamed D, Arava Y. Genome-wide analysis of mRNA polysomal profiles with spotted DNA microarrays. Methods Enzymol 431: 177–201, 2007 [DOI] [PubMed] [Google Scholar]
- 38. Miller RL, Lucero OM, Riemondy KA, Baumgartner BK, Brown D, Breton S, Nelson RD. The V-ATPase B1-subunit promoter drives expression of Cre recombinase in intercalated cells of the kidney. Kidney Int 75: 435–439, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Miller RL, Zhang P, Chen T, Rohrwasser A, Nelson RD. Automated method for the isolation of collecting ducts. Am J Physiol Renal Physiol 291: F236–F245, 2006 [DOI] [PubMed] [Google Scholar]
- 40. Moeller MJ, Sanden SK, Soofi A, Wiggins RC, Holzman LB. Podocyte-specific expression of Cre recombinase in transgenic mice. Genesis 35: 39–42, 2003 [DOI] [PubMed] [Google Scholar]
- 41. Mueller KL, Hoon MA, Erlenbach I, Chandrashekar J, Zuker CS, Ryba NJ. The receptors and coding logic for bitter taste. Nature 434: 225–229, 2005 [DOI] [PubMed] [Google Scholar]
- 42. Nelson RD, Stricklett P, Gustafson C, Stevens A, Ausiello D, Brown D, Kohan DE. Expression of an AQP2 Cre recombinase transgene in kidney and male reproductive system of transgenic mice. Am J Physiol Cell Physiol 275: C216–C226, 1998 [DOI] [PubMed] [Google Scholar]
- 43. Okabe M, Ikawa M, Kominami K, Nakanishi T, Nishimune Y. “Green mice” as a source of ubiquitous green cells. FEBS Lett 407: 313–319, 1997 [DOI] [PubMed] [Google Scholar]
- 44. Peng J, Bencsik M, Louie A, Lu W, Millard S, Nguyen P, Burghardt A, Majumdar S, Wronski TJ, Halloran B, Conklin BR, Nissenson RA. Conditional expression of a Gi-coupled receptor in osteoblasts results in trabecular osteopenia. Endocrinology 149: 1329–1337, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Poche RA, Larina IV, Scott ML, Saik JE, West JL, Dickinson ME. The Flk1-myr::mCherry mouse as a useful reporter to characterize multiple aspects of ocular blood vessel development and disease. Dev Dyn 238: 2318–2326, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Porrero C, Rubio-Garrido P, Avendano C, Clasca F. Mapping of fluorescent protein-expressing neurons and axon pathways in adult and developing Thy1-eYFP-H transgenic mice. Brain Res 1345: 59–72, ???? [DOI] [PubMed] [Google Scholar]
- 47. Redfern CH, Coward P, Degtyarev MY, Lee EK, Kwa AT, Hennighausen L, Bujard H, Fishman GI, Conklin BR. Conditional expression and signaling of a specifically designed Gi-coupled receptor in transgenic mice. Nat Biotechnol 17: 165–169, 1999 [DOI] [PubMed] [Google Scholar]
- 48. Redfern CH, Degtyarev MY, Kwa AT, Salomonis N, Cotte N, Nanevicz T, Fidelman N, Desai K, Vranizan K, Lee EK, Coward P, Shah N, Warrington JA, Fishman GI, Bernstein D, Baker AJ, Conklin BR. Conditional expression of a Gi-coupled receptor causes ventricular conduction delay and a lethal cardiomyopathy. Proc Natl Acad Sci USA 97: 4826–4831, 2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Rhee JM, Pirity MK, Lackan CS, Long JZ, Kondoh G, Takeda J, Hadjantonakis AK. In vivo imaging and differential localization of lipid-modified GFP-variant fusions in embryonic stem cells and mice. Genesis 44: 202–218, 2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Sakaue-Sawano A, Kurokawa H, Morimura T, Hanyu A, Hama H, Osawa H, Kashiwagi S, Fukami K, Miyata T, Miyoshi H, Imamura T, Ogawa M, Masai H, Miyawaki A. Visualizing spatiotemporal dynamics of multicellular cell-cycle progression. Cell 132: 487–498, 2008 [DOI] [PubMed] [Google Scholar]
- 51. Sakaue-Sawano A, Ohtawa K, Hama H, Kawano M, Ogawa M, Miyawaki A. Tracing the silhouette of individual cells in S/G2/M phases with fluorescence. Chem Biol 15: 1243–1248, 2008 [DOI] [PubMed] [Google Scholar]
- 52. Scearce-Levie K, Coward P, Redfern CH, Conklin BR. Engineering receptors activated solely by synthetic ligands (RASSLs). Trends Pharmacol Sci 22: 414–420, 2001 [DOI] [PubMed] [Google Scholar]
- 53. Srinivas S, Watanabe T, Lin CS, William CM, Tanabe Y, Jessell TM, Costantini F. Cre reporter strains produced by targeted insertion of EYFP and ECFP into the ROSA26 locus. BMC Dev Biol 1: 4, 2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Sweger EJ, Casper KB, Scearce-Levie K, Conklin BR, McCarthy KD. Development of hydrocephalus in mice expressing the Gi-coupled GPCR Ro1 RASSL receptor in astrocytes. J Neurosci 27: 2309–2317, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Tanaka D, Nakaya Y, Yanagawa Y, Obata K, Murakami F. Multimodal tangential migration of neocortical GABAergic neurons independent of GPI-anchored proteins. Development 130: 5803–5813, 2003 [DOI] [PubMed] [Google Scholar]
- 56. Terada M, Nobori K, Munehisa Y, Kakizaki M, Ohba T, Takahashi Y, Koyama T, Terata Y, Ishida M, Iino K, Kosaka T, Watanabe H, Hasegawa H, Ito H. Double transgenic mice crossed GFP-LC3 transgenic mice with α-MyHC-mCherry-LC3 transgenic mice are a new and useful tool to examine the role of autophagy in the heart. Circ J 74: 203–206, 2010 [DOI] [PubMed] [Google Scholar]
- 57. Veizis EI, Carlin CR, Cotton CU. Decreased amiloride-sensitive Na+ absorption in collecting duct principal cells isolated from BPK ARPKD mice. Am J Physiol Renal Physiol 286: F244–F254, 2004 [DOI] [PubMed] [Google Scholar]
- 58. Vigneau C, Polgar K, Striker G, Elliott J, Hyink D, Weber O, Fehling HJ, Keller G, Burrow C, Wilson P. Mouse embryonic stem cell-derived embryoid bodies generate progenitors that integrate long term into renal proximal tubules in vivo. J Am Soc Nephrol 18: 1709–1720, 2007 [DOI] [PubMed] [Google Scholar]
- 59. Webster NL, Forni M, Bacci ML, Giovannoni R, Razzini R, Fantinati P, Zannoni A, Fusetti L, Dalpra L, Bianco MR, Papa M, Seren E, Sandrin MS, McKenzie IF, Lavitrano M. Multi-transgenic pigs expressing three fluorescent proteins produced with high efficiency by sperm mediated gene transfer. Mol Reprod Dev 72: 68–76, 2005 [DOI] [PubMed] [Google Scholar]
- 60. Yu J, Carroll TJ, McMahon AP. Sonic hedgehog regulates proliferation and differentiation of mesenchymal cells in the mouse metanephric kidney. Development 129: 5301–5312, 2002 [DOI] [PubMed] [Google Scholar]
- 61. Zhao GQ, Zhang Y, Hoon MA, Chandrashekar J, Erlenbach I, Ryba NJ, Zuker CS. The receptors for mammalian sweet and umami taste. Cell 115: 255–266, 2003 [DOI] [PubMed] [Google Scholar]
- 62. Zharkikh L, Zhu X, Stricklett PK, Kohan DE, Chipman G, Breton S, Brown D, Nelson RD. Renal principal cell-specific expression of green fluorescent protein in transgenic mice. Am J Physiol Renal Physiol 283: F1351–F1364, 2002 [DOI] [PubMed] [Google Scholar]