Abstract
The potential of metabolites to contribute to drug-drug interactions is not well defined. The aim of this study was to determine the quantitative role of circulating metabolites in inhibitory drug-drug interactions in vivo. The AUC data for at least one circulating metabolite was available for 71% of the 102 inhibitors identified. 78% of the 80 metabolites characterized at steady state had AUCs greater than 10% of the parent drug. A comparison of the metabolite’s and parent’s [I]/Ki showed that 17 of the 21 (80%) reversible inhibitors studied had metabolites that are likely to contribute to in vivo drug-drug interactions, with some metabolites predicted to be the major inhibitors. The in vivo drug interaction risk of amiodarone, bupropion and sertraline could only be identified from in vitro data when metabolites were included into predictions. In conclusion, circulating metabolites are common with CYP inhibitors and do contribute to clinically observed CYP inhibition.
INTRODUCTION
It has long been known that metabolites may contribute to drug action, toxicity, act as inhibitors of drug elimination or displace the drug from plasma or tissue binding sites. However, the quantitative importance of metabolites in drug therapy is not well characterized. Classical examples of drugs that have pharmacologically important (active) metabolites include for example morphine as a metabolite of codeine, phenobarbital as a metabolite of primidone, enalaprilat as a metabolite of enalapril and desipramine as a metabolite of imipramine.
In some cases, metabolites might be a risk factor for drug toxicity. To acknowledge this, the US Food and Drug Administration (FDA) issued the so called MIST (metabolites in safety testing) guidelines (www.fda.gov/downloads/Drugs/GuidanceComplianceRegulatoryInformation/Guidances/ucm079266.pdf) that establish a strategy for toxicology evaluation of circulating metabolites. Attention is drawn to metabolites that circulate in concentrations greater than 10% of the parent at steady state. The recently issued ICH M3 guideline by the European Medicines Agency, on the other hand, focuses on metabolites that circulate at 10% of total drug related material. The MIST and ICH guidelines are, however, specific for toxicity testing and do not recommend study of the metabolites as inhibitors of metabolic enzymes or as binding displacers.
Despite the fact that metabolites have received consistent attention regarding their role in pharmacological action and in drug toxicity, their role in inhibitory drug-drug interactions has not been well characterized. This is perhaps due to the fact that there are no established examples of clinical interactions which are caused solely by an inhibitory metabolite with a lack of any inhibitory effect from the parent drug. Several clinically important inhibitors have recently been recognized to have circulating metabolites that are inhibitory. For example, hydroxy-itraconazole as a metabolite of itraconazole, N-desmethyldiltiazem as a metabolite of diltiazem and norfluoxetine as a metabolite of fluoxetine appear to contribute to in vivo DDIs [1–4]. Whether these three drugs are the exceptions or examples of more prevalent phenomena of multiple inhibitors being responsible for a single interaction is currently unknown. To evaluate the role of inhibitory metabolites in clinically important drug-drug interactions, we recently determined that majority (82%) of clinically used cytochrome P450 inhibitors possessed circulating metabolites, and that all known potent P450 inhibitors had circulating metabolites [5]. The current work was undertaken to quantitatively analyze the exposures to these metabolites after administration of a parent compound and to analyse the relative importance of the circulating inhibitors in clinical drug-drug interactions. The University of Washington Metabolism and Transport Drug Interaction Database™ was used to collect the relevant in vitro enzyme inhibition data and clinical interaction data, and in vivo interaction potential was analyzed in accordance to FDA guidelines.
RESULTS
Prevalence of circulating metabolites
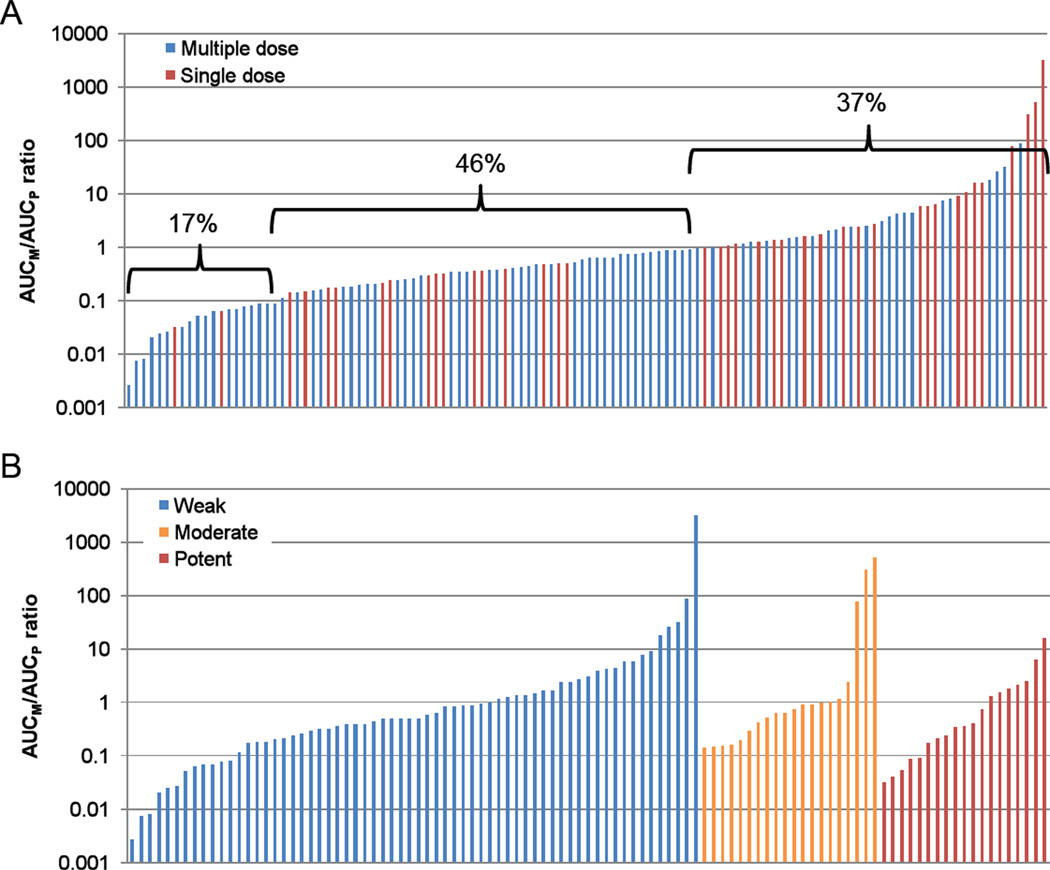
The metabolite to parent AUC ratios were calculated for 72 of the 102 inhibitors previously identified [5] based on availability of data. As some of the inhibitors have multiple circulating metabolites, this resulted in 120 datasets of AUCM/AUCP-ratios (Figure 1). Within these data, 37% of the metabolites had an AUC that was greater than the AUC of the parent and an additional 46% had an AUC that was between 10% and 100% of the parent AUC. Only 17% of all the metabolites were determined to be insignificant in terms of their AUC in relation to the parent drug (AUCM/AUCP<0.1). Figure 1 depicts the distribution of the inhibitors based on metabolite to parent AUC ratio with steady state and single dose data. The individual metabolite to parent pairs and the AUC values used for the analysis are summarized in Appendix 1 (steady state data) and 2 (single dose data). Appendix 3 shows the list of circulating metabolites for which no pharmacokinetic characterization was available. When the data was analyzed separately for weak, moderate and potent inhibitors the distribution of AUCM/AUCP ratios was similar as that shown for all the inhibitors together.
Figure 1.
Distribution of metabolite to parent AUC ratios amongst all analyzed inhibitors. Panel A shows all the inhibitors analyzed in Appendices 1 and 2 with each bar representing a specific metabolite-parent pair. Blue bars refer to steady-state data and red bars to single dose data. The percentages above the bars indicate the fraction of inhibitors in each group (AUCM/AUCP≤ 0.1; 0.1<AUCM/AUCP >1; AUCM/AUCP ≥1). Panel B shows the distribution of AUCM/AUCP ratios for the inhibitors that had available in vivo interaction data including clearance or AUC data from marker substrates and were classified according to the FDA-recommended system (www.fda.gov/cder/drug/drugInteractions/) as potent (> 5-fold increase in AUC), moderate (> 2- but < 5-fold increase in AUC), or weak (> 1.25- but < 2-fold increase in AUC) inhibitors of the target enzyme.
Prediction of relative contribution of metabolites to in vivo drug-drug interactions
Quantitative prediction of the importance of metabolites in in vivo drug-drug interactions was conducted by calculating the IPM/IPP-ratios (inhibitory potency of the metabolite as [I]/Ki, over the inhibitory potency of the parent as [I]/Ki) for all inhibitors for which circulating concentrations of the metabolite and parent were available, and in vitro Ki-values had been determined. Table 1 shows a summary of the calculated IPM/IPP-ratios. The metabolites were predicted to be most significant in vivo inhibitors for 11 of the 21 inhibitors (52%) analyzed whereas 5 metabolites belonging to 4 inhibitors (19%) were predicted not to contribute to the in vivo interaction (IPM/IPP-ratio<0.1). Notably, the two inhibitors that are commonly recognized to have inhibitory metabolites, itraconazole and fluoxetine, had IPM/IPP-ratios less than 1. For three analyzed potent or moderate inhibitors, amiodarone, amodiaquine and bupropion, the IPM/IPP-ratios ranged from 5 to 25 demonstrating that the metabolites are predicted to be the main contributors to interactions. On the other hand, omeprazole sulfone, quinidine N-oxide and 3-hydroxy quinidine had IPM/IPP-ratios <0.01 resulting in a prediction of insignificant contribution of metabolites to in vivo interactions.
Table 1.
Determination of the relative importance of the metabolites in comparison to parent drug using the IPM/IPP ratios.
| Parent (P) | Metabolite (M) | CYP | P | M | IPM/IPP | Reference(s) | ||||
|---|---|---|---|---|---|---|---|---|---|---|
| [I] (µM) |
Ki (µM) |
[I]/Ki | [I] (µM) |
Ki (µM) |
[I]/Ki | |||||
| amiodarone | N-desethylamiodarone | 2C9 | 2.5 | 94.6* | 0.026 | 1.5 | 2.3* | 0.67 | 25.3 | [10], [11]* |
| bupropion | threobupropion | 2D6 | 0.3 | 21 | 0.014 | 1.9 | 5.4 | 0.36 | 24.8 | [12] |
| hydroxybupropion | 2D6 | 3.2 | 13 | 0.24 | 17.0 | [12] | ||||
| erythrobupropion | 2D6 | 0.37 | 1.7 | 0.22 | 15.3 | [12] | ||||
| sulfinpyrazone | sulfinpyrazone sulphide | 2C9 | 17.8 | 229 | 0.031 | 5.0 | 27 | 0.19 | 6.0 | [13] |
| venlafaxine | N-desmethylvenlafaxine | 2D6 | 0.23 | 30 | 0.0078 | 0.93 | 20 | 0.047 | 6.0 | [14] |
| atorvastatin | atorvastatin lactone | 3A | 0.06* | 90 | 0.0007 | 0.003* | 0.9 | 0.004 | 5.5 | [15], [16]* |
| amodiaquine | desethylamodiaquine | 2D6 | 0.08 | 2.1 | 0.039 | 0.82 | 4.1 | 0.20 | 5.1 | [17] |
| clomipramine | N-desmethylclomipramine | 2D6 | 0.24 | 16 | 0.014 | 0.49 | 7.9 | 0.062 | 4.2 | [18] |
| sertraline | N-desmethylsertraline | 2D6 | 0.086 | 23 | 0.0038 | 0.14 | 16 | 0.009 | 2.4 | [19] |
| risperidone | paliperidone | 3A | 0.020 | 67 | 0.0003 | 0.046 | 80 | 0.001 | 2.0 | [20] |
| sertraline | N-desmethylsertraline | 3A4 | 0.33 | 3.5 | 0.094 | 0.53 | 3.5 | 0.15 | 1.6 | [21], [22] |
| haloperidol | reduced haloperidol | 2D6 | 0.023 | 0.89 | 0.026 | 0.008 | 0.24 | 0.033 | 1.3 | [23] |
| risperidone | paliperidone | 2D6 | 0.020 | 6.9 | 0.0028 | 0.046 | 16 | 0.003 | 1.0 | [20] |
| fluoxetine | norfluoxetine | 2D6 | 0.37 | 0.2 | 2.2 | 0.38 | 0.2 | 2.0 | 0.92 | [24] |
| citalopram | didesmethylcitalopram | 2C19 | 0.20 | 87.3 | 0.002 | 0.015 | 7.7 | 0.002 | 0.85 | [25] |
| desmethylcitalopram | 2C19 | 0.24 | 87.3 | 0.003 | 0.13 | 56 | 0.002 | 0.85 | [25] | |
| propranolol | 4-hydroxypropranolol | 2D6 | 0.42 | 4.5* | 0.094 | 0.046 | 1.0 | 0.046 | 0.49 | [26], [27]* |
| sulfinpyrazone | sulfinpyrazone sulfone | 2C9 | 17.8 | 229 | 0.078 | 2.3 | 73 | 0.031 | 0.40 | [13] |
| itraconazole | hydroxyitraconazole | 3A4 | 0.94 | 8.5 | 0.11 | 1.09 | 26. | 0.042 | 0.38 | [3] |
| ranolazine | O-demethyl ranolazine | 3A4 | 2.2 | 90.0 | 0.025 | 0.89 | 104. | 0.009 | 0.35 | a |
| chloroquine | monodesethylchloroquine | 2D6 | 0.62 | 15.3 | 0.040 | 0.20 | 15.5 | 0.013 | 0.32 | [28] |
| imatinib | N-desmethylimatinib | 2C9 | 8.6* | 59.0 | 0.15 | 1.4* | 40.3 | 0.034 | 0.23 | b, [29]* |
| 3A | 8 | 1.1 | 13.7 | 0.099 | 0.092 | b, [29]* | ||||
| 2D6 | 7.5 | 1.1 | 13.5 | 0.10 | 0.088 | b, [29]* | ||||
| carbamazepine | carbamazepine-10,11-epoxide | 2C19 | 30.5 | 29.6 | 1.0 | 4.8 | 53.5 | 0.089 | 0.086 | [30] |
| atomoxetine | N-desmethylatomoxetine | 3A | 0.59 | 34.0 | 0.017 | 0.019 | 16.0 | 0.001 | 0.069 | [31] |
| omeprazole | omeprazole sulfone | 2C | 1.6 | 2.0 | 0.80 | 0.38 | 8.2 | 0.046 | 0.058 | [32] |
| omeprazole sulfone | 3A | 201.0 | 0.008 | 188 | 0.002 | 0.0025 | [33] | |||
| quinidine | 3-hydroxyquinidine | 2D6 | 1.9 | 0.03 | 70 | 0.30 | 2.3 | 0.13 | 0.0019 | [34] |
| quinidine N-oxide | 2D6 | 0.60 | 43.5 | 0.014 | 0.0002 | [34] | ||||
U.S. Food and Drug Administration. Clinical Reviews- Ranolazine. Drugs@FDA 2007
U.S. Food and Drug Administration. Clinical Review- Imatinib. Drugs@FDA 2002.
Role of circulating metabolites in risk analysis of inhibitory interactions
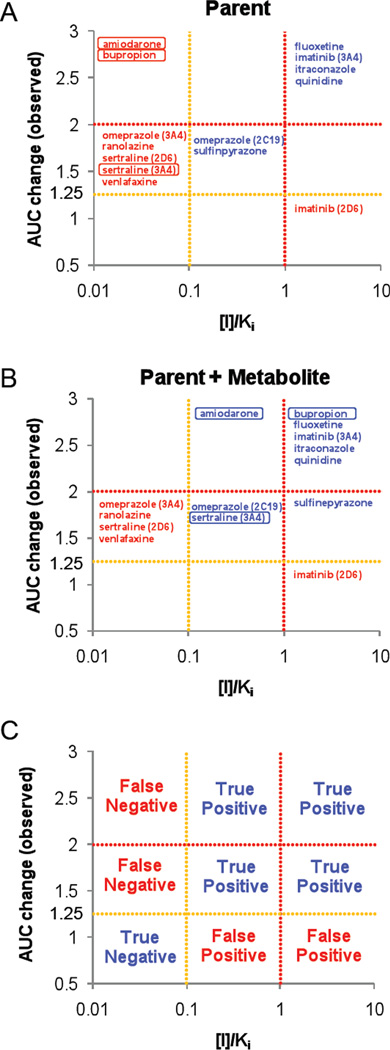
As metabolites were predicted to quantitatively contribute to selected in vivo drug-drug interactions, the effect of circulating metabolites on preclinical risk analysis of potential drug-drug interactions was determined. Only inhibitors that had in vivo interaction data with accepted probes, allowing establishment of in vivo interaction risk were included in the analysis and the [I]/Ki ratios for the inhibitors and metabolites that had sufficient data available were calculated. Six (54%) of the eleven inhibitors included in the analysis were classified into “possible” or “likely” interactions based on parent drug only (Table 2). Metabolites of six of the eleven compounds were qualitatively predicted to have equal or greater risk to cause drug-drug interactions than the parent inhibitor. Five compounds, amiodarone, bupropion, ranolazine, sertraline and venlafaxine, were classified as “remote risk” for in vivo interactions when only parent drug was considered. Two of these in vivo inhibitors, amiodarone (CYP 2C9) and sertraline (CYP3A4), ended as “possible” and one, bupropion (CYP2D6), as “likely” risk when inhibition by known circulating metabolites was considered, whereas the in vivo interaction caused by ranolazine (CYP3A4) and venlafaxine (CYP2D6) was not predicted with the available in vitro inhibition data. Figure 2 shows the correlation between in vitro risk assessment and the magnitude of in vivo drug-drug interactions when only parent drug is accounted for and when parent and metabolites are analyzed together. As shown in Figure 2, the moderate inhibitor amiodarone and potent inhibitor bupropion, were predicted to be in vivo inhibitors only when metabolites as well as parent drug were accounted for. It is noteworthy that for sertraline, the interaction with CYP3A4 could be predicted with the metabolite taken into account whereas the CYP2D6 interaction could not.
Table 2.
Evaluation of Potential DDI risk using parent and metabolite [I]/Ki data according to the FDA guidance.
| Parent | Metabolite | CYP | Inhibitor dose |
Victim drug |
AUC change of victim drug (observed) |
Parent (P) | Metabolite (M) | P+M | FDA guidanceb |
Reference | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| [I] (µM) |
[I]/Ki | [I] (µM) |
[I]/Ki | [I]/Ki | P | M | P+M | AUC | [I] | ||||||
| amiodarone | N-desethylamiodarone | 2C9 | 200 mg, b.i.d., 10 d | (s)-warfarin | 2.1 | 1.8 | 0.019 | 1.2 | 0.51 | 0.53 | R | P | P | [35] | [36] |
| bupropion | erythrohydrobupropion | 2D6 | 150 mg, b.i.d., 12 d | desipramine | 5.2 | 0.67 | 0.032 | 0.43 | 0.25 | 1.02 | R | P | L | [12] | [12] |
| hydroxybupropion | 4.0 | 0.30 | P | ||||||||||||
| threohydrobupropion | 2.3 | 0.43 | P | ||||||||||||
| fluoxetine | norfluoxetine | 2D6 | 60 mg, q.d., 8 d | desipramine | 7.8 | 1.4a | 7.9a | 1.6a | 8.1 | 16 | L | L | L | [37] | [38] |
| imatinib | N-desmethylimanitib | 2D6 | 400 mg, b.i.d., 9 d | metoprolol | 1.2 | 8.6 | 1.1 | 1.4 | 0.10 | 1.2 | L | P | L | [29] | [29] |
| 3A | 400 mg, q.d., 7 d | simvastatin | 2.9 | 8.6 | 1.0 | 1.4 | 0.099 | 1.2 | L | R | L | c | [29] | ||
| itraconazole | hydroxyitraconazole | 3A4 | 200 mg, q.d., 4 d | lovastatin | 36.4 | 0.94 | 60 | 1.1 | 29 | 89 | L | L | L | [39] | [39] |
| omeprazole | omeprazole sulfone | 2C19 | 40 mg, q.d., 7 d | proguanil | 1.5 | 1.2a | 0.62 | 0.44a | 0.054 | 0.67 | P | R | P | [40] | [41] |
| 3A4 | 20 mg, q.d., 8 d | nifedipine | 1.3 | 0.62 | 0.003 | 0.22 | 0.001 | 0.004 | R | R | R | [42] | [41] | ||
| quinidine | quinidine N-oxide | 2D6 | 50 mg, single | dextromethorphan | 42.6 | 0.57a | 19 | 0.18a | 0.004 | 19 | L | R | L | [43] | [44] |
| 3-hydroxyquinidine | 0.09 | 0.039 | R | ||||||||||||
| ranolazine | O-desmethylranolazine | 3A4 | 1000 mg, b.i.d., 7 d | simvastatin | 1.9 | 5.0 | 0.056 | 2.7 | 0.026 | 0.082 | R | R | R | d | d |
| sertraline | N-desmethylsertraline | 2D6 | 150 mg, q.d., 8 d | imipramine | 1.7 | 0.25a | 0.011 | 0.32a | 0.020 | 0.031 | R | R | R | [45] | [46] |
| 3A4 | 200 mg, q.d., 29 d | pimozide | 1.4 | 0.33 | 0.095 | 0.53 | 0.15 | 0.25 | R | P | P | [22] | [22] | ||
| sulfinpyrazone | sulfinpyrazone sulfide | 2C9 | 200 mg, b.i.d., 13 d | (s)-warfarin | 1.9 | 51 | 0.223 | 36 | 2.1 | 2.4 | P | L | L | [47] | [48] |
| sulfinpyrazone sulfone | 8.1 | 0.11 | P | ||||||||||||
| venlafaxine | N-desmethylvenlafaxine | 2D6 | 50 mg, t.i.d., 3 d | imipramine | 1.3 | 0.25 | 0.008 | 0.027 | 0.001 | 0.010 | R | R | R | [49] | [50] |
The inhibitor and its metabolites concentration were calculated with dose normalization.
R; remote, P; possible, L; likely
U.S. Food and Drug Administration. Clinical Reviews- Imatinib. Drugs@FDA 2002
U.S. Food and Drug Administration. Clinical Review- Ranolazine. Drugs@FDA 2007
The Ki-values used were the same in Table 1.
Figure 2.
Evaluation of DDI risk with [I]/Ki data for inhibitors presented in Table 2. Panels A and B show the correlation between in vitro risk assessment and the magnitude of in vivo DDIs when only parent drug is accounted for (A) and when parent and metabolites are analyzed together (B). Panel C shows the 9 zones for prediction in accordance to panels A and B, including true negative, true positive, false negative, and false positive.
Quantitative in vitro to in vivo extrapolation of drug-drug interactions
In vivo drug-drug interactions are commonly rationalized by the use of static or dynamic models that allow extrapolation of in vitro inhibition potency to in vivo interactions. The existence of circulating inhibitory metabolites could cause a significant underprediction of the existing interactions and, if metabolites are not accounted for, lead to poor understanding of the characteristics of specific interactions. To determine if circulating metabolites lead to significant underpredictions in known, well characterized drug-drug interactions, the effect of inhibitory metabolites in the accuracy of in vitro to in vivo predictions of interaction magnitude was tested using classic static prediction methods. Table 3 shows the predicted and observed interactions for the 10 inhibitors analyzed. Interactions were included in the analysis, if in vivo data with a well characterized probe was available and [I]/Ki ratios could be calculated for both the parent drug and metabolite. Both steady state Cmax and steady state Cav were used for the analysis and no apparent differences were observed between the predictions with these two values. This might be because at steady state there is minimum fluctuation in the inhibitor concentrations over the dosing interval and hence Cmax and Cav are similar. For amiodarone, bupropion, fluoxetine and sulfinpyrazone, an improved quantitative prediction of the in vivo interaction was obtained when metabolites were accounted for whereas for the remaining six inhibitors no significant differences in prediction accuracy were observed. The fact that quantitative predictions were possible only after accounting for the metabolites for four of ten analyzed inhibitors, suggests that knowledge of circulating metabolites and their inhibitory potency should be obtained prior to quantitative modeling of the observed interaction.
Table 3.
Predicted versus observed AUC values calculated with Cmax and Cav.
| Parent (P) Metabolite (M) |
CYP | Inhibitor dose |
Victim drug |
AUC change of victim drug | ||||
|---|---|---|---|---|---|---|---|---|
| observed | predicted (Cmax) | predicted (Cav) | ||||||
| P | P+M | P | P+M | |||||
|
amiodarone N-desethylamiodarone |
2C9 | 200 mg, q.d., 6.5 w | phenytoin | 1.4 | 1.01 | 1.35 | 1.01 | 1.31 |
|
bupropion erythrohydrobupropion hydroxybupropion threohydrobupropion |
2D6 | 150 mg, b.i.d., 12 d | desipramine | 5.2 | 1.03 | 1.8 | 1.01 | 1.5 |
|
fluoxetine norfluoxetine |
2D6 | 20 mg, q.d., 21 d | desipramine | 4.4 | 2.41 | 3.35 | 2.47 | 3.61 |
|
imatinib N-desmethylimanitib |
2D6 | 400 mg, b.i.d., 9 d | metoprolol | 1.2 | 1.79 | 1.85 | — | — |
|
itraconazole hydroxyitraconazole |
3A4 | 200 mg, q.d., 4 d | midazolam | 10.8 | 10.9 | 12.5 | 8.1 | 10.4 |
|
omeprazole omeprazole sulfone |
3A4 | 20 mg, q.d., 8 d | nifedipine | 1.3 | 1.0 | 1.0 | 1.0 | 1.0 |
|
quinidine quinidine N-oxide 3-hydroxyquinidine |
2D6 | 50 mg, single | desipramine | 3.2 | 6.0* | 6.0* | — | — |
|
ranolazine O-desmethylranolazine |
3A4 | 1000 mg, b.i.d., 7 d | simvastatin | 1.9 | 1.06 | 1.08 | 1.07 | 1.1 |
|
sertraline N-desmethylsertraline |
2D6 | 50 mg, q.d., 10 d | desipramine | 1.4 | 1.0 | 1.01 | 1.0 | 1.01 |
|
sulfinpyrazone sulfinpyrazone sulfide sulfinpyrazone sulfone |
2C9 | 200 mg, b.i.d., 13 d | (s)-warfarin | 1.9 | 1.19 | 2.61 | — | — |
: The Cav was not available.
The inhibitor and it’s metabolites concentration were calculated with dose normalization.
DISCUSSION
This study aimed at determining to what extent circulating metabolites contribute to in vivo drug-drug interactions in context of preclinical risk assessment and of development of quantitative methods of predicting in vivo drug-drug interactions. The analysis shows that a majority of the clinically important P450 inhibitors have metabolites that circulate at appreciable concentrations and a third of the known metabolites are more abundant than the parent drug. However, the P450 inhibition potential has been published only for a fraction of the metabolites. It is possible that unpublished negative inhibition data exists but was not available for this analysis.
The existing metabolite data was evaluated to determine whether the known metabolites would alter the preclinical risk assessment of known clinical P450 inhibitors. Surprisingly, for three of the 11 inhibitors evaluated, the risk assessment changed from remote likelihood of DDIs to possible or likely with the inclusion of metabolites. This suggests that determination of Ki values for quantitatively important metabolites can be useful when interactions are not predicted or only weak interactions are predicted with parent drug. On the other hand, testing for inhibitory potency for metabolites might be of limited usefulness when parent drug on its own has an identified risk at the likely category, since the true magnitude of the in vivo interaction will be determined in relevant in vivo DDI studies. Interestingly, all the compounds that were “missed” in the risk analysis (amiodarone, bupropion, ranolazine, sertraline and venlafaxine) when only parent inhibitor data was included, had metabolites with IPM/IPP-ratios>0.1, suggesting that metabolites of these compounds may be critical in in vivo drug-drug interactions. On the other hand omeprazole sulfone, quinidine N-oxide and 3-hydroxy quinidine which were predicted in the risk analysis to only have remote potential to cause drug-drug interactions had IPM/IPP-ratios <0.1.
A possible explanation for the incorrect assignment of risk for the ranolazine-simvastatin interaction could be the involvement of transporters such as P-glycoprotein (P-gp) in the in vivo interaction. Ranolazine has been shown to cause in a 1.5-fold increase in in vivo digoxin concentrations, suggesting that it is a P-gp inhibitor (http://www.accessdata.fda.gov/drugsatfda_docs/nda/2006/021526_s000_Ranexa.cfm).
Since simvastatin has been shown to be a P-gp substrate [6], it is possible that the in vivo interaction is due to P-gp inhibition during simvastatin absorption. Although data for the other probes, inhibitors and metabolites is not available, it is possible that transporter mediated interactions contribute to the other underpredicitons as well. In addition, metabolites may be sequestered in the hepatocytes and hence plasma concentrations do not accurately reflect the intracellular free inhibitor concentrations. Such sequestration is not accounted for in the current analysis.
A special consideration may be warranted with compounds that are multi-CYP inhibitors in vitro. These compounds are typically analyzed using a rank-order approach [7]. As inhibitor depletion will prevent formation of significant amounts of metabolites in in vitro inhibition assays, rank order analyses in vitro are likely to reflect the potency of the parent drug. If metabolites do not follow the same rank order, or inhibit only a subset of the CYPs that the parent compound inhibits, rank order approach may fail. For the three multi-CYP inhibitors included in this analysis, the rank order for the metabolite was similar to that observed for the parent drug when reversible inhibition was considered, but the dataset is not representative of the overall multi-CYP inhibitors.
The recently published MIST guidance recommends toxicology review of metabolites that circulate in excess of 10% of the parent drug. Although the MIST guidance does not address metabolites in drug-drug interactions, the current data was analyzed in light of the 10% cutoff value of the MIST guidance. Based on the [I]/Ki ratios of the metabolites in relation to the parent compounds, with the exception of 4-hydroxypropranolol, all the metabolites that were predicted to contribute to in vivo interactions had an AUC greater than 10% of the parent at steady state. Interestingly, there was no correlation between the contribution of the metabolites to interactions (IPM/IPP-ratio) and the relative abundance of the metabolites in plasma, as many metabolites were more potent inhibitors than the parent drug while they circulated at lower concentrations than the parent drug. Hence, the 10% cut-off AUC value appears appropriate for inhibition testing of metabolites. Based on these analyses, predicting the [I]/Ki-ratio for abundant metabolites (as well as for the parent inhibitor) can be helpful in drug discovery and development process even when the parent compound is not inhibitory. This also requires that the methodology predicting [I] not only for parent but also metabolite should be established.
The inhibitory metabolites included in this analysis may not be the sole circulating metabolites of that given inhibitor but were the only ones with published in vitro inhibition data and in vivo AUC values. For example 5-hydroxyomeprazole is more abundant than omeprazole sulfone after omeprazole administration. However, no in vitro inhibition data was available for 5-hydroxyomeprazole and hence this metabolite could not be analyzed. Similarly, atorvastatin has several metabolites that circulate at similar concentrations as atorvastatin itself but the inhibition potency of these metabolites has not been determined. On the other hand, itraconazole has two other metabolites, keto-itraconazole and N-desalkyl-itraconazole that inhibit CYP3A4 potently but their steady state concentrations have not been determined. The existence of multiple inhibitory metabolites would further increase the relative contribution of metabolites as a whole in comparison to the parent drug.
The role of inhibitory metabolites in quantitatively predicting the magnitude of drug-drug interactions may be less than would be expected from the abundance of the metabolites. In the analysis of quantitative predictions, inclusion of metabolites did not always improve the accuracy of the prediction and the increase in average predictive accuracy was only modest. However, for some of the inhibitors significant improvement in predictions was observed. As plasma and microsomal unbound fractions are not available for majority of these compounds, the quantitative prediction may not accurately reflect the true relative importance of all of the metabolites analyzed. Despite this, the fact that so many inhibitors have circulating inhibitory metabolites may have implications to overall correlation analyses of in vitro to in vivo extrapolations, as an accurate model should incorporate all inhibitory species.
In conclusion, these results suggest that metabolites or candidates for major circulating metabolites in vivo, should be investigated in addition to parent drug for inhibition potential in vitro unless clinical interaction studies are already warranted due to the parent compound. Although the available literature data does not allow for comprehensive evaluation, these results suggest that circulating metabolites contribute to in vivo drug-drug interactions and further study will be required for in-depth understanding of their overall clinical significance.
METHODS
Identification of the inhibitors and their metabolites
The University of Washington Metabolism and Transport Drug Interaction Database™ (MTDI database: http://www.druginteractioninfo.org) was searched to identify known P450 inhibitors. All inhibitors analyzed had to be drugs currently available in the US market, and herbal products, combination therapies and oral contraceptives were excluded from the analysis. The complete list of identified inhibitors and their P450 inhibition characteristics was previously published [5].
The circulating metabolites and the AUC values for the parent drug and metabolites of these inhibitors were extracted from the MTDI database by searching these compounds as objects in DDI studies in which their metabolites were also measured. If no metabolite data was available in the MTDI database, a Pubmed search was conducted to extract circulating metabolite data. Finally, the product labels and Micromedex were used as a reference for data on circulating metabolites. To calculate metabolite to parent AUC ratios the following criteria were used:
The metabolite and the parent must have been measured in the same study and complete (dosing interval at steady state or 0-infinity after single dose) AUC’s for both had to be available
Steady state values for the parent drug and metabolite were used if available. If no steady state pharmacokinetic data was available single dose kinetics were accepted. The ratio between the metabolite and parent AUC was calculated either by using the steady state dosing interval AUC or by using the AUC to infinity after single dose.
For compounds that had data from multiple dosing levels the highest AUCM/AUCP-ratio was used.
All AUC values were converted to µmol∙h/mL units using the molecular weight of the parent drug and the metabolite, and the ratio between the metabolite and parent drug AUCs were calculated.
Determination of in vivo drug-drug interaction magnitude and the inhibitory potency for parent and metabolites
The MTDI database was queried to retrieve all reported in vivo interactions (defined as resulting in a ≥ 20% increase in the AUC or decrease in clearance of the victim drug) for the inhibitors that were previously identified to have circulating metabolites. From the resulting list of in vivo interaction studies, the ones conducted with a known marker substrates (www.fda.gov/downloads/Drugs/GuidanceComplianceRegulatoryInformation/Guidances/ucm072101.pdf) were kept for further analysis. All data for in vivo AUC change using marker substrates were extracted and the change in object AUC was recorded for analysis. The inhibitor and its metabolite concentrations measured in the interaction study were recorded if available. For studies that did not measure the plasma concentrations of the inhibitor and its metabolites, literature data using the same dosing regimen was used to obtain steady state Cmax and Cav values for the inhibitor and its metabolites. If data for the inhibitor and its metabolite was not available at the dose level used in the in vivo interaction studies, the concentrations were dose normalized to allow the risk analysis.
The MTDI database was queried to extract the in vitro Ki-values for the identified metabolites and the parent inhibitors. Inhibitors that were known inactivators of a given enzyme were excluded from further analysis for the enzyme that was irreversibly inactivated in vitro by the parent drug or the metabolite. In case there were multiple reversible Ki-values for the parent inhibitor or its metabolites, the Ki-value used for analysis was chosen according to the following criteria: 1) The metabolite and the parent drug were tested in the same study against the same P450 and same probe; 2) if the metabolite and parent were not available in the same study two studies that used the same probe were chosen; 3) in the absence of such data two studies that used the same microsomal protein concentration were used. For equivalent data, the study with the lowest microsomal protein concentration was always used.
Relative importance of circulating metabolites
To predict the relative contribution of the metabolites in in vivo drug-drug interactions, the [I]/Ki ratios were calculated for all inhibitors and metabolites that had sufficient in vitro data. The clinically relevant steady state average total concentrations of the drug and its metabolite (measured in the same study and dose level) were used as [I] and the Ki values were collected as described above. The [I]/Ki ratio was calculated for the metabolite and parent drug separately, and then the ratio between the [I]/Ki of the metabolite to parent (IPM/IPP ratio) was calculated. As the [I]/Ki ratio for the metabolite (IPM) and for the parent (IPP) is indicative of their in vivo inhibitory potential (IP), the ratio of the two can be used as a predictor of the relative in vivo contribution of metabolites to CYP inhibition. The predicted contribution of the metabolites was classified as follows: 1) If the IPM/IPP ratio was ≤0.1 the metabolites were not predicted to contribute to in vivo interactions; 2) if IPM/IPP ratio was ≥1 the metabolite was predicted to be the main inhibitory species causing the in vivo interaction; and 3) if 0.1< IPM/IPP-ratio<1 metabolite contribution was predicted to be likely in the interaction but the parent drug was predicted to be the main inhibitor causing the interaction.
Risk analysis using metabolite and parent drug data
A risk analysis for inhibitory interactions was conducted according to the FDA guidance for testing parent drugs (www.fda.gov/downloads/Drugs/GuidanceComplianceRegulatoryInformation/Guidances/ucm079266.pdf). Only inhibitors for which in vivo interaction data with a known probe was available were included in this analysis. The steady state inhibitor concentrations (total Cmax) and in vitro Ki values were used to compute the [I]/Ki ratio for the inhibitors and their metabolites. In order to evaluate the highest risk, the in vivo studies with the most sensitive probes for the inhibited P450 and highest in vivo interactions were included in the analysis. Based on the [I]/Ki ratio, the potential of the drugs to cause in vivo interactions was classified as likely ([I]/Ki>1), possible (1>[I]/Ki>0.1) or remote (0.1>[I]/Ki) for the parent drug and the metabolite separately. To predict the combined risk of metabolites and parent drug, the [I]/Ki ratios were summed together and the sum was ranked similar to the individual compounds.
Quantitative prediction of in vivo drug-drug interactions
Ten inhibitors with in vivo DDI data with a probe substrate with a known fmCYP were used to estimate the role of metabolites in quantitative in vitro to in vivo extrapolations (IVIVE) of inhibitory interactions. Studies were included in the quantitative analysis if the in vivo study was conducted with a reliable probe with a defined fm and the Cmax and Cav were available in the DDI study or in another study with similar dosing regimen. If multiple DDI studies were available, the study with the highest AUC change was selected to consider possible worst case scenario. Both the total Cmax and Cav values for the inhibitor and its metabolite were used in the predictions as [I]. IVIVE was done according to Equation 1:
| (1) |
in which the fmCYP for each marker substrate was included according to the following list. fmCYP3A4: midazolam 0.94, nifedipine 0.71, simvastatin 0.99; fmCYP2D6: desipramine 0.877, metoprolol 0.828; fmCYP2C9: phenytoin 0.75, (S)-warfarin 0.87 ([8, 9]). The effect of multiple inhibitors was accounted for by summing the [I]/Ki ratios.
Supplementary Material
ACKNOWLEDGEMENTS
This study was supported in part by an NIH grant P01 GM32165 and the Elmer M. and Joy B. Plein Fellowship for Excellence in Pharmacy Education (CKY) at the School of Pharmacy University of Washington.
Abbreviations
- CYP
cytochrome P450
- Cmax
maximum plasma concentrations
- Cav
average steady state plasma concentration
- fm
fraction metabolized
- fmCYP
fraction metabolized via specific CYP
- AUC
area under plasma concentration time curve
- AUCi
area under plasma concentration time curve in the presence of an inhibitor
- DDI
drug-drug interaction
- [I]
inhibitor concentration
- Ki
inhibition constant
Footnotes
Conflict of Interest/Disclosure
None
REFERENCES
- 1.Zhang X, Jones DR, Hall SD. Prediction of the effect of erythromycin, diltiazem, and their metabolites, alone and in combination, on CYP3A4 inhibition. Drug Metab Dispos. 2009 Jan;37(1):150–160. doi: 10.1124/dmd.108.022178. [DOI] [PubMed] [Google Scholar]
- 2.Zhang X, Quinney SK, Gorski JC, Jones DR, Hall SD. Semiphysiologically based pharmacokinetic models for the inhibition of midazolam clearance by diltiazem and its major metabolite. Drug Metab Dispos. 2009 Aug;37(8):1587–1597. doi: 10.1124/dmd.109.026658. [DOI] [PubMed] [Google Scholar]
- 3.Isoherranen N, Kunze KL, Allen KE, Nelson WL, Thummel KE. Role of itraconazole metabolites in CYP3A4 inhibition. Drug Metab Dispos. 2004 Oct;32(10):1121–1131. doi: 10.1124/dmd.104.000315. [DOI] [PubMed] [Google Scholar]
- 4.Stevens JC, Wrighton SA. Interaction of the enantiomers of fluoxetine and norfluoxetine with human liver cytochromes P450. J Pharmacol Exp Ther. 1993 Aug;266(2):964–971. [PubMed] [Google Scholar]
- 5.Isoherranen N, Hachad H, Yeung CK, Levy RH. Qualitative analysis of the role of metabolites in inhibitory drug-drug interactions: literature evaluation based on the metabolism and transport drug interaction database. Chem Res Toxicol. 2009 Feb;22(2):294–298. doi: 10.1021/tx800491e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hochman JH, Pudvah N, Qiu J, Yamazaki M, Tang C, Lin JH, et al. Interactions of human P-glycoprotein with simvastatin, simvastatin acid, and atorvastatin. Pharm Res. 2004 Sep;21(9):1686–1691. doi: 10.1023/b:pham.0000041466.84653.8c. [DOI] [PubMed] [Google Scholar]
- 7.Obach RS, Walsky RL, Venkatakrishnan K, Houston JB, Tremaine LM. In vitro cytochrome P450 inhibition data and the prediction of drug-drug interactions: qualitative relationships, quantitative predictions, and the rank-order approach. Clin Pharmacol Ther. 2005 Dec;78(6):582–592. doi: 10.1016/j.clpt.2005.09.004. [DOI] [PubMed] [Google Scholar]
- 8.Brown HS, Ito K, Galetin A, Houston JB. Prediction of in vivo drug-drug interactions from in vitro data: impact of incorporating parallel pathways of drug elimination and inhibitor absorption rate constant. Br J Clin Pharmacol. 2005 Nov;60(5):508–518. doi: 10.1111/j.1365-2125.2005.02483.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ito K, Hallifax D, Obach RS, Houston JB. Impact of parallel pathways of drug elimination and multiple cytochrome P450 involvement on drug-drug interactions: CYP2D6 paradigm. Drug Metab Dispos. 2005 Jun;33(6):837–844. doi: 10.1124/dmd.104.003715. [DOI] [PubMed] [Google Scholar]
- 10.Robinson K, Johnston A, Walker S, Mulrow JP, McKenna WJ, Holt DW. The digoxin-amiodarone interaction. Cardiovasc Drugs Ther. 1989 Mar;3(1):25–28. doi: 10.1007/BF01881526. [DOI] [PubMed] [Google Scholar]
- 11.Ohyama K, Nakajima M, Suzuki M, Shimada N, Yamazaki H, Yokoi T. Inhibitory effects of amiodarone and its N-deethylated metabolite on human cytochrome P450 activities: prediction of in vivo drug interactions. Br J Clin Pharmacol. 2000 Mar;49(3):244–253. doi: 10.1046/j.1365-2125.2000.00134.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Reese MJ, Wurm RM, Muir KT, Generaux GT, St John-Williams L, McConn DJ. An in vitro mechanistic study to elucidate the desipramine/bupropion clinical drug-drug interaction. Drug Metab Dispos. 2008 Jul;36(7):1198–1201. doi: 10.1124/dmd.107.020198. [DOI] [PubMed] [Google Scholar]
- 13.He M, Kunze KL, Trager WF. Inhibition of (S)-warfarin metabolism by sulfinpyrazone and its metabolites. Drug Metab Dispos. 1995 Jun;23(6):659–663. [PubMed] [Google Scholar]
- 14.Otton SV, Ball SE, Cheung SW, Inaba T, Rudolph RL, Sellers EM. Venlafaxine oxidation in vitro is catalysed by CYP2D6. Br J Clin Pharmacol. 1996 Feb;41(2):149–156. doi: 10.1111/j.1365-2125.1996.tb00173.x. [DOI] [PubMed] [Google Scholar]
- 15.Jacobsen W, Kuhn B, Soldner A, Kirchner G, Sewing KF, Kollman PA, et al. Lactonization is the critical first step in the disposition of the 3-hydroxy-3- methylglutaryl-CoA reductase inhibitor atorvastatin. Drug Metab Dispos. 2000 Nov;28(11):1369–1378. [PubMed] [Google Scholar]
- 16.Keskitalo JE, Kurkinen KJ, Neuvoneni PJ, Niemi M. ABCB1 haplotypes differentially affect the pharmacokinetics of the acid and lactone forms of simvastatin and atorvastatin. Clin Pharmacol Ther. 2008 Oct;84(4):457–461. doi: 10.1038/clpt.2008.25. [DOI] [PubMed] [Google Scholar]
- 17.Bapiro TE, Egnell AC, Hasler JA, Masimirembwa CM. Application of higher throughput screening (HTS) inhibition assays to evaluate the interaction of antiparasitic drugs with cytochrome P450s. Drug Metab Dispos. 2001 Jan;29(1):30–35. [PubMed] [Google Scholar]
- 18.Skjelbo E, Brosen K. Inhibitors of imipramine metabolism by human liver microsomes. Br J Clin Pharmacol. 1992 Sep;34(3):256–261. doi: 10.1111/j.1365-2125.1992.tb04133.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.von Moltke LL, Greenblatt DJ, Cotreau-Bibbo MM, Duan SX, Harmatz JS, Shader RI. Inhibition of desipramine hydroxylation in vitro by serotonin-reuptakeinhibitor antidepressants, and by quinidine and ketoconazole: a model system to predict drug interactions in vivo. J Pharmacol Exp Ther. 1994 Mar;268(3):1278–1283. [PubMed] [Google Scholar]
- 20.Prakash C, Kamel A, Cui D, Whalen RD, Miceli JJ, Tweedie D. Identification of the major human liver cytochrome P450 isoform(s) responsible for the formation of the primary metabolites of ziprasidone and prediction of possible drug interactions. Br J Clin Pharmacol. 2000;49(Suppl 1):35S–42S. doi: 10.1046/j.1365-2125.2000.00151.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.von Moltke LL, Greenblatt DJ, Harmatz JS, Duan SX, Harrel LM, Cotreau-Bibbo MM, et al. Triazolam biotransformation by human liver microsomes in vitro: effects of metabolic inhibitors and clinical confirmation of a predicted interaction with ketoconazole. J Pharmacol Exp Ther. 1996 Feb;276(2):370–379. [PubMed] [Google Scholar]
- 22.Alderman J. Coadministration of sertraline with cisapride or pimozide: an openlabel, nonrandomized examination of pharmacokinetics and corrected QT intervals in healthy adult volunteers. Clin Ther. 2005 Jul;27(7):1050–1063. doi: 10.1016/j.clinthera.2005.07.013. [DOI] [PubMed] [Google Scholar]
- 23.Shin JG, Kane K, Flockhart DA. Potent inhibition of CYP2D6 by haloperidol metabolites: stereoselective inhibition by reduced haloperidol. Br J Clin Pharmacol. 2001 Jan;51(1):45–52. doi: 10.1046/j.1365-2125.2001.01313.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Otton SV, Wu D, Joffe RT, Cheung SW, Sellers EM. Inhibition by fluoxetine of cytochrome P450 2D6 activity. Clin Pharmacol Ther. 1993 Apr;53(4):401–409. doi: 10.1038/clpt.1993.43. [DOI] [PubMed] [Google Scholar]
- 25.Kobayashi K, Yamamoto T, Chiba K, Tani M, Ishizaki T, Kuroiwa Y. The effects of selective serotonin reuptake inhibitors and their metabolites on S-mephenytoin 4'- hydroxylase activity in human liver microsomes. Br J Clin Pharmacol. 1995 Nov;40(5):481–485. doi: 10.1111/j.1365-2125.1995.tb05793.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rowland K, Yeo WW, Ellis SW, Chadwick IG, Haq I, Lennard MS, et al. Inhibition of CYP2D6 activity by treatment with propranolol and the role of 4-hydroxy propranolol. Br J Clin Pharmacol. 1994 Jul;38(1):9–14. doi: 10.1111/j.1365-2125.1994.tb04315.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Crespi CL, Penman BW. Use of cDNA-expressed human cytochrome P450 enzymes to study potential drug-drug interactions. Adv Pharmacol. 1997;43:171–188. doi: 10.1016/s1054-3589(08)60205-7. [DOI] [PubMed] [Google Scholar]
- 28.Masimirembwa CM, Hasler JA, Johansson I. Inhibitory effects of antiparasitic drugs on cytochrome P450 2D6. Eur J Clin Pharmacol. 1995;48(1):35–38. doi: 10.1007/BF00202169. [DOI] [PubMed] [Google Scholar]
- 29.Wang Y, Zhou L, Dutreix C, Leroy E, Yin Q, Sethuraman V, et al. Effects of imatinib (Glivec) on the pharmacokinetics of metoprolol, a CYP2D6 substrate, in Chinese patients with chronic myelogenous leukaemia. Br J Clin Pharmacol. 2008 Jun;65(6):885–892. doi: 10.1111/j.1365-2125.2008.03150.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lakehal F, Wurden CJ, Kalhorn TF, Levy RH. Carbamazepine and oxcarbazepine decrease phenytoin metabolism through inhibition of CYP2C19. Epilepsy Res. 2002 Dec;52(2):79–83. doi: 10.1016/s0920-1211(02)00188-2. [DOI] [PubMed] [Google Scholar]
- 31.Sauer JM, Long AJ, Ring B, Gillespie JS, Sanburn NP, DeSante KA, et al. Atomoxetine hydrochloride: clinical drug-drug interaction prediction and outcome. J Pharmacol Exp Ther. 2004 Feb;308(2):410–418. doi: 10.1124/jpet.103.058727. [DOI] [PubMed] [Google Scholar]
- 32.Chiba K, Kobayashi K, Manabe K, Tani M, Kamataki T, Ishizaki T. Oxidative metabolism of omeprazole in human liver microsomes: cosegregation with Smephenytoin 4'-hydroxylation. J Pharmacol Exp Ther. 1993 Jul;266(1):52–59. [PubMed] [Google Scholar]
- 33.Zomorodi K, Houston JB. Diazepam-omeprazole inhibition interaction: an in vitro investigation using human liver microsomes. Br J Clin Pharmacol. 1996 Aug;42(2):157–162. doi: 10.1046/j.1365-2125.1996.03563.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ching MS, Blake CL, Ghabrial H, Ellis SW, Lennard MS, Tucker GT, et al. Potent inhibition of yeast-expressed CYP2D6 by dihydroquinidine, quinidine, and its metabolites. Biochem Pharmacol. 1995 Sep 7;50(6):833–837. doi: 10.1016/0006-2952(95)00207-g. [DOI] [PubMed] [Google Scholar]
- 35.O'Reilly RA, Trager WF, Rettie AE, Goulart DA. Interaction of amiodarone with racemic warfarin and its separated enantiomorphs in humans. Clin Pharmacol Ther. 1987 Sep;42(3):290–294. doi: 10.1038/clpt.1987.149. [DOI] [PubMed] [Google Scholar]
- 36.Shoaf SE, Elizari MV, Wang Z, Sekar K, Grinfeld LR, Barbagelata NA, et al. Tolvaptan administration does not affect steady state amiodarone concentrations in patients with cardiac arrhythmias. J Cardiovasc Pharmacol Ther. 2005 Sep;10(3):165–171. doi: 10.1177/107424840501000304. [DOI] [PubMed] [Google Scholar]
- 37.Bergstrom RF, Peyton AL, Lemberger L. Quantification and mechanism of the fluoxetine and tricyclic antidepressant interaction. Clin Pharmacol Ther. 1992 Mar;51(3):239–248. doi: 10.1038/clpt.1992.18. [DOI] [PubMed] [Google Scholar]
- 38.Harvey AT, Preskorn SH. Fluoxetine pharmacokinetics and effect on CYP2C19 in young and elderly volunteers. J Clin Psychopharmacol. 2001 Apr;21(2):161–166. doi: 10.1097/00004714-200104000-00007. [DOI] [PubMed] [Google Scholar]
- 39.Neuvonen PJ, Jalava KM. Itraconazole drastically increases plasma concentrations of lovastatin and lovastatin acid. Clin Pharmacol Ther. 1996 Jul;60(1):54–61. doi: 10.1016/S0009-9236(96)90167-8. [DOI] [PubMed] [Google Scholar]
- 40.Funck-Brentano C, Becquemont L, Lenevu A, Roux A, Jaillon P, Beaune P. Inhibition by omeprazole of proguanil metabolism: mechanism of the interaction in vitro and prediction of in vivo results from the in vitro experiments. J Pharmacol Exp Ther. 1997 Feb;280(2):730–738. [PubMed] [Google Scholar]
- 41.Stuht H, Lode H, Koeppe P, Rost KL, Schaberg T. Interaction study of lomefloxacin and ciprofloxacin with omeprazole and comparative pharmacokinetics. Antimicrob Agents Chemother. 1995 May;39(5):1045–1049. doi: 10.1128/aac.39.5.1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Soons PA, van den Berg G, Danhof M, van Brummelen P, Jansen JB, Lamers CB, et al. Influence of single- and multiple-dose omeprazole treatment on nifedipine pharmacokinetics and effects in healthy subjects. Eur J Clin Pharmacol. 1992;42(3):319–324. doi: 10.1007/BF00266355. [DOI] [PubMed] [Google Scholar]
- 43.Capon DA, Bochner F, Kerry N, Mikus G, Danz C, Somogyi AA. The influence of CYP2D6 polymorphism and quinidine on the disposition and antitussive effect of dextromethorphan in humans. Clin Pharmacol Ther. 1996 Sep;60(3):295–307. doi: 10.1016/S0009-9236(96)90056-9. [DOI] [PubMed] [Google Scholar]
- 44.Damkier P, Hansen LL, Brosen K. Effect of diclofenac, disulfiram, itraconazole, grapefruit juice and erythromycin on the pharmacokinetics of quinidine. Br J Clin Pharmacol. 1999 Dec;48(6):829–838. doi: 10.1046/j.1365-2125.1999.00099.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kurtz DL, Bergstrom RF, Goldberg MJ, Cerimele BJ. The effect of sertraline on the pharmacokinetics of desipramine and imipramine. Clin Pharmacol Ther. 1997 Aug;62(2):145–156. doi: 10.1016/S0009-9236(97)90062-X. [DOI] [PubMed] [Google Scholar]
- 46.Alderman J, Preskorn SH, Greenblatt DJ, Harrison W, Penenberg D, Allison J, et al. Desipramine pharmacokinetics when coadministered with paroxetine or sertraline in extensive metabolizers. J Clin Psychopharmacol. 1997 Aug;17(4):284–291. doi: 10.1097/00004714-199708000-00008. [DOI] [PubMed] [Google Scholar]
- 47.O'Reilly RA. Stereoselective interaction of sulfinpyrazone with racemic warfarin and its separated enantiomorphs in man. Circulation. 1982 Jan;65(1):202–207. doi: 10.1161/01.cir.65.1.202. [DOI] [PubMed] [Google Scholar]
- 48.Rosenkranz B, Fischer C, Jakobsen P, Kirstein Pedersen A, Frolich JC. Plasma levels of sulfinpyrazone and of two of its metabolites after a single dose and during the steady state. Eur J Clin Pharmacol. 1983;24(2):231–235. doi: 10.1007/BF00613823. [DOI] [PubMed] [Google Scholar]
- 49.Albers LJ, Reist C, Vu RL, Fujimoto K, Ozdemir V, Helmeste D, et al. Effect of venlafaxine on imipramine metabolism. Psychiatry Res. 2000 Nov 20;96(3):235–243. doi: 10.1016/s0165-1781(00)00213-4. [DOI] [PubMed] [Google Scholar]
- 50.Lindh JD, Annas A, Meurling L, Dahl ML, A AL-S Effect of ketoconazole on venlafaxine plasma concentrations in extensive and poor metabolisers of debrisoquine. Eur J Clin Pharmacol. 2003 Sep;59(5–6):401–406. doi: 10.1007/s00228-003-0627-x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.