Abstract
The plasma membrane sodium/proton exchanger Salt-Overly-Sensitive 1 (SOS1) is a critical salt tolerance determinant in plants. The SOS2–SOS3 calcium-dependent protein kinase complex up-regulates SOS1 activity, but the mechanistic details of this crucial event remain unresolved. Here we show that SOS1 is maintained in a resting state by a C-terminal auto-inhibitory domain that is the target of SOS2–SOS3. The auto-inhibitory domain interacts intramolecularly with an adjacent domain of SOS1 that is essential for activity. SOS1 is relieved from auto-inhibition upon phosphorylation of the auto-inhibitory domain by SOS2–SOS3. Mutation of the SOS2 phosphorylation and recognition site impeded the activation of SOS1 in vivo and in vitro. Additional amino acid residues critically important for SOS1 activity and regulation were identified in a genetic screen for hypermorphic alleles.
Keywords: ion transport, salinity, sodium tolerance
Salinity is a major problem in agriculture because the total area of salt-affected soils, including saline and sodic soils, exceeds 900 million ha (1). Salt-affected soils reduce both the ability of crops to take up water and the availability of mineral nutrients. Often, the high sodium (Na) content relative to other cations is the main factor affecting plant growth by causing a set of metabolic derangements (2). Because most crop species have only very limited capacities to cope with excess Na, the elucidation of Na tolerance mechanisms in plants is of paramount importance (2). Plant ion transporters mediating Na fluxes have recently been cloned and characterized, and the knowledge of the regulatory mechanisms of transporter abundance and activity in response to environmental, hormonal, and developmental signals is critical for understanding salinity tolerance (3).
The plasma membrane Na/H antiporter SOS1 is essential for the salt tolerance of various model plants, including Arabidopsis thaliana (4) and its halophytic relative Thellungiella salsuginea (5), tomato (6), and the moss Physcomitrella patens (7). SOS1 is thought to mediate Na efflux at the root epidermis and long-distance transport from roots to shoots (4, 6) while protecting individual cells from Na toxicity (7–9). SOS1 is also indirectly required for the uptake of potassium (K) in the presence of Na, although the mechanistic basis is not fully understood (7, 8, 10). Both the protein kinase SOS2 and its associated calcium-sensor subunit SOS3 are required for the posttranslational activation of SOS1 Na/H exchange activity in Arabidopsis (11, 12), and a similar regulatory module operates also in cereals (13).
To understand further the mechanism(s) of SOS1 regulation, we identified the SOS2-dependent phosphorylation site and began to dissect the structure–function relationship in the SOS1 protein. Our results indicate that the SOS1 C-terminal domain comprises an auto-inhibitory domain the activity of which is counteracted by SOS2-dependent phosphorylation upon salinity stress.
Results
SOS1 Residues Phosphorylated by the SOS2 Protein Kinase.
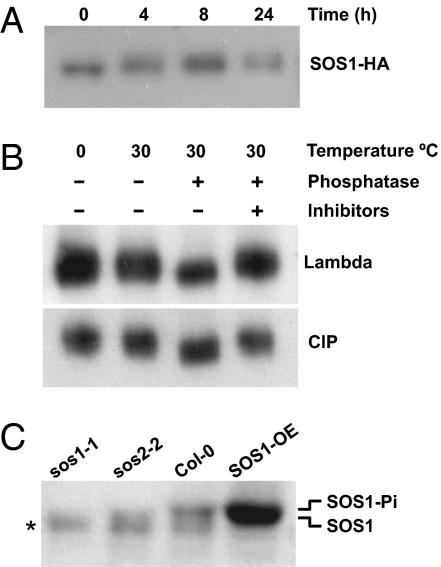
We have previously shown that the plasma membrane Na/H antiporter SOS1 of Arabidopsis is regulated positively by the protein kinase complex comprising SOS2 and SOS3 (11, 12). To demonstrate that SOS1 becomes phosphorylated in planta upon salinity stress, the relative mobility of HA-tagged SOS1 was determined in Columbia (Col-0) plants treated or not with 100 mM NaCl. Salt treatment elicited a mobility shift of SOS1-HA (Fig. 1A) that was reversed by incubating the samples with either λ-phosphatase or alkaline phosphatase (Fig. 1B). The phosphorylation-dependent in-gel retardation of SOS1 appeared small due to the large size of the protein (127 kDa). To test whether the phosphorylation of SOS1 was SOS2-dependent, the relative mobility of SOS1 in SDS/PAGE was also analyzed in the sos2-2 mutant. Because polyclonal antibodies against SOS1 were used in this experiment for protein detection, the sos1-1 mutant and wild-type plants over-expressing SOS1 were included as controls. The sos1-1 allele contains a 14-bp deletion causing a frameshift that truncates the SOS1 protein (∼49 kDa vs. 127 kDa) (14). The truncated protein does not cross-react with the SOS1 antibodies generated against the C terminus of the SOS1 protein. As shown in Fig. 1C, no in-gel retardation of the SOS1 band was observed in the sos2-2 mutant compared with that in the wild-type plants. Together these data strongly indicate that SOS2 phosphorylates SOS1 in vivo in response to salinity stress.
Fig. 1.
Phosphorylation of SOS1 in planta. (A) Gel mobility shift of SOS1 elicited by salt treatment. Arabidopsis plants were treated with 100 mM NaCl for the time indicated in hours; the protein extracts were resolved by SDS/PAGE, and the HA-tagged SOS1 protein was detected with anti-HA antibodies. (B) Reversal of gel mobility shift by phosphatase treatment. Protein extracts from plants treated with NaCl for 24 h were kept on ice or treated at 30 °C with λ-phosphatase or calf intestinal (CIP) phosphatase with and without the addition of the phosphatase inhibitors; HA-tagged SOS1 protein was detected with anti-HA antibodies. (C) SOS2-dependent mobility shift of SOS1. Protein extracts from mutants sos1-1, sos2-2, wild-type Col-0 gl1 plants, and the latter transformed to over-express the SOS1 protein were resolved by SDS/PAGE and the SOS1 protein was immunodetected with anti-SOS1 antibodies; plants were treated with 100 mM NaCl for 24 h. The identity of the lower band cross-reacting in all samples (asterisk), including the deletion mutant sos1-1, is unknown.
Next, the amino acid residue(s) of the SOS1 polypeptide that are phosphorylated by SOS2 were determined. There are three distinct modules in SOS1 on the basis of sequence comparisons with other proteins, hydrophobicity, and predicted globular domains by GLOBPLOT (Fig. 2A). The N-terminal module comprises the hydrophobic transmembrane domain encompassing amino acid residues 1–440 that shows sequence homology to other ion exchangers of the CPA1 family from plants and that is likely to act as the pore domain for ion transport (15). Next is an ∼300 amino-acid-long stretch that is predicted to comprise a separate globular domain and that, together with the pore domain, presents extensive homology to AtNHX8, a member of the CPA1 family that functions in lithium tolerance in Arabidopsis (16). From amino acid 740 to 1146, the SOS1 sequence is unique and does not show significant homology to any other protein except other SOS1-like proteins in plants. The second and third modules (amino acids 441–1146) are predicted to be cytosolic. Therefore, nested deletions of the SOS1 hydrophilic tail spanning residues 441 to 1146 were fused to the C-terminal end of GST, affinity purified with GST-Sepharose, and subjected to SOS2-dependent phosphorylation assays (Fig. 2B). The SOS2 derivative SOS2T168D/Δ308 was used because this mutant protein has much higher protein kinase activity than wild-type SOS2 and is independent of SOS3 (17). The SOS1 peptide comprising amino acids 1072–1146 was the smallest fragment phosphorylated by SOS2, indicating that SOS2 phosphorylates SOS1 at the very end of this large protein. In keeping with this, full-length SOS2 interacted with the SOS1998-1145 fragment, but not with the SOS1451-997 fragment in the yeast two-hybrid system (Fig. 2C).
Fig. 2.
Phosphorylation of the SOS1 C terminus. (A) Schematic structure and deletion mapping of SOS1; fragments F1–F4 were subjected to the SOS2 phosphorylation assay (B). Purified proteins corresponding to fragments F1–F4 were incubated with SOS2T168D/Δ308 in the presence of [γ-32P]ATP, resolved in SDS/PAGE (Left), and exposed to X-ray film (Right). (C) Yeast two-hybrid assay demonstrating the interaction of SOS2 with the last 147 amino acids of SOS1.
To identify the actual phosphorylated residues, fragment SOS1998-1146 was incubated with the SOS2T168D/Δ308 kinase in the presence of ATP and then subjected to nano-liquid chromatography tandem mass-spectrometry analysis (nLC-MS/MS). An untreated SOS1998-1146 sample was used as control to discard phosphorylation events that may have taken place in the yeast cells before protein purification. The phosphopeptide of sequence IDSPpSKIVFR, corresponding to amino acids 1134–1143 and in which the serine in position 1138 appeared phosphorylated, was specifically identified by nLC-MS/MS in the SOS1 sample incubated with SOS2T168D/Δ308 but not in the control sample. Prior biochemical characterization of SOS2 indicated a phosphorylation recognition site similar to those of SNF1/AMPK/SnRK protein kinases with the consensus H-X-B-X2-(S/T)-X3-H, where “H” indicates hydrophobic residues, “B” is a basic residue, and “S/T” is the phosphorylation site (18). Thus, S1136, which is embedded in the sequence IVVRIDSPSKIV (serines 1136 and 1138 are underlined), is a better match with the consensus than S1138. To validate nLC-MS/MS results and to check whether S1136 could be also involved in the phosphorylation of SOS1, serine-to-alanine mutations were introduced in positions S1136 and S1138 either individually or in combination. The entire cytosolic region of SOS1 (amino acids 441–1146), with and without S1136A/S1138A mutations, was tested in SOS2-dependent phosphorylation assays. Both mutations S1136A and S1138A prevented phosphorylation by SOS2 (Fig. 3A). Thus, it appears that S1138 is phosphorylated by SOS2, whereas S1136 is essential for substrate recognition by the protein kinase. No other residues of SOS1 appeared to be phosphorylated by SOS2 in the SOS1441-1146 fragment as efficiently as S1138. Background phosphorylation in mutant proteins likely occurred at places in the cytosolic region of SOS1 weakly conforming to SNF1/AMPK/SnRK recognition sites. A survey of all SOS1-like sequences in public databases showed that the SOS2 phosphorylation site identified here is conserved in nearly all plant sequences available, with the consensus [V,I]VR[V,I]DSPS (Fig. S1C). There are few motifs that depart from this consensus among the SOS1 sequences available in protein databases. The most dissimilar is the tomato SOS1 protein where the DSPS motif is interrupted by two intervening amino acids.
Fig. 3.
Serines 1136 and 1138 in the SOS2 phosphorylation site are essential for SOS1 activation. (A) GST fusions encompassing the SOS1 cytosolic tail (amino acids 441–1146) from the wild-type protein (DSPS), mutant S1136A (DAPS), mutant S1138A (DSPA), and mutant S1136A/S1138A (DAPA) were subjected to phosphorylation by SOS2 in vitro, resolved by SDS/PAGE (Upper) and exposed to X-ray film (Lower). (B) Full-length wild-type or mutant SOS1 proteins were expressed in the yeast strain AXT3K with (Right) or without (Left) the coexpression of SOS2T168D/Δ308. Decimal dilutions of saturated cultures were plated in AP medium supplemented with 1 mM KCl and with the indicated concentration of NaCl. Growth of all transformants was indistinguishable in plates without NaCl.
Residues S1136 and S1138 Are Essential for the Activation of SOS1 in Vivo.
To assess the requirement of the phosphorylation by SOS2 on the activation of SOS1, proteins carrying single or combined S1136A and S1138A mutations were expressed in the salt-sensitive yeast strain AXT3K, with and without coexpression of SOS2T168D/Δ308, and the capacity to grow on media with NaCl was determined. As depicted in Fig. 3B, the mutant proteins conferred the same degree of salt tolerance as the wild-type SOS1 in the absence of SOS2, indicating that the basal activity of the transporter had not been affected by the S1136A and/or S1138A mutations. Wild-type SOS1 coexpressed with SOS2T168D/Δ308 produced a remarkable increase of salt tolerance, allowing the growth of yeast cells in the presence of 200 mM NaCl. By contrast, no increase in salt tolerance was observed when SOS2T168D/Δ308 was coexpressed with either one of the SOS1 proteins harboring mutations S1136A and/or S1138A. As expected, the kinase-dead mutant SOS2K40N/T168D/Δ308 (17) failed to activate wild-type SOS1 or S-to-A mutants in yeast cells, implying that phosphorylation by SOS2 is essential for SOS1 activation. The SOS1 variants with mutations S1136A and/or S1138A were also tested for complementation of the knock-out allele sos1-1 of Arabidopsis. Five-day-old T2 transgenic seedlings expressing the SOS1S1136A/S1138A mutant allele under the 35S cauliflower mosaic virus (CaMV) promoter were germinated on Murashige–Skoog (MS) medium and then transferred to plates with 50 or 100 mM NaCl for 15 d. The nonphosphorylable SOS1 mutant protein failed to complement sos1-1 under 100 mM NaCl treatment. However, growth was partially restored in 50 mM NaCl compared with plants transformed with the empty vector, but these lines did not reach the growth rate produced by wild-type protein (Fig. S2 A and B). This partial complementation in low salt is likely due to the basal activity of SOS1, in keeping with the residual growth of yeast cells expressing the mutant SOS1 proteins. Indeed, the salt tolerance of the sos1-1 transgenic lines expressing SOS1S1136A/S1138A was equivalent to that of the sos2-2 mutant (Fig. S2C), in which the wild-type SOS1 protein remains in a state with basal activity (11, 12). Similar results were obtained with the single-site mutant protein SOS1S1136A (Fig. S2D). Together, these results demonstrate that phosphorylation at S1138 by SOS2 is an essential step for the activation of SOS1 in response to salt stress. To further demonstrate that SOS2-dependent phosphorylation is also sufficient for the activation of ion transport, a His-tagged SOS1 protein was purified from yeast plasma membrane and reconstituted in artificial proteoliposomes. Efflux of luminal protons by Na/H exchange was monitored by pyranine fluorescence recovery after the addition of 50 mM Na2SO4 (Fig. S3A). As depicted in Fig. S3B, the addition of SOS2T168D/Δ308 in the presence of ATP stimulated Na+ transport by SOS1.
Auto-Inhibitory Domain in SOS1.
To gain further insights into the structure–function relationships of SOS1, a mutant screen for gain-of-function alleles of SOS1 was performed. Expression of wild-type SOS1 in the yeast strain AXT3K improves growth in medium containing moderate concentrations of NaCl (up to 50 mM NaCl), whereas coexpression of SOS2 and SOS3 allow yeast cells to grow in 200 mM salt concentration (11). Hence, mutations locking SOS1 in the activated state in the absence of SOS2–SOS3 could be selected on medium containing the appropriate NaCl concentration. A randomly mutagenized population (∼2 × 105 independent clones) of SOS1 was created using the Escherichia coli hypermutable strain XL1-Red. The mutagenized plasmids were recovered in six independent pools and then transformed into the yeast strain AXT3K. Transformants were selected in arginine-phosphate (AP) medium supplemented with 1 mM KCl and 150 mM NaCl, a salt concentration that completely inhibits the growth of yeast cells expressing wild-type SOS1. Forty-four independent, salt-tolerant clones were isolated that were further classified into two groups according to their relative salt tolerance and their response to SOS2–SOS3 (Table S1). Transformants of class 1 tolerated 200 mM NaCl in the absence of SOS2–SOS3, but salt tolerance increased to 400 mM NaCl when the mutant SOS1 protein was coexpressed with the SOS2–SOS3 protein complex (Fig. 4A). Transformants of class 2 showed extreme resistance to salt and were able to grow up to 800 mM NaCl (in AP medium with 1 mM KCl), but their halotolerance was not further increased by coexpression of SOS2–SOS3. Incremental levels of NaCl tolerance strictly correlated with reduced net uptake of Na+ in yeast transformants. A disproportionate amount of individual class 2 clones were isolated relative to class 1 clones. Several clones shared the same mutation even though they were isolated from different pools of mutagenized plasmids, indicating that they were redundant mutations that originated from independent events. In total, 11 different mutations were identified, 6 of them belonging to class 1 and 5 to class 2 mutants (Table S1). Class 1 mutations E281K, A399V, and A399T mapped at the putative 7th and 11th transmembrane segments (TMs). Mutation R551G is in the central part of the cytosolic domain, and mutations S742L and V743I cluster together at the junction between the SOS1 segment that is highly homologous to AtNHX8 and the SOS1-specific C-terminal tail. Interestingly, class 2 mutations were all clustered (residues P985, Q998, Q1000, K1005, and W1013), and all but mutation P985S produced premature stop codons, thereby creating C-terminal deletions.
Fig. 4.
Functional domains of SOS1. (A) Representative class 1 and class 2 mutants were transformed in strain AXT3K, with and without the coexpression of the SOS2–SOS3 kinase complex, and compared with wild-type SOS1 in AP medium (1 mM KCl) with the indicated concentrations of NaCl. All transformants had identical growth in plates without NaCl. (B) The yeast mutant cdc25-2, transformed with the reporter protein hSos:SOS1998-1146, was further transformed to express wild-type SOS1, or mutant proteins truncated at residues 745 and 998, or with an empty vector. Decimal dilutions of liquid cultures were plated and incubated at the permissive (23 °C) or restrictive (37 °C) temperature to identify protein interactions at the plasma membrane. (C) Salt-tolerance test of AXT3K cells expressing the indicated SOS1 mutant proteins with and without the coexpression of the SOS2–SOS3 kinase complex.
The finding that class 2 mutants showed maximal SOS1 activity and were impervious to the SOS2–SOS3 kinase complex (Fig. 4A) suggested that the last ∼130 residues of the SOS1 protein act as an auto-inhibitory domain. Also, because no other class 2 mutations were isolated farther downstream from the residue W1013 despite the redundancy of isolates with class 2 mutations (Table S1), the putative auto-inhibitory domain was likely downstream from and adjacent to the cluster of class 2 mutations (see diagram in Fig. S4A). To confirm this, stop codons were introduced in residues L1030, N1047, and L1072 by site-directed mutagenesis. Also, the loss-of-function allele sos1-11 isolated in Arabidopsis (14), consisting of a frameshift mutation in residue L1106, was created in the SOS1 cDNA. Mutant proteins with these nested deletions were expressed in yeast with and without SOS2T168D/Δ308. The relative salt tolerance of these transformants showed that mutant SOS1Δ1030 had a basal activity greater than that of wild-type SOS1 (Fig. S4B) but much lower than that of class 2 mutants (Fig. 4A), suggesting that the auto-inhibitory domain was partly active in this mutant. By contrast, mutants with truncations at N1047, L1072, and L1106 showed low basal activity equivalent to the wild-type protein demonstrating that the auto-inhibitory domain was present and fully functional. Neither of these mutants could be activated by SOS2T168D/Δ308, as expected from the removal of the phosphorylation site by SOS2. Together with the derepressed state of the class 2 mutant SOS1W1013Z, these results indicate that the auto-inhibitory domain spans from amino acid W1013 to N1047. There are two motifs within this stretch that are conserved in SOS1-like proteins and that may comprise the auto-inhibitory domain: [K,R][D,E]HxGLxSWPE (residues K1005 and W1013 mutated in class 2 mutants are underlined) and Sx[R,K]Axx[L,I,V]S[I,M][F,Y]GS (amino acids 1033–1044) (Fig. S1B). The finding that both truncation at W1013 within the first conserved motif and truncation at L1030 in the intervening sequence between the two conserved motifs conveyed lower constitutive salt tolerance than other class 2 mutant proteins strongly indicate that the auto-inhibitory domain was partly active in these truncated proteins and that the two conserved motifs identified here together constitute the auto-inhibitory domain of SOS1.
Auto-Inhibitory Domain Interacts with a Conserved Domain That Is Essential for SOS1 Activity.
An activation mechanism of SOS1 consisting of counteracting auto-inhibition predicts that the inhibitory domain interacts with another domain of SOS1 to keep the transporter in a resting state with basal activity. To test this idea, the human Son-of-Sevenless (hSos) recruitment system (SRS) was used to monitor in yeast the in vivo interaction between wild-type and truncated SOS1 proteins residing on the plasma membrane (bait) and a cytosolic reporter protein (prey) consisting of the auto-inhibitory domain SOS1998-1146 fused to the human hSos protein. Upon interaction of pray and bait proteins, recruitment of the hSos to the plasma membrane restored Ras guanyl nucleotide exchange factor activity and growth at 37 °C of the thermosensitive yeast mutant cdc25-2 (19). The interaction of the reporter protein (hSos:SOS1998-1146) was tested using as bait the full-length SOS1 protein and the SOS1 proteins truncated at position 998 (SOS1Δ998) or 742 (SOS1Δ742), i.e., lacking only the auto-inhibitory domain or the entire module that is specific to SOS1-like proteins, respectively (Fig. 2A). As depicted in Fig. 4B, the reporter protein hSos:SOS1998-1146 was recruited to the plasma membrane upon coexpression of SOS1Δ998. Interaction of the reporter protein with the wild-type SOS1 is probably hindered because the docking site is already occupied by its own auto-inhibitory domain acting in cis. Interaction was lost with the truncated protein SOS1Δ742 indicating that the target sequence of the auto-inhibitory domain is located between positions 742 and 998. Within this region there is a highly conserved stretch, from residue 764 to 849, that shows homology with cyclic nucleotide-binding domains (CNBDs; Prosite pf00027; Fig. S1A) and that is known to play an essential yet undetermined role in SOS1 function (14). In Arabidopsis, loss-of-function alleles sos1-8 and sos1-9 consist of single-point mutations that are translated to amino acid changes G777D and G784E, respectively, affecting two fully conserved G residues in this putative CNBD (14). To evaluate the role of this domain in the activity and regulation of SOS1, the cDNAs encoding SOS1Δ742, which had the CNBD removed, and the mutant alleles sos1-8 and sos1-9 were expressed in yeast, and the salt tolerance that they conveyed was determined. As shown in Fig. 4C, the absence of the 742–998 region produced a great reduction of salt tolerance compared with the wild-type SOS1 activated by coexpression of SOS2–SOS3 and with the SOS1Δ998 lacking the auto-inhibitory domain. However, the SOS1Δ742 protein demonstrated greater salt tolerance than wild-type SOS1 alone, likely as the result of having the auto-inhibitory domain removed. It is worth noting that this protein would be equivalent to AtNHX8 (16). On the other hand, mutant proteins SOS1G777D (sos1-8 allele) and SOS1G784E (sos1-9) retained a basal activity similar to that of wild-type SOS1, but contrary to the latter they could not be activated by SOS2–SOS3. Accordingly, the SOS1G777D protein (sos1-8 allele) could not be activated by SOS2 in transport assays (Fig. S3). Deletion of the auto-inhibitory domain in SOS1G777D (sos1-8) produced only a marginal increase in salt tolerance, indicating that the mutation G777D is epistatic over the derepressed state of SOS1Δ998 (Fig. S4C).
Discussion
Here we show that phosphorylation by SOS2 is both necessary and sufficient to activate SOS1. Serine 1138 within the highly conserved consensus motif [V,I]VR[V,I]DSPS (serine 1138 underlined) was identified as the SOS2 phosphorylation site. However, serine 1136 fitted better the hypothetical recognition site H-X-B-X2-(S/T)-X3-H (18). Mutations S1136A or S1138A prevented SOS1 phosphorylation by SOS2. Moreover, mutation of S1136 to aspartate, which mimics the negative charge of a phosphorylated residue, yielded an inactive SOS1 protein (Fig. S2D). The simplest explanation for these observations is that S1138 is phosphorylated by SOS2, whereas S1136 is essential for substrate recognition by the protein kinase. However, while this work was being completed, Yu et al. (20) reported that the MAP kinase MPK6 phosphorylated the C-terminal fragment of SOS1, although neither the precise phosphorylated residue nor the effect on SOS1 activity was determined. The motif RIDSPSK of SOS1 also conforms to a proline-directed MAPK site in which S1136 preceding the proline residue would be phosphorylated. Thus, the possibility exists that this conserved motif is targeted by both SOS2 and MPK6. If so, the putative phosphorylation of S1136 by MPK6 would prevent activation by SOS2 because the mutation S1136D abrogated SOS1 activity in planta (Fig. S2D). Like SOS1, the C terminus of the plasma membrane H+-ATPase includes an auto-inhibitory domain to inhibit the activity of the pump (21). PKS5, a SnRK3 kinase similar to SOS2, phosphorylates the H+-ATPase isoform AHA2 at S931 in the C-terminal regulatory domain. Phosphorylation at this site inhibits interaction between the H+-ATPase and an activating 14–3-3 protein binding the neighboring phosphorylated residue T947, thereby preventing the activation of the pump (21). The motif RIDSPSK within the recognition site of SOS2 is also a putative 14–3-3–binding site, but it remains to be determined whether SOS1 interacts with 14–3-3 proteins. The possible interplay between SOS2- and MPK6-dependent phosphorylation of SOS1 deserves future research.
Through the selection of hyperactive alleles of SOS1 we have identified critical residues for SOS1 activity. Class 1 mutations appeared to enhance both the basal activity (i.e., without SOS2–SOS3) and the SOS1 activity upon up-regulation by SOS2–SOS3. All these mutations affected either fully conserved amino acids or residues embedded in conserved motifs. Mutation E261K lies in a hydrophobic region predicted to be TM7 and affects a glutamic acid residue fully conserved among SOS1 proteins. Likewise, mutations A399V and A399T affect a fully conserved alanine residue in TM11. Mutation R551G affects an invariant arginine that is embedded in the conserved sequence DxRxRxLNGVQAAYWxMLDEGRI (R551 underlined) in the cytosolic module shared by SOS1 and AtNHX8. In contrast, mutations S742L and V743I change residues that are not well conserved, although they lie in a short intervening sequence between two fully conserved P740 and P746 residues (sequence PLSVALP in Arabidopsis) at the boundary between the homologous regions of SOS1 and AtNHX8 and the C-terminal extension that is unique to SOS1 proteins. The significance of these amino acid substitutions for SOS1 activity or regulation is presently unclear. All but one class 2 mutation rendered C-terminally truncated SOS1 proteins with maximal activity that was independent of SOS2–SOS3, strongly indicating that the last 130 amino acids of SOS1 include an auto-inhibitory domain. By comparing the relative salt tolerance of yeast expressing serial truncations created downstream of K1005 (the most downstream class 2 mutation with full SOS1 activity) this inhibitory domain was mapped to the bipartite conserved sequences [K,R][D,E]HxGLxSWPE (amino acids 1005–1015) and Sx[R,K]Axx[L,I,V]S[I,M][F,Y]GS (amino acids 1033–1044) (Fig. S1B). Truncations downstream from these motifs produced SOS1 proteins locked in the basal state because they retained the auto-inhibitory domain but lacked the C-terminal phosphorylation site by SOS2 that acts to relieve SOS1 from auto-inhibition (Fig. S4B). On the other hand, mutations sos1-8 (G777D) and sos1-9 (G784E) created proteins that were still largely inactive even after removal of the auto-inhibitory domain (Fig. S4C). These two glycine residues are embedded in a very-well-conserved domain that shows weak similarity to a CNBD (Prosite pf00027). The null allele sos1-7 creates a frameshift mutation starting in residue E759, thereby eliminating this domain and producing an inactive SOS1 protein in planta (14). Together, these data strongly suggest that this putative CNBD plays an important, yet undetermined, role in the activation of SOS1. Because a fragment comprising the auto-inhibitory domain interacted in the yeast system with SOS1Δ998 but not with SOS1Δ742, we suggest that the auto-inhibitory and the CNBDs interact to keep SOS1 inactive (Fig. S5). Upon salinity stress, the SOS2–SOS3 kinase complex phosphorylates the auto-inhibitory domain, thereby relieving SOS1 auto-inhibition. In this regard, it is worth noting that the class 2 mutation P985S lies in the intervening region between the CNBD and the auto-inhibitory domains. This mutation may remove a critical turn in the polypeptide secondary structure and disrupt the interaction between these two domains, thereby preventing SOS1 auto-inhibition.
Materials and Methods
Plasmid Constructs and Protein Production.
GST-tagged fusion proteins used in phosphorylation assays and site-directed mutagenesis of serine 1136 and 1138 in SOS1 were produced according to standard methods, which are detailed in SI Materials and Methods, together with primer sequences in Table S2. For expression in Arabidopsis, SOS1 mutant alleles were subcloned in pBISOS1 (22) or pCAMBIA2300, both containing the 35S CaMV promoter.
Phosphorylation Assays.
For in vitro assays with SOS2, different GST:SOS1 peptides were purified from yeast cells. Cells were collected by centrifugation and lysed with glass beads in PBS buffer (10 mM Na2HPO4, 2 mM KH2PO4, 2.7 mM KCl, 137 mM NaCl, pH 7.4) supplemented with 1% Triton X-100. SOS2T168D/Δ308 was expressed as a GST-tagged fusion protein in E. coli (17). Substrate recombinant proteins (∼100 ng) were subjected to phosphorylation by the SOS2T168D/Δ308 protein kinase (∼100 ng) in 30 μL of buffer (20 mM Tris·HCl, pH 7.5, 5 mM MgCl2, 1 mM DTT). Reactions were started by adding ATP (0.2 mM with 1 μCi of [γ-32P]ATP), which was incubated at 30 °C for 30 min, and stopped with 10 μL of 4× SDS/PAGE sample buffer. Aliquots were then resolved by SDS/PAGE and the gel was exposed to X-ray films. To determine the phosphorylated residues, 20 μg of GST:SOS1998-1146 incubated with and without SOS2T168D/Δ308 were purified by 12% SDS/PAGE and subjected to nLC-MS/MS. For identification of protein modification, fragmentation spectra were searched against the Mass Spectrometry Protein Sequence DataBase using the Mascot software (Matrix Science). For SOS1 phosphorylation in planta, 2- to 3-wk-old plants were treated with 100 mM NaCl. Grinded leaves were extracted in 1 vol of extraction buffer (50 mM Tris·HCl, pH 7.5, 150 mM NaCl, 0.5% Nonidet P-40, 1 mM EDTA, 3 mM DTT, 2 mM Na3VO4, 2 mM NaF, 2 mM β-glycerophosphate) supplemented with a proteinase inhibitor mixture (1 mM phenylmethylsulfonyl fluoride, 5 μg/mL leupeptin, 1 μg /mL aprotinin, 1 μg/mL pepstatin, 5 μg /mL antipain, 5 μg/mL chymostatin, 50 μM MG132, 50 μM MG115, 50 μM μM ALLN). After separation in 6% SDS/PAGE, proteins were transferred to polyvinylidene difluoride membrane and detected with anti-HA (1:2,000) or anti-SOS1 (1:250) monoclonal antibodies. For phosphatase treatments, protein extracts were prepared either in λ-phosphatase buffer (50 mM Hepes, pH 7.5, 100 mM NaCl, 2.5 mM MnCl2, 2 mM DTT, 0.01% Brij-35, 0.5% Triton X-100, 0.4% Nonidet P-40), or in calf intestinal phosphatase (CIP) buffer (50 mM Tris·HCl, pH 7.9, 100 mM NaCl, 10 mM MgCl2, 1 mM DTT) supplemented with the proteinase inhibitor mixture described above. Fifty-microliter aliquots of protein extracts were incubated with 400 units of λ-protein phosphatase or with 10 units of CIP, with or without phosphatase inhibitors (2 mM NaF, 2 mM Na3VO4) at 30 °C for 5 min.
Ion Transport.
The wild-type and SOS1G777D (sos1-8) proteins were His-tagged, purified from yeast plasma membrane by Ni2+ affinity chromatography, and reconstituted in artificial proteoliposomes as described (23). After imposing the internal acidification of vesicles, 50 mM Na2SO4 was added to the incubation buffer and Na/H exchange was monitored by changes in pyranine fluorescence (23). ATP (1.5 mM) was added to all samples and kinase SOS2T168D/Δ308 (1 μg) only when indicated.
Yeast Methods.
Saccharomyces cerevisiae AXT3K strain (Δena1::HIS3::ena4, Δnha1::LEU2, Δnhx1::KanMX4) has been described elsewhere (24). Sodium tolerance tests were performed in the alkali cation-free medium AP (25) supplemented with 1 mM KCl and with NaCl as indicated for each experiment. For yeast two-hybrid, the SOS2 cDNA was amplified by PCR with oligos P14 and P15 and cloned in-frame between the NdeI and PstI sites of pAS2.1 (Clontech). The SOS1 fragments encompassing residues 441–997 and 998–1146 were amplified with the oligos P16 and P17 and P18 and P19, respectively, and cloned in-frame between NcoI and BamHI sites of pACT2 (Clontech). All PCR products were sequenced. For the SRS, a 450-pb fragment extending from 2,993 bp downstream of the ATG to the end of the SOS1 ORF was amplified with primers P20 and P21 and inserted into plasmid pADNS to produce a translational fusion downstream from the hSos protein (19). The bait proteins were expressed from vector pYPGE15. All plasmids used for SRS were transformed in the S. cerevisiae strain cdc25-2 (19).
Isolation of Hyperactive SOS1 Mutants.
The plasmid pSOS1-1 (11), harboring the SOS1 cDNA, was transformed into the mutator strain Epicurian E. coli XL1-Red (Stratagene). Transformants were selected on LB plates containing ampicillin (100 μg/mL), pooled in six groups, and grown overnight at 37 °C in LB broth. Randomly mutated plasmid DNA extracted from individual pools was used to transform the yeast strain AXT3K. Mutations leading to increased salt tolerance were selected by plating transformants on AP medium containing 1 mM KCl and 150 mM NaCl. Plasmids were extracted from salt-resistant clones and rechecked in fresh transformants, and the SOS1 cDNA insert was fully sequenced to identify the mutations.
Antibody Production and Immunoblotting.
Antibodies against SOS1 were produced according to modifications of standard techniques, detailed in SI Materials and Methods. Leaf extracts were immunoblotted following standard techniques.
Supplementary Material
Acknowledgments
We are indebted to Unidad de Proteomica, Centro Nacional de Investigaciones Cardiovasculares Carlos III, for mass-spectrometric analysis. This work was supported by Grants BIO2009-08641 and CSD2007-00057 from the Ministerio de Ciencia e Innovacion (cofinanced by Fondo Europeo de Desarrollo Regional) (to J.M.P. and F.J.Q.), World Class University Program Grant R32-10148 (to D.-J.Y. and W.Y.K.), and National Institutes of Health Grants R01GM070795 and R01GM059138 (to J.-K.Z.).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1018921108/-/DCSupplemental.
References
- 1.Abrol IP, Yadav JSP, Massoud FI. FAO Soils Bull. Food and Agriculture Organization (Rome); 1988. Salt-affected soils and their management. [Google Scholar]
- 2.Tester M, Davenport R. Na+ tolerance and Na+ transport in higher plants. Ann Bot. 2003;91:503–527. doi: 10.1093/aob/mcg058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Munns R, Tester M. Mechanisms of salinity tolerance. Annu Rev Plant Biol. 2008;59:651–681. doi: 10.1146/annurev.arplant.59.032607.092911. [DOI] [PubMed] [Google Scholar]
- 4.Shi HZ, Quintero FJ, Pardo JM, Zhu JK. The putative plasma membrane Na(+)/H(+) antiporter SOS1 controls long-distance Na(+) transport in plants. Plant Cell. 2002;14:465–477. doi: 10.1105/tpc.010371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Oh D-H, et al. Loss of halophytism by interference with SOS1 expression. Plant Physiol. 2009;151:210–222. doi: 10.1104/pp.109.137802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Olías R, et al. The plasma membrane Na+/H+ antiporter SOS1 is essential for salt tolerance in tomato and affects the partitioning of Na+ between plant organs. Plant Cell Environ. 2009;32:904–916. doi: 10.1111/j.1365-3040.2009.01971.x. [DOI] [PubMed] [Google Scholar]
- 7.Fraile-Escanciano A, Kamisugi Y, Cuming AC, Rodríguez-Navarro A, Benito B. The SOS1 transporter of Physcomitrella patens mediates sodium efflux in planta. New Phytol. 2010;188:750–761. doi: 10.1111/j.1469-8137.2010.03405.x. [DOI] [PubMed] [Google Scholar]
- 8.Wu SJ, Ding L, Zhu JK. SOS1, a genetic locus essential for salt tolerance and potassium acquisition. Plant Cell. 1996;8:617–627. doi: 10.1105/tpc.8.4.617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhu Z, Wu R. Regeneration of transgenic rice plants using high salt for selection without the need for antibiotics or herbicides. Plant Sci. 2008;174:519–523. [Google Scholar]
- 10.Qi Z, Spalding EP. Protection of plasma membrane K+ transport by the salt overly sensitive1 Na+-H+ antiporter during salinity stress. Plant Physiol. 2004;136:2548–2555. doi: 10.1104/pp.104.049213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Quintero FJ, Ohta M, Shi HZ, Zhu JK, Pardo JM. Reconstitution in yeast of the Arabidopsis SOS signaling pathway for Na+ homeostasis. Proc Natl Acad Sci USA. 2002;99:9061–9066. doi: 10.1073/pnas.132092099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Qiu QS, Guo Y, Dietrich MA, Schumaker KS, Zhu JK. Regulation of SOS1, a plasma membrane Na+/H+ exchanger in Arabidopsis thaliana, by SOS2 and SOS3. Proc Natl Acad Sci USA. 2002;99:8436–8441. doi: 10.1073/pnas.122224699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Martínez-Atienza J, et al. Conservation of the salt overly sensitive pathway in rice. Plant Physiol. 2007;143:1001–1012. doi: 10.1104/pp.106.092635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shi HZ, Ishitani M, Kim CS, Zhu JK. The Arabidopsis thaliana salt tolerance gene SOS1 encodes a putative Na+/H+ antiporter. Proc Natl Acad Sci USA. 2000;97:6896–6901. doi: 10.1073/pnas.120170197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mäser P, et al. Phylogenetic relationships within cation transporter families of Arabidopsis. Plant Physiol. 2001;126:1646–1667. doi: 10.1104/pp.126.4.1646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.An R, et al. AtNHX8, a member of the monovalent cation: Proton antiporter-1 family in Arabidopsis thaliana, encodes a putative Li/H antiporter. Plant J. 2007;49:718–728. doi: 10.1111/j.1365-313X.2006.02990.x. [DOI] [PubMed] [Google Scholar]
- 17.Guo Y, et al. Transgenic evaluation of activated mutant alleles of SOS2 reveals a critical requirement for its kinase activity and C-terminal regulatory domain for salt tolerance in Arabidopsis thaliana. Plant Cell. 2004;16:435–449. doi: 10.1105/tpc.019174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gong DM, Guo Y, Jagendorf AT, Zhu JK. Biochemical characterization of the Arabidopsis protein kinase SOS2 that functions in salt tolerance. Plant Physiol. 2002;130:256–264. doi: 10.1104/pp.004507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Aronheim A. Improved efficiency sos recruitment system: Expression of the mammalian GAP reduces isolation of Ras GTPase false positives. Nucleic Acids Res. 1997;25:3373–3374. doi: 10.1093/nar/25.16.3373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yu L, et al. Phosphatidic acid mediates salt stress response by regulation of MPK6 in Arabidopsis thaliana. New Phytol. 2010;188:762–773. doi: 10.1111/j.1469-8137.2010.03422.x. [DOI] [PubMed] [Google Scholar]
- 21.Fuglsang AT, et al. Arabidopsis protein kinase PKS5 inhibits the plasma membrane H+ -ATPase by preventing interaction with 14-3-3 protein. Plant Cell. 2007;19:1617–1634. doi: 10.1105/tpc.105.035626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shi HZ, Lee BH, Wu SJ, Zhu JK. Overexpression of a plasma membrane Na+/H+ antiporter gene improves salt tolerance in Arabidopsis thaliana. Nat Biotechnol. 2003;21:81–85. doi: 10.1038/nbt766. [DOI] [PubMed] [Google Scholar]
- 23.Venema K, Quintero FJ, Pardo JM, Donaire JP. The Arabidopsis Na+/H+ exchanger AtNHX1 catalyzes low affinity Na+ and K+ transport in reconstituted liposomes. J Biol Chem. 2002;277:2413–2418. doi: 10.1074/jbc.M105043200. [DOI] [PubMed] [Google Scholar]
- 24.Quintero FJ, Blatt MR, Pardo JM. Functional conservation between yeast and plant endosomal Na(+)/H(+) antiporters. FEBS Lett. 2000;471:224–228. doi: 10.1016/s0014-5793(00)01412-5. [DOI] [PubMed] [Google Scholar]
- 25.Rodríguez-Navarro A, Ramos J. Dual system for potassium transport in Saccharomyces cerevisiae. J Bacteriol. 1984;159:940–945. doi: 10.1128/jb.159.3.940-945.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.