Abstract
Cyclin-dependent kinases (CDKs) are key regulatory enzymes in cell cycle progression and transcription. Aberrant activity of CDKs has been implicated in a number of medical conditions, and numerous small molecule CDK inhibitors have been reported as potential drug leads. However, these inhibitors exclusively bind to the ATP site, which is largely conserved among protein kinases, and clinical trials have not resulted in viable drug candidates, attributed in part to the lack of target selectivity. CDKs are unique among protein kinases, as their functionality strictly depends on association with their partner proteins, the cyclins. In an effort to identify potential target sites for disruption of the CDK-cyclin interaction, we probed the extrinsic fluorophore 8-anilino-1-naphthalene sulfonate (ANS) with human CDK2 and cyclin A using fluorescence spectroscopy and protein crystallography. ANS interacts with free CDK2 in a saturation-dependent manner with an apparent Kd of 37 μM, and cyclin A displaced ANS from CDK2 with an EC50 value of 0.6 μM. Co-crystal structures with ANS alone and in ternary complex with ATP site-directed inhibitors revealed two ANS molecules bound adjacent to one another, away from the ATP site, in a large pocket that extends from the DFG region above the C-helix. Binding of ANS is accompanied by substantial structural changes in CDK2, resulting in a C-helix conformation that is incompatible for cyclin A association. These findings indicate the potential of the ANS binding pocket as a new target site for allosteric inhibitors disrupting the interaction of CDKs and cyclins.
Keywords: protein kinases, cell cycle, anticancer therapeutics, contraceptive agents, structure-based drug design
INTRODUCTION
Cyclin-dependent kinases (CDKs) belong to the CMGC class of serine/threonine protein kinases which act as key regulatory elements in cell cycle progression. Deregulation of CDKs has been implicated in a number of diseases (1–4). Both the activity and the substrate specificity of CDKs are critically dependent on the presence of their partner proteins, the cyclins (5–7). The CDK2-cyclin A complex predominantly controls the G1- to S-phase checkpoint (8, 9) and, therefore, represents an attractive target for therapeutics designed to arrest or recover control of the cell cycle in dividing cells (10, 11). Although several CDK2 inhibitors are in clinical trials, none are currently approved for clinical use (12). The main hurdle encountered in the development of drugs selectively targeting CDK2 is the topology of the ATP binding site, which is well conserved among protein kinases. To date, more than 140 crystal structures of CDK2 liganded with small molecule inhibitors have been deposited in the Protein Data Bank (PDB). These inhibitors exclusively bind to the ATP site through interactions with the hinge or linker region (residues 81–83) and, therefore, belong to the type I family of protein kinase inhibitors (13). Little progress has been made towards the design of noncompetitive, allosteric inhibitors of CDKs, particularly those able to disrupt the interaction with cyclins.
A key structural change observed during the association between CDK2 and cyclin A is the migration of the PSTAIRE helix (the C-helix), the sequence of which is highly conserved among CDKs. One method for targeting the CDK2-cyclin interaction has been proposed, based on cellular inhibition by the Kip/Cip family of proteins (14). The cyclin-binding motif identified within these proteins was used to design peptidomimetics able to disrupt the CDK2-cyclin A complex (15–18). However, this approach has not yielded promising lead molecules for drug discovery to date.
In contrast to CDKs, protein kinases such as Abl, p38, and MEK1 have been successfully targeted by small molecule inhibitors with allosteric properties (19–21). Type II inhibitors, such as imatinib (Gleevec) for Abl, extend from the ATP site into a neighboring hydrophobic pocket, whereas type III inhibitors of MEK1 are purely allosteric. The binding of allosteric inhibitors is often accompanied by large structural changes within the highly conserved DFG motif. For many kinases, the DFG region assumes distinct conformations in the active (DFG-in) and inactive (DFG-out) enzyme states, with the majority of allosteric inhibitors appearing to stabilize the DFG-out conformation (22, 23). However, the DFG region in CDKs does not seem to undergo this structural transition, with both the active and inactive enzyme exhibiting DFG-in conformation, thereby rendering the nearby hydrophobic pocket inaccessible from the ATP site.
In an attempt to identify potential allosteric binding sites in CDK2, we probed the extrinsic fluorophore 8-anilino-1-naphthalene sulfonate (ANS). Previously, we utilized ANS for the characterization of ligand-induced conformational changes in the bacterial enzyme MurA (24) Here, we demonstrate that ANS specifically interacts with free CDK2, but not with cyclin A, to produce a characteristic fluorescence spectrum. Fluorescence intensity decreased in a dose-dependent manner upon interaction with cyclin A but not with substrate ATP. High-resolution crystal structures revealed two ANS molecules bound adjacent to one another, away from the ATP site, and in the vicinity of the C-helix. Binding of ANS induced large conformational changes to the C-helix, indicating the potential of this site to accommodate compounds able to disrupt the interaction of CDK2 with cyclins. Structures of ternary complexes of CDK2 liganded with ANS and selected ATP-competitive inhibitors were determined as well, demonstrating that the ANS site is independent of the ATP site and allosteric in nature.
RESULTS AND DISCUSSION
Fluorescence characterization of the CDK2-ANS interaction
We probed the extrinsic fluorophore ANS (Figure 1) for interaction with free CDK2 and free cyclin A. ANS alone exhibits a low fluorescence yield in aqueous solutions, which increases substantially upon binding to certain proteins (24, 25). Incubation of free CDK2 with ANS resulted in a characteristic emission spectrum with a single maximum around 460 nm (Figure 2a). The fluorescence intensity increased in a saturation-dependent manner, yielding an apparent dissociation constant of Kd = 37 μM. Free cyclin A reacted poorly with ANS, and saturation was not observed, up to 200 μM ANS (data not shown). Next, we evaluated the ability of diverse, known CDK2 ligands to displace ANS from the CDK2-ANS complex. Ligands interfering with the binding of ANS were expected to quench the CDK2-ANS fluorescence spectrum either directly, by competing with ANS for binding, or indirectly, by inducing long-range conformational changes to the ANS site. Cyclin A, ATP, and selected CDK2 inhibitors were among the ligands tested (Figure 1). For these experiments, the ANS concentration was maintained at 50 μM. Titration of cyclin A into the CDK2-ANS complex resulted in dose-dependent quenching of the emission spectrum with an EC50 value of 0.6 μM (Figure 2b). The equilibrium constant of the CDK2-cyclin A complex was previously assessed using mant-ATP (26, 27), and it was concluded that an initial rapid equilibrium (Kd = 1.3 μM) is followed by a rate-determining conformational change, giving rise to an overall dissociation constant of 48 nM. The difference in assay signals between CDK2-ANS and CDK2-mant-ATP likely accounts for the higher apparent dissociation constant observed here. Furthermore, ANS inhibited the catalytic activity of the phosphorylated CDK2-cyclin A2 complex with an IC50 value of 91 μM, indicating that the presence of ANS obstructs formation of the complex between CDK2 and cyclins. (Supplementary Figure 1).
Figure 1.
ANS and the ATP site-directed inhibitors JWS648 (IC50 = 5.9 μM) and SU9516 (IC50 = 0.13 μM).
Figure 2.
Fluorescence measurements of CDK2-ANS interaction, the effect of selected CDK2 ligands, and the efficency of ANS interaction with other protein kinases. a) Emission spectra of CDK2 (1.6 μM) were recorded for increasing concentrations of ANS. Fitting the emission maximum at 460 nm to equation (3) yielded an apparent Kd of 37 μM (inset). b) Emission spectra for the CDK2-ANS complex were recorded at 50 μM ANS and 1.6 μM CDK2 in the presence of increasing concentrations of cyclin A2. Cyclin A2 quenched the fluorescence yield in a dose-dependent manner, indicating the displacement of ANS with an EC50 value of 0.6 μM (inset). c) Spectra were recorded for 50 μM ANS and 1.6 μM CDK2 in the presence of small molecule ligands and the fluorescence maximum at 460 nm analyzed as a function of compound concentration. ATP site-directed ligands exhibited differential ANS displacement potential: SU9515 (■) readily quenched the fluorescence signal (EC50 = 0.3 μM), while concentrations as high as 300 μM of JWS648 (▼) and ATP (●) only marginally affected the fluorescence yield. Note that the displacement potential of SU9515 decreased in the presence of 50 μM JWS648 (□) to EC50 = 1.2 μM. Spectra for a-c were recorded with emission and excitation slit widths 5/5. d) ANS titrations were also performed for Aurora A (5.8 μM) (■), Rock1 (4.3 μM) (◇) and Akt 1 (4.1 μM) (▼) in parallel with CDK2 (1.5 μM) (●). Plotted is the maximum fluorescence yield observed per μM respective kinase in the presence of increasing ANS concentrations. Spectra for d were recorded with slit widths 10/10. Data were fitted to equation (3) yielding Kd values of 37 μM for CDK2, 54 μM for Aurora A, 450 μM for Akt and 1900 μM for Rock 1.
Compounds JWS648 and SU9516 are ATP-competitive inhibitors of CDK2 with IC50 values of 5.9 and 0.13 μM, respectively (Supplementary Figure 1). Titration of JWS648 into the CDK2-ANS complex caused only slight quenching of the fluorescence signal (EC50 > 300 μM), suggesting that this inhibitor does not compete with ANS for binding (Figure 2c). In contrast, titration of SU9516 resulted in significant quenching of the fluorescence signal in a dose-dependent manner, with an EC50 value of 0.3 μM, comparable to its IC50 value. Notably, the presence of JWS648 led to a substantial decrease in the potential of SU9516 to displace ANS, as reflected by a shift of the dose-response curve towards higher inhibitor concentrations. It appears that the interaction of ANS with CDK2 is unaffected by smaller inhibitors such as JWS648, whereas ANS is readily displaced by larger and more potent inhibitors such as SU9516. ATP did not appear to affect the CDK2-ANS fluorescence at concentrations up to 100 μM. Combined the data indicate that ANS binds immediately adjacent to the ATP site in a region susceptible to the structural changes that occur upon interaction with cyclin A.
We compared the fluorescence efficiency of the ANS-CDK2 interaction with that of other Ser/Thr protein kinases, namely Akt, Aurora A and Rho associated kinase 1 (Rock 1) (Figure 2d). Akt and Rock 1 reacted poorly with ANS and saturation was not reached up to 100 μM ANS. The fluorescence yield of ANS with Aurora A was about 5 times less efficient than with CDK2, and the apparent Kd of 54 μM indicates less binding potential. The results suggest that the interaction of ANS with CDK2 is relatively specific among the protein kinases tested.
Structural characterization of the ANS binding pocket
To understand the interaction between ANS and CDK2 on a molecular level, we determined the crystal structures of CDK2 in binary complex with ANS alone and in ternary complexes with ANS and inhibitors (Table 1). Initial attempts to co-crystallize the enzyme with ANS using well-known crystallization conditions for free and liganded CDK2 (28, 29) failed, indicating that conformational changes upon binding of ANS are incompatible with crystal formation under these conditions. Therefore, we first applied in-diffusion (“soaking”) experiments, in which preformed crystals of free or inhibitor-liganded CDK2 were incubated with ANS incrementally over a period of 1–5 days. This allowed the crystals to adapt to the structural changes induced by ANS, resulting in a ~ 2 Å decrease of the unit cell b-axis without loss of diffraction quality. Simultaneously, new crystallization conditions suitable for the co-crystallization of CDK2 with ANS were established. Notably, although grown from different precipitants (Jeffamine ED-2001 vs. PEG3350), the crystals obtained by co-crystallization or in-diffusion were of identical space group and possessed the same unit cell dimensions, with the refined structures displaying the same conformational changes unique to the binding of ANS.
Table 1.
Summary of crystallographic data collection and structure refinement. Values in parentheses refer to the highest resolution shell.
| Structure (PDB ID) | 2 ANS MOLECULES (3PXF) | ANS+JWS648 (3PXZ) | ANS+SU9516 (3PY1) |
|---|---|---|---|
|
Crystallization | |||
| Precipitant | Jeffamine ED-2001 | PEG 3350 | PEG 3350 |
| Co-crystallization | ANS | JWS648 | SU9516 |
| Soaking | - | ANS | ANS |
|
Data Collection | |||
| Space group | P212121 | P212121 | P212121 |
| Unit cell dimensions (Å) | a=53.0 b=69.4 c=72.3 |
a=53.1 b=69.6 c=72.6 |
a=53.3 b=70.2 c=71.5 |
| Resolution range | 20–1.80 (1.85–1.80) | 20–1.70 (1.75–1.70) | 20–2.05 (2.10–2.05) |
| Unique reflections | 24998 (1872) | 30104 (2434) | 17189 (1183) |
| Completeness (%) | 98.5 (96.6) | 99.3 (98.3) | 98.7 (99.5) |
| I/σI | 21 (3.8) | 27 (4.4) | 23 (4.8) |
| Rmergea (%) | 3.5 (19.9) | 2.9 (22.7) | 4.4 (31.4) |
|
Structure refinement | |||
| Protein atoms | 2438 | 2431 | 2448 |
| Average B-factor (Å2) | 25 | 24 | 31 |
| Ligand atoms | 2 ANS (2 × 21 atoms) | 2 ANS (2 × 21 atoms) 1 JWS (1 × 17 atoms) |
2 ANS (2 × 21 atoms) 1 SU9516 (18 atoms) |
| Average B-factor (Å2) | 22 | 19 | 33 |
| Solvent molecules | 203 | 187 | 128 |
| Average B-factor (Å2) | 31 | 30 | 35 |
| Rmsdb bonds (Å) | 0.010 | 0.009 | 0.010 |
| Rmsd angles (°) | 1.4 | 1.4 | 1.4 |
| Rcrystc (%) | 19.8 | 21.4 | 19.9 |
| Rfreed (%) | 22.9 | 25.8 | 25.1 |
| Rfree reflection set size | 1100 (4.4 %) | 1114 (3.7 %) | 860 (5.0 %) |
| Cross-validated coordinate error: | |||
| - From Luzzati plot (Å) | 0.24 | 0.24 | 0.30 |
| - From SigmaA (Å) | 0.12 | 0.16 | 0.15 |
Rmerge = 100 × ΣhΣi|Ihi − Ih|/ΣhiIhi where h are unique reflection indices.
Rmsd = root mean square deviation from ideal values.
Rcryst = 100 x Σ|Fobs−Fmodel|/Fobs where Fobs and Fmodel are observed and calculated structure factor amplitudes, respectively.
Rfree is Rcryst calculated for randomly chosen unique reflections, which were excluded from the refinement.
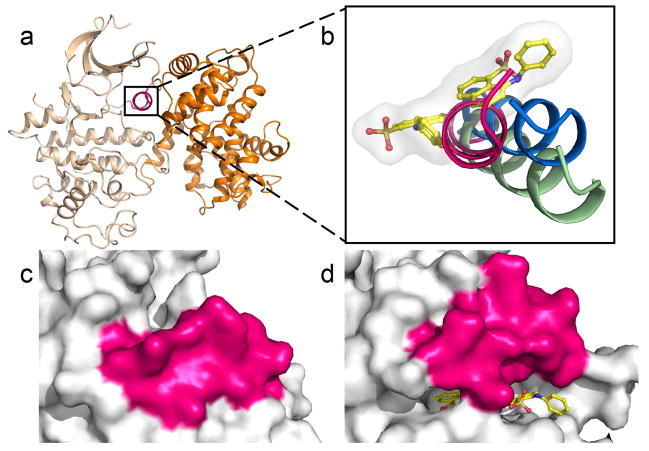
The crystal structure of the CDK2-ANS complex revealed two distinct ANS molecules bound adjacent to one another, away from the ATP site and in the vicinity of the C-helix (Figure 3a). The ANS site that is located approximately midway between the ATP site and the C-helix likely binds ANS with the highest affinity, as indicated by the elaborate network of hydrogen bonding and van der Waals carbon-carbon (hydrophobic) interactions, and the significantly lower temperature factors (B-factors) of the ANS molecule (Supplementary Figure 2a). Therefore, this site was defined as the primary ANS site. The naphthalene ring is positioned between C-helix residues Leu55 and Lys56 and gatekeeper residue Phe80. The sulfonate group is within hydrogen bonding distance of the main chain amides of Asp145 and Phe146, which constitute the DFG motif, and appears to establish a salt bridge with the ε-amino group of the conserved Lys33 residue (Figures 3b and c). The second ANS molecule binds adjacent to the primary site, within van der Waals distance of the first ANS molecule (Figure 2a). This secondary site is also comprised of C-helix residues. The naphthalene is located between residues Ile52 and Leu76, and the sulfonate group interacts with residues Lys56 and His71, while the aniline moiety is largely solvent exposed. Notably, upon incubation of apo-CDK2 crystals with high concentrations of ANS (> 10 mM), a third ANS molecule appeared in the ATP site (Supplementary Figure 3). However, the weak electron density and absence of interactions with the hinge region indicate a low binding potential of ANS for the ATP site.
Figure 3.
Molecular mode of action of ANS binding to CDK2. a) Two ANS molecules (yellow) bind adjacent to one another, away from the ATP site (hinge region shown in cyan) and in the vicinity of the C-helix (magenta). Displayed in blue is the 2Fo-Fc electron density, contoured at 1σ around both ANS molecules. The Fo-Fc electron density map with the ANS molecules omitted during refinement is shown in Fig. S7a. b) Schematic representation of potential hydrogen bonding and van der Waals (hydrophobic) interactions between ANS and enzyme residues. c) Stereoview of binding interactions within the ANS pocket. Black and green dotted lines indicate hydrogen bonding and hydrophobic interactions, respectively. Water molecules are shown as blue spheres.
ANS induces large structural changes in CDK2 not previously observed with small molecule ligands of this enzyme. The C-helix is locked in a conformation substantially different from that of CDK2 alone or in complex with cyclin A (Figures 4a and b). Interaction of cyclin A with the CDK2-ANS complex requires repositioning of the C-helix, which effectively narrows the ANS pocket. As a result, ANS dissociates from the enzyme, accompanied by quenching of the fluorescence signal (Figure 2b). The activation loop (residues 152–171) and the DFG region (residues 145–147) undergo substantial structural changes as well, but Asp145 and Phe146 remain in the DFG-in conformation. Since the gatekeeper residue Phe80 shifts only slightly, the ANS pocket remains largely shielded from the ATP site. Notably, binding of ANS also causes a main chain conformational change of the hinge region around His84, indicating an increase in conformational flexibility of the ATP site (Supplementary Figure 4). When occupied by ANS, the pocket becomes partially solvent accessible, while it is buried in free CDK2 (Figures 4c and d). Entry of ANS to the pocket likely occurs at the interface between the N-terminus of the C-helix and the activation loop surrounding residue Phe152 (Supplementary Figure 5). These regions exhibit high conformational flexibility and are likely to expose the hydrophobic pocket during fluctuating transitions between the various conformational states of CDK2 (30).
Figure 4.
Binding of ANS induces large conformational changes in the C-helix. a) Overall view of the CDK2-cyclin A complex structure (PDB entry 2CCI) (45). CDK2 is shown in wheat, the C-helix in magenta, and cyclin A in orange. b) Superimposition reveals the distinct conformation of the C-helix in the CDK2-ANS complex (green) and in free CDK2 (blue) as compared to the CDK2-cyclin A complex (magenta). Surface representations of the C-helix and surrounding residues for free CDK2 (c) and the CDK2-ANS complex (d) reveal the partial opening of the ANS pocket towards solvent.
To further evaluate the allosteric characteristics of the ANS binding pocket, we assessed the ability of inhibitors to form ternary complexes with CDK2-ANS. Crystal structures of CDK2-ANS-JWS648 and CDK2-ANS-SU9516 were obtained both by co-crystallization and in-diffusion (Table 1 and Supplementary Table 1). The ternary CDK2-ANS-JWS648 complex (Figures 5a and c) is nearly identical to the binary CDK2-ANS complex (rmsd = 0.35 Å, Figure S2b), suggesting that binding of JWS648 does not impact the allosteric site, in agreement with the low fluorescence quenching potential of this inhibitor (Figure 2c). Unexpectedly, we were also able to obtain the structure of CDK2 in complex with ANS and SU9516 (Figures 5b and d), although this inhibitor readily displaced ANS in the fluorescence assay. The CDK2-ANS-SU9516 complex, however, differs considerably from the ternary complex with JWS648 (rmsd = 1.7 Å, Supplementary Figure 2b), and the volume of the binding pocket appears significantly smaller. To fit the altered pocket, the two ANS molecules adopt new conformations by rotating about the aniline moiety, with increased B-factor values indicating reduced binding potential (Supplementary Figure 6). It appears that SU9516 induces a conformational change in the side chain of Lys33, which is located at the interface between the ATP site and the allosteric pocket. This causes the activation loop around Val156 and the glycine-rich loop around Tyr15 to change structure, in turn affecting the integrity of the ANS pocket. Thus, the crystal structures of the ternary complexes provide an explanation for the differential effects of JWS648 and SU9516 on CDK2-ANS fluorescence and demonstrate the allosteric nature of the ANS binding pocket.
Figure 5.
Allosteric nature of the ANS binding pocket. Crystal structures of ternary complexes with ATP site-directed inhibitors were determined for CDK2-ANS-JWS648 (a) and CDK2-ANS-SU9516 (b). The 2Fo-Fc electron density maps contoured at 1σ around the respective ligands are displayed in blue, with inhibitors shown in orange and ANS in yellow. The Fo-Fc electron density maps with the ligands omitted during refinement are presented in the supplemental material (Fig. S7). (c) and (d) represent stereo views of the binding interactions of ligands in the CDK2-ANS-JWS648 and CDK2-ANS-SU9516 complexes, respectively.
Implications for the design of allosteric CDK inhibitors
The majority of protein kinase inhibitors are ATP-competitive (type I), and comparatively few allosteric (noncompetitive) inhibitors have been reported to date. Abl, p38, and MEK1 kinases (19–21) are prime examples for the successful design of high affinity inhibitors with distinct allosteric binding characteristics (type II–IV) (23, 31, 32). For these enzymes, the allosteric site extends along the DFG region, with the inhibitor located under the C-helix (Figure 6). In contrast, the allosteric pocket in CDK2 points away from the DFG region and extends above the helix. These substantial architectural differences may provide an opportunity for the design of a new generation of inhibitors selectively targeting CDK2 and possibly other CDKs as well, since the residues interacting with ANS are largely conserved (Figure 7). The structural changes induced by ANS indicate the potential of the allosteric site to modulate complex formation between CDKs and cyclins by locking the C-helix in a conformation incompatible with this protein-protein interaction. However, the tight interaction of the CDK-cyclin complex requires allosteric inhibitors to be exceptionally potent. With an apparent Kd of 37 μM, ANS is readily displaced from CDK2 upon interaction with cyclin A and, consequently, exhibits relatively weak inhibitory potency against the activated CDK2-cyclin A complex with an IC50 of 91 μM. However, it is conceivable that larger compounds occupying both ANS sites simultaneously could interact more efficiently with CDK2, thereby inhibiting complex formation with cyclins. For example, aryl groups introduced at the 7-position of the naphthalene ring via alkyl or alkylamine linkers could extend into the secondary ANS site. The 4-position is oriented towards a solvent exposed region suitable for the introduction of bulky polar substituents to increase solubility, and the sulfonate group could be replaced by a sulfonamide to improve pharmacological properties (Figure 8a). However, defining an allosteric pharmacophore based on the binding interactions of ANS alone is challenging, and chemical scaffolds other than ANS are needed to comprehensively evaluate the potential of this pocket as a viable drug target. The fluorescence assay used herein is suitable for high-throughput screening (HTS) to identify compounds selectively competing with ANS for binding (Figure 8b). It is readily adaptable to 384-well format, robust in the presence of DMSO, and cost effective due to the use of only free, non-phosphorylated CDK2. The assay could be conducted in the presence of inhibitors, such as JWS648, to effectively block the ATP site from compounds that may impact the binding of ANS indirectly, as seen with SU9516. Combined, the findings indicate the potential of the ANS binding pocket as a new target site for allosteric inhibitors disrupting the interactions between CDKs and cyclins.
Figure 6.
Comparison of the ANS pocket in CDK2 with allosteric sites in other protein kinases. a) In CDK2, the allosteric site extends away from the DFG motif (green) and above the C-helix (magenta). In contrast, the allosteric sites in the Abl-Imatinib complex (b, PDB entry 2HYY) (19), the MEK1-PD318088-ATP complex (c, PDB entry 1S9J) (20) and the P38-BIRB796 complex (d, PDB entry 1KV2) (21) extend along the DFG region so that ligands are underneath the C-helix. Allosteric ligands are shown in yellow, ATP site-binders in orange, and the hinge region in cyan.
Figure 7.

Residues comprising the ANS binding pocket are largely conserved among CDKs. Pairwise sequence alignments were performed for CDK2 and CDKs1-9, and their overall sequence similarity and identity values are indicated. Identical residues are highlighted in grey, with conservative and non-conservative substitutions in blue and red, respectively.
Figure 8.
Potential of the ANS pocket for the discovery of novel CDK2 inhibitors. a) Based on the binding interactions of ANS with CDK2, potential allosteric inhibitors spanning both ANS sites could be designed by attaching aryl groups via rotatable linkers to the 7-position of the naphthalene. Introduction of polar moieties at the 4-position could serve to increase solubility, and the sulfonate group could be replaced by a sulfonamide to improve pharmacological properties. b) HTS using the CDK2-ANS fluorescence assay, in which compounds that compete for binding to the ANS site induce quenching of the fluorescence signal, could lead to the discovery of new chemical scaffolds with allosteric binding potential. The assay could be performed in the presence of ATP site-directed inhibitors such as JWS648 to protect the CDK2-ANS complex from compounds which indirectly affect the ANS pocket.
METHODS
Reagents and compounds
Reagents and compounds for biochemical and crystallization experiments were purchased from Sigma–Aldrich and Hampton Research unless otherwise indicated. ANS ammonium salt (purity ≥ 97.0% by HPLC) was from Sigma–Aldrich, the peptide substrate for activity assays from Biomatik and the CDK2 inhibitor SU9516 from Tocris Bioscience. During protein purification, the protein concentration was determined using the Coomassie reagent from BioRad with bovine serum albumin as a standard. The concentration of crystallization grade proteins was determined by A280 molar absorbance using a nanodrop ND-1000 spectrophotometer (Nanodrop Technologies). Non-linear regression analysis was performed using SigmaPlot (Systat Software).
Cloning and expression
The gene for human CDK2 (comprising residues 1–298) was custom-synthesized (GeneArt), cloned into the pGEX6P-1 vector to provide an N-terminal GST-tag, and transformed into E. coli Tuner(DE3) cells (Novagen). The pGEX4T-2 plasmid used for expression of yeast CAK1 and the pGEX6P-1 used for expression of human cyclin A2 (comprising residues 174–432) were generously provided by Dr. Philip Kaldis (National Cancer Institute) and transformed into E. coli Tuner(DE3) cells. Cultures were grown for 2–3 h at 37°C, then the temperature was decreased to 16 °C prior to induction with 0.1 mM IPTG at OD600 = 0.9. The cultures were allowed to grow for an additional 20–24 h at 16 °C, and were harvested by centrifugation (20 min at 6000 x g).
Enzyme purification
Harvested cells were resuspended in 50 mM HEPES buffer (pH 7.5) containing 150 mM NaCl, 10 mM MgCl2, 2 mM dithiothreitol (DTT), 1 mM ethylene glycol tetra-acetic acid (EGTA), 0.5 mg mL−1 lysosyme, and 0.01 % Triton X-100 at 4 °C for 1 h. After sonication and centrifugation (1 h at 29000 x g), the supernatant was purified by GST-affinity column chromatography (GE LifeSciences). After incubation of peak fractions with GST-PreScission protease (20:1) at 4°C, the cleaved GST tag was removed by a second GST-affinity column. CDK2 peak fractions were loaded onto a Superdex 200 (26/60) column, eluting with 50 mM HEPES buffer (pH 7.5) containing 150 mM NaCl, 10 mM MgCl2, 2 mM DTT, 2 mM EGTA, and 0.01 mM ADP. Purified CDK2 was concentrated to ~ 10 mg mL−1 and stored at −80 °C. Cyclin A2 was purified by GST-affinity chromatography as described above, adding 100 mM MgCl2 to fractions to prevent protein aggregation. After PreScission cleavage, the protein solution was applied to a QFF ion-exchange column and eluted with 50 mM HEPES buffer (pH 7.5) containing 2 mM EDTA and 2 mM DTT over a 0–500 mM NaCl gradient. Fractions containing cyclin A were pooled, concentrated to ~ 20 mg mL−1, and stored at −80 °C. CAK1 was also purified using GST-affinity chromatography, concentrated to ~ 10 mg mL−1, and stored at −80 °C. Activation of the CDK2-cyclin A complex was achieved in vitro. Briefly, 6 mg of CDK2 was added to a 15 mL solution of 30 mM Tris/HCl (pH 7.5), 10 mM MgCl2, and 3 mM DTT containing 3 mg GST-CAK1 and 10 mM ATP. The mixture was incubated for 4 h at room temperature, then overnight at 4°C, prior to addition of cyclin A (6 mg) to yield the activated CDK2-cyclin A complex. Purification of the activated CDK2-cyclin A complex included GST-affinity chromatography to remove the GST–CAK1 fusion protein, followed by buffer exchange into 50 mM sodium phosphate buffer (pH 7.5) containing 150 mM NaCl, 2 mM DTT, and 1 mM MgCl2. The specific activity of this complex was ~ 6 μmol min−1 mg−1. The comparative analysis with other Ser/Thr protein kinases (Figure. 2d) was performed for the kinase domains of human Aurora A (residues 123–380, overexpressed in E. coli Tuner DE3) and Rock1 (residues 5–415, overexpressed in SF9 insect cells) and full-length Akt1 (residues 1–480, overexpressed in SF9 insect cells), purified to crystallization grade using similar purification protocols as described for CDK2.
Enzyme assays
The synthetic peptide PKTPKKAKKL (33) served as a substrate for the activated CDK2-cyclin A complex, and the formation of ADP from ATP was coupled to the oxidation of NADH using pyruvate kinase (PK) and lactate dehydrogenase (LDH), monitored at 340 nm (34, 35). Reactions were carried out at room temperature in 50 mM Tris/HCl buffer (pH 7.5) containing 10 mM MgCl2, 0.24 mM NADH, 5 mM DTT, 6 U mL−1 LDH, 10 U mL−1 PK, 1 mM phosphoenolpyruvate, 5 % (v/v) DMSO, and 0.01 mg mL−1 activated CDK-cyclin A complex. Inhibitor was added to the mixture, and the reaction was started by the addition of ATP and peptide substrate. Assays were performed in 384-well plates using a SpectraMax 340PC plate reader (Molecular Devices). Since ANS absorbs strongly at 340 nm, the inhibitory potential of ANS was determined using a different coupled enzyme assay (DiscoverX), in which a fluorescent resorufin dye is generated from the interaction of ADP with hydrogen peroxide and acetyl dihydroxy phenoxazine (measured with excitation and emission wavelengths of 540 and 590 nm, respectively). Reactions were carried out at 20 °C in 15 mM HEPES buffer (pH 7.4) containing 20 mM NaCl, 1 mM EGDTA, 0.02% Tween 20, 10 mM MgCl2, 5 % (v/v) DMSO, and 45 ng mL−1 activated CDK-cyclin A complex. Inhibitor was added to the mixture, and the reaction was initiated by the addition of 50 μM ATP and 200 μM peptide substrate. Assays were performed in 384-well plates using a Envision 2102 plate reader (Perkin Elmer). IC50 values for were obtained by fitting the data to equation (1),
| Equation (1) |
where A is the remaining activity, [I] is the concentration of the inhibitor, and n is the Hill slope coefficient.
Fluorescence spectroscopy
Fluorescence spectra were recorded with a Varian Eclipse fluorescence spectrophotometer using 1 × 1 cm quartz cells in 1 ml reaction mixtures at 20 °C Excitation was at 366 nm and emission spectra were recorded between 400 and 600 nm. The concentration of ANS was determined spectrophotometrically from the molar absorbance of 5000 M−1 cm−1 at 350 nm (36). Emission spectra were corrected for the inner filter effect caused by the absorption of ANS using equation (2) (37),
| Equation (2) |
where Fobs is the observed fluorescence intensity, and ODex and ODem the optical density at the extinction and emission wavelengths, respectively. Under the experimental conditions tested here absorption of ANS between 450 and 600 nm was negligible. Titrations were performed by progressive addition of ANS to CDK2 or ligands to the CDK2-ANS complex into the sample solution (50 mM HEPES/NaOH pH 7.5, 100 mM NaCl, 2 mM DTT and 1.6 μM (55 μg/ml) CDK2. CDK2-ANS spectra were subtracted from those recorded in the absence of CDK2, and the emission maximum at 460 nm was plotted as a function of ligand concentration. The apparent Kd of ANS was obtained by fitting data to equation (3),
| Equation (3) |
where Fmax is the maximum fluorescence yield.. EC50 values for the quenching potential of ligands were obtained by fitting F466 values to equation (4),
| Equation (4) |
where [L] is the concentration of ligand and n is the Hill slope coefficient.
Protein crystallography
For crystallization, CDK2 was transferred into 100 mM Na/K phosphate buffer (pH 6.2) including 2 mM DTT via PD10 columns (GE Lifesciences) and concentrated to 10 mg mL−1 using Amicon Ultra-4 10K centrifugal devices (Millipore). Crystallization of CDK2 was performed at 19 °C using the hanging drop vapor-diffusion method. Initially, crystals of free CDK2 and of the enzyme in the presence of JWS648, SU9516, and ANS were grown from 5 % (v/v) PEG 3350 in 50 mM HEPES/NaOH (pH 7.5) at a CDK2 concentration of ~ 5 mg mL−1. While free CDK2 and complexes with inhibitors yielded crystals after two days, the CDK2-ANS crystals appeared after several weeks. Therefore, alternative crystallization conditions using 15 % (v/v) Jeffamine ED-2001 and 50 mM HEPES were established, yielding co-crystals with ANS alone and in ternary complex with ANS and JWS648 after two days (1.5–5 mM of compound was added to the crystallization solution). In parallel, in-diffusion experiments with ANS were performed for free CDK2 and complexes of CDK2-JWS648 and CDK2-SU9516. Crystals were incubated for 24 h in 50 mM HEPES (pH 7.5), 50 mM phosphate (Na/K, pH 7.5), 7.5 % (v/v) PEG 3350, and 5–15 mM aqueous ANS, with or without 2 mM of compound JWS648 or SU9516. All crystals were harvested in cryoprotectant prior to data collection: 50 mM HEPES (pH 7.5), 50 mM phosphate (Na/K, pH 7.5), 7.5 % (v/v) PEG 3350, and 25 % (v/v) ethylene glycol, with 0.5–2 mM inhibitor.
X-ray diffraction data were recorded at –180 °C in the Moffitt Cancer Center Structural Biology Core using CuKα X-rays generated by a Rigaku Micro-Max 007-HF X-ray generator, focused by mirror optics and equipped with a Rigaku R-AXIS HTC imager and a Rigaku CCD Saturn 944 system on single crystals frozen in liquid N2. Data were reduced with XDS (38). CNS (39) was employed for phasing and refinement (minimization and simulated annealing), and model building was performed using Coot (40) or O (41). Initial models for the small molecule ligands were generated using Chem3DPro (CambridgeSoft), with parameters and topology files from the program xplo2d (42). Figures were drawn with PyMol (Schrodinger) (43).
B-factor analysis
The frequency of B-factor values for all protein atoms, rounded to the nearest integer, were fitted to the logarithmic gaussian distribution equation (5),
| Equation (5) |
where A is the amplitude (height) of the center, Bo is the B-factor for the peak, and c is the width of the distribution, respectively. The Bo value was then used to normalize the atomic B-factors of the ANS molecules bound to the binary and ternary complexes.
Synthesis of CDK2 inhibitor JWS648
JWS648 was prepared in two steps (Scheme 1) by the reaction of biguanide with methyl 2-hydroxy-5-methoxybenzoate to form the corresponding monoacylated biguanide, which was then subjected to microwave irradiation to form the triazine (44):
Scheme 1.
Synthesis of JWS648
Small pieces of sodium metal (108 mg, 4.96 mmol) were added to dry ethanol (40 mL, cooled to 0 °C). After stirring for 30 min, the sodium was completely dissolved, and biguanide hydrochloride (1.05 g, 4.96 mmol) was added to the solution. The resulting suspension was stirred at room temperature for 30 min, then the precipitated sodium chloride was collected by suction filtration and washed with dry ethanol (20 mL). The filtrate was collected, and methyl 2-hydroxy-5-methoxybenzoate (903 mg, 4.96 mmol) was added at once. The suspension was stirred at reflux for 4 h, then the solvent was removed under reduced pressure. The residue was dissolved in dry DMSO (15 mL), placed into a 20 mL microwave tube, and irradiated at 190 °C for 15 min. The reaction was quenched with water and extracted with ethyl acetate (3 × 50 mL). The combined organic layers were washed with brine, dried over sodium sulfate, and filtered. The solvent was removed under reduced pressure, and the crude residue was purified by flash column chromatography over silica gel using an ethyl acetate/hexanes gradient (1:1 to 2:1), to afford 150 mg (14 %) of JWS648 as a light yellow solid. 1H NMR (400 MHz, DMSO-d6): δ=13.35 (s, 1H), 7.71 (d, J = 3.2 Hz, 1H), 7.18 (br s, 2H), 6.98 (m, 3H), 6.79 (d, J = 8.9 Hz, 1H), 3.70 (s, 3H). HR-MS calcd for C10H11N5O2 (M+H) 234.0991, found 234.0993.
Supplementary Material
Acknowledgments
We thank the Moffitt Structural Biology Core for use of the X-ray crystallography facility. This work was supported in part by National Institutes of Health Grants N01-HD53400 and U54-HD055763.
Footnotes
Accession codes. The atomic coordinates and structure factors of the crystal structures determined as part of this work have been deposited in the Protein Data Bank (entries 3PXF, 3PXZ, 3PY1, 3PXR, 3PXQ, 3PXY, 3PY0).
Supporting Information Available. Additional kinetic and structural analysis is provided in Supplementary Table 1 and Supplementary Figures 1–7. This material is available free of charge via the Internet.
References
- 1.Hall M, Peters G. Genetic alterations of cyclins, cyclin-dependent kinases, and Cdk inhibitors in human cancer. Adv Cancer Res. 1996;68:67–108. doi: 10.1016/s0065-230x(08)60352-8. [DOI] [PubMed] [Google Scholar]
- 2.Johnson LN. Protein kinase inhibitors: contributions from structure to clinical compounds. Q Rev Biophys. 2009;42:1–40. doi: 10.1017/S0033583508004745. [DOI] [PubMed] [Google Scholar]
- 3.Zhang J, Yang PL, Gray NS. Targeting cancer with small molecule kinase inhibitors. Nat Rev Cancer. 2009;9:28–39. doi: 10.1038/nrc2559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bain J, Plater L, Elliott M, Shpiro N, Hastie CJ, McLauchlan H, Klevernic I, Arthur JS, Alessi DR, Cohen P. The selectivity of protein kinase inhibitors: a further update. Biochem J. 2007;408:297–315. doi: 10.1042/BJ20070797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sherr CJ. Cancer cell cycles. Science. 1996;274:1672–1677. doi: 10.1126/science.274.5293.1672. [DOI] [PubMed] [Google Scholar]
- 6.Grana X, Reddy EP. Cell cycle control in mammalian cells: role of cyclins, cyclin dependent kinases (CDKs), growth suppressor genes and cyclin-dependent kinase inhibitors (CKIs) Oncogene. 1995;11:211–219. [PubMed] [Google Scholar]
- 7.Morgan DO. Principles of CDK regulation. Nature. 1995;374:131–134. doi: 10.1038/374131a0. [DOI] [PubMed] [Google Scholar]
- 8.Ortega S, Malumbres M, Barbacid M. Cyclin D-dependent kinases, INK4 inhibitors and cancer. Biochim Biophys Acta. 2002;1602:73–87. doi: 10.1016/s0304-419x(02)00037-9. [DOI] [PubMed] [Google Scholar]
- 9.Weinberg RA. The retinoblastoma protein and cell cycle control. Cell. 1995;81:323–330. doi: 10.1016/0092-8674(95)90385-2. [DOI] [PubMed] [Google Scholar]
- 10.van den Heuvel S, Harlow E. Distinct roles for cyclin-dependent kinases in cell cycle control. Science. 1993;262:2050–2054. doi: 10.1126/science.8266103. [DOI] [PubMed] [Google Scholar]
- 11.Malumbres M, Barbacid M. To cycle or not to cycle: a critical decision in cancer. Nat Rev Cancer. 2001;1:222–231. doi: 10.1038/35106065. [DOI] [PubMed] [Google Scholar]
- 12.Echalier A, Endicott JA, Noble ME. Recent developments in cyclin-dependent kinase biochemical and structural studies. Biochim Biophys Acta. 2010;1804:511–519. doi: 10.1016/j.bbapap.2009.10.002. [DOI] [PubMed] [Google Scholar]
- 13.Liu Y, Gray NS. Rational design of inhibitors that bind to inactive kinase conformations. Nat Chem Biol. 2006;2:358–364. doi: 10.1038/nchembio799. [DOI] [PubMed] [Google Scholar]
- 14.Russo AA, Jeffrey PD, Patten AK, Massague J, Pavletich NP. Crystal structure of the p27Kip1 cyclin-dependent-kinase inhibitor bound to the cyclin A-Cdk2 complex. Nature. 1996;382:325–331. doi: 10.1038/382325a0. [DOI] [PubMed] [Google Scholar]
- 15.Gondeau C, Gerbal-Chaloin S, Bello P, Aldrian-Herrada G, Morris MC, Divita G. Design of a novel class of peptide inhibitors of cyclin-dependent kinase/cyclin activation. J Biol Chem. 2005;280:13793–13800. doi: 10.1074/jbc.M413690200. [DOI] [PubMed] [Google Scholar]
- 16.McInnes C, Andrews MJ, Zheleva DI, Lane DP, Fischer PM. Peptidomimetic design of CDK inhibitors targeting the recruitment site of the cyclin subunit. Curr Med Chem Anticancer Agents. 2003;3:57–69. doi: 10.2174/1568011033353506. [DOI] [PubMed] [Google Scholar]
- 17.Kontopidis G, Andrews MJ, McInnes C, Plater A, Innes L, Renachowski S, Cowan A, Fischer PM. Truncation and optimisation of peptide inhibitors of cyclin-dependent kinase 2-cyclin a through structure-guided design. Chem Med Chem. 2009;4:1120–1128. doi: 10.1002/cmdc.200900093. [DOI] [PubMed] [Google Scholar]
- 18.Mendoza N, Fong S, Marsters J, Koeppen H, Schwall R, Wickramasinghe D. Selective cyclin-dependent kinase 2/cyclin A antagonists that differ from ATP site inhibitors block tumor growth. Cancer Res. 2003;63:1020–1024. [PubMed] [Google Scholar]
- 19.Cowan-Jacob SW, Fendrich G, Floersheimer A, Furet P, Liebetanz J, Rummel G, Rheinberger P, Centeleghe M, Fabbro D, Manley PW. Structural biology contributions to the discovery of drugs to treat chronic myelogenous leukaemia. Acta Crystallogr D Biol Crystallogr. 2007;63:80–93. doi: 10.1107/S0907444906047287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ohren JF, Chen H, Pavlovsky A, Whitehead C, Zhang E, Kuffa P, Yan C, McConnell P, Spessard C, Banotai C, Mueller WT, Delaney A, Omer C, Sebolt-Leopold J, Dudley DT, Leung IK, Flamme C, Warmus J, Kaufman M, Barrett S, Tecle H, Hasemann CA. Structures of human MAP kinase kinase 1 (MEK1) and MEK2 describe novel noncompetitive kinase inhibition. Nat Struct Mol Biol. 2004;11:1192–1197. doi: 10.1038/nsmb859. [DOI] [PubMed] [Google Scholar]
- 21.Pargellis C, Tong L, Churchill L, Cirillo PF, Gilmore T, Graham AG, Grob PM, Hickey ER, Moss N, Pav S, Regan J. Inhibition of p38 MAP kinase by utilizing a novel allosteric binding site. Nat Struct Biol. 2002;9:268–72. doi: 10.1038/nsb770. [DOI] [PubMed] [Google Scholar]
- 22.Zuccotto F, Ardini E, Casale E, Angiolini M. Through the “gatekeeper door”: exploiting the active kinase conformation. J Med Chem. 2010;53:2681–2694. doi: 10.1021/jm901443h. [DOI] [PubMed] [Google Scholar]
- 23.Kluter S, Grutter C, Naqvi T, Rabiller M, Simard JR, Pawar V, Getlik M, Rauh D. Displacement assay for the detection of stabilizers of inactive kinase conformations. J Med Chem. 2010;53:357–367. doi: 10.1021/jm901297e. [DOI] [PubMed] [Google Scholar]
- 24.Stryer L. The interaction of a naphthalene dye with apomyoglobin and apohemoglobin. A fluorescence probe of non-polar binding sites. J Mol Biol. 1965;13:482–495. doi: 10.1016/s0022-2836(65)80111-5. [DOI] [PubMed] [Google Scholar]
- 25.Schonbrunn E, Eschenburg S, Luger K, Kabsch W, Amrhein N. Structural basis for the interaction of the fluorescence probe 8-anilino-1-naphthalene sulfonate (ANS) with the antibiotic target MurA. Proc Natl Acad Sci USA. 2000;97:6345–6349. doi: 10.1073/pnas.120120397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Heitz F, Morris MC, Fesquet D, Cavadore JC, Doree M, Divita G. Interactions of cyclins with cyclin-dependent kinases: a common interactive mechanism. Biochemistry. 1997;36:4995–5003. doi: 10.1021/bi962349y. [DOI] [PubMed] [Google Scholar]
- 27.Morris MC, Gondeau C, Tainer JA, Divita G. Kinetic mechanism of activation of the Cdk2/cyclin A complex. Key role of the C-lobe of the Cdk. J Biol Chem. 2002;277:23847–23853. doi: 10.1074/jbc.M107890200. [DOI] [PubMed] [Google Scholar]
- 28.Lawrie AM, Noble ME, Tunnah P, Brown NR, Johnson LN, Endicott JA. Protein kinase inhibition by staurosporine revealed in details of the molecular interaction with CDK2. Nat Struct Biol. 1997;4:796–801. doi: 10.1038/nsb1097-796. [DOI] [PubMed] [Google Scholar]
- 29.Brown NR, Noble ME, Lawrie AM, Morris MC, Tunnah P, Divita G, Johnson LN, Endicott JA. Effects of phosphorylation of threonine 160 on cyclin-dependent kinase 2 structure and activity. J Biol Chem. 1999;274:8746–8756. doi: 10.1074/jbc.274.13.8746. [DOI] [PubMed] [Google Scholar]
- 30.Gu J, Bourne PE. Identifying allosteric fluctuation transitions between different protein conformational states as applied to Cyclin Dependent Kinase 2. BMC Bioinformatics. 2007;8:45. doi: 10.1186/1471-2105-8-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Simard JR, Getlik M, Grutter C, Schneider R, Wulfert S, Rauh D. Fluorophore labeling of the glycine-rich loop as a method of identifying inhibitors that bind to active and inactive kinase conformations. J Am Chem Soc. 2010;132:4152–4160. doi: 10.1021/ja908083e. [DOI] [PubMed] [Google Scholar]
- 32.Cox KJ, Shomin CD, Ghosh I. Tinkering outside the kinase ATP box: allosteric (type IV) and bivalent (type V) inhibitors of protein kinases. Future Med Chem. 2011;3:29–43. doi: 10.4155/fmc.10.272. [DOI] [PubMed] [Google Scholar]
- 33.Stevenson-Lindert LM, Fowler P, Lew J. Substrate specificity of CDK2-cyclin A. What is optimal? J Biol Chem. 2003;278:50956–50960. doi: 10.1074/jbc.M306546200. [DOI] [PubMed] [Google Scholar]
- 34.Adams JA, McGlone ML, Gibson R, Taylor SS. Phosphorylation modulates catalytic function and regulation in the cAMP-dependent protein kinase. Biochemistry. 1995;34:2447–2454. doi: 10.1021/bi00008a007. [DOI] [PubMed] [Google Scholar]
- 35.Cook PF, Neville ME, Jr, Vrana KE, Hartl FT, Roskoski R., Jr Adenosine cyclic 3′,5′-monophosphate dependent protein kinase: kinetic mechanism for the bovine skeletal muscle catalytic subunit. Biochemistry. 1982;21:5794–5799. doi: 10.1021/bi00266a011. [DOI] [PubMed] [Google Scholar]
- 36.Weber G, Young LB. Fragmentation of bovine serum albumin by pepsin. I. The origin of the acid expansion of the albumin molecule. J Biol Chem. 1964;239:1415–1423. [PubMed] [Google Scholar]
- 37.Puchaslki MM, Morra MJ, von Wandruszka R. Assessment of inner filter effect corrections in fluorimetry. Fresenius J Anal Chem. 1991;340:341–344. (and cited herein: Lakowicz, L. (1983) Principles of fluorescence spectroscopy, Plenum Press, NY) [Google Scholar]
- 38.Kabsch W. Automatic processing of rotation diffraction data from crystals of initially unknown symmetry and cell constants. J Appl Crystallogr. 1993;26:795–800. [Google Scholar]
- 39.Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, Read RJ, Rice LM, Simonson T, Warren GL. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr D Biol Crystallogr. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 40.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 41.Jones TA, Zou JY, Cowan SW, Kjeldgaard M. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr. 1991;A47(Pt 2):110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- 42.Kleywegt GJ, Zou J-Y, Kjeldgaard M, Jones TA. Around O in International Tables for Crystallography. International Union of Crystallography; Chester: 2001. pp. 353–356. [Google Scholar]
- 43.DeLano WL. PyMOL Molecular Graphics System. DeLano Scientific LLC; Palo Alto, CA, USA: 2008. http://www.pymol.org. [Google Scholar]
- 44.Jaroslav S, Jaroslav N, Zdenek B. 2,4-Diamino-6-phenyl-1,3,5-triazines. Coll Czech Chem Commun. 1978;43:1639–1646. [Google Scholar]
- 45.Cheng KY, Noble ME, Skamnaki V, Brown NR, Lowe ED, Kontogiannis L, Shen K, Cole PA, Siligardi G, Johnson LN. The role of the phospho-CDK2/cyclin A recruitment site in substrate recognition. J Biol Chem. 2006;281:23167–23179. doi: 10.1074/jbc.M600480200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.