Abstract
The nematode Caenorhabditis elegans is a model organism best known for its powerful genetics. There is an increasing need in the worm community to couple genetics with biochemistry. Isolation of functionally-active proteins, or nucleic acids without the use of strong, oxidizing denaturants, or of sub-cellular compartments from C. elegans has, however, been challenging because of the worms' thick surrounding cuticle. The Balch Homogenizer is a tool that has found much use in mammalian cell culture biology. The interchangeable, single ball bearing design of this instrument permits rapid permeabilization, or homogenization, of cells. Here we demonstrate the utility of the Balch Homogenizer for studies with C. elegans. We describe procedures for the efficient breakage and homogenization of every larval stage, including dauers, and show that the Balch Homogenizer can be used to extract functionally-active proteins. Enzymatic assays for catalase and dihydrolipoamide dehydrogenase show that sample preparation using the Balch Homogenizer equals or out-performs conventional methods employing boiling, sonication or Dounce homogenization. We also describe phenol-free techniques for isolation of genomic DNA and RNA. Finally, we use the tool to isolate coupled mitochondria and polysomes. The reuseable Balch Homogenizer represents a quick and convenient solution for undertaking biochemical studies on C. elegans.
Keywords: Balch Homogenizer, Caenorhabditis elegans, dihydrolipoamide dehydrogenase, catalase, genomic DNA, mitochondria, polysomal profiling
Introduction
Caenorhabditis elegans is a free-living, soil nematode that is used extensively as a model organism to address fundamental questions in biology [1]. Adult hermaphrodites measure ∼100 × 1500 μm in length [2] and are comprised of 959 somatic cells [1]. The body plan of C. elegans is organized into a primitive nervous system, a gut connected anteriorly to a pharynx and posteriorly to an anus, a surrounding epithelium comprised of syncytial hypodermal cells that secrete a protective cuticle, several circumferential body wall muscles that are used for locomotion, and a simple excretory system. C. elegans hermaphrodites also contain a gonad that is connected to the outside via a uterus. During the course of an animals' life (∼20 days at 20°C) up to 350 self-progeny, or 1000 progeny if fertilized by a male, can be produced.
The developmental cycle of C. elegans is comprised of six life stages: embryogenesis (egg stage), four larval stages (L1 to L4), and an adult stage. Animals hatch from eggs as L1 larvae comprised of 560 cells. Growth during each larval stage occurs by cell division and by cell hypertrophy [3]. Cuticular molting punctuates each larval stage [4]. If environmental cues signal to the developing animal that conditions are unlikely to support adult fertility, C. elegans can alter its development and form an alternate L3 larval stage called a dauer. In this state, animals are stress-tolerant and long-lived (surviving three to nine months). Dauers isolate themselves from adversity by sealing both their buccal and anal cavities, shrinking their gut, and turning on a daf-16/FOXO-dependent genetic program that, among other things, leads to expression of a dauer-specific cuticle [5].
The C. elegans cuticle is synthesized five times during the course of development - once during embryogenesis, and subsequently at the end of each larval stage, just prior to each molt. This flexible, but resilient, exoskeleton serves several functions, including acting as a site of anchorage for the bodywall musculature, environmental protection, and the permittance of growth. The cuticle is composed of a highly-structured extracellular matrix comprised predominantly of cross-linked collagens (80%) as well as other insoluble proteins (cuticulins). The outer surface of the cuticle is coated in a lipid rich layer and this layer is, in turn, covered by a glycoprotein-rich layer [4]. The cuticle layer of all stages is resilient enough to interfere with most standard cell-solubilization protocols. Indeed, the cuticle confers significant resistance to a variety of detergents. Dauer larvae are particularly resilient; they are able to survive several hours of exposure to the strong anionic detergent 1% SDS. The resilence of the dauer cuticle is a result of extensive tyrosine cross-linking between cuticulins [4]. The cuticle of all stages also displays significant mechanical resistance. Moreover, worms can withstand pressures up to 30,000 psi (LD50 of 7400 psi); eggs can withstand double this pressure [6]. Recovery of live animals from the space shuttle Columbia crash debris demonstrates the resilience of C. elegans[7].
The ideal worm homogenization technique should be cheap, easy to apply, reproducible, safe, gentle so as to permit recovery of target molecule activity or subcellular identity, applicable to all developmental stages of C. elegans, unlimited by sample size, and finally, temperature-controllable. Surprisingly, no method that fulfills all these criteria exists. In the present study we have adapted an old tool – the Balch Homogenizer [8], to address this limitation of C. elegans research. This instrument has found widespread use in sub-cellular fractionation studies of mammalian cells [8; 9; 10; 11; 12; 13]. We present procedures that are sufficiently gentle to isolate functional protein, PCR-quality nucleic acids, coupled mitochondria and ribosomal polysomes from C. elegans. The Balch Homogenizer opens the way for the establishment of more refined C. elegans procedures, such as sub-cellular fractionation and deep sequencing of ribosomal sub-fractions.
Materials & Methods
C. elegans Growth & Maintenance
The following C. elegans strains were employed: N2 (Bristol); CB1370 [daf-2(e1 370)III], PD4251 [ccIs4251[pSAK2(Pmyo-3∷NLSGFP-LacZ) + pSAK4(Pmyo-3∷MITOGFP) + dpy-20(wt)]I; dpy-20(e1282ts)IV], and SD1241 [gaIs153[pPRSK29(F25B3.3:FLAG∷PAB-1); TG96(sur-5∷GFP)]. Strains were maintained at 20°C on NGM agar plates seeded with E. coli (OP50), as described [1]. Synchronous worm cultures were obtained by allowing gravid adults to lay eggs onto NGM/OP50 plates for 2-6 hours. Eggs were then collected in S-basal (100 mM NaCl, 50 mM KH2PO4, pH 6.0), counted using a dissecting microscope, then seeded onto BNGM agar plates (NGM + an additional 7.5 g peptone per liter) at a density of 2500 eggs/plate (7500 eggs/plate were used for polysomal profiling). Dauer larvae were generated using temperature-sensitive daf-2(e1370) mutants, as described [14]. Briefly, synchronized eggs (2500/plate) were allowed to hatch on NGM/OP50 plates at 20°C (∼18 hours). Plates were then sealed with parafilm and placed at 25°C for 2 weeks to allow dauer formation.
Balch homogenizer
Construction of the original Balch Homogenizer has been described [8]. The model we used was purchased from Isobiotec (Heidelberg, Germany), and is described under the German patent #202 09 547.9. We tested tungsten carbide ball bearings ranging in size from 7.982 mm to 7.999 mm, corresponding to wall-to-ball bearing clearances of 18 μm down to 1 μm. Pictures of the instrument and its assembly are presented in Figure 1.
Figure 1. Balch Homogenizer.
(A) Instrument construction - stainless steel chamber (1) with a hollow barrel into which a single tungsten carbide ball bearing (2) of defined tolerance (+/− 0.0001 mm) is inserted. The barrel is sealed with bolted steel caps (3). Glass syringes (4) force worm samples around the ball bearing. (B) Assembled instrument - placement of the Balch homogenizer on an ice-cold metal platform is ideal for maintaining the metal chamber at 4°C.
Worm Homogenization & Protein Extraction Protocols
For each of the following procedures, animals of the relevant stage (30,000 per sample) were collected and washed in ice-cold S-basal, concentrated by centrifugation (1500 rpm, 2 min), washed six times with ice-cold S-basal to remove residual bacteria, then stored on ice until ready for use. For protein extraction studies, gravid-adult worms were allowed to form a compact pellet after the last S-basal wash. Worm pellets were then resuspended in 1 ml of ice-cold Extraction Buffer [20 mM potassium phosphate, pH 7.4, 2mM EDTA, 1% Triton-X-100, protease inhibitors (Sigma P2714)] and processed immediately.
(i) Balch Homogenization
For studies monitoring worm breakage, worm pellets (30,000 worms for each larval stage) were resuspended in 2 ml ice-cold S-basal, then transferred to a 2 ml glass syringe fitted with a metal luer lock (ICO Interchangeable –this item has been discontinued but a similar item is obtainable from Sigma - Z314528. We have also used disposable plastic syringes without concern). Animals were passed though the Balch Homogenizer for a defined number of times. Worm breakage was monitored using a dissecting microscope. For extraction of protein, gravid-adult worms were resuspended in 1 ml of cold Extraction Buffer then homogenized using a 16 μm ball clearance and 25 passes. The homogenate was kept on ice for 30 minutes, then centrifuged at 16,470 × g for 15 minutes (4°C). The supernatant was retained (referred to as ‘supernatant’) and the pellet was resuspended in 100 μl of 5% SDS, boiled for 5 minutes, then re-centrifuged. The resulting supernatant was retained (referred to as ‘pellet’). An identical procedure was employed for L1 larvae except worms were homogenized using a 4 μm ball clearance and 14 passes.
(ii) Sonication
∼30,000 gravid-adult worms (corresponding to ∼100 mg wet weight), were sonicated on ice using a VirSonic 60 sonicator with microtip probe (VirTis, Gardiner, NY), for three minutes at 4 W power, in 1 ml of ice-cold Extraction Buffer. The homogenate resulting from this method (and likewise in the following two protein extraction methods), was processed exactly as described for the Balch Homogenizer, resulting in ‘supernatant’ and ‘pellet’ fractions.
(iii) Dounce Homogenization
Gravid-adult worms (∼30,000) were resuspended in 1 ml of cold Extraction Buffer and homogenized on ice using 25 stokes of a 15 ml Duall® 21, Dounce Homogenizer (Kontes Glass Co.).
(iv) Boiling
1 ml of boiling extraction buffer was added to gravid-adult worm pellets (∼30,000 worms) and further boiled for 5 minutes. Samples were vortex-mixed for 5 seconds, every minute.
Protein quantification
The amount of protein in each homogenization extract was quantified using the bicinchoninic acid (BCA) method (Thermo Fisher Scientific, Rockford, IL).
Western Analysis
Protein extracts (25 μg) were separated by 10% SDS-PAGE, transferred to PVDF membranes, blocked with 5% non-fat milk in PBS containing 0.1% Tween-20 (1 hr), then analyzed for pyruvate dehydrogenase (PDH) E1α subunit integrity using a mouse anti-PDH E1α antibody (cat# MSP07, Mito Sciences, 1:2000). This antibody cross-reacts with the worm protein. Bands were visualized using a HRP-conjugated donkey anti-mouse IgG secondary antibody (cat# 2314, Santa Cruz, 1:10,000) in conjunction with Amersham ECL plus reagent (cat# RPN 2132, GE Healthcare, Piscataway, NJ).
Enzymatic Assays
(i) Catalase Activity
The catalatic activity (H2O2 consumption) of catalase in whole-worm extracts was measured spectrophotometrically by following the loss of exogenously added H2O2, using an adaptation of a procedure described by Aebi [15] for human tissue. Briefly, extracts from adult wild type (N2) worms (30,000 per sample), were prepared in 1ml of ice-cold Extraction Buffer using each of the four homogenization procedures described above. After removal of insoluble material by centrifugation (16,470 × g, 15 minutes, 4°C), catalatic activity was measured by adding 30 μl of extract to 1 ml (final volume) of Assay Buffer (50mM potassium phosphate, pH 7.0, 25°C) containing 8.2 mM H2O2 (Sigma, H3410). Loss of H2O2 was monitored spectrophotometrically (A240nm) for three minutes (Spectra Max, NY, USA). All experiments were independently replicated. Catalatic activity is expressed as a first-order rate constant normalized to extract protein amount (sec−1 . mg−1).
(ii) Dihydrolipoamide Dehydrogenase (DLD) Activity
DLD activity in whole worm extracts was monitored spectrophotometrically following the addition of dihydrolipoamide and NAD+. Briefly, dihydrolipoamide was synthesized in-house via NaBH4 reduction of D,L-lipoamide [16; 17]. Whole-worm extracts from 30,000 adult worms (per sample) were prepared in 1 ml of Extraction Buffer supplemented with 2mM EGTA, using each of the four extraction techniques described above. One unit (U) of DLD activity is defined as the rate of production of 1 μmol of NADH in 1 minute at 25°C. NADH production was followed spectrophotometrically at A340nm. Assays were performed in triplicate using 250 μL aliquots per reaction. All experiments were replicated and are expressed in U per mg of protein extract.
Purification of gDNA from egg nuclei
Isolation of nuclei from N2 and PD4251 eggs was carried out using an adaptation of a procedure described by Dixon et al (19) with the following modifications. One-day-old gravid adults (40,000) were allowed to lay ∼500,000 eggs on NGM agar plates, over two consecutive 4 hour intervals (20°C). Cuticle synthesis in embryonic larvae begins at ∼10 hrs post-fertilization. Embryonic cell number reaches a maximum at ∼ 5 hrs post-fertilization (eggs are laid ∼4 hours after fertilization [1]). Eggs were collected off plates using S-Basal, then freed of bacteria and debris on a 50% sucrose cushion (2000 × g for 3 minutes at 4°C) as described [18]. Egg pellicles were pooled, diluted with cold S-Basal and centrifuged at 12,000 × g for 1 minute. Egg pellets were then snap frozen in liquid nitrogen and either stored at −80C or thawed and processed immediately (a freeze-thaw cycle substantially increases the genomic DNA (gDNA) yield and quality - see Results and Figure 5A and B). Thawed eggs were resuspended in buffer A [1M sucrose, 100 mM Tris HCl (pH 8),100 mM MgCl2 100 mM EGTA, 100 mM PMSF and 100 mM β-mercaptoethanol] then homogenized on ice using the Balch homogenizer (10μm clearance, 20 passes). The resulting homogenate was centrifuged at 4000 × g (5 minutes), and the gDNA-containing pellet was re-suspended in buffer A containing 0.25 % NP-40 and 0.1% Triton -X-100. The homogenate was centrifuged at 50×g for 5 minutes, then the gDNA-containing supernatant set aside and the pellet was re-extracted twice with buffer A containing detergents (0.25% NP40 and 0.1% Triton-X-100). The three supernatant fractions were pooled, centrifuged at 4000 × g for 5 minutes and the gDNA-containing pellet retained. The pellet was subsequently washed once in buffer A without PMSF, pelleted again at 4000 ×g for 5 minutes, then finally resuspended in nuclease free water. gDNA was then purified using a Gentra Puregene Tissue Kit (Qiagen, cat. # 158622). The manufacturer would not provide details on the composition of the isolation buffers, however the methodology uses a proprietary anionic detergent, proteinase K and RNase A to disrupt cells, digest proteins and remove RNA, respectively. Finally, the resulting gDNA was ethanol precipitated.
Figure 5. Isolation of high quality gDNA using the Balch Homogenizer.
(A-C) SD1241 worms contain a sur-5∷GFP transgene that is genomic localized. Different stages of egg development are shown (A). A single round of freeze-thawing (-80°C) is sufficient to disrupt lipid membrane integrity (B). Balch Homogenization results in almost complete dissolution of eggs. A rare piece of debris is shown (C). For panes A-C: left panels, DIC images; right panels, GFP fluorescence. (D) Agarose gel showing phenol-free gDNA purified from either freeze-thawed nuclei or fresh eggs. For both extractions, eggs were homogenized with the Balch homogenizer. Size marker: HindIII-restricted λ DNA (left lane). (E) Purified gDNA isolated from egg nuclei is free of restriction enzyme-inhibiting factors. gDNA smear following EcoR1 digestion (1.5μg gDNA in 30μL, 40U EcoR1, 6hrs, 37°C), indicates sample is highly enriched in gDNA (compare right two lanes). Size markers: Gibco 1kb ladder and HindIII-restricted λ DNA (left two lanes). (F) Q-PCR using primers specific for gDNA (daf-12), mtDNA, or E. coli DNA confirms gDNA purified from egg nuclei is highly pure. Q-PCR amplicon abundance (Sytox stain) is plotted against PCR cycle number. Line marks cycle threshold cutoff (Ct). Genome sizes are C. elegans mtDNA: 13,794 bp, C. elegans genomic DNA: 1×109 bp, and E. coli genomic DNA: 4.6 ×106 bp.
RNA Extraction
Two methods were employed to isolate RNA from C. elegans. In both procedures, use of the Balch Homogenizer (4°C) to homogenize worms preceded the addition of either chaotrope or denaturant. The instrument was pre-rinsed in RNase-free water before use.
(i) Guanididium Isothiocyanate Method
20,000 L4 N2 larvae were homogenized in 1 ml S-Basal using the Balch Homogenizer (14 μm clearance, 20 passes). Homogenates were centrifuged at 13,000 rpm (16,470g) at 4°C for 2 minutes then the cleared supernatant was processed for RNA using a Nucleospin RNA Isolation kit (cat# 740-933-50, Clontech, CA).
(ii) Phenol-based Extraction Procedure
50,000 N2 adult worms were homogenized in 1ml S-Basal using the Balch homogenizer (14 mm clearance, 20 passes). 2ml TRIZOL® Reagent (Gibco cat# 15596-018) was then added to the homogenate and the RNA was extracted following the manufacturer's protocol.
For both procedures, RNA concentration and purity was determined using a Nanodrop Spectrophotometer (Thermo Scientific) and RNA integrity was confirmed by gel electrophoresis.
Quantitative Reverse Transcription-PCR (qRT-PCR)
qRT-PCR was used to assess mRNA quality. cDNA was generated from 2 μg purified RNA using a Vilo cDNA Synthesis Kit (Invitrogen). qRT-PCR was performed using an ABI 7900HT (Applied Biosystems) and reaction mixtures contained 1:50 diluted cDNA, 2 μM each forward and reverse primer, 10 μl 2×SYBR Green PCR Master Mix (Applied Biosystems) in a final reaction volume of 20 μl. cDNA targets were amplified using 40 PCR cycles (95° C for 15 seconds and 60° C for 45 seconds). We tested several target genes including icd-1, pmp-3, csq-1, atp-3 and Y45F10D.4, as described [19; 20]. Primers were designed using PrimerQuestSM (Integrated DNA Technologies). For both qRT-PCR and Q-PCR (see next), all primer pairs were verified by agarose gel electrophoresis to amplify only a single product. Primer amplification efficiencies were determined prior to data collection.
Quantitative PCR (Q-PCR)
Contamination of gDNA preparations by mitochondrial DNA(mtDNA) and E. coli genomic DNA was measured by Q-PCR with an ABI 7900HT instrument (Applied Biosystems). Reaction mixtures contained gDNA template (15ng), 2μM each forward and reverse primer, 10 μl 2×SYBR Green PCR Master Mix (Applied Biosystems) in a final reaction volume of 20 μl. Targets were amplified using 40 PCR cycles (95° C for 15 seconds and 60°C for 45 seconds). Primer pairs were: C. elegans mtDNA – (Forward) 5′-GTAATTGCTGAACTTAACCGGGG-3′, (Reverse) 5′-AGCTACTCTGGCAAACTCCACA-3′; daf-12 gDNA – (Forward) 5′-ATGAGACCACGGTCCAAGTATCAC-3′, (Reverse) 5′-TGGAAAGTTCATCCTCCTCAGACG-3′; E. coli genomic DNA - (Forward) 5′-AGCTTCACCGACTGTTCCGGTTAT-3′ and (Reverse) 5′-CCTGCGTGTCTTCACGGCAATTA-3′.
Immunofluorescence
Eggs were mounted for GFP immunofluorescence on 2% agarose pads as described previously [20]. Images were collected using an Axiovert 200M (Zeiss) inverted fluorescence microscope. Images were processed using ImageJ (1.41, NIH).
Preparation of Mitochondria-enriched Fractions
(i) C. elegans
∼100,000 synchronized N2 or PD4251 eggs were hatched on BNGM agar plates (2500 worms/plates) and allowed to develop into L3 larvae at 20oC (∼28 hours). Animals were then collected and washed in S-Basal, pooled, transferred into a baffled flask containing S-basal (200 worms/ml) supplemented with 1×109 cfu/ml OP50 + 200ug/ml cholesterol, and allowed to complete development at 20°C with shaking (100 rpm). Growth of larval stages on solid media facilitated subsequent development into well-fed adults in liquid media. One-day-old adult worms (∼1g wet weight) were collected in a glass separatory funnel, then washed several times with S-basal. To separate bacteria, dead worms and debris, the washed worm pellet was suspended in 35% sucrose solution and centrifuged at 3000 rpm for 3 minutes. The upper, worm-containing layer was collected and washed three times with S-basal to remove residual sucrose. Worms were subsequently resuspended in ice-cold isolation buffer (10mM Tris-MOPS, 1mM EGTA-Tris, 0.2 M Sucrose pH7.4) and gently fractured on ice using the Balch homogenizer (18 μm clearance, 3 passes). The mitochondria-containing homogenate was centrifuged at 600 ×g for 10 minutes at 4°C (Avanti J-20 XP, Beckman Coulter). The supernatant was collected, centrifuged at 7,000×g for 10 minutes at 4°C, and the pellet retained. The pellet was washed with 5ml of isolation buffer then re-centrifuged at 7000 ×g for 10 minutes, 4°C. The crude mitochondria-containing pellet was resuspended in isolation buffer and protein concentration quantified using the bicinchoninic acid (BCA) method (Thermo Fisher Scientific, Rockford, IL).
(ii) Mouse Liver
Mitochondria were isolated from 6 month old BALBc mice exactly as described by Frezza et al [21].
Mitochondrial Bioenergetics
Mitochondrial oxygen consumption and mitochondrial membrane potential were measured using a Clarke-type oxygen electrode and a tetraphenyl phosphonium (TPP+)-selective electrode, respectively, (Hansatech Oxygraph System, Norfolk, England) exactly as described previously [21]. Briefly, mitochondria (250 μg for worms, and 500 μg for mouse liver) were resuspended in 0.5 ml Experimental Buffer (125 mM KCl, 10 mM Tris- MOPS, 10 μM EGTA, 1 mM potassium phosphate, pH 7.4) and both oxygen and [TPP+] changes recorded immediately at 25°C, with stirring (60 rpm). All substrates and inhibitors were purchased from Sigma and prepared as described [21]. Final working concentrations were – Malate (2.5 mM)/Glutamate (5 mM); Succinate (5 mM); ADP (100μM); TMPD (300 μM)/Ascorbat (6mM); Rotenone (2 μM); Oligomycin (0.32 nM), Antimycin A(0.25 μg/ml) FCCP (100 nM).
Polysomal Profiling
Polysomal fractions, containing ribosomes from cytosolic and endoplasmic reticulum-bound origin, were isolated from whole-worm samples using the Balch Homogenizer, in conjunction with sucrose density gradient centrifugation. Briefly, 78,000 two day-old gravid adult worms, or 6 million L1 larvae, were collected from BNGM agar plates and washed using cold S-Basal supplemented with 100 μg/ml cycloheximide. To each packed worm pellet (∼500 (μl) an equal volume of 2X Lysis Buffer (80 mM Tris-HCl (pH 8.0), 470 mM KCl, 16 mM MgCl2, 1.6 mM EGTA (pH 8.0), 0.32 mg/ml heparin, 0.32 mg/ml cycloheximide, 3.9 mM PMSF, 640 U/ml RNasin, 1.6% Triton X-100, 0.16% Na Sodium deoxycholate) was added and samples immediately homogenized on ice (16 μM clearance, 25 passes, for adults; 6 μM clearance, 25 passes for L1 larvae). Cuticular fragments were cleared by centrifugation (13,200 rpm, 18 mins, 4°C). In the case of adult worms, a second centrifugation was necessary to remove residues of intestinal fat (13,200 rpm, 10mins 4°C). 20-30 OD260nm units of homogenate, in a final volume of 750 μl 1X Lysis Buffer, was loaded on top of a chilled 7% - 47% continuous sucrose gradient, then tubes were centrifuged in a Beckman SW-41 Ti rotor (38,000 rpm, 2 hours, 4°C). Gradients were prepared beforehand using 5.5 ml each of 7% and 47% sucrose solutions (RT, 3 hours), according to the method of Abe & Davies [22]. At the end of gradient centrifugation 24 × 500 μl fractions were collected from the top of the gradient and immediately placed into 250 ul phenol/chloroform/isoamyl alcohol (25:24:1) containing 20 μl of 1% SDS. Following re-extraction with chloroform (200 μl), total RNA from each fraction was isolated by ethanol precipitation, then analyzed by gel electrophoresis.
Results
Construction of the Balch Homogenizer is shown in Figure 1A. The instrument consists of a hollow metal chamber into which a ball bearing of defined diameter is inserted. Screw-held caps physically limit lateral movement of the bearing. Worms are disrupted by the shearing generated when they are sequentially forced around the ball bearing by two syringes. During homogenization, fluid entering the chamber is dispersed evenly over the surface of the bearing. This process ensures formation of a gap equal to one-half the difference in diameter between the chamber wall and the ball bearing. In the present study, we used ball bearings ranging in size from 7.999 mm down to 7.982 mm - generating clearances ranging from 1 to 18 μm. By varying the clearance inside the chamber we controllably fragmented or homogenized C. elegans of all stages, including dauer larvae (Table I and Figure 2). We found both re-useable, metal-reinforced glass syringes or disposable plastic syringes worked equally well for homogenization. Caution should of course be exercised when using either type of syringe since plunger resistance is inversely proportional to ball-bearing clearance, and directly proportional to worm density. During fractionations, we routinely maintained the Balch Homogenizer in contact with a large, ice-cooled metal plate in order to minimize temperature fluctuations (Figure 1B). In Table I we have provided a comprehensive analysis of the number of syringe passes required to disrupt each developmental stage of C. elegans with respect to various ball-bearing clearances. By selecting the appropriate combination of ball bearing and pass number, one can choose to fracture or homogenize worm samples of any stage.
Table I.
Relationship Between Number of Syringe Passes, Ball Bearing Clearance, Developmental Stage and Sample Integrity.
| Developmental Stage | Egg | L1 | L2 | L3 | |||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
| |||||||||||||||||||||
| Pass Number‡ | 5 | 10 | 15 | 20 | 25 | 5 | 10 | 15 | 20 | 25 | 5 | 10 | 15 | 20 | 25 | 5 | 10 | 15 | 20 | 25 | |
| Clearance (μm) | 2 | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * |
| 4 | * | * | * | * | * | H | * | * | * | * | * | * | * | * | * | * | * | * | * | * | |
| 6 | * | * | * | * | * | F | F | F/H | H | * | N/F | N/F | N/F | N/F | N/F | * | * | * | * | * | |
| 8 | * | * | * | * | * | * | * | * | * | * | N/F | F | H/F | H | H | F/H | F/H | ||||
| 10 | H | * | * | * | * | * | * | * | * | * | F/H | H | H | * | * | F | F/H | F/H | F/H | H | |
| 12 | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | N/F | F | F/H | F/H | F/H | |
| 14 | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | |
| 16 | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | |
| 18 | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | |
| Developmental Stage | L4 | Young Adult | Gravid Adult | daf-2 dauers‡ | |||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
| |||||||||||||||||||||
| Pass Number | 5 | 10 | 15 | 20 | 25 | 5 | 10 | 15 | 20 | 25 | 5 | 10 | 15 | 20 | 25 | 5 | 10 | 15 | 20 | 25 | |
| Clearance (μm) | 2 | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | F | F | F | F | H/F |
| 4 | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | |
| 6 | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | |
| 8 | N/F | F | F | H | H | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | |
| 10 | N/F | N/F | F | F/H | F/H | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | |
| 12 | F | F | F | F | F | F | F/H | F/H | F/H | F/H | * | * | * | * | * | * | * | * | * | * | |
| 14 | * | * | * | * | * | * | * | * | * | * | F/H | F/H | F/H | F/H | H | * | * | * | * | * | |
| 16 | * | * | * | * | * | N/F | N/F | F | F | F/H | F/H | F/N | F/H | F/H | F/H | * | * | * | * | * | |
| 18 | * | * | * | * | * | N/F | F | F | F | F | F | F | F/H | F | F/N | * | * | * | * | * | |
Key: H – ‘Homogenized’, speckles and small worm fragments present; F – ‘Fractured’, worms are cut in halves or thirds; N – ‘Not Effective’, worms are intact (motile or non-motile) and/or worms are in the process of fragmentation (cut in half but still held together);
Condition not tested.
We also tested a ball bearing with a 1 μm clearance but were unable to even move buffer through the chamber effectively. Clearances ≥ 4 μm were ineffective at fracturing dauers.
Figure 2. Fragmentation of staged, wild type C. elegans using the Balch homogenizer.
Left panels: before homogenization; right panels, after homogenization. (A, B) L2 larvae – 10 μm clearance, 25 passes. (C, D) L4 larvae – 14 μm clearance, 25 passes. (E, F) Gravid adults - 18 μm clearance, 25 passes. (G, H) daf-2(e1370) dauer larvae - 2 μm clearance, 25 passes. In all cases >90% of animals were disrupted. Scale bar: 500 μm.
Many biochemical procedures require isolation of large amounts of intact macromolecules in their native state. We tested the ability of the Balch Homogenizer to leave soluble proteins in their functional state. We also compared the performance of the Balch Homogenizer against three other protein extraction procedures – Boiling, Dounce Homogenization and Sonication (refer to Materials & Methods). We observed that the amount of protein retained in the soluble state after homogenization with the Balch Homogenizer was comparable to, or exceeded, that obtained using either Sonication or the Dounce Homogenizer (Fig. 3A). Boiling drastically reduced the yield of soluble protein. When we checked protein integrity, in this case by western analysis against the E1α subunit of mitochondrial pyruvate dehydrogenase, we observed that almost all protein remained intact following Balch Homogenization and Sonication. Dounce Homogenization resulted in marked E1 α degradation (Figure 3B-D). Activity measurements of the mitochondrial enzyme dihydrolipoamide dehydrogenase (DLD), as well as of the peroxisomal enzyme catalase, revealed similar findings – that the Balch Homogenizer either equaled the best performing alternate method (Sonication), or performed only slightly less better (Table II). The ability of the Balch Homogenizer to extract soluble protein from worm samples was not limited to adult animals. Even larger amounts of protein could be obtained from L1 larval samples that were volume matched against adult worm samples (compare rows 1 and 3 of Table III).
Figure 3. Quality of C. elegans protein after various homogenization techniques.
Adult worms (wild type) were separated into four equal volumes then total protein extracted using one of four techniques – Balch homogenization, boiling, Dounce homogenization or sonication. (A) Extracted protein was separated into supernatant (S) and pellet (P) fractions as described under Materials and Methods, then protein concentration in each fraction determined. (B) Equal quantities of protein (25μg) were analyzed by 10% SDS-PAGE. Simply Blue Stain (Invitrogen) was used to assess relative protein abundance. (C, D) Western analysis was used to assess relative protein stability during each of the four extraction techniques. The E1α subunit of pyruvate dehydrogenase (PDH) was used as a marker protein (arrowhead). (D) is a longer exposure of (C).
Table II. Enzymatic Activities - Measure of Protein Extract Integrity.
| Protein Extraction Method | DLD Activity (Units/mg extract) | Catalase Activity (k/mg extract)* |
|---|---|---|
|
| ||
| Balch | 0.759 | 1.59 |
| Dounce | 0.620 | 1.26 |
| Sonication | 0.976 | 1.59 |
| Boiling | 0.068 | 8.37** |
| No Homogenization | - | - |
The first order rate constant, k (sec−1), for catalysis of H2O2 is used as a direct measure of catalase concentration.
This value is unexpectedly high. The same protein extract samples were utilized for both enzymatic assays.
Table III. Comparison of Protein Extraction Efficiency from L1 and Adult Animals.
| Protein Extraction Method | Stage (n††) | Soluble Protein (μg) | Pellet Protein (5% SDS Soluble) (μg) | Net Yield (μg) |
|---|---|---|---|---|
|
| ||||
| Balch* | Adult (11,000) | 595 | 500 | 1095 |
| Sonication** | Adult (11,000) | 420 | 770 | 1190 |
|
| ||||
| Balch† | L1 (1 million) | 900 | 265 | 1165 # |
| Sonication** | L1 (1 million) | 1700 | 120 | 1820 |
Balch Homogenizer: 25 passes, 16 μM clearance,
VirSonic 60 with microtip probe (4Watt power, 3 minutes, on ice),
Balch Homogenizer: 14 passes, 4 μM clearance,
n = number of worms. The packed volume of worms was 50 μl for each sample.
Higher yields are expected if a larger number of passes are employed.
Sonication cannot be used to extract nucleic acids without significant structural damage. Isolation of mRNA from tissue extracts is often undertaken following the direct addition of phenol-based mixtures such as Trizol Reagent, or chaotropes such as sodium iodide or guanididium isothiocyanate. The metal construction of the Balch Homogenizer is not compatible with acidic or corrosive chemicals. We therefore tested the quality of mRNA prepared using Trizol and guanididium isothiocyanate added after disruption of worms by the Balch Homogenizer. Figure 4 reveals that even in the absence of either chemical during the homogenization procedure, it is possible to isolate high quality mRNA from worm samples. Our homogenization buffer lacked RNase inhibitors, illustrating that the rapid tissue disruption caused by the Balch Homogenizer, in conjunction with ice-cold temperatures, is sufficient to isolate quality RNA. The functionality of our purified mRNA was confirmed using RT-PCR, against multiple genetic loci (refer to Materials & Methods for details, data not shown).
Figure 4. RNA integrity following Balch Homogenization.
Two methods were employed to extract RNA from C. elegans following sample disruption with the Balch Homogenizer. (A) Guanididium isothiocyanate method employing a Nucleospin RNA Isolation Kit. (B) Trizol (phenol-based) method. In both instances, worm samples were homogenized before molecular disruptants were added. No obvious compromise in RNA integrity was observed in either method. Listed in each panel are - DNA marker sizes (base pairs, bp), and RNA species (28S, 18S, and 5S rRNA, transfer RNA).
We next tested the ability of the Balch Homogenizer to isolate intact genomic DNA (gDNA) from C. elegans. We were particularly interested in developing a phenol-free procedure for future measurement of 8-oxo-deoxyguanosine [23]. One peculiar finding that we have consistently observed with C. elegans is that gDNA prepared from whole-worm lysates in the absence of phenol contains an unidentified inhibitory molecule that prevents endonuclease restriction and efficient PCR amplification. We therefore spent significant effort establishing a procedure using the Balch Homogenizer, a proprietary DNA purification kit, and nuclei isolated from freeze-fractured eggs (Materials & Methods) to isolate high-quality gDNA that functioned even in sensitive qPCR assays (Figure 5). gDNA isolated using this procedure contained ∼1% mtDNA, ∼0.3% E. coli genomic DNA, and was comprised of fragments in excess of 25 kbp (Figure 5E & F). We found that use of freeze-fractured nuclei greatly enhanced the purity of gDNA over that obtained when using fresh, whole eggs (Figure 5D). This phenomenon is most likely due to enhanced permeability of the nuclei to the non-ionic detergents used in the extraction procedure (see Figure A-C).
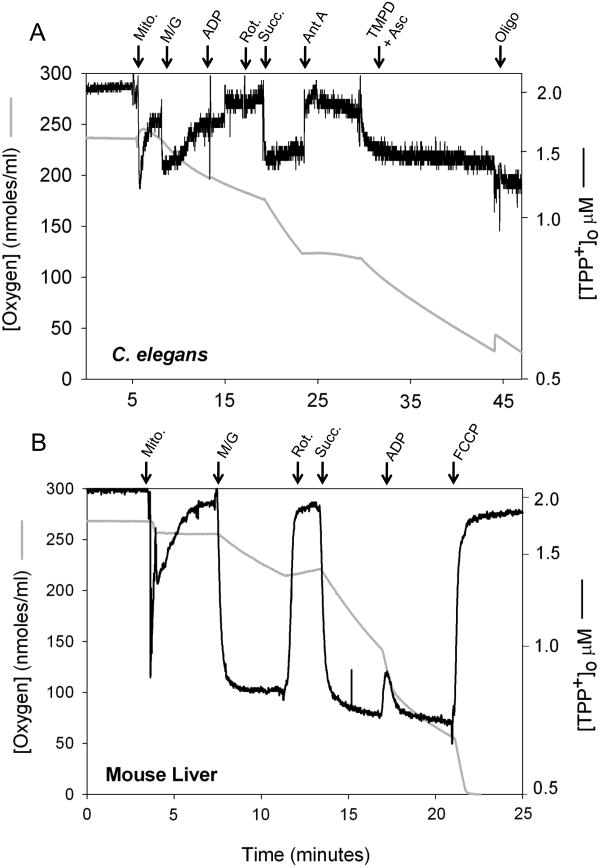
Next, we sought to determine if the Balch Homogenizer could be used as the starting point for isolation of sub-cellular organelles from C. elegans. As a proof of principle we performed a crude assay showing coupled mitochondria can be isolated from adult worms with as few as three strokes of the Balch Homogenizer (Materials & Methods). Mitochondrial isolation buffers differ depending on the type of tissue from which mitochondria are being isolated [21]. In the study shown in Figure 6A we fractionated worms in the presence of an isolation buffer typically employed for collection of mouse liver mitochondria (Figure 6B). Even in this sub-optimal buffer it is apparent that we have isolated coupled mitochondria. Oxygen consumption and reduction of TPP+ concentration following addition of TMPD plus ascorbate illustrates cytochrome c is present and that the outer mitochondrial membrane is intact. Interestingly, complex I activity in the mitochondria isolated from worms is quickly lost under these assay conditions. It is probable that a protease is responsible for this effect, because only EGTA was present in our sample buffer.
Figure 6. Isolation of coupled mitochondria from C. elegans using the Balch Homogenizer.
(A) Adult, wild type worms were gently sheared using the Balch homogenizer (18 μm clearance, 3 passes) and then a mitochondria-enriched fraction isolated. Mitochondrial bioenergetics were assessed using a Hansatech oxygen electrode in conjunction with a tetraphenylphosphonium (TPP+)-selective electrode. (B) Functional mitochondria were also isolated from mouse liver following established procedures (Frezza et al [21]). Data in both panels have been normalized to 500 μg protein (1μg/μl). Arrows mark points of substrate or inhibitor addition: mitochondria (Mito.); malate/glutamate (M/G); adenosine diphosphate (ADP); rotenone (Rot.); antimycin A (Ant A); tetramethyl phenylene diamine in conjunction with ascorbate (TMPD + Asc); oligomycin (Oligo) and p-trifluoromethoxy carbonyl cyanide phenyl hydrozone (FCCP).
Finally, we tested whether the Balch Homogenizer could be used to isolate polysome-bound mRNA from crude adult or L1 larval extracts (Materials & Methods). Cycloheximide inhibits the translocation step of ribosome elongation [24]. Ribosomes effectively become clamped to their host mRNA. This effect is restricted to eucaryotic ribosomes [25]. When whole-worm extracts were prepared using the Balch Homogenizer in the presence of cycloheximide, then separated on a continuous sucrose density gradient [26; 27], intact polysomes could be easily isolated from both stages (Figure 7, and data not shown).
Figure 7.
Polysomal Profiling using the Balch Homogenizer. A whole-cell extract from adult worms was prepared using the Balch Homogenizer (16 μm clearance, 25 passes). Samples were fractionated into polysomes using continuous density gradient centrifugation (7% - 47% sucrose). Total RNA was extracted from each fraction then separated on a 1% agarose gel. Fraction number is shown under lanes (labeled from least to most dense). Identity of various RNA species is marked on right; corresponding ribosomal species are marked at the top of each panel [34]. Size marker (M): HindIII-restricted λ DNA.
Discussion
Although C. elegans has proven to be an exceptionally powerful tool for genetic analyses, its use in biochemical studies has been hindered by the inability to efficiently and rapidly break its resilient outer cuticle in a gentle, temperature-controlled manner. Several techniques have been described to ‘crack’ worms, but each suffers from one or more limitations. Pressure Cycling Technology (PCT) uses expensive equipment to expose samples to multiple rounds of combined freezing, mechanical shearing and elevated hydrostatic pressure (40 cycles 35,000 psi) [6; 28]. Complete disruption of all worm stages, including dauers, can be accomplished using this procedure but it requires mixing of animals with a SiC abrasive [29]. High pressure alone disrupted fewer than 15% of worms [6].
Another way to homogenize worms is by the ‘bead beating method’ [30]. In this procedure microscopic glass beads are used to mechanically disrupt the cuticle. While cheap and fast, a drawback of this method is that protein recovery is variable. Also, proteins are prone to denaturation as samples heat up, leading to protein aggregation and gradual precipitation of normally-soluble proteins [6]. A variation of this procedure is employed in the Precellys (Bertin) range of instruments [31; 32]. Worm samples are homogenized by a single ball bearing vortexed in a figure eight motion. Cooling is possible in this system, but requires additional equipment, including a source of liquid nitrogen or dry ice. Sample volumes are also restricted in this system.
Grinding worms after flash cooling is yet another technique to crack C. elegans [33]. In this procedure worm samples are snap frozen with liquid nitrogen and then ground to a powder with a mortar and pestle. Although, cheap, rapid and efficient, small sample volumes are impractical to grind and are potentially easily lost. Sonication and Dounce Homogenization have also been used to break C. elegans. Sonication of small volumes on ice is an effective method to isolate functional protein extracts, as shown in this study, but this technique cannot be used to isolate intact nucleic acids. Great care also has to be taken to avoid over-heating samples. Dounce homogenization led to unacceptable protein instability in our hands.
Here we have shown that the Balch Homogenizer can be used to disrupt C. elegans of any stage, including dauers. The instrument is gentle enough to allow recovery of functional proteins, RT-PCR quality mRNA, contaminant-free gDNA, as well as coupled mitochondria and intact polysomes. Worms can be fragmented or homogenized, simply by altering the number of syringe passes and ball bearing clearance inside the chamber. It is re-useable, safe, reproducible and sample size is only limited by the syringe volume. Relative to other pieces of laboratory equipment, the Balch Homogenizer is affordable (US $2,000). Novel application of the Balch Homogenizer to C. elegans research now opens the way for a number of studies that in the past have been difficult, if not impossible, to perform. This simple tool is set to become a standard piece of equipment in every worm laboratory.
Acknowledgments
We thank Yvonne Penrod, Bill Swan, and Adwitiya Kar for technical assistance; and Dr. Shivani Maffi for help with the confocal image. We also thank Drs. Vivian MacKay and Brian Kennedy (University of Washington, Seattle) for providing training and the methodology we used as the basis of our polysomal profiling protocol. Funding was provided by NIA (R21-AG025207-01A1), and the Ellison Medical Foundation (AG-NS-0519-08).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Wood WB, editor. The Nematode Caenorhabditis elegans. Cold Spring Harbor Laboratory; New York: 1988. [Google Scholar]
- 2.Knight CG, Patel MN, Azevedo RB, Leroi AM. A novel mode of ecdysozoan growth in Caenorhabditis elegans. Evol Dev. 2002;4:16–27. doi: 10.1046/j.1525-142x.2002.01058.x. [DOI] [PubMed] [Google Scholar]
- 3.Flemming AJ, Shen ZZ, Cunha A, Emmons SW, Leroi AM. Somatic polyploidization and cellular proliferation drive body size evolution in nematodes. Proc Natl Acad Sci U S A. 2000:97. 5285–90. doi: 10.1073/pnas.97.10.5285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Page AP, Johnstone IL. The cuticle. WormBook. 2007:1–15. doi: 10.1895/wormbook.1.138.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Henderson ST, Bonafe M, Johnson TE. daf-16 protects the nematode Caenorhabditis elegans during food deprivation. J Gerontol A Biol Sci Med Sci. 2006;61:444–60. doi: 10.1093/gerona/61.5.444. [DOI] [PubMed] [Google Scholar]
- 6.Smejkal GB, Anderson J, Thomas WK, Robinson MH, Kwan AT, Carlson G, Gross V, Schumacher RT, Lazarev AV. Pressure Cycling Technology (PCT) mediated sample preparation schemes for isolating proteins from the nematode Caenorhabditis elegans; 2008 HUPO World Congress (Aug 16-20); Amsterdam. 2008. [Google Scholar]
- 7.Szewczyk NJ, Mancinelli RL, McLamb W, Reed D, Blumberg BS, Conley CA. Caenorhabditis elegans survives atmospheric breakup of STS-107, space shuttle Columbia. Astrobiology. 2005;5:690–705. doi: 10.1089/ast.2005.5.690. [DOI] [PubMed] [Google Scholar]
- 8.Balch WE, Rothman JE. Characterization of protein transport between successive compartments of the Golgi apparatus: Asymmetric properties of donor and acceptor activities in a cell-free system. Archives of Biochemistry and Biophysics. 1985;240:413–425. doi: 10.1016/0003-9861(85)90046-3. [DOI] [PubMed] [Google Scholar]
- 9.Rea S, Martin LB, McIntosh S, Macaulay SL, Ramsdale T, Baldini G, James DE. Syndet, an adipocyte target SNARE involved in the insulin-induced translocation of GLUT4 to the cell surface. J Biol Chem. 1998;273:18784–92. doi: 10.1074/jbc.273.30.18784. [DOI] [PubMed] [Google Scholar]
- 10.Widberg CH, Bryant NJ, Girotti M, Rea S, James DE. Tomosyn interacts with the t-SNAREs syntaxin4 and SNAP23 and plays a role in insulin-stimulated GLUT4 translocation. J Biol Chem. 2003;278:35093–101. doi: 10.1074/jbc.M304261200. [DOI] [PubMed] [Google Scholar]
- 11.Clark SF, Molero JC, James DE. Release of insulin receptor substrate proteins from an intracellular complex coincides with the development of insulin resistance. J Biol Chem. 2000;275:3819–26. doi: 10.1074/jbc.275.6.3819. [DOI] [PubMed] [Google Scholar]
- 12.Hill MM, Clark SF, Tucker DF, Birnbaum MJ, James DE, Macaulay SL. A role for protein kinase Bbeta/Akt2 in insulin-stimulated GLUT4 translocation in adipocytes. Mol Cell Biol. 1999;19:7771–81. doi: 10.1128/mcb.19.11.7771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Barysch SV, Jahn R, Rizzoli SO. A fluorescence-based in vitro assay for investigating early endosome dynamics. Nat Protocols. 2010;5:1127–1137. doi: 10.1038/nprot.2010.84. [DOI] [PubMed] [Google Scholar]
- 14.Houthoofd K, Braeckman BP, Lenaerts I, Brys K, Matthijssens F, Vreese AD, Eygen SV, Vanfleteren JR. DAF-2 pathway mutations and food restriction in aging Caenorhabditis elegans differentially affect metabolism. Neurobiol Aging. 2005;26:689–96. doi: 10.1016/j.neurobiolaging.2004.06.011. [DOI] [PubMed] [Google Scholar]
- 15.Aebi H. Catalase in vitro. Methods Enzymol. 1984;105:121–6. doi: 10.1016/s0076-6879(84)05016-3. [DOI] [PubMed] [Google Scholar]
- 16.Patel MS, Vettakkorumakankav NN, Liu TC. Dihydrolipoamide dehydrogenase: v activity assays. Methods Enzymol. 1995;252:186–95. doi: 10.1016/0076-6879(95)52022-8. [DOI] [PubMed] [Google Scholar]
- 17.Argyrou A, Sun G, Palfey BA, Blanchard JS. Catalysis of Diaphorase Reactions by Mycobacterium tuberculosis Lipoamide Dehydrogenase Occurs at the EH4 Level. Biochemistry. 2003;42:2218–2228. doi: 10.1021/bi020654f. [DOI] [PubMed] [Google Scholar]
- 18.Foll RL, Pleyers A, Lewandovski GJ, Wermter C, Hegemann V, Paul RJ. Anaerobiosis in the nematode Caenorhabditis elegans. Comp Biochem Physiol B Biochem Mol Biol. 1999;124:269–80. doi: 10.1016/s0305-0491(99)00130-3. [DOI] [PubMed] [Google Scholar]
- 19.Hoogewijs D, Houthoofd K, Matthijssens F, Vandesompele J, Vanfleteren JR. Selection and validation of a set of reliable reference genes for quantitative sod gene expression analysis in C. elegans. BMC Mol Biol. 2008;9:9. doi: 10.1186/1471-2199-9-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rea SL, Ventura N, Johnson TE. Relationship Between Mitochondrial Electron Transport Chain Dysfunction, Development, and Life Extension in Caenorhabditis elegans. PLoS Biol. 2007;5:e259. doi: 10.1371/journal.pbio.0050259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Frezza C, Cipolat S, Scorrano L. Organelle isolation: functional mitochondria from mouse liver, muscle and cultured fibroblasts. Nat Protoc. 2007;2:287–95. doi: 10.1038/nprot.2006.478. [DOI] [PubMed] [Google Scholar]
- 22.Abe S, Davies E. Memoirs of the College of Agriculture. Vol. 31. Ehime University; 1986. Quantitative analysis of polysomes using a baseline from uncentrifuged blank gradients; pp. 187–199. [Google Scholar]
- 23.Helbock HJ, Beckman KB, Shigenaga MK, Walter PB, Woodall AA, Yeo HC, Ames BN. DNA oxidation matters: the HPLC-electrochemical detection assay of 8-oxo- deoxyguanosine and 8-oxo-guanine. Proc Natl Acad Sci U S A. 1998;95:288–93. doi: 10.1073/pnas.95.1.288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Felicetti L, Colombo B, Baglioni C. Inhibition of protein synthesis in reticulocytes by antibiotics. II. The site of action of cycloheximide, streptovitacin A and pactamycin. Biochim Biophys Acta. 1966;119:120–9. [PubMed] [Google Scholar]
- 25.Garden GA, Canady KS, Lurie DI, Bothwell M, Rubel EW. A biphasic change in ribosomal conformation during transneuronal degeneration is altered by inhibition of mitochondrial, but not cytoplasmic protein synthesis. J Neurosci. 1994;14:1994–2008. doi: 10.1523/JNEUROSCI.14-04-01994.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sparkuhl J, Gare RL, Setterfield G. Metabolism of Free and Membrane-bound Ribosomes during Aging of Jerusalem Artichoke Tuber Slices. Planta (Berl) 1976;129:97–104. doi: 10.1007/BF00390014. [DOI] [PubMed] [Google Scholar]
- 27.Zid BM, Rogers AN, Katewa SD, Vargas MA, Kolipinski MC, Lu TA, Benzer S, Kapahi P. 4E-BP extends lifespan upon dietary restriction by enhancing mitochondrial activity in Drosophila. Cell. 2009;139:149–60. doi: 10.1016/j.cell.2009.07.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ringham H, Bell RL, Smejkal GB, Behnke J, Witzmann FA. Application of pressure cycling technology to tissue sample preparation for 2-DE. Electrophoresis. 2007;28:1022–4. doi: 10.1002/elps.200600434. [DOI] [PubMed] [Google Scholar]
- 29.I. Pressure BioSciences. Complete Disruption of Caenorhabditis elegans Under Non- denaturing Conditions Using Pressure Cycling Technology (PCT) BioTechniques Protocol Guide. 2009;2009:53. [Google Scholar]
- 30.Grad LI, Sayles LC, Lemire BD. Isolation and functional analysis of mitochondria from the nematode Caenorhabditis elegans. Methods Mol Biol. 2007;372:51–66. doi: 10.1007/978-1-59745-365-3_4. [DOI] [PubMed] [Google Scholar]
- 31.Tazir Y, Steisslinger V, Soblik H, Younis AE, Beckmann S, Grevelding CG, Steen H, Brattig NW, Erttmann KD. Molecular and functional characterisation of the heat shock protein 10 of Strongyloides ratti. Mol Biochem Parasitol. 2009;168:149–57. doi: 10.1016/j.molbiopara.2009.07.007. [DOI] [PubMed] [Google Scholar]
- 32.Hoerauf A, Specht S, Buttner M, Pfarr K, Mand S, Fimmers R, Marfo-Debrekyei Y, Konadu P, Debrah AY, Bandi C, Brattig N, Albers A, Larbi J, Batsa L, Taylor MJ, Adjei O, Buttner DW. Wolbachia endobacteria depletion by doxycycline as antifilarial therapy has macrofilaricidal activity in onchocerciasis: a randomized placebo-controlled study. Med Microbiol Immunol. 2008;197:295–311. doi: 10.1007/s00430-007-0062-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Johnstone IL. Molecular Biology. In: Hope IA, editor. C elegans: A Practical Approach. Oxford University Press; New York: 1999. pp. 201–225. [Google Scholar]
- 34.Ding XC, Grosshans H. Repression of C. elegans microRNA targets at the initiation level of translation requires GW182 proteins. EMBO J. 2009;28:213–22. doi: 10.1038/emboj.2008.275. [DOI] [PMC free article] [PubMed] [Google Scholar]