Abstract
Using a diverse collection of small molecules we recently found that compound sets from different sources (commercial; academic; natural) have different protein-binding behaviors, and these behaviors correlate with trends in stereochemical complexity for these compound sets. These results lend insight into structural features that synthetic chemists might target when synthesizing screening collections for biological discovery. We report extensive characterization of structural properties and diversity of biological performance for these compounds and expand comparative analyses to include physicochemical properties and three-dimensional shapes of predicted conformers. The results highlight additional similarities and differences between the sets, but also the dependence of such comparisons on the choice of molecular descriptors. Using a protein-binding dataset, we introduce an information-theoretic measure to assess diversity of performance with a constraint on specificity. Rather than relying on finding individual active compounds, this measure allows rational judgment of compound subsets as groups. We also apply this measure to publicly available data from ChemBank for the same compound sets across a diverse group of functional assays. We find that performance diversity of compound sets is relatively stable across a range of property values as judged by this measure, both in protein-binding studies and functional assays. Because building screening collections with improved performance depends on efficient use of synthetic organic chemistry resources, these studies illustrate an important quantitative framework to help prioritize choices made in building such collections.
Keywords: chemical diversity, cheminformatics, natural products, small-molecule probes, small-molecule profiling
A central theme in applying cheminformatics to discovery chemistry is to relate synthetic decisions to consequences on both chemical structure and biological assay performance. Historically, such efforts focused on small sets of similar compounds, and single performance measurements (1–3), providing guidance to chemists in compound optimization against single-target proteins or processes (4). However, additional methods are needed to judge large sets of compounds, such as those used in small-molecule screening. Progress toward more valuable screening collections (5) requires unbiased methods to evaluate diversity of assay performance for compound sets rather than performance of individual members.
A widely used method to judge compounds for drug discovery is the “rule of 5” (RO5) (6), which predicts poor absorption or permeation for compounds that deviate from property-value constraints: H-bond donors (Hd) and acceptors (Ha), molecular weight (MW), and calculated partition coefficients (cLogP). Recent studies have attempted to refine such rules (7–9) and extend them to other goals (10–13), such as making leads or probes. Such property filters have been debated and reviewed (14–16), and their long-term impact on pharmaceutical research is starting to be analyzed (17, 18). Importantly, exceptions to these rules, including natural products (19–21), are well-noted and suggest that previously undescribed types of chemistry might access property distributions acceptable for certain goals despite nonadherence to established rules.
Comparative analyses of compound sets usually use computed properties (19, 22, 23) or historical assay results (24, 25). Significant progress has been made quantifying and visualizing properties of compound sets (26), including methods that relate structure to intuitive notions of shape (27–29), and similarity fusion methods (30–33) that describe relationships between sets. Moreover, chemical similarity and diversity analyses continue to progress (34–37), including studies using Shannon entropy (38) as a measure of structure information among compounds (39–41), addressing reagent selection (42), database similarity searches (43), and scaffold diversity (44). Entropy-based methods have also been used on assay data to distinguish single-target compounds from those with multitarget effects (45), and to quantify relationships between targets based on Ki profiles among sets of common inhibitors (46).
Despite advances in cheminformatics, methods to measure assay performance of compound collections remain underexplored. One important study focused on compounds from different sources, including drugs (19). Other studies focused on molecular complexity, suggesting intermediate complexity is preferable for drug leads (17, 47). Recently, we investigated relationships between intermediate stereochemical complexity and binding specificity (48). What these previous studies did not address is set-based behavior of compound collections. In screening collections, the value of chemistry investment needs to be measured in terms of overall collection performance, rather than anecdotes about the best performers. An assumption often made is that diverse structures will result in diverse outcomes across many assays, but few studies address this question directly (49–53). Likewise, our recent analysis (48) did not account for the distinction between individual and groupwise compound performance. The availability of data from large-scale experiments (48) and public databases provides an opportunity to measure set-based performance quantitatively, rather than measuring success by finding a few “special” compounds.
We recently analyzed a large compound collection in 100 parallel protein-binding assays (48) and found both protein-binding frequencies and specificities are increased among compounds having intermediate stereochemical complexity. Here, we extend analysis of the same compounds to physicochemical properties (6, 7, 16, 54) and shape-based descriptors (27, 29), revealing additional similarities and differences between sets. We use principal component analysis (PCA) (55) of chemically intuitive properties to analyze sources of variation among the sets. Different descriptors report different aspects of chemical structure (36), and we aim to illustrate for synthetic chemists how quantitative relationships can be reconciled with chemical intuition. We also provide a framework to evaluate performance diversity of compound sets using Shannon entropy (38) on profiles of assay measurements. In this context, entropy measures how evenly a compound set is distributed over all possible patterns of performance accessible from a given set of measurements. We apply this method to both protein-binding profiles (48) and diverse profiles of functional assays extracted from ChemBank (56). These methods provide a quantitative measure for performance evaluation of small-molecule collections (5, 48) and encourage exploration of emerging relationships between performance diversity and molecular property distributions.
Results
We characterized compounds from three sources that were exposed to 100 parallel protein-binding assays (48). The compound collection consists of (i) 6,152 compounds representative of screening collections (commercial compounds; CC); (ii) 2,477 naturally occurring compounds (natural products; NP); and (iii) 5,963 compounds from the academic synthetic chemistry community (diverse compounds; DC′). These sets provide an opportunity to compare properties and performance of compounds from different origins (cf. ref. 19), including one group (DC′) whose property distributions differ from other compounds, and whose properties and performance have not thoroughly been investigated.
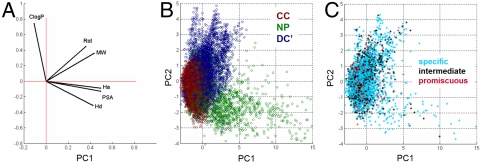
Not surprisingly, when applying established filtering criteria (16) nearly all CC will “pass” RO5 (99.9%) (6, 16) or an alternative based on polar surface area (PSA) and rotatable bonds (Rot) (99.7%) (7, 16). In contrast, up to approximately 1/3 of DC′ (73.0% RO5, 66.5% alternative) and up to approximately 2/5 of NP (71.1% RO5, 60.7% alternative) would “fail.” Because differences between natural products and typical screening compounds are established (16, 19, 21, 57), we sought to compare DC′ with each of CC and NP to determine which set DC′ more resembles for each of six common properties (16). For five of the six properties, DC′ is more similarly distributed to either CC or NP than the latter two are to each other, and for three of these, the values for DC′ are intermediate between those of CC and NP (Table 1). That DC′ is heavier and more lipophilic than CC or NP is evident by inspecting the Table 1 and visualizing MW versus cLogP (Fig. 1A). In a PCA “chemical space” composed of these six properties (Fig. 1B), both NP and DC′ overlap substantially with the more compact CC, but DC′ is more similar to NP along one dimension (Fig. 1C) and more different in another (Fig. 1D). These results show that some members of DC′ access part of the space not accessed by either CC or NP, suggesting properties and performance for DC′ should be evaluated in their own right, rather than being presumed similar to either CC or NP.
Table 1.
Distributions of properties typically used for filtering (e.g., Lipinski RO5)
| Molecular weight (Da) | Calculated logP | Polar surface area (Å2) | |||||||
| Median | Mean | SD | Median | Mean | SD | Median | Mean | SD | |
| CC | 311 | 314 | 75 | 3.2 | 3.2 | 1.6 | 68 | 72 | 31 |
| NP | 386 | 457 | 232 | 1.9 | 1.8 | 2.2 | 104 | 136 | 93 |
| DC′ | 496 | 509 | 157 | 3.9 | 3.9 | 2.3 | 96 | 100 | 41 |
| H-bond donors |
H-bond acceptors |
Rotatable bonds |
|||||||
| Median | Mean | SD | Median | Mean | SD | Median | Mean | SD | |
| CC | 1 | 0.9 | 0.9 | 4 | 3.7 | 1.6 | 4 | 3.8 | 1.9 |
| NP | 3 | 4.1 | 3.6 | 7 | 8.5 | 5.9 | 5 | 6.0 | 4.6 |
| DC′ | 1 | 1.6 | 1.2 | 6 | 6.0 | 2.5 | 8 | 8.9 | 4.3 |
Bold pairs of values for each property indicate more similarly distributed pairs of sets. Also indicated is whether the distribution for DC′ is centered between (underline) or outside (italics) those of CC and NP.
Fig. 1.
DC′ occupies a distinct part of the property space from CC and NP. (A) Scatterplot of MW versus cLogP, omitting three compounds (all from NP) with MW > 1,500 and three compounds (all from DC′) with cLogP > 15. (B) Top three principal components (PCs) (93% of total variance) using six properties. (c) PC1 versus PC3, illustrating dimension in which DC′ is similar to NP. (D) PC2 versus PC3, illustrating dimension in which DC′ is distinct from NP (CC: dark red; NP: dark green; DC′: dark blue).
To refine our chemical intuition about similarities and differences among CC, NP, and DC′, we analyzed PCA coefficients to learn which properties are correlated (Fig. 2A), and how these properties vary in each set (Fig. 2B). Not surprisingly, PSA is correlated with Ha and Hd, and NP has high variation in this direction, consistent with other studies (19, 21). Similarly, Rot is correlated with MW, and these properties, along with cLogP, account for much of the variation in DC′. Previously, we classified these compounds for binding specificity in protein-binding profiles (48). Using similarly defined specificity groups (“specific”: bound 1 protein; “intermediate”: bound 2–5 proteins; “promiscuous”: bound 6+ proteins), we asked whether compounds in different groups are concentrated or distributed in the property space (Fig. 2C). We found that specific and intermediate compounds, regardless of source, are well-distributed throughout the space, whereas promiscuous compounds are significantly concentrated in the center (p < 0.0099). Because the center of the space corresponds to common property filters, this result suggests that our binding experiments accessed a greater number of desirable outcomes (specific binding) than had we restricted ourselves to compounds passing common filters.
Fig. 2.
Specific binders occur throughout the property space. (A) PCA coefficient map of six properties onto first two PCs, showing correlations between properties and interpretation of dimensions; horizontal and vertical scales are relative to unit vectors along the PCs. (B) PC1 versus PC2 showing compound sets (CC: dark red; NP: dark green; DC′: dark blue); scales are unit standard deviations. (C) PC1 versus PC2 showing distributions of protein-binding specificity groups (cf. figure 7 of ref. 48); (specific: 1 protein, cyan; intermediate: 2–5 proteins, black; promiscuous: 6 + proteins, red); scales are the same as in B. Promiscuous compounds are significantly concentrated (p < 0.0099) in the center of the space.
Although the above properties allow rapid application of filters, they tend to oversimplify relationships between compounds and therefore offer only partial guidance to chemists planning syntheses. To illustrate additional possibilities for guidance, we characterized relationships among CC, NP, and DC′, using three chemical spaces that shed light on different aspects of structure variation (Fig. 3). First, we considered atom counts: PCA of this chemical space reveals that carbon, nitrogen, and oxygen are the dominant contributors to variation (Fig. 3A). Notably, NP achieves most of its variation with oxygen and carbon, with little variation in nitrogen composition (Fig. 3B). In contrast, both CC and DC′ get most compositional variation from nitrogen and carbon. Second, to examine compounds in terms of ring and side-chain content, we used descriptors counting rings, side chains, and branches (Fig. 3 C and D). Here, CC varies primarily in number of aromatic rings, as does DC′ (with higher variation in total rings). In contrast, NP has high variation in side-chain number (unconnected fragments after removing ring atoms), but less variation in number of aromatic rings. Third, to extend our previous analysis (48) of electronic character of carbon atoms, we used electrotopological-state (E-state) descriptors for nine carbon environments (Fig. 3E). These descriptors measure variation in electronic environment (54) for these carbon types, rather than simply the number of each type. Comparing variation of position in each set (Fig. 3F) to the coefficient map allows interpretation of the relative importance of varying carbon environments to the diversity of CC, NP, and DC′. For example, more variation in NP than in CC or DC′ corresponds to variation in electronic character of sp3 carbon atoms connected to three or four other heavy atoms (ssssC, sssCH). In contrast, CC and DC′ express more of their variation around less-connected sp3 carbons (sCH3, ssCH2) or sp2 carbons (aaCH, aaaC). Often, descriptors such as E states are used to build predictors of compound performance using statistical learning methods (58, 59). Here, these examples illustrate how quantitative, yet chemically intuitive, information can be used to guide chemists building discovery collections.
Fig. 3.
Different chemical spaces provide intuitive comparisons between collections. (A) PCA coefficient map of select atom counts onto first two PCs, showing interpretation of PCA dimensions. (B) PC1 versus PC2 showing compound sets in the space of A (CC: dark red; NP: dark green; DC′: dark blue). (C) PCA coefficient map of select ring and chain counts onto first two PCs, showing interpretation of PCA dimensions. (D) PC1 versus PC2 showing compound sets in the space of C. (E) PCA coefficient map of E-state sums (54) (reporting electronic environments of different carbon atom types) onto first two PCs, showing interpretation of PCA dimensions (s: single bond; d: double bond; a: aromatic bond). (F) PC1 versus PC2 showing compound sets in the space of E. Scale units for coefficient maps and PCA plots are the same as Fig. 2.
Recently, we used principal moment-of-inertia (PMI) descriptors (27) of 3D shape to analyze chemist decisions during diversity-oriented syntheses, including relationships between skeletal and stereochemical diversity (60, 61) and a unique fragment library (62). PMI descriptors provide an intuitive notion of molecular shape, occupying a triangular map bounded by canonical shapes (rod, disk, and sphere). We mapped the three sets (CC, NP, and DC) and the specificity groups (promiscuous, intermediate, and specific) to PMI plots (Fig. 4A). Consistent with earlier observations, CC is dominated by flat and linear compounds. NP covers more of the PMI map, its distribution centered up and to the right relative to CC. Notably, NP has less coverage in the disk-like (bottom center) region of the map than either CC or DC′. DC′ covers more of the shape space than NP, with its distribution centered down and to the right of CC. DC′ also contains low density on the extreme left diagonal of the space, indicating few very flat or linear compounds. The specificity groups also follow a trend, though with less difference between them than the compound sets.
Fig. 4.
Different compound sets and specificity groups are quantitatively different in shape distributions. (A) PMI maps showing compounds from each set (Top; CC: dark red; NP: dark green; DC′: dark blue) and from each specificity group (specific: 1 protein, cyan; intermediate: 2–5 proteins, black; promiscuous: 6 + proteins, red). Canonical PMI shapes are shown on the bottom-left map. (B) Cumulative distributions of distances from canonical sphere shape using PMI descriptors. (C) Cumulative distributions of distances from canonical flat shape using alpha-shape descriptors (29). Color-coding of distributions is the same as in A.
To quantify shifts in PMI density, we measured distance distributions for each set or group relative to vertices of PMI space. Cumulative distance distributions allow us to quantify the statistical significance of these differences, with all three of CC, NP, and DC′ significantly different relative to the sphere shape (Fig. 4B), and DC′ less rod-like and more disk-like than either CC or NP. Importantly, more specific binders (regardless of set) are significantly more sphere-like than promiscuous ones. To refine this analysis, we also computed distance distributions from canonical flat and spherical shapes based on alpha-shape descriptors (29). The results for sphere likeness exactly match those with PMI distance to the sphere vertex, and for flat shapes significantly discriminate CC, NP, and DC′. Moreover, promiscuous compounds are significantly flatter in their distance distribution (Fig. 4C).
The foregoing analyses treat properties as distributions, but until now we have considered assay performance for each compound separately, for example, assigning each to a specificity group. An important consideration for compound collections is how well they access a diversity of assay outcomes. Given 100 specific compounds (each binds exactly one of 100 proteins), compare the case where each compound binds the same protein to the case where each compound binds a different protein: “Hit” rates and specificity groups are not sufficient. To evaluate compound sets, we need to distinguish these cases. Shannon entropy (38) and related measures (45) provide a framework to do so when applied to matrices of assay outcomes (cf. figure 4 in ref. 48). Entropy applied to profiles (profile entropy) of small-molecule performance allows us to distinguish the possibilities above, providing a higher score for more diverse sets of assay outcomes, a maximal score for uniform coverage of all possible outcomes, and penalties for missing outcomes or “dilution” with inactive compounds. By itself, however, profile entropy does not distinguish profiles based on their selectivity. A profile with a single assay activity is equivalent to one with a single assay inactivity (i.e., a promiscuous compound). To address this concern we define a weighted measure (weighted profile entropy) to reward more specific profiles over less specific ones.
Using profile entropy, we analyzed the performance of CC, NP, and DC′ using profiles of binding to 100 proteins (Fig. 5A) (48). By this measure, CC exhibits higher performance diversity than DC′, and substantially higher than NP, possibly due to lower hit rates in NP (48). However, considering hit compounds only (bound at least one protein) does not change relative scores; NP, and to a lesser extent DC′, “concentrates” its compounds on particular patterns of protein binding (which entropy penalizes). Weighted profile entropies show similar relationships between sets, as expected because most hit compounds bind one (65.3%) or two (13.2%) proteins. As an independent test of performance diversity for the compound sets, we extracted functional assay data from ChemBank corresponding to all 14,592 compounds. Each compound had been exposed to between 51 and 154 different functional assay readouts (median = 71, mean = 85). Using these data, we computed profile entropies for CC, NP, and DC′ (Fig. 5B). The functional assay results exhibit higher overall entropies (a smaller fraction is never called active; 30.4% versus 78.4%), but performance diversity remains highest for CC and lowest for NP, except in weighted profile entropies, where DC′ drops below NP, suggesting that either nonspecific effects or correlations between assay readouts are more common for DC′ than for NP.
Fig. 5.
Shannon entropy measures performance diversity for sets of compounds across many assays. (A) Performance diversity of CC, NP, and DC′ in 100 protein-binding assays, including profile entropy for all compounds (red bars), hits only (black bars), and weighted profile entropy (cyan bars). (B) Performance diversity of CC, NP, and DC′ in ChemBank assay data; color coding is the same as A. (c) Trend lines in relative performance diversity for all compounds in protein-binding assays as a function of increasing ranked cLogP values, including both profile entropy (black line) and weighted profile entropy (cyan line). (D) Trend lines in relative performance diversity for all compounds in ChemBank functional assays as a function of increasing ranked cLogP (solid lines) or MW (dashed lines) values, including both profile entropies (black) and weighted profile entropies (cyan). In C and D, entropy values are normalized by subtraction to the first compound set considered (i.e., lowest values of cLogP or MW).
Finally, we revisited the idea that computed property distributions will impact compound set performance: specifically the ability to produce diverse activities in primary assay formats (binding or functional). We computed profile entropies for subsets of compounds with similar property values. We took sets of profiles of equal size, centered on a compound with the (ranked) property value of interest. We found that increasing cLogP increased performance diversity in protein-binding profiles using both unweighted and weighted profile entropy measures (Fig. 5C). In contrast, though increasing both MW and cLogP produced increased performance diversity in unweighted functional assay profiles, both properties showed stable performance diversity when specificity-weighted profile entropies were considered (Fig. 5D), suggesting increases in the unweighted case may stem from nonspecific effects in assays. Similar observations were observed with several descriptors (see SI Datasets D1–D8); for example, we observed that increasing PSA to extreme values decreased performance diversity in functional assays, consistent with the inability of compounds to penetrate cells. Clearly, other such comparisons are warranted, but these preliminary results illustrate a framework to evaluate set-based performance in terms of calculated property distributions.
Discussion
Quantifying properties and behavior of compound sets has implications for organic synthesis toward small-molecule probes and drugs. In particular, it is valuable to make quantitative decisions about choices: which compounds to buy, which to synthesize, which to include in a screening collection, which to retire from screening. Decisions regarding individual compounds typically dominate such discussions: Individual compounds pass or fail filtering rules, individual compounds hit or not in assays, etc. In this study, we use real-world compounds and data to illustrate methods that can guide decisions about sets of compounds.
Our results reflect the properties a particular set of compounds (48), but the message of this study is not a statement about these particular compounds. Indeed, our current discovery collection at the Broad Institute reflects many considerations (26, 63–67) that were never applied to the compounds in this study. Rather, we aim to promote a way of thinking about compound sets that is unbiased, quantitative, and decision-oriented, not to prescribe which particular decisions should be made. Depending on the goal of any particular study, attention to different properties or performance is warranted. For example, Figs. 1 and 2 remind us that although for certain applications (e.g., drug discovery) additional property constraints might be applied, we should be cautious against overapplication of constraints that might obscure previously undescribed insights into small-molecule behavior—lack of “drug-likeness” does not mean that a compound’s profile of interactions with 100 different proteins is not valuable information.
Compounds are often judged by whether they add “diversity” to a compound collection. Fig. 3 reminds us that diversity is a relative concept, depending on the sets being compared and properties used to describe them. Consistent with earlier studies (60, 61), visualizing molecular shape relative to canonical shapes (27–29) provides a powerful stimulus to chemists interested in “escaping flatland” (68) by making more “globular” compounds. Importantly, we show in Fig. 4 how these relationships can be quantified and their significance assessed, but our results do not support a causative relationship between globularity and specificity. DC′ is more globular (sphere-like) than other sets, and the group of specific binders is more globular than promiscuous compounds. However, this is primarily because DC′ is enriched in specific binders (48), not because globularity “imparts” specificity. Specific members of CC and NP are not statistically enriched for globularity relative to their promiscuous counterparts, suggesting more complicated relationships that warrant investigation with different compounds.
An important advance in this study is extending Shannon entropy (38) to profiles of assay performance for compound sets. Entropy has been used extensively for chemical structure analysis (39–44), but has been underutilized in the area of measuring diversity of assay performance (45, 46). Here, we apply entropy broadly to multiassay performance profiles (51, 53) for large collections and observe that CC accesses 654 unique protein-binding profiles with 1,415 active compounds, NP accesses 112 profiles with 324 compounds, and DC′ accesses 422 profiles with 1,406 compounds. Entropy is sensitive precisely to these relationships—how uniformly are compounds distributed over different patterns of performance? Weighted profile entropy addresses an important concern about specificity among binding profiles, and future studies should reconcile weighting with further analysis of on-target and off-target effects in phenotypic assays and provide improved handling of missing data (45). As suggested by Fig. 5, profile entropy can be sensitive to the size of compound sets and the number of assays. Moreover, our nonparametric analysis of entropy relative to property distributions is likely sensitive to the shapes of these distributions. Additional studies with other datasets are needed to refine the methodology, but the speed of profile entropy calculations, and their generality to any small-molecule profiling study (69), are indicators of future promise.
Overall, this study focuses attention on the need of computational methods to adapt to the changing requirements of high-throughput chemistry and screening. It provides a framework for analysis of compound collections that focuses on overall collection performance rather than performance of individual members (as with more conventional structure-activity studies). We intend this study as a follow-up to our prior work (48), in that it provides informative additional analysis of compounds from different sources. However, we also intend this work to advance methods for analysis that can be applied to previously undescribed compounds and profiling datasets in the future. Such large-scale computational analysis of compound sets in the context of screening data can influence synthetic (or acquisition) decisions leading toward the assembly of improved screening collections.
Materials and Methods
Chemical structures, categories, sources, and ChemBank (56) identifiers for each compound are already published (48), and we used this information as the source of structures and binary protein-binding data. We calculated molecular property counts (SI Datasets D1 and D2) and E-state descriptors using Pipeline Pilot (Accelrys, Inc.). We also considered these descriptors as a function of protein-binding specificity (SI Dataset D3). For 3D descriptors, we used a ChemAxon module employing DREIDING force field (70) in Pipeline Pilot to generate up to 16 3D conformers per molecule, retaining those within 3 kcal/mol of the lowest energy conformer. We computed PMI (27) and alpha-shape (29) descriptors using MATLAB code (The MathWorks, Inc.) that follows published methods. Median PMI values and alpha-shape-based distances were taken across retained conformers. In PCA visualizations, small numbers of outliers are not shown for graphical clarity, but all compounds were included in calculations. Significance of spread or concentration in PCA was performed between distributions of Hotelling’s T2 values, and in PMI and alpha-shape plots between distance distributions (SI Dataset D4), each using Kolmogorov–Smirnov tests (71). Correlations between 2D and PMI descriptors are also provided (SI Dataset D5).
For each compound we constructed a performance profile assigning binary ([0,1], binding data) or discrete ([-1,0,1], functional data) values representing activity for a compound in a given assay, collecting such values across all assays into a vector x. Discretization of binding data are described elsewhere (48), and ChemBank data were handled similarly, except both high- and low-signal outlier values (56) were accepted (SI Datasets D6 and D7). All distinct performance profile vectors x for compounds were collected to set S, and Shannon entropy (H) was computed by calculating relative frequencies p(x) and summing frequency terms over x∈S: H(S) = -Σp(x) log 2[p(x)] (profile entropy). To calculate entropy for a set of profiles weighted by a specificity constraint, we first computed H separately for each subset Sm of profiles sharing the same number m of nonzero profile features, then computed a weighted sum of these entropies with weighting factor wm = exp[- ln(2)m], to give Hw(S) = ΣwmH(Sm) (weighted profile entropy). For ChemBank profiles, missing data were censored to zero before entropy calculations (SI Dataset D8). Statistical analyses, visualizations, and entropy calculations were performed in MATLAB.
Supplementary Material
Acknowledgments.
The authors thank past and current members of the Broad Institute Chemical Biology community, especially Joshua Bittker, Nicole Bodycombe, Julia Lamenzo Fox, Annaliese Franz, Daniel Kahne, Young-kwon Kim, Justin Lamb, Lisa Marcaurelle, Alykhan Shamji, Nicola Tolliday, and Damian Young. This work was supported by National Institute of General Medical Sciences (P50-GM069721), the National Institutes of Health RoadMap (P20-HG003895), and the National Cancer Institute (N01-CO-12400). S.L.S. is an investigator at the Howard Hughes Medical Institute.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1015024108/-/DCSupplemental.
References
- 1.Iwasa J, Fujita T, Hansch C. Substituent constants for aliphatic functions obtained from partition coefficients. J Med Chem. 1965;8:150–153. doi: 10.1021/jm00326a002. [DOI] [PubMed] [Google Scholar]
- 2.Fujita T, Hansch C. Analysis of the structure-activity relationship of the sulfonamide drugs using substituent constants. J Med Chem. 1967;10:991–1000. doi: 10.1021/jm00318a001. [DOI] [PubMed] [Google Scholar]
- 3.Hansch C. A quantitative approach to biochemical structure-activity relationships. Acc Chem Res. 1969;2:232–239. [Google Scholar]
- 4.Clemons PA. Chemical Informatics. In: Schreiber SL, Kapoor TM, Wess G, editors. Chemical Biology: From Small Molecules to Systems Biology and Drug Design. Vol 2. Weinheim Germany: Wiley-VCH; 2007. pp. 723–759. [Google Scholar]
- 5.Drewry DH, Macarron R. Enhancements of screening collections to address areas of unmet medical need: An industry perspective. Curr Opin Chem Biol. 2010;14:289–298. doi: 10.1016/j.cbpa.2010.03.024. [DOI] [PubMed] [Google Scholar]
- 6.Lipinski CA, Lombardo F, Dominy BW, Feeney PJ. Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv Drug Deliv Rev. 2001;46:3–26. doi: 10.1016/s0169-409x(00)00129-0. [DOI] [PubMed] [Google Scholar]
- 7.Veber DF, et al. Molecular properties that influence the oral bioavailability of drug candidates. J Med Chem. 2002;45:2615–2623. doi: 10.1021/jm020017n. [DOI] [PubMed] [Google Scholar]
- 8.Fichert T, Yazdanian M, Proudfoot JR. A structure-permeability study of small drug-like molecules. Bioorg Med Chem Lett. 2003;13:719–722. doi: 10.1016/s0960-894x(02)01035-1. [DOI] [PubMed] [Google Scholar]
- 9.Wenlock MC, Austin RP, Barton P, Davis AM, Leeson PD. A comparison of physiochemical property profiles of development and marketed oral drugs. J Med Chem. 2003;46:1250–1256. doi: 10.1021/jm021053p. [DOI] [PubMed] [Google Scholar]
- 10.Bleicher KH, Bohm HJ, Muller K, Alanine AI. Hit and lead generation: beyond high-throughput screening. Nat Rev Drug Discov. 2003;2:369–378. doi: 10.1038/nrd1086. [DOI] [PubMed] [Google Scholar]
- 11.Wunberg T, et al. Improving the hit-to-lead process: Data-driven assessment of drug-like and lead-like screening hits. Drug Discov Today. 2006;11:175–180. doi: 10.1016/S1359-6446(05)03700-1. [DOI] [PubMed] [Google Scholar]
- 12.Oprea TI, et al. Lead-like, drug-like or “Pub-like”: How different are they? J Comput Aided Mol Des. 2007;21:113–119. doi: 10.1007/s10822-007-9105-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wager TT, Hou X, Verhoest PR, Villalobos A. Moving beyond rules: The development of a central nervous system multiparameter optimization (CNS MPO) approach to enable alignment of druglike properties. ACS Chem Neurosci. 2010;1:435–449. doi: 10.1021/cn100008c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Muegge I. Selection criteria for drug-like compounds. Med Res Rev. 2003;23:302–321. doi: 10.1002/med.10041. [DOI] [PubMed] [Google Scholar]
- 15.Lajiness MS, Vieth M, Erickson J. Molecular properties that influence oral drug-like behavior. Curr Opin Drug Disc. 2004;7:470–477. [PubMed] [Google Scholar]
- 16.Keller TH, Pichota A, Yin Z. A practical view of ‘druggability’. Curr Opin Chem Biol. 2006;10:357–361. doi: 10.1016/j.cbpa.2006.06.014. [DOI] [PubMed] [Google Scholar]
- 17.Selzer P, Roth HJ, Ertl P, Schuffenhauer A. Complex molecules: Do they add value? Curr Opin Chem Biol. 2005;9:310–316. doi: 10.1016/j.cbpa.2005.04.001. [DOI] [PubMed] [Google Scholar]
- 18.Leeson PD, Springthorpe B. The influence of drug-like concepts on decision-making in medicinal chemistry. Nat Rev Drug Discov. 2007;6:881–890. doi: 10.1038/nrd2445. [DOI] [PubMed] [Google Scholar]
- 19.Feher M, Schmidt JM. Property distributions: Differences between drugs, natural products, and molecules from combinatorial chemistry. J Chem Inf Comput Sci. 2003;43:218–227. doi: 10.1021/ci0200467. [DOI] [PubMed] [Google Scholar]
- 20.Clardy J, Walsh C. Lessons from natural molecules. Nature. 2004;432:829–837. doi: 10.1038/nature03194. [DOI] [PubMed] [Google Scholar]
- 21.Ganesan A. The impact of natural products upon modern drug discovery. Curr Opin Chem Biol. 2008;12:306–317. doi: 10.1016/j.cbpa.2008.03.016. [DOI] [PubMed] [Google Scholar]
- 22.Ertl P, Schuffenhauer A. Cheminformatics analysis of natural products: Lessons from nature inspiring the design of new drugs. Prog Drug Res. 2008;66:217, 219–235. doi: 10.1007/978-3-7643-8595-8_4. [DOI] [PubMed] [Google Scholar]
- 23.Singh N, et al. Chemoinformatic analysis of combinatorial libraries, drugs, natural products, and molecular libraries small molecule repository. J Chem Inf Model. 2009;49:1010–1024. doi: 10.1021/ci800426u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schuffenhauer A, Brown N, Selzer P, Ertl P, Jacoby E. Relationships between molecular complexity, biological activity, and structural diversity. J Chem Inf Model. 2006;46:525–535. doi: 10.1021/ci0503558. [DOI] [PubMed] [Google Scholar]
- 25.Muchmore SW, et al. Application of belief theory to similarity data fusion for use in analog searching and lead hopping. J Chem Inf Model. 2008;48:941–948. doi: 10.1021/ci7004498. [DOI] [PubMed] [Google Scholar]
- 26.Akella LB, DeCaprio D. Cheminformatics approaches to analyze diversity in compound screening libraries. Curr Opin Chem Biol. 2010;14:325–330. doi: 10.1016/j.cbpa.2010.03.017. [DOI] [PubMed] [Google Scholar]
- 27.Sauer WH, Schwarz MK. Molecular shape diversity of combinatorial libraries: A prerequisite for broad bioactivity. J Chem Inf Comput Sci. 2003;43:987–1003. doi: 10.1021/ci025599w. [DOI] [PubMed] [Google Scholar]
- 28.Ballester PJ, Richards WG. Ultrafast shape recognition to search compound databases for similar molecular shapes. J Comput Chem. 2007;28:1711–1723. doi: 10.1002/jcc.20681. [DOI] [PubMed] [Google Scholar]
- 29.Wilson JA, Bender A, Kaya T, Clemons PA. Alpha shapes applied to molecular shape characterization exhibit novel properties compared to established shape descriptors. J Chem Inf Model. 2009;49:2231–2241. doi: 10.1021/ci900190z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ginn C, Willett P, Bradshaw J. Combination of molecular similarity measures using data fusion. Perspect Drug Discov. 2000;20:1–16. [Google Scholar]
- 31.Salim N, Holliday J, Willett P. Combination of fingerprint-based similarity coefficients using data fusion. J Chem Inf Comput Sci. 2003;43:435–442. doi: 10.1021/ci025596j. [DOI] [PubMed] [Google Scholar]
- 32.Whittle M, Gillet VJ, Willett P, Loesel J. Analysis of data fusion methods in virtual screening: similarity and group fusion. J Chem Inf Model. 2006;46:2206–2219. doi: 10.1021/ci0496144. [DOI] [PubMed] [Google Scholar]
- 33.Medina-Franco JL, Maggiora GM, Giulianotti MA, Pinilla C, Houghten RA. A similarity-based data-fusion approach to the visual characterization and comparison of compound databases. Chem Biol Drug Des. 2007;70:393–412. doi: 10.1111/j.1747-0285.2007.00579.x. [DOI] [PubMed] [Google Scholar]
- 34.Horvath D, Jeandenans C. Neighborhood behavior of in silico structural spaces with respect to in vitro activity spaces—A benchmark for neighborhood behavior assessment of different in silico similarity metrics. J Chem Inf Comput Sci. 2003;43:691–698. doi: 10.1021/ci025635r. [DOI] [PubMed] [Google Scholar]
- 35.Gillet VJ. New directions in library design and analysis. Curr Opin Chem Biol. 2008;12:372–378. doi: 10.1016/j.cbpa.2008.02.015. [DOI] [PubMed] [Google Scholar]
- 36.Bender A, et al. How similar are similarity searching methods? A principal component analysis of molecular descriptor space. J Chem Inf Model. 2009;49:108–119. doi: 10.1021/ci800249s. [DOI] [PubMed] [Google Scholar]
- 37.Khanna V, Ranganathan S. Molecular similarity and diversity approaches in chemoinformatics. Drug Develop Res. 2011;72:74–84. [Google Scholar]
- 38.Shannon CE. The mathematical theory of communication. 1963. MD Comput. 1997;14:306–317. [PubMed] [Google Scholar]
- 39.Godden JW, Stahura FL, Bajorath J. Variability of molecular descriptors in compound databases revealed by Shannon entropy calculations. J Chem Inf Comput Sci. 2000;40:796–800. doi: 10.1021/ci000321u. [DOI] [PubMed] [Google Scholar]
- 40.Godden JW, Bajorath J. Differential Shannon entropy as a sensitive measure of differences in database variability of molecular descriptors. J Chem Inf Comput Sci. 2001;41:1060–1066. doi: 10.1021/ci0102867. [DOI] [PubMed] [Google Scholar]
- 41.Stahura FL, Godden JW, Bajorath J. Differential Shannon entropy analysis identifies molecular property descriptors that predict aqueous solubility of synthetic compounds with high accuracy in binary QSAR calculations. J Chem Inf Comput Sci. 2002;42:550–558. doi: 10.1021/ci010243q. [DOI] [PubMed] [Google Scholar]
- 42.Chen H, et al. ProSAR: A new methodology for combinatorial library design. J Chem Inf Model. 2009;49:603–614. doi: 10.1021/ci800231d. [DOI] [PubMed] [Google Scholar]
- 43.Wang Y, Geppert H, Bajorath J. Shannon entropy-based fingerprint similarity search strategy. J Chem Inf Model. 2009;49:1687–1691. doi: 10.1021/ci900159f. [DOI] [PubMed] [Google Scholar]
- 44.Medina-Franco JL, Martínez-Mayorga K, Bender A, Scior T. Scaffold diversity analysis of compound data sets using an entropy-based measure. QSAR Comb Sci. 2009;28:1551–1560. [Google Scholar]
- 45.Klekota J, Brauner E, Roth FP, Schreiber SL. Using high-throughput screening data to discriminate compounds with single-target effects from those with side effects. J Chem Inf Model. 2006;46:1549–1562. doi: 10.1021/ci050495h. [DOI] [PubMed] [Google Scholar]
- 46.Metz JT, et al. Navigating the kinome. Nat Chem Biol. 2011;7:200–202. doi: 10.1038/nchembio.530. [DOI] [PubMed] [Google Scholar]
- 47.Hann MM, Leach AR, Harper G. Molecular complexity and its impact on the probability of finding leads for drug discovery. J Chem Inf Comput Sci. 2001;41:856–864. doi: 10.1021/ci000403i. [DOI] [PubMed] [Google Scholar]
- 48.Clemons PA, et al. Small molecules of different origins have distinct distributions of structural complexity that correlate with protein-binding profiles. Proc Natl Acad Sci USA. 2010;107:18787–18792. doi: 10.1073/pnas.1012741107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Potter T, Matter H. Random or rational design? Evaluation of diverse compound subsets from chemical structure databases. J Med Chem. 1998;41:478–488. doi: 10.1021/jm9700878. [DOI] [PubMed] [Google Scholar]
- 50.Harper G, Pickett SD, Green DV. Design of a compound screening collection for use in high throughput screening. Comb Chem High T Scr. 2004;7:63–70. doi: 10.2174/138620704772884832. [DOI] [PubMed] [Google Scholar]
- 51.Kim YK, et al. Relationship of stereochemical and skeletal diversity of small molecules to cellular measurement space. J Am Chem Soc. 2004;126:14740–14745. doi: 10.1021/ja048170p. [DOI] [PubMed] [Google Scholar]
- 52.Brown N, et al. A chemoinformatics analysis of hit lists obtained from high-throughput affinity-selection screening. J Biomol Screen. 2006;11:123–130. doi: 10.1177/1087057105283579. [DOI] [PubMed] [Google Scholar]
- 53.Tanikawa T, et al. Using biological performance similarity to inform disaccharide library design. J Am Chem Soc. 2009;131:5075–5083. doi: 10.1021/ja806583y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hall LH, Kier LB. The E-state as the basis for molecular structure space definition and structure similarity. J Chem Inf Comput Sci. 2000;40:784–791. doi: 10.1021/ci990140w. [DOI] [PubMed] [Google Scholar]
- 55.Jolliffe IT. Principal Component Analysis. New York: Springer; 2002. [Google Scholar]
- 56.Seiler KP, et al. ChemBank: A small-molecule screening and cheminformatics resource database. Nucleic Acids Res. 2008;36(Database issue):D351–359. doi: 10.1093/nar/gkm843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Harvey AL. Natural products in drug discovery. Drug Discov Today. 2008;13:894–901. doi: 10.1016/j.drudis.2008.07.004. [DOI] [PubMed] [Google Scholar]
- 58.Li H, et al. Prediction of genotoxicity of chemical compounds by statistical learning methods. Chem Res Toxicol. 2005;18:1071–1080. doi: 10.1021/tx049652h. [DOI] [PubMed] [Google Scholar]
- 59.Li H, et al. Effect of selection of molecular descriptors on the prediction of blood-brain barrier penetrating and nonpenetrating agents by statistical learning methods. J Chem Inf Model. 2005;45:1376–1384. doi: 10.1021/ci050135u. [DOI] [PubMed] [Google Scholar]
- 60.Muncipinto G, et al. Expanding stereochemical and skeletal diversity using petasis reactions and 1,3-dipolar cycloadditions. Org Lett. 2010;12:5230–5233. doi: 10.1021/ol102266j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Pizzirani D, Kaya T, Clemons PA, Schreiber SL. Stereochemical and skeletal diversity arising from amino propargylic alcohols. Org Lett. 2010;12:2822–2825. doi: 10.1021/ol100914b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hung AW, et al. Route to three-dimensional fragments using diversity-oriented synthesis. Proc Natl Acad Sci USA. 2011;108:6799–6804. doi: 10.1073/pnas.1015271108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Dandapani S, Marcaurelle LA. Current strategies for diversity-oriented synthesis. Curr Opin Chem Biol. 2010;14:362–370. doi: 10.1016/j.cbpa.2010.03.018. [DOI] [PubMed] [Google Scholar]
- 64.Dandapani S, Marcaurelle LA. Grand challenge commentary: Accessing new chemical space for ‘undruggable’ targets. Nat Chem Biol. 2010;6:861–863. doi: 10.1038/nchembio.479. [DOI] [PubMed] [Google Scholar]
- 65.Marcaurelle LA, et al. An aldol-based build/couple/pair strategy for the synthesis of medium- and large-sized rings: Discovery of macrocyclic histone deacetylase inhibitors. J Am Chem Soc. 2010;132:16962–16976. doi: 10.1021/ja105119r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Schreiber SL, et al. Towards patient-based cancer therapeutics. Nat Biotechnol. 2010;28:904–906. doi: 10.1038/nbt0910-904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Comer E, et al. Fragment-based domain shuffling approach for the synthesis of pyran-based macrocycles. Proc Natl Acad Sci USA. 2011;108:6751–6756. doi: 10.1073/pnas.1015255108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lovering F, Bikker J, Humblet C. Escape from flatland: Increasing saturation as an approach to improving clinical success. J Med Chem. 2009;52:6752–6756. doi: 10.1021/jm901241e. [DOI] [PubMed] [Google Scholar]
- 69.Wagner BK, Clemons PA. Connecting synthetic chemistry decisions to cell and genome biology using small-molecule phenotypic profiling. Curr Opin Chem Biol. 2009;13:539–548. doi: 10.1016/j.cbpa.2009.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Mayo SL, Olafson BD, Goddard WA. DREIDING: A generic force field for molecular simulations. J Phys Chem. 1990;94:8897–8909. [Google Scholar]
- 71.Sheshkin DJ. Handbook of Parametric and Nonparametric Statistical Procedures. 2nd Ed. Boca Raton, FL: Chapman & Hall/CRC; 2004. p. 1016. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.