Abstract
Serine/threonine protein kinases (STPKs) are the major participants in intracellular signal transduction in eukaryotes, such as yeasts, fungi, plants, and animals. Genome sequences indicate that these kinases are also present in prokaryotes, such as cyanobacteria. However, their roles in signal transduction in prokaryotes remain poorly understood. We have attempted to identify the roles of STPKs in response to heat stress in the prokaryotic cyanobacterium Synechocystis sp. PCC 6803, which has 12 genes for STPKs. Each gene was individually inactivated to generate a gene-knockout library of STPKs. We applied in vitro Ser/Thr protein phosphorylation and phosphoproteomics and identified the methionyl-tRNA synthetase, large subunit of RuBisCO, 6-phosphogluconate dehydrogenase, translation elongation factor Tu, heat-shock protein GrpE, and small chaperonin GroES as the putative targets for Ser/Thr phosphorylation. The expressed and purified GroES was used as an external substrate to screen the protein extracts of the individual mutants for their Ser/Thr kinase activities. The mutants that lack one of the three protein kinases, SpkC, SpkF, and SpkK, were unable to phosphorylate GroES in vitro, suggesting possible interactions between them towards their substrate. Complementation of the mutated SpkC, SpkF, and SpkK leads to the restoration of the ability of cells to phosphorylate the GroES. This suggests that these three STPKs are organized in a sequential order or a cascade and they work one after another to finally phosphorylate the GroES.
Keywords: cyanobacteria, GroES, heat stress, phosphorylation, serine–threonine protein kinase
1. Introduction
Phosphorylation is a reversible post-translational modification of proteins, and, in many cases, substrate proteins are involved in signal transduction in both eukaryotic and prokaryotic cells. Protein kinases transfer phosphate groups, mostly from ATP, to proteins, with resultant changes in the enzymatic activities and/or properties of the substrate proteins. Prokaryotes widely employ two-component regulatory systems composed of histidine kinase and its cognate response regulator to control various regulatory functions that require fine-tuning of gene expression.1,2 Most protein kinases in eukaryotes are tyrosine or serine/threonine protein kinases (2.7.1.37; the latter are abbreviated as STPKs). They catalyse phosphorylation of the hydroxyl group of serine or threonine residues of their substrate proteins.3 All STPKs analysed to date include a conserved domain of 250–300 amino acids, which binds first to ATP (and, in very few cases, like casein kinase 2, to GTP) and then binds to a substrate protein for transfer of the phosphate group derived from ATP (GTP) to the substrate protein.3
The complete sequences of the genomes of Arabidopsis thaliana, rice, human, and rat reveal the presence of hundreds of genes for STPKs. Homologues of eukaryotic-like STPKs have been found in many bacterial species.4–6 For example, Bacillus subtilis has three and Mycobacterium tuberculosis has 11 genes for STPKs. Myxococcus xanthus carries nearly 100 genes for eukaryotic-like STPKs.5 In contrast, Escherichia coli has no such genes; however, Ser/Thr protein kinase activity was found in E. coli cells.7 Substrate proteins for Ser/Thr phosphorylation were identified in E. coli by phosphoproteomics,8 and the protein YihE, which has no homology to eukaryotic STPKs, but possesses Ser/Thr protein kinase activity, has been identified and crystallized.9
In bacteria, STPKs are involved in central metabolism and regulate the activity of enzymes such as enolase, pyruvate dehydrogenase, inorganic pyrophosphatase, cell division proteins, FtsZ, and others,10 or globally control the gene expression.11,12 Usually, mutation of genes that encode STPKs in bacteria leads to multiple changes in overall metabolism and/or gene expression, and identification of the original targets for Ser/Thr phosphorylation is not easy.
Among cyanobacteria, unicellular freshwater Synechocystis sp. PCC 6803 (hereafter abbreviated as Synechocystis) has 12 genes for putative STPKs,13–15 whereas filamentous nitrogen-fixing Anabaena sp. PCC 7120 has 52 such genes.16,17 Zhang18 demonstrated that one of these genes in Anabaena, pknA, is essential for normal cell growth and the differentiation of heterocysts. Another STPK, Pkn22, is involved in the acclimation of Anabaena cells to iron starvation and oxidative stress, and it regulates the expression of the isiA gene for chlorophyll-binding CP43′ protein.19
Of the 12 putative genes for Ser/Thr kinases in Synechocystis, seven of these genes encode proteins that belong to the PKN2 subfamily of STPKs and five encode proteins that belong to the ABC1 subfamily.15,20 The genes for kinases of the PKN2 type are designated spkA, spkB, spkC, spkD, spkE, spkF, and spkG and those for kinases of the ABC1 type are designated spkH, spkI, spkJ, spkK, and spkL (http://genome.kazusa.or.jp/cyanobase/). The spkA21,22 and spkB23 genes appear to encode proteins that participate in the formation of thick pili and the control of cell motility, whereas the spkC gene encodes a kinase that was reported to be involved in the regulation of nitrogen metabolism.24 The involvement of SpkE in the control of cell motility has been proposed but not yet confirmed.25 Until today, the protein substrates, which are the targets for Ser/Thr phosphorylation in Synechocystis, have not yet been identified.
In this study, we identified several proteins that change the extent of their Ser/Thr phosphorylation upon heat stress. GroES co-chaperonin was one of the targets for such phosphorylation. Here we introduced mutations into 11 genes for STPKs, from spkB to spkL. The phenotype of the mutation in spkA has been described earlier by Panichkin et al.22 Analysis of their capacity to phosphorylate the externally provided GroES demonstrated the involvement of three STPKs into phosphorylation of this co-chaperonin in Synechocystis. Inactivation of either spkC, spkF, or spkK, leads to inability of cells to phosphorylate GroES, suggesting the existence of a regulatory circuit or cascade, in which these three STPKs interact with each other.
2. Materials and methods
2.1. Strains and culture conditions
Synechocystis sp. PCC 6803, a glucose-sensitive strain (GS), was obtained from Department of Genetics, Moscow State University.26 It served as a wild-type strain for construction of the gene knock-out library of individual spk genes. This strain already possesses a spontaneous frame-shift mutation in the spkA gene.21 All cyanobacterial cells were grown in a liquid or on agar-solidified BG11 medium27 that had been buffered to pH 7.6 with 20 mM HEPES-NaOH. Cells in 50 ml liquid cultures were grown photoautotrophically in 100 ml glass tubes in a water bath at 32°C with continuous illumination at 50 µmol photons m−2 s−1, which was provided by incandescent lamps, and with continuous bubbling with air that contained 1% CO2 (v/v) (hereafter referred to as normal growth conditions). The concentration of cells in liquid cultures was estimated from the optical density at 750 nm (A750), as determined with a spectrophotometer (Genesis 10UV; Thermo Fisher Scientific, MA, USA). Cultures for experiments were inoculated with cells from liquid precultures at a concentration of cells that corresponded to A750 = 0.1 and were grown under normal growth conditions. Cells at a density that corresponded to A750 = 0.4–0.6 were used for analysis. Mutant strains were maintained on a agar-solidified or liquid medium that contained 50 µg ml−1 kanamycin, 1 µg ml−1 gentamycin, 20 µg ml−1 chloramphenicol, or 20 µg ml−1 spectinomycin, as appropriate. No antibiotics were added to the final cultures used for analysis to avoid the possibility of phenotypic changes due to the presence of an antibiotic.
For heat-shock experiments, both GS and mutant cultures were withdrawn form 32°C and placed to 44°C and incubated at the same light intensity for 30 min. After treatment, the cells were collected by centrifugation (5000g, 10 min, 4°C), washed twice with 50 mM Tris–HCl (pH 7.6), 10 mM MgCl2, and washed pellets were used immediately for protein isolation.
Competent cells of E. coli strains JM109, DH5α, or BL21 (DE3 pLysS) were obtained with the standard CaCl2 protocol28 and were used for the cloning and isolation of plasmids. Cells were grown overnight at 37°C in Luria–Bertani medium supplemented with 50 µg ml−1 ampicillin.
2.2. Construction of the plasmid library of mutated genes for Ser/Thr kinases
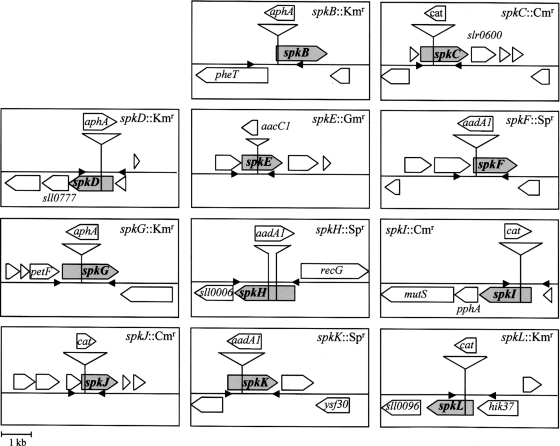
Partial sequences of individual spk genes have been amplified by PCR with the primers listed in Supplementary Table S1. The amplified DNA fragments were ligated into pGEM®-T or pGEM®-T Easy (Promega Inc., Madison, WI, USA), or into pT7Blue-T, or pUC18.29 Antibiotic-resistance gene cassettes,30–33 a transposon EZ::TN™ <KAN-2> (Epicentre, Madison, WI, USA), or pGPS2.1 (New England BioLabs, Beverly, MA, USA) have been inserted into the cloned DNA fragments. Each of the resultant plasmids was used for transformation of Synechocystis cells. The transformed cells were selected on a agar-solidified BG11 medium that had been supplemented with an appropriate antibiotic, as described by Grigorieva and Shestakov.26 Each cyanobacterial cell contains several copies of the cyanobacterial chromosome.34,35 In order to mutate every copy of the chromosome, we transferred the transformed cells to several plates of the agar-solidified BG11 medium with increasing concentrations of the antibiotic. We confirmed the complete replacement of native copies of the chromosome by mutated copies by amplification by PCR (Supplementary Fig. S1) with the pairs of primers listed in Supplementary Table S2. Positions of insertions are shown in Fig. 1.
Figure 1.
A schematic representation of the mutation of individual spk genes in Synechocystis by insertion of antibiotic-resistance gene cassettes. Open arrows represent the open reading frames (ORFs). Shaded arrows represent the target spk genes. Small black triangles indicate sites for binding of primers (see Supplementary Table S2) and directions of elongation of primers. The position and orientation of insertion of each cassette and the size and orientation of each respective gene are indicated in Supplementary Table S1.
2.3. Construction of the ΔhrcA (Δsll1670) mutant
Synechocystis sp. PCC 6803, a GS, was used for disruption of the hrcA (sll1670) gene that encodes a transcription factor, which negatively controls transcription of some heat stress genes.36 The complete nucleotide sequence of the hrcA gene36 was amplified with forward (5′-TTGGACTGGAATTCTAAGATG) and reverse (5′-TTAATTTTTGCTCAATGCTTCCG) primers, agarose-purified, and cloned into pT7-Blue. The HincII Kmr-cassette derived from pUC4K30 was inserted into blunted Eco91I site of the hrcA to substitute a part of a gene (positions from 223 to 939 of the gene sequence). Complete segregation of the mutated chromosomes was confirmed by PCR with the primers described above.
2.4. Complementation of mutations in spkC, spkF, and spkK genes
The spkC, spkF, and spkK genes together with their upstream regions were amplified with Pfu DNA polymerase (Fermentas) from the genomic DNA of Synechocystis GS with synthetic nucleotides (Supplementary Table S3) used as primers for PCR. The amplified DNA fragments, which contained the spkF and spkK, were ligated into the SmaI site of pVZ321.37 The amplified DNA fragment that contained the spkC was ligated into the EcoRI site of pVZ321 blunted with Klenow exo− enzyme. Mutant cells of Synechocystis GS defective in spkC, spkF, and spkK genes were transformed with recombinant pVZ-spkC, pVZ-spkF, and pVZ-spkK plasmids, respectively. The clones that carried the spkF and spkK genes were selected on spectinomycin (30 μg ml−1) and chloramphenicol (5 μg ml−1). The clones that carried the spkC gene were selected on kanamycin (10 μg ml−1) and chloramphenicol (12.5 μg ml−1).
2.5. Other manipulations of DNA
DNA was isolated from Synechocystis cells with lysozyme and purified with chloroform as described previously.37 Restriction analysis with restriction enzymes (Fermentas), isolation of plasmids, transformation of E. coli, and ligation of DNA fragments were performed by standard methods.27
2.6. Protein isolation
All chemicals were purchased from Sigma. All procedures were carried out at 4°C. To isolate the soluble protein fraction, the cell pellets were suspended in ice-cold buffer which contained 50 mM Tris–HCl (pH 7.6), 10 mM MgCl2, 2 mM EDTA, 1 mM dithiothreitol (DTT), 1 mM phenylmethylsulfonyl fluoride, 1 mM EGTA, 2 mM Na3VO4, 1 mM benzamidine, 10 mM NaF, 50 mM β-glycerophosphate, and 250 mM sucrose. Disruption was achieved using French Press (2.8 MPa; SLM, American Instruments Co., Urbana, IL, USA). The crude extracts were centrifuged stepwise, first at 7500g to remove unbroken cells and afterwards at 16000g for 20 min. The final supernatants were transferred into an appropriate buffer by gel filtration on NAP-5 columns (GE Healthcare) and used for biochemical assays. Protein concentration was determined using the BCA protein assay.
2.7. Protein phosphorylation in vitro and 2D PAGE
Protein phosphorylation in vitro was performed in a mixture that contained 20 mM MES-KOH (pH 6.5), 10 mM MgCl2, 10 µM ATP, and 740 kBq [γ-32P]ATP (148 TBq mmol−1). The reaction was initiated by the addition of 50 µg protein and allowed to proceed for 15 min at 30°C. After labelling, the proteins were precipitated at –20°C with 80% (v/v) acetone in the presence of 15% TCA and 1% 2-mercaptoethanol and pelleted by centrifugation. The pellets were washed twice with 80% (v/v) acetone.
Labelled proteins were dissolved in rehydration buffer (130 µl) contained 6 M urea, 1.5 M thiourea, 3% CHAPS, 66 mM DTT, 0.2% Bio-Lytes (pH 3–10, Bio-Rad), and traces of bromophenol blue. The samples (125 µl each) were loaded on IPG strips (7 cm, pH 4–7; ReadyStrips, Bio-Rad) for passive rehydration at 20°C for 12 h. The strips were then run using a Bio-Rad Protean IEF Cell at 20°C as follows: 200 V constant voltage for 15 min, 500 V constant voltage for 15 min, linear increase up to 1000 V for 30 min, low increase up to 5000 V for 30 min, and 5000 V constant voltage until a total of 10 000 V h−1 was reached. After IEF, the strips were equilibrated for 15 min with the buffer (50 mM Tris–HCl, pH 8.8, 6 M urea, 30% glycerol, 2% SDS) containing 10 mg ml−1 DTT followed by equilibration for 15 min with the same buffer, which contained 25 mg ml−1 iodoacetamide instead of DTT. The strips were then placed on the top of 12.5% polyacrylamide mini gels (0.75 mm thick) and subjected to SDS–PAGE at 200 V in a Bio-Rad MiniProtean III. After electrophoresis, the gels were fixed in 50% methanol with 3% phosphoric acid to remove the phosphate groups associated with His residues. After acidic treatment, the gels were stained with colloidal CBB G-250, dried, and subjected to autoradiography on a Kodak Biomax MR film.
The resultant autoradiographs were aligned with the corresponding images of the CBB G-250-stained gels using Image Quant 5.2 (Molecular Dynamics) gel analysis software. Selected protein spots were excised, digested in-gel with trypsin, and subsequently analysed by MALDI-TOF-MS at the service centre of Postgenomic Technology at the Institute of Biomedical Chemistry (Russian Academy of Medical Sciences, Moscow, Russia).
2.8. Western blotting
After 2DE separation, phosphoproteins were transferred onto 45 µm nitrocellulose filters (Hybond-C Extra) for western blotting using a Trans-Blot SD Electrophoretic Semi-Dry Transfer Cell (Bio-Rad) at 0.8 mA cm−2 for 2 h at room temperature. Air-dried membranes were subjected to autoradiography on Kodak Biomax MR films. After exposure to X-ray films, nitrocellulose blots were treated with rabbit antisera that had been raised against a mixture of Hsp60s (homologous GroEL and Cpn60) of Synechococcus vulcanus.38 Protein bands were visualized with secondary antibodies coupled with horseradish peroxidase.
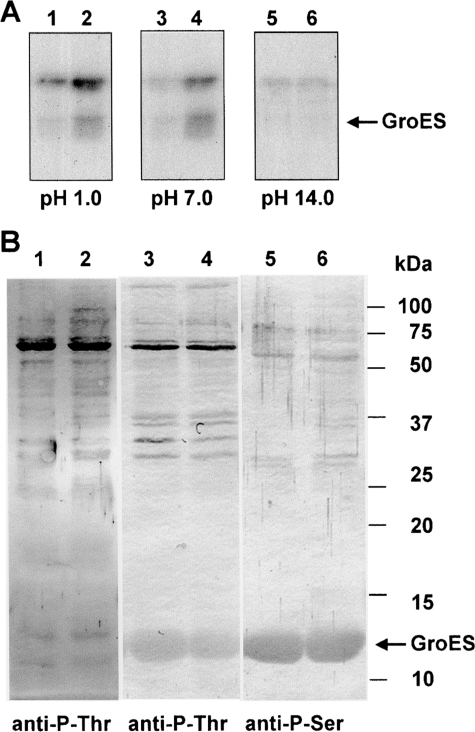
Phosphoserines and phosphothreonines have been determined with the corresponding anti-P-Ser and anti-P-Thr monoclonal antibodies produced in mouse purchased form Sigma-Aldrich (Product Nos P 3430 and P 3555 with dilution of 1:1000 and 1:500, respectively). Anti-mouse secondary antibodies conjugated with horseradish peroxidase and raised in rabbits have been used for visualization.
2.9. Expression and purification of the recombinant GroES
The recombinant GroES protein was expressed in the E. coli strain BL21(DE3 pLysS) harbouring the recombinant plasmid pET22b(+) (Novagen) with His-tagged groES. A DNA fragment carrying the groES gene was amplified from the genomic DNA of Synechocystis sp. PCC 6803 by PCR using synthetic oligonucleotides (5′-CATATGGCCGCTATTTCC and 5′-CTCGAGGGCAACGGAGGC) as forward and reverse primers, respectively. The amplified DNA fragment was digested with NdeI and XhoI restriction endonucleases and cloned into the corresponding sites of the pET22b(+). The resultant plasmid, pET-groES, was used for transformation of E. coli cells. The transformed cells were grown in LB-medium until the OD750 of suspension reached 0.5. Overproduction of the recombinant protein was initiated by the addition of isopropyl thio-β-d-galactoside to a final concentration of 1 mM and continued for 3 h at 37°C. Cells were harvested by centrifugation, disrupted by French Press, and the expressed protein was purified from the soluble fraction by affinity chromatography on a Ni2+-resin with a Protino® Ni-IDA Kit (Macherey-Nagel Gmbh & Co, Düren, Germany) according to the manufacturer's protocol. Alternatively, the recombinant GroES was purified by FPLC on a Mono Q column (GE Healthcare). Expression and purification protein profiles are shown in Supplementary Fig. S2.
2.10. Ser/Thr protein kinase assay on GroES
Supernatant protein factions (5–10 µg of protein), obtained from Synechocystis wild-type and its 11 mutant (ΔspkB-L) cells, were incubated with purified GroES (0.04 mg ml−1) for 15 min at 30°C in a final volume of 25 µl of buffer that contained 20 mM MES-KOH (pH 6.5), 10 mM MgCl2, 10 nM ATP, and 74 kBq [γ-32P]ATP (148 TBq mmol−1). The reaction was terminated by adding half of the reaction volume of three times concentrated Laemmli SDS sample buffer,27 and samples were boiled for 5 min. Phosphorylated GroES was resolved using 18% SDS–PAGE, stained with CBB R-250 in 40% (v/v) methanol with 10% (v/v) acetic acid, dried, and autoradiographed as described above. The same kinase assay was performed with the protein fractions obtained from the Synechocystis mutants defective in spkC, spkF, and spkK, which have been complemented with the pVz-spkC, pVZ-spkF, and pVZ-spkK, respectively.
2.11. pH-dependence of the phosphorylation of the GroES protein in vitro
Proteins were isolated from control and heat-stressed wild-type cells and phosphorylated with [γ-32P]ATP. Phosphorylated proteins were loaded on a 15% SDS–PAGE and blotted onto the PVDF membrane after electrophoresis. The membrane was then stained with Ponceau red, and the portion of the membrane containing the GroES was incubated at a different pH in the following buffers: 50 mM KCl (pH 1.0), 0.1 M Tris–HCl (pH 7.0), and 1 M KOH (pH 14.0). After incubation for 2 h at 45°C, membranes were washed thoroughly with deionized water, air dried, and exposed onto a Kodak X-ray film.
3. Results and discussion
3.1. Complete replacement of wild-type copies of individual spk genes by mutated copies
We inactivated 11 spk genes (spkB through spkL) in the GS strain of Synechocystis by insertional mutations (Fig. 1). In the GS strain, the spkA gene has already been inactivated by a frame-shift mutation. Therefore, we previously mutated the spkA gene in the Pasteur strain, in which the spkA gene is intact and the corresponding kinase is active.21
We examined the extent of replacement of wild-type copies of individual genes after each mutagenesis by PCR, using DNA isolated from wild-type and mutant cells as templates and appropriate forward and reverse primers (Fig. 1 and Supplementary Table S2). Analysis of each mutant (Supplementary Fig. S1) confirmed that the PCR-amplified fragment from mutant's DNA was longer than that amplified from the wild-type genomic DNA. The increase in the length of PCR-amplified fragments in the mutants was due to the insertion of the antibiotic-resistance gene cassette, except the spkF::Spr and spkH::Spr mutants, where both insertion and deletion occurred (Fig. 1). No mutant cells yielded an amplified fragment of the same length as the parental wild-type gene (Supplementary Fig. S1). This fact indicates that there were no copies of the respective wild-type genes in any of the 11 mutants.
Phenotype of all mutants was similar to wild-type cells in terms of size, colour, and the shape of colonies during cultivation on solid medium and in terms of the growth rate in liquid bubbling cultures under standard growth conditions.
3.2. Ser/Thr phosphorylation of the soluble proteins in relation to high temperature treatment
Phosphorylation of proteins catalysed by the sensory histidine kinases and STPKs is the key regulatory mechanism that ensures the perception and transmission of the intracellular and intercellular signals in eukaryotic39 and prokaryotic cells.40 The evidence exists that typical prokaryotic two-component signalling systems may work in cooperation with eukaryotic-like STPKs.6 In Synechocystis, sensory histidine kinase Hik34 is a repressor of heat-shock genes at normal growth temperatures.41 The involvement of STPKs into temperature signal transduction pathways was not yet demonstrated.
Here we tried to visualize the substrates of Ser/Thr phosphorylation in Synechocystis upon a rise in temperature. Cells that have been grown at 32°C were subjected to high temperature treatment (44°C) for 30 min. The influence of heat stress on Ser/Thr protein phosphorylation has been studied in soluble protein fractions using in vitro phosphorylation technique (see Experimental procedures). It is generally accepted that phospho-Ser/phospho-Thr are stable at low pH but labile at alkaline conditions. On the contrary, phospho-His is both acidic and heat labile. PAGs have been fixed in a 50% methanol in the presence of 3% phosphoric acid to remove the phosphate groups associated with His residues. Therefore, we are confident that under phosphorylation conditions described above, we were dealing with Ser/Thr phosphorylation.
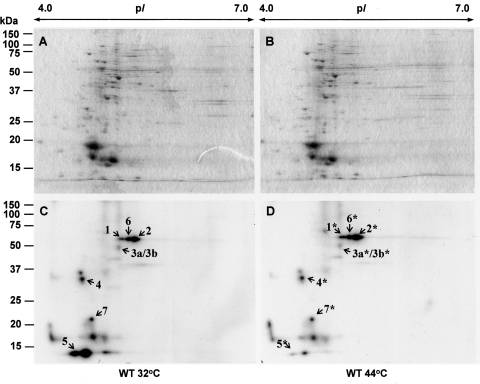
Short-term heat stress had no significant influence on soluble protein composition as demonstrated by 2D gels stained with colloidal CBB G-250 (Fig. 2A and B). However, the extent of phosphorylation of the individual polypeptides changed upon heat treatment (Fig. 2C and D). The phosphorylation signals from polypeptides presented by spots 1 and 2 increased twice, whereas the phosphorylation signal from spots 3а/3b decreased 1.5 times. Phosphopeptide 4 did not show any temperature-dependent change in phosphorylation. The high-intensity signal from phosphopeptide 5 dropped significantly upon an increase in temperature from 32 to 44°C (Fig. 2C and D).
Figure 2.
Proteome of soluble fractions of Synechocystis sp. PCC 6803. Soluble proteins (50 μg) from wild-type GS cells grown at 32°C (A) and wild-type GS cells treated for 30 min at 44°C (B) were separated by 2DE and stained with Colloidal Coomassie Briliant Blue G-250. Corresponding autoradiographs (C and D). Dried gels were exposed to X-ray films for 30 h at –70°C. Spot numbers of the identified proteins correspond to those presented in Table 1.
Thus, one can see that the increase in temperature causes significant changes in the intensity of Ser/Thr phosphorylation of proteins in Synechocystis. These changes might be due to autophosphorylation of the individual polypeptides, or due to temperature-dependent changes in the activity of different STPKs. For example, high temperature may inactivate certain STPKs. Protein kinases may phosphorylate either the same or a similar set of protein, or, alternatively, each individual STPK may phosphorylate its own substrate in vitro. To discriminate between these two possibilities, it is necessary (i) to identify the proteins, which correspond to the phosphorylated spots, and (ii) to clarify which of the 11 STPKs is able to phosphorylate the identified substrate in vitro.
3.3. MALDI-TOF MS identification of phosphorylated proteins
Seven proteins that have been identified as phosphoproteins phosphorylated by STPKs were subjected to MALDI-TOF MS analysis. We analysed similar spots that were visible in soluble protein extracts obtained from cells that had been grown at optimal growth temperature and subjected to heat treatment for 30 min. This was done to ensure that the visible temperature-dependent changes in Ser/Thr phosphorylation referred to the same protein(s). Thus, all together, 14 spots have been subjected to mass-spectrometry analysis. These spots were easily and reproducibly separated by 2DE and isolated with sufficient amounts for MALDI-TOF MS analysis (Table 1). The phosphoproteins observed at 32°C (Fig. 1C) were identical to those observed at 44°C (Fig. 1D). It means that, indeed, the raise in temperature causes changes in Ser/Thr phosphorylation.
Table 1.
List of Synechocystis phosphoproteins detected by in vitro assays and identified by MS analysis
| Spot # | Protein | ORF | MW (kDa) Dt/Calc | pI Dt/Calc | No. of peptides | Coverage, % |
|---|---|---|---|---|---|---|
| 1 | MetS, methionyl-tRNA synthetase | slr0649 | 58.7/61.442 | 5.0/5.0 | 10 | 26 |
| 2 | RbcL, large subunit of RuBisCO | slr0009 | 58.7/52.490 | 5.2/5.73 | 11 | 23 |
| 3a | 6-phosphogluconate dehydrogenase | sll0329 | 47.3/52.873 | 5.0/4.93 | 18 | 55 |
| 3b | TufA, elongation factor Tu | sll1099 | 47.3/43.733 | 5.0/4.99 | 26 | 71 |
| 4 | GrpE, heat-shock protein | sll0057 | 33.1/27.567 | 4.5/4.43 | 13 | 57 |
| 5 | GroES, co-chaperonin | slr2075 | 11.7/10.859 | 4.3/4.8 | 8 | 83 |
| 6 | tRNA δ2-isopentenylpyro-phosphate (IPP) transferase | sll0817 | 58.7/33.561 | 5.1/5.87 | 5 | 12 |
| 7 | Unknown protein | slr1270 | 21.1/57.735 | 4.6/4.62 | 5 | 10 |
Spot numbers correspond to those in Fig. 2. Spots in Fig. 2D marked with asterisk have been also analysed by MS, and they found to be identical to those listed in this table and marked in Fig. 2C. MW Dt/Calc, molecular weight experimentally determined vs. calculated from the amino acid sequence; pI Dt/Calc, pI experimentally determined vs. calculated from the amino acid sequence. No. of peptides, number of peptides determined by MS analysis. Coverage %, the per cent of MS-determined peptides to the complete amino acid sequence of the corresponding protein. The table summarizes the results of three independent experiments.
In general, we obtained rather high compliance with calculated and experimental data concerning molecular mass, pI, and peptide location in 2D gels and autoradiograms. It should be noted that the results of MS analysis were compared with the database of Synechocystis proteins, which, among others, includes polypeptides phosphorylated at His and Asp residues. In our case, however, such proteins were never found, since in our case phosphoproteins have been exposed to low pH at several stages of sample preparation (precipitation after phosphorylation in vitro, fixation of 2D gels at pH 1.0 after 2DE).
The identified phosphoproteins belong to several functional groups: (i) the enzymes for basic metabolism (nos 1, 2, 3a, 3b, and 6); (ii) proteins for general stress response (nos 4 and 5); and (iii) proteins with unknown function (no. 7). Among the first group, there is a protein that participates in the Calvin cycle (no. 2, RuBisCo large subunit), and another protein is involved in protein biosynthesis (no. 1, methionyl-tRNA synthetase). It might be that an increase in phosphorylation of these proteins correlates with an increase in their enzymatic activity—the fact, which was demonstrated for some enzymes of the cycle of tricarbonic acids.42
6-Phosphogluconate dehydrogenase (no. 3a), which also belongs to the first group of identified proteins, participates in the first phase of pentose phosphate pathway. Phosphorylated status for proteins that are involved in this pathway has been demonstrated in B. subtilis.43
Elongation factor Tu (no. 3b) exists in its phosphorylated form in all organisms, from archaeabacteria to humans, and it is characterized by the conserved sites of phosphorylation.7 Phosphorylation of factor Tu is, probably, required for optimization of the rate of translation. Here, we identified this protein with relatively high level of probability, higher than 70% (Table 1).
It was not surprising that we have found the so-called heat-shock proteins among the identified phosphoproteins (nos 4 and 5). It should be noted, however, that these proteins, GrpE and GroES, were highly labelled with phosphate in the extracts obtained from cells grown at the normal temperature. Heat stress had no effect on the phosphorylation of GrpE, and it caused a decrease in phosphorylation of GroES (Fig. 2C and D). These changes might reflect temperature-dependent changes in the rate of translation of these proteins, in the activity of phosphatases, and/or changes in the activity of STPKs that use these proteins as the substrates.
Our further efforts have been concentrated on the co-chaperonin GroES of 10 kDa as a putative substrate for STPKs activity. Identification of this polypeptide met the stringent criteria of analysis: in all our samples, this protein (no. 5) was digested with trypsin into five polypeptides with the 83% coverage of GroES amino acid sequence of Synechocystis (Table 1).
It was earlier demonstrated that the expression of the groESL operon and the amount of GroES protein increase after 60 min of heat stress.44,45 Our experimental setup (44°C for 30 min) did not reveal any significant changes in the amount of GroES (Fig. 2B). However, we observed distinct changes in its Ser/Thr phosphorylation in vitro (Fig. 2D) under the heat stress. To our knowledge, phosphorylated form of GroES had been recently reported in E. coli,8 although the functional significance of such phosphorylation is still unknown. Much work has been done on GroEL phosphorylation and its importance for correct assembly of the GroESL chaperonin machinery.46–48 Here we show that (i) GroES is phosphorylated by STPKs, (ii) its Ser/Thr phosphorylation runs even at low temperature, and (iii) its Ser/Thr phosphorylation is reduced upon heat stress.
3.4. GroES is a substrate for in vitro phosphorylation
To confirm that GroES might be a substrate for STPKs in Synechocystis and to identify the individual STPK, which participate in phosphorylation of GroES, we applied in vitro phosphorylation with the purified recombinant GroES (Supplementary Fig. S2) as a substrate for the protein kinase activity.
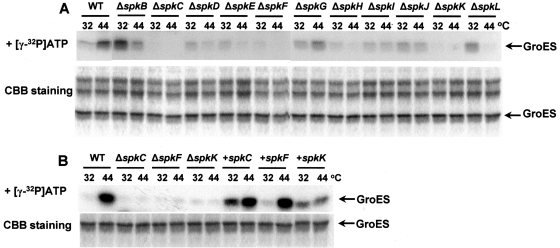
Wild-type and 11 mutant (ΔspkB-L) cells were treated at 44°C for 30 min, and soluble protein fractions were isolated from each strain. Twelve protein fractions isolated from those cells were equalized by protein content and used for phosphorylation of equal amounts of the purified recombinant GroES in the presence of [γ-32P]ATP. The products of reaction were resolved with SDS–PAGE, and phosphorylated GroES was visualized by autoradiography (Fig. 3A). Since equal amounts of isolated proteins were incubated with equal amounts of the recombinant GroES in the presence of equal amounts of [γ-32P]ATP, we can reliably validate the data on GroES phosphorylation presented in Fig. 3A.
Figure 3.
Phosphorylation in vitro of recombinant GroES by soluble protein fractions isolated from Synechocystis sp. PCC6803 (GS), spk mutants, and some of the complemented mutants. (A) GroES was phosphorylated in vitro with protein extracts obtained from the wild-type cells of Synechocystis (GS) and spk mutants grown at 32°C, and incubated for 30 min at 44°C. Autoradiographs and Coomassie R-250-stained gel are presented. (B) Similar reactions have been performed with wild-type cells of Synechocystis (GS), and with the mutant strains defective in SpkF, SpkC, and SpkK, which have been complemented with pVZ-spkC, pVZ-spkF, and pVZ-spkK (designated as +spkC, +spkF, and +spkK, respectively).
Phosphorylation of GroES was relatively low with the protein extracts isolated from wild-type cells and the mutant defective in spkG that had been grown at 32°C. Phosphorylation, however, increased in the extracts obtained from heat-stressed cells. Other mutants could be divided into the following groups according to their character of GroES phosphorylation. Mutants defective in spkB and spkL displayed relatively high levels of GroES phosphorylation at 32°C when compared with wild-type cells grown at 32°C. Heat stress at 44°C caused a severe decrease in the level of GroES phosphorylation. Mutants defective in spkD, spkE, spkH, spkI, and spkJ phosphorylated GroES less intensively than ΔspkG-mutant or wild-type cells at 32°C. Heat stress at 44°C also caused a decrease in GroES phosphorylation in those mutants.
Finally, three mutants defective in spkC, spkF, and spkK displayed nearly null level of GroES phosphorylation either at normal or at high temperatures.
Thus, one can conclude that, in Synechocystis, indeed, small co-chaperonin GroES is a target of phosphorylation by STPKs. It is unlikely that SpkC, SpkF, and SpkK all phosphorylate GroES. If this would be the case, the other two kinases should successfully phosphorylate GroES in vitro when the third kinase is knocked out. However, this does not happen. We suggest that these three protein kinases compose a chain or a cascade, and they phosphorylate one another with the final target of GroES. The place of each kinase in this cascade and the order of their functioning are still unclear, but it could be suggested that they might work in the following order: SpkF (transmembrane) → SpkC (membrane associated)→ SpkK (soluble). Anyway, only such mode of action explains the fact that phosphorylation of the GroES is lost, when any of the three kinases is knocked out.
To prove such a sequential (one after another) way of action, we complemented these three mutations in the mutants defective in SpkF, SpkC, and SpkK, with the corresponding genes expressed in the autonomously replicating plasmid pVZ321 under control of their own promoters. One can see that complementation of any of the STPK restores the ability of cells to phosphorylate GroES (Fig. 3B). This confirms the suggestion that the SpkF, SpkC, and SpkK might comprise a cascade to phosphorylate the GroES.
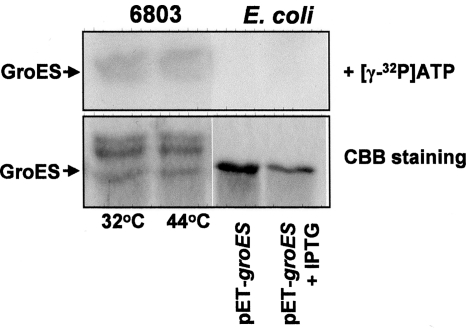
To ensure that the recombinant GroES is not phosphorylated by E. coli cells, we compared the patterns of phosphorylation obtained with Synechocystis and E. coli-soluble protein extracts. Figure 4 demonstrates that E. coli cells did not phosphorylate expressed and/or externally provided recombinant GroES protein. Thus, we conclude that phosphorylation of GroES demonstrated in Fig. 3 entirely results from the activity of the cyanobacterial STPKs.
Figure 4.
Phosphorylation of the recombinant GroES protein by soluble protein fractions obtained from Synechocystis and E. coli. (A) Soluble protein fraction was obtained from disrupted cells of Synechocystis GS, which had been grown at 32°C and transferred to 44°C for 30 min, by centrifugation at 100 000g for 1 h. Soluble protein fraction of E. coli BL21cells transformed with pET-GroES was obtained from disrupted cells by centrifugation at 16 000g for 20 min. Cells were grown at 37°C, and the protein extracts were obtained from non-induced cells, and from cells, in which the expression of GroES was induced by 50 mM IPTG for 3 h. About 1.8 μg of the purified recombinant GroES was added to each protein fraction (7.5 μg) in the presence of 1.5 μCi of [γ-32P]ATP. The reaction of phosphorylation was carried out for 15 min at 30°C. Upper panel represents the results of phosphorylation of externally provided GroES. Lower panel shows parts of gels stained with CBB.
To make sure that we are dealing with Ser/Thr-type of protein phosphorylation, the stability of the in vitro phosphorylated proteins in the presence of different pH was examined.
Proteins were isolated from control and heat-stressed wild-type cells, phosphorylated with [γ-32P]ATP, separated by electrophoresis, blotted onto PVDF, and stained with Ponceau red. The portions of the membrane containing the GroES were incubated in solutions with different pH: 1.0, 7.0, and 14.0. A majority of phosphate incorporated into GroES was released when the incubation was carried out at 45°C for 2 h at pH 14 (Fig. 5A). N-Phosphate amino acids (phoshohistidine, phospholysine, and phosphoarginine) are known to be stable at pH of 14.0 but not at pH of 1.0, while O-phosphate amino acids (phosphoserine and phosphothreonine) are known to be stable at pH of 1.0 but not at pH of 14.0.49 On the other hand, acyl-phosphate amino acids (phosphoaspartate and phosphoglutamate) are labile at both pH of 1.0 and 14.0, whereas the phosphotyrosine and phosphocysteine are stable at both pH. We observed a disappearance of the radioactive signal from the blots at pH of 14.0. Therefore, the stability profile for phosphorylated GroES is in agreement with Ser/Thr nature of protein phosphorylation (Fig. 5A).
Figure 5.
Stability of phosphorylated GroES at different pHs. (A) Autoradiographs of the phosphorylated GroES. Purified GroES (2.5 μg) mixed with either protein extracts derived from control (32°C) wild-type cells (10 μg) (lanes 1, 3, and 5) or with heat-treated (44°C for 30 min) cells (10 μg) (lanes 2, 4, and 6) was phosphorylated in vitro with [γ-32P]ATP. The reaction was terminated with 3× concentrated SDS–PAGE sample buffer and immediately subjected to SDS–PAGE (15% PAG). After electrophoresis, the proteins were transferred onto PVDF membrane. The membranes were stained with Ponceau red and incubated at 45°C for 2 h in 50 mM KCl–HCl (pH 1.0), 0.1 M Tris–HCl (pH 7), or 1 M KOH (pH 14). The radioactivity remaining in the membrane was revealed after its exposure onto a X-ray film. (B). Immunoblots of proteins from the wild-type cells of Synechocystis probed with monoclonal antibodies against phosphorylated Ser and Thr (anit-P-Thr and anti-P-Ser). Left panel: Protein extracts (25 μg) isolated from control (lane 1) and heat-treated (lane 2) wild-type cells were probed with anti-P-Thr antibodies. Central panel: Protein extracts (25 μg) isolated from control (lanes 3 and 5) and heat-treated (lanes 4 and 6) wild-type cells were probed with anti-P-Thr antibodies after phosphorylation in vitro with exogenously added recombinant GroES (2.5 μg). Right panel: The same as the middle panel but probed with anti-P-Ser antibodies.
Additional confirmation of the presence of phosphoserines and phosphothreonines in the protein was obtained by western blotting with the use of the anti-P-Ser and anti-P-Thr specific monoclonal antibodies (Fig. 5B). This experiment confirmed the presence of the Ser/Thr phosphorylation in the protein extract obtained from the wild-type cells of Synechocystis (Fig. 5B, left panel), and the ability of this extract to phosphorylate Ser and Thr residues of the externally added GroES (Fig. 5, central and right panels).
3.5. Ser/Thr phosphorylation of the soluble proteins isolated from the ▵hrcA mutant
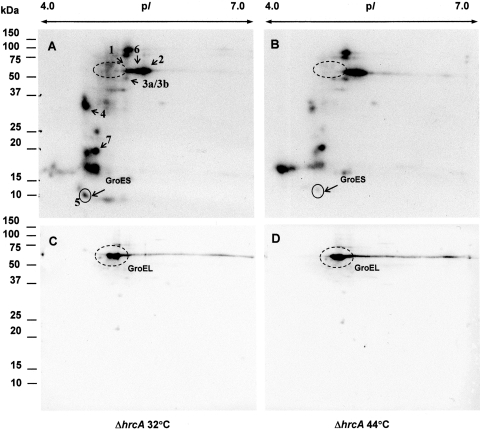
Up to date, it is known that a number of heat-inducible genes are under negative control of the histidine kinase Hik34,41 and some of them (like groESL operon) are under negative control of the transcription factor called HrcA (Sll1670).36 Taking into account the results described above, it is reasonable to suggest the involvement of STPKs into the temperature signal transduction pathway(s). If this is the case, it would be predicted that the mutants defective in HrcA and/or Hik34 actively express heat-shock genes and demonstrate the maximum level of Ser/Thr phosphorylation at normal temperature, such as 32°C.
Indeed, total Ser/Thr phosphorylation was much higher in ▵hrcA cells than in wild-type cells (Fig. 6). Among individual polypeptides, those with nos 1, 2, 4, 6, and 7 were phosphorylated most intensively. In ▵hrcA cells grown at 32°C, the phosphorylation level of polypeptides 1, 2, and 6 was higher (Fig. 6A) than in wild-type cells exposed to 44°C (Fig. 2D).
Figure 6.
Phosphorylation in vitro of soluble protein fraction isolated from Synechocystis GS ΔHrcA mutant grown at 32°C and treated at 44°C for 30 min. Autoradiographs of phosphoprotein patterns at 32°C (A) and 44°C (B) and the corresponding western blots (C and D) developed with anti-Hsp60 (GroEL) antibodies are presented. Dashed circles designate the positions of GroEL. Dried gels were exposed to X-ray films for 20 h at –70°C.
When ▵hrcA cells that had been grown at 32°C were exposed at 44°C, the extent of phosphorylation of these polypeptides increased, at least, two times, when compared with 32°C-grown mutant cells (Fig. 6A and B), whereas that of the polypeptide no. 4 decreased more than three times. Heat treatment at 44°C did not affect phosphorylation of phosphoprotein 7, resembling its temperature-independent behaviour in wild-type cells (Fig. 2C and D). The observed patterns of phosphorylation are in agreement with the repressive nature of HrcA regulation. Our results suggest that most of genes for Ser/Thr-phosphorylated proteins (Fig. 6) might be under the control of transcription factor HrcA.
The most striking difference between wild-type and ▵hrcA cells was that the latter displayed phosphorylated protein, which was similar to 60 kDa chaperonin GroEL, in terms of its molecular weight and pI characteristics. This protein in its phosphorylated form was visible only in ▵hrcA cells grown at 32°C (Fig. 6A), and it was not visible in cells exposed to 44°C (Fig. 6B). Cross-reaction of this polypeptide with antibodies against GroEL (Fig. 6C and D) revealed the presence of GroEL in protein extract obtained from both, 32°C-grown and 44°C-treated cells.
Since in vitro phosphorylation demonstrates only de novo incorporation of exogenously provided [γ-32P]ATP into polypeptides due to intrinsic protein kinase activities, it did not display any phosphorylation that had already occurred in cells or cell extract before the addition of radioactively labelled ATP. In the ▵hrcA cells, GroEL should be expressed constitutively at 32 and 44°C. It is very likely that GroEL was actively phosphorylated at 32°C and, therefore, the exposure of cells to 44°C did not contribute significantly to Ser/Thr phosphorylation in vitro of GroEL, as shown in Fig. 6. It is also possible that phosphorylation of GroEL occurs within few minutes under heat stress conditions, and additional incubation (30–60 min) at 44°C did not further affect its phosphorylation status (Fig. 6B).
It is known that chaperonin GroEL can prevent misfolding of polypeptides and promote the refolding and proper assembly of unfolded proteins generated that had been generated under heat stress. It is generally accepted that in E. coli cells, heat stress promotes reversible phosphorylation of GroEL,46,47 which is necessary for the formation of an active oligomeric GroESL complex. However, in Synechocystis mutant cells that lack transcription regulator HrcA, GroEL is produced and, perhaps, phosphorylated by STPKs constitutively.
Phosphorylation of GroES (phosphoprotein no. 5) was lower in the ▵hrcA mutant (Fig. 6A) than in wild-type cells at 32°С (Fig. 2C). After heat treatment at 44°C for 30 min, the signal from GroES in ▵hrcA mutant cells (Fig. 6B) became comparable with that of wild-type cells exposed to 44°C (Fig. 2D). Therefore, it seems that phosphorylation of GroES also ran constitutively at 32°C, and a decrease in its phosphorylation at 44°C reflected a decrease in the activity of the corresponding STPK(s).
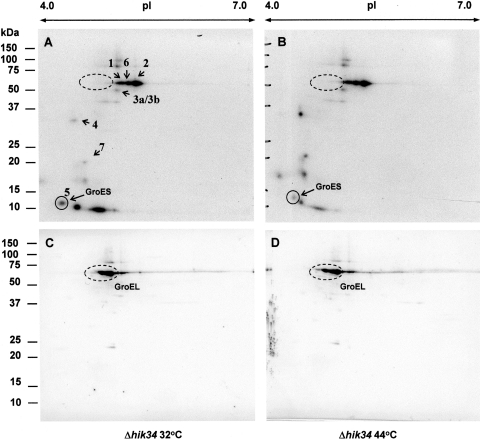
3.6. Ser/Thr phosphorylation of the soluble proteins isolated from the ▵hik34 mutant
Histidine protein kinase Hik34 (Slr1285) is a negative regulator for the expression of heat-shock genes. Disruption of the hik34 gene causes constitutive expression of a number of genes for heat-shock proteins at normal temperature.41 This allows mutant cells to survive under severe heat stress, such as exposure to 48°C for 3 h. Inactivation of hik34 up-regulates transcription of groESL, htpG, hspA, dnaK2, and slr1634,41 whereas inactivation of hrcA up-regulates only groESL and cpn60.36 DNA microarray analysis demonstrated that mutation in hik34 does not affect transcription of the hrcA gene at 34 or 44°C.41 Possible interaction of Hik34 and HrcA has been suggested,50 although experimental evidence for that is unavailable.
Here we examined the phosphorylation patterns in wild-type cells and in mutant cells that lack histidine kinase Hik34. It should be noted that the parental strain for the Hik34 mutant possessed the glucose-tolerant (GT) phenotype, whereas all other mutants were generated with a parental strain, which had GS phenotype.51 The intensity of Ser/Thr phosphorylation in the protein extracts of GT cells was, in general, lower than that of GS cells (Fig. 7A; Supplementary Fig. S3).
Figure 7.
Phosphorylation in vitro of soluble protein fraction isolated from Synechocystis GT ΔHik34 mutant grown at 32°C and treated at 44°C for 30 min. Autoradiographs of phosphoprotein patterns at 32°C (A) and 44°C (B) and the corresponding western blots (C and D) developed with anti-Hsp60 (GroEL) antibodies are presented. Dashed circles designate the positions of GroEL. Dried gels were exposed to X-ray films for 30 h at −70°C.
Heat stress caused alterations in the phosphorylation pattern in ▵hik34 cells that differed from the alterations observed in GS cells. There was no effect of temperature on phosphoproteins nos 1 and 3а/3b. Phosphorylation of phosphoproteins 2 and 6 increased, although to a less extent than it was observed in GS cells. In ▵hik34 cells, constitutive phosphorylation of phosphoprotein 7 was lower than in GS cells. Heat stress, however, caused the increase in its phosphorylation (Fig. 7B).
In ▵hik34 cells grown at 32°С, phosphorylation of GroES was lower than in GS cells, but higher than in ▵hrcA cells. Heat stress for 30 min diminished GroES phosphorylation (Fig. 6B), although to a less extent than it was observed in GS and ▵hrcA cells (Figs 2 and 6).
The GroEL chaperonin appears in equal amounts in ▵hik34 cells at both 32 and 44°C, as revealed by western blotting (Fig. 7C and D). However, we could not find even the traces of its phosphorylation in vitro. This implies that GroEL might be rapidly and intensively phosphorylated already at 32°C. Slabas et al.44 identified five spots that corresponded to GroEL1 and three spots that corresponded to GroEL2 (Cpn60) by proteomics. The antibodies used in our assay were raised against a mixture of GroEL1 and GroEL2, and they certainly recognize both forms of this chaperonin. There is no doubt about the constitutive expression of GroEL in ▵hrcA and ▵hik34 cells, but there is a serious doubt about its temperature-dependent phosphorylation. Also, comparison of the phosphorylation patterns for GroEL in ▵hrcA and ▵hik34 mutant cells leads to the suggestion that phosphorylation of GroEL somehow depends on the activity of HrcA, but not on the activity of Hik34.
Phosphoproteome of E. coli also did not demonstrate any phosphorylated GroEL at normal growth temperature,8 although the enrichment of phosphorylated proteins was carried out in that work. However, the phosphorylated GroES was apparent in E. coli cells. Since GroES and GroEL are encoded in one operon in E. coli and Synechocystis (the latter organism has an additional gene for GroEL-like protein, called groEL2 or cpn60), it is reasonable to expect their co-transcription, co-translation, and, finally, 1:1 ratio in cells. Differences in their Ser/Thr phosphorylation observed in Synechocystis imply that either (i) they have different affinity to similar or same STPKs or (ii) they are phosphorylated by different protein kinases.
Anyhow, it is evident that the expression of groESL operon is regulated by HrcA and Hik34, but phosphorylation of GroEL and GroES is regulated differentially with possible participation of HrcA, but not Hik34.
The data obtained here allow us to conclude that STPKs are involved in phosphorylation of several proteins during routine growth and under heat stress in Synechocystis. We found that the small co-chaperonin GroES was phosphorylated by STPKs, as it was previously found for 60 kDa chaperonin GroEL.46,47 We also found that the SpkC, SpkF, and SpkK might comprise a regulatory cascade that ensures phosphorylation of the GroES. Phosphorylation in vitro of the recombinant GroES demonstrated that heat stress at 44°C caused an increase in its phosphorylation by STPKs. However, we could not observe such an increase in our in vitro assays when no GroES was provided externally. Our data imply that Ser/Thr phosphorylation in Synechocystis runs intensively and constitutively at normal growth temperature.
Supplementary Data
Supplementary Data are available at www.dnaresearch.oxfordjournals.org.
Funding
The work was supported by grants from the Russian Foundation for Basic Research (nos 09-04-01074-a, 10-04-00840-а, 11-04-01006, and 11-04-01509) to D.A.L., V.V.Z., G.V.N., and I.E.M., and by a grant from the ‘Molecular and Cell Biology Program' of the Russian Academy of Sciences, and by a grant from Russian Ministry of Education and Science (no. 16.740.11.0176) to D.A.L.
Supplementary Material
References
- 1.Dutta R., Quin L., Inouye M. Histidine kinases: diversity of domain organization. Mol. Microbiol. 1999;34:633–40. doi: 10.1046/j.1365-2958.1999.01646.x. doi:10.1046/j.1365-2958.1999.01646.x. [DOI] [PubMed] [Google Scholar]
- 2.Los D.A., Zorina A., Sinetova M., Kryazhov S., Mironov K., Zinchenko V.V. Stress sensors and signal transducers in cyanobacteria. Sensors. 2010;10:2386–415. doi: 10.3390/s100302386. doi:10.3390/s100302386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hanks S.K., Hunter T. The eukaryotic protein kinase superfamily: kinase (catalytic) domain structure and classification. FASEB J. 1995;9:576–96. [PubMed] [Google Scholar]
- 4.Kennelly P.J. Protein kinases and protein phosphatases in prokaryotes: a genomic perspective. FEMS Microbiol. Lett. 2002;206:1–8. doi: 10.1111/j.1574-6968.2002.tb10978.x. doi:10.1111/j.1574-6968.2002.tb10978.x. [DOI] [PubMed] [Google Scholar]
- 5.Krupa A., Abhinandan K.R., Srinivasan N. KinG: a database of protein kinases in genomes. Nucleic Acids Res. 2004;32:D153–5. doi: 10.1093/nar/gkh019. http://hodgkin.mbu.iisc.ernet.in/king2/cgi-bin/index. doi:10.1093/nar/gkh019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pérez J., Castaneda-García A., Jenke-Kodama H., Müller R., Munoz-Dorado J. Eukaryotic-like protein kinases in the prokaryotes and the myxobacterial kinome. Proc. Natl Acad. Sci. USA. 2008;105:15950–5. doi: 10.1073/pnas.0806851105. doi:10.1073/pnas.0806851105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Enami M., Ishihama A. Protein phosphorylation in Escherichia coli and purification of a protein kinase. J. Biol. Chem. 1984;259:526–33. [PubMed] [Google Scholar]
- 8.Macek B., Gnad F., Soufi B., et al. Phosphoproteome analysis of E. coli reveals evolutionary conservation of bacterial Ser/Thr/Tyr phosphorylation. Mol. Cell. Proteomics. 2008;7:299–307. doi: 10.1074/mcp.M700311-MCP200. [DOI] [PubMed] [Google Scholar]
- 9.Zheng J., He C., Singh V.K., Martin N.L., Jia Z. Crystal structure of a novel prokaryotic Ser/Thr kinase and its implication in the Cpx stress response pathway. Mol. Microbiol. 2007;63:1360–71. doi: 10.1111/j.1365-2958.2007.05611.x. doi:10.1111/j.1365-2958.2007.05611.x. [DOI] [PubMed] [Google Scholar]
- 10.Nováková L., Bezousková S., Pompach P., et al. Identification of multiple substrates of the StkP Ser/Thr protein kinase in Streptococcus pneumoniae. J. Bacteriol. 2010;192:3629–38. doi: 10.1128/JB.01564-09. doi:10.1128/JB.01564-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sasková L., Nováková L., Basler M., Branny P. Eukaryotic-type serine/threonine protein kinase StkP is a global regulator of gene expression in Streptococcus pneumoniae. J. Bacteriol. 2007;189:4168–79. doi: 10.1128/JB.01616-06. doi:10.1128/JB.01616-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Donat S., Streker K., Schirmeister T., et al. Transcriptome and functional analysis of the eukaryotic-type serine/threonine kinase PknB in Staphylococcus aureus. J. Bacteriol. 2009;191:4056–69. doi: 10.1128/JB.00117-09. doi:10.1128/JB.00117-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kaneko T., Sato S., Kotani H., et al. Sequence analysis of the genome of the unicellular cyanobacterium Synechocystis sp. strain PCC 6803. II. Sequence determination of the entire genome and assignment of potential protein-coding regions. DNA Res. 1996;3:109–36. doi: 10.1093/dnares/3.3.109. doi:10.1093/dnares/3.3.109. [DOI] [PubMed] [Google Scholar]
- 14.Kaneko T., Nakamura Y., Sasamoto S., et al. Structural analysis of four large plasmids harboring in a unicellular cyanobacterium, Synechocystis sp. PCC 6803. DNA Res. 2003;10:221–8. doi: 10.1093/dnares/10.5.221. doi:10.1093/dnares/10.5.221. [DOI] [PubMed] [Google Scholar]
- 15.Leonard C.J., Aravind L., Koonin E.V. Novel families of putative protein kinases in bacteria and archaea: evolution of the ‘eukaryotic' protein kinase superfamily. Genome Res. 1998;8:1038–47. doi: 10.1101/gr.8.10.1038. [DOI] [PubMed] [Google Scholar]
- 16.Kaneko T., Nakamura Y., Wolk C.P., et al. Complete genomic sequence of the filamentous nitrogen-fixing cyanobacterium Anabaena sp. strain PCC 7120. DNA Res. 2001;8:205–13. doi: 10.1093/dnares/8.5.205. doi:10.1093/dnares/8.5.205. [DOI] [PubMed] [Google Scholar]
- 17.Ohmori M., Ikeuchi M., Sato N., et al. Characterization of genes encoding multi-domain proteins in the genome of the filamentous nitrogen-fixing cyanobacterium Anabaena sp. strain PCC 7120. DNA Res. 2001;8:271–84. doi: 10.1093/dnares/8.6.271. doi:10.1093/dnares/8.6.271. [DOI] [PubMed] [Google Scholar]
- 18.Zhang C.C. A gene encoding a protein related to eukaryotic protein kinases from the filamentous heterocystous cyanobacterium Anabaena PCC 7120. Proc. Natl Acad. Sci. USA. 1993;90:11840–4. doi: 10.1073/pnas.90.24.11840. doi:10.1073/pnas.90.24.11840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Xu W.L., Jeanjean R., Liu Y.D., Zhang C.C. pkn22 (alr2502) encoding a putative Ser/Thr kinase in the cyanobacterium Anabaena sp. PCC 7120 is induced by both iron starvation and oxidative stress and regulates the expression of isiA. FEBS Lett. 2003;553:179–82. doi: 10.1016/s0014-5793(03)01019-6. doi:10.1016/S0014-5793(03)01019-6. [DOI] [PubMed] [Google Scholar]
- 20.Shi L., Potts M., Kennelly P.J. The serine, threonine, and/or tyrosine-specific protein kinases and protein phosphatases of prokaryotic organisms. A family portrait. FEMS Microbiol. Rev. 1998;22:229–53. doi: 10.1111/j.1574-6976.1998.tb00369.x. doi:10.1111/j.1574-6976.1998.tb00369.x. [DOI] [PubMed] [Google Scholar]
- 21.Kamei A., Yuasa T., Orikawa K., Geng X.X., Ikeuchi M. A eukaryotic-type protein kinase, SpkA, is required for normal motility of the unicellular cyanobacterium Synechocystis sp. strain PCC 6803. J. Bacteriol. 2001;183:1505–10. doi: 10.1128/JB.183.5.1505-1510.2001. doi:10.1128/JB.183.5.1505-1510.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Panichkin V.B., Arakawa-Kobayashi S., Kanaseki T., et al. Serine/Threonine protein kinase SpkA in Synechocystis sp. PCC 6803 is a regulator of expression of three putative pilA operons, formation of thick pili, and cell motility. J. Bacteriol. 2006;188:7696–9. doi: 10.1128/JB.00838-06. doi:10.1128/JB.00838-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kamei A., Yoshihara S., Yuasa T., Geng X., Ikeuchi M. Biochemical and functional characterization of a eukaryotic-type protein kinase, SpkB, in the cyanobacterium, Synechocystis sp. PCC 6803. Curr. Microbiol. 2003;46:296–301. doi: 10.1007/s00284-002-3887-2. doi:10.1007/s00284-002-3887-2. [DOI] [PubMed] [Google Scholar]
- 24.Galkin A.N., Mikheeva L.E., Shestakov S.V. Insertional inactivation of genes encoding eukaryotic-type serine/threonine protein kinases in the cyanobacterium Synechocystis sp. PCC 6803. Mikrobiologiia. 2003;72:64–9. [PubMed] [Google Scholar]
- 25.Kamei A., Yuasa T., Geng X., Ikeuchi M. Biochemical examination of the potential eukaryotic-type protein kinase genes in the complete genome of the unicellular cyanobacterium Synechocystis sp. PCC 6803. DNA Res. 2002;9:71–8. doi: 10.1093/dnares/9.3.71. doi:10.1093/dnares/9.3.71. [DOI] [PubMed] [Google Scholar]
- 26.Grigorieva G.A., Shestakov S.V. Transformation in the cyanobacterium Synechocystis sp. PCC 6803. FEMS Microbiol. Lett. 1982;13:367–70. doi:10.1111/j.1574-6968.1982.tb08289.x. [Google Scholar]
- 27.Rippka R. Isolation and purification of cyanobacteria. Methods Enzymol. 1988;167:3–27. doi: 10.1016/0076-6879(88)67004-2. doi:10.1016/0076-6879(88)67004-2. [DOI] [PubMed] [Google Scholar]
- 28.Sambrook J., Frietsch E.F., Maniatis T. Molecular Cloning: A Laboratory Manual. 2nd edition. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 29.Yanisch-Perron C., Vieira J., Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33:103–119. doi: 10.1016/0378-1119(85)90120-9. doi:10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]
- 30.Vieira J., Messing J. The pUC plasmids, an M13mp7-derived system for insertion mutagenesis and sequencing with synthetic universal primers. Gene. 1982;19:259–68. doi: 10.1016/0378-1119(82)90015-4. doi:10.1016/0378-1119(82)90015-4. [DOI] [PubMed] [Google Scholar]
- 31.Rose R.E. The nucleotide sequence of pACYC184. Nucleic Acids Res. 1988;16:35. doi: 10.1093/nar/16.1.355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Prentki P., Binda A., Epstein A. Plasmid vectors for selecting IS1-promoted deletions in cloned DNA: sequence analysis of the omega interposon. Gene. 1991;103:17–23. doi: 10.1016/0378-1119(91)90385-o. doi:10.1016/0378-1119(91)90385-O. [DOI] [PubMed] [Google Scholar]
- 33.Schweizer H.D. Small broad-host-range gentamycin resistance gene cassettes for site-specific insertion and deletion mutagenesis. Biotechniques. 1993;15:831–4. [PubMed] [Google Scholar]
- 34.Labarre J., Chauvat F., Thuriaux P. Insertional mutagenesis by random cloning of antibiotic resistance genes into the genome of the cyanobacterium Synechocystis strain PCC 6803. J. Bacteriol. 1989;171:3449–57. doi: 10.1128/jb.171.6.3449-3457.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mann N., Carr N.G. Control of macromolecular composition and cell division in the blue-green alga Anacystis nidulans. J. Gen. Microbiol. 1974;83:399–405. doi: 10.1099/00221287-83-2-399. [DOI] [PubMed] [Google Scholar]
- 36.Nakamoto H., Suzuki M., Kojima K. Targeted inactivation of the hrcA repressor gene in cyanobacteria. FEBS Lett. 2003;549:57–62. doi: 10.1016/s0014-5793(03)00768-3. doi:10.1016/S0014-5793(03)00768-3. [DOI] [PubMed] [Google Scholar]
- 37.Zinchenko V.V., Piven I.V., Melnik V.A., Shestakov S.V. Vectors for the complementation analysis of cyanobacterial mutants. Genetika. 1999;35:228–32. [Google Scholar]
- 38.Tanaka N., Hiyama T., Nakamoto H. Cloning, characterization and functional analysis of groESL operon from thermophilic cyanobacterium Synechococcus vulcanus. Biochim. Biophys. Acta. 1997;1343:335–48. doi: 10.1016/s0167-4838(97)00159-3. doi:10.1016/S0167-4838(97)00159-3. [DOI] [PubMed] [Google Scholar]
- 39.Hunter T. Protein kinases and phosphatases: the yin and yang of protein phosphorylation and signaling. Cell. 1995;80:225–36. doi: 10.1016/0092-8674(95)90405-0. doi:10.1016/0092-8674(95)90405-0. [DOI] [PubMed] [Google Scholar]
- 40.Cozzone A.J. Post-translational modification of proteins by reversible phosphorylation in prokaryotes. Biochimie. 1998;80:43–8. doi: 10.1016/s0300-9084(98)80055-2. doi:10.1016/S0300-9084(98)80055-2. [DOI] [PubMed] [Google Scholar]
- 41.Suzuki I., Kanesaki Y., Hayashi H., et al. The histidine kinase Hik34 is involved in thermotolerance by regulating the expression of heat shock genes in Synechocystis. Plant Physiol. 2005;138:1409–21. doi: 10.1104/pp.104.059097. doi:10.1104/pp.104.059097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lévine A., Vannier F., Absalon C., et al. Analysis of the dynamic Bacillus subtilis Ser/Thr/Tyr phosphoproteome implicated in a wide variety of cellular processes. Proteomics. 2006;6:2157–73. doi: 10.1002/pmic.200500352. doi:10.1002/pmic.200500352. [DOI] [PubMed] [Google Scholar]
- 43.Eymann C., Becher D., Bernhardt J., Gronau K., Klutzny A., Hecker M. Dynamics of protein phosphorylation on Ser/Thr/Tyr in Bacillus subtilis. Proteomics. 2007;7:3509–26. doi: 10.1002/pmic.200700232. doi:10.1002/pmic.200700232. [DOI] [PubMed] [Google Scholar]
- 44.Slabas A.R., Suzuki I., Murata N., Simon W.J., Hall J.J. Proteomic analysis of the heat shock response in Synechocystis PCC6803 and a thermally tolerant knockout strain lacking the histidine kinase 34 gene. Proteomics. 2006;6:845–64. doi: 10.1002/pmic.200500196. doi:10.1002/pmic.200500196. [DOI] [PubMed] [Google Scholar]
- 45.Suzuki I., Simon W.J., Slabas A.R. The heat shock response of Synechocystis sp. PCC 6803 analysed by transcriptomics and proteomics. J. Exp. Bot. 2006;57:1573–8. doi: 10.1093/jxb/erj148. doi:10.1093/jxb/erj148. [DOI] [PubMed] [Google Scholar]
- 46.Sherman M.Yu., Goldberg A.L. Heat shock in Escherichia coli alters the protein-binding properties of the chaperonin groEL by inducing its phosphorylation. Nature. 1992;357:167–9. doi: 10.1038/357167a0. doi:10.1038/357167a0. [DOI] [PubMed] [Google Scholar]
- 47.Sherman M.Yu., Goldberg A.L. Heat shock-induced phosphorylation of GroEL alters its binding and dissociation from unfolded proteins. J. Biol. Chem. 1994;269:31479–83. [PubMed] [Google Scholar]
- 48.Kumar C.M., Khare G., Srikanth C.V., Tyagi A.K., Sardesai A.A., Mande S.C. Facilitated oligomerization of mycobacterial GroEL: evidence for phosphorylation-mediated oligomerization. J. Bacteriol. 2009;191:6525–38. doi: 10.1128/JB.00652-09. doi:10.1128/JB.00652-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Duclos B., Marcandier S., Cozzone A.J. Chemical properties and separation of phospho-amino acids by thin-layer chromatography and/or electrophoresis. Methods Enzymol. 1991;201:10–21. doi: 10.1016/0076-6879(91)01004-l. doi:10.1016/0076-6879(91)01004-L. [DOI] [PubMed] [Google Scholar]
- 50.Singh A.K., Summerfield T.C., Li H., Sherman L.A. The heat shock response in the cyanobacterium Synechocystis sp. Strain PCC 6803 and regulation of gene expression by HrcA and SigB. Arch. Microbiol. 2006;186:273–86. doi: 10.1007/s00203-006-0138-0. doi:10.1007/s00203-006-0138-0. [DOI] [PubMed] [Google Scholar]
- 51.Ikeuchi M., Tabata S. Synechocystis sp. PCC 6803—a useful tool in the study of the genetics of cyanobacteria. Photosynth. Res. 2001;70:73–83. doi: 10.1023/A:1013887908680. doi:10.1023/A:1013887908680. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.