Abstract
Ochratoxin A (OTA) is one of the most potent rodent renal carcinogens studied to date. Although controversial results regarding OTA genotoxicity have been published, it is now widely accepted that OTA is not a mutagenic, DNA-reactive carcinogen. Instead, increasing evidence from both in vivo and in vitro studies suggests that OTA may promote genomic instability and tumorigenesis through interference with cell division. The aim of the present study was to provide further support for disruption of mitosis as a key event in OTA toxicity and to understand how OTA mediates these effects. Immortalized human kidney epithelial cells (IHKE) were treated with OTA and monitored by differential interference contrast microscopy for 15 h. Image analysis confirmed that OTA at concentrations ≥ 5μM, which correlate with plasma concentrations in rats under conditions of carcinogenesis, causes sustained mitotic arrest and exit from mitosis without nuclear or cellular division. Mitotic chromosomes were characterized by aberrant condensation and premature sister chromatid separation associated with altered phosphorylation and acetylation of core histones. To test if OTA directly interferes with histone acetyltransferases (HATs) which regulate lysine acetylation of histones and nonhistone proteins, a cell-free HAT activity assay was conducted using total nuclear extracts of IHKE cells. In this assay, OTA significantly blocked HAT activity in a concentration-dependent manner Overall, results from this study provide further support for a mechanism of OTA carcinogenicity involving interference with the mitotic machinery and suggest HATs as a primary cellular target of OTA.
Keywords: ochratoxin A, carcinogenesis, mitosis, histone, histone acetyltransferase, Haspin
Due to fungal infection of crops in the field or postharvest, ochratoxin A (OTA) (N-{[(3R)-5-chloro-8-hydroxy-3-methyl-1-oxo-7-isochromanyl]-carbonyl}-3-phenyl-L-alanine), a mycotoxin produced by strains of Aspergillus and Penicillium species, is frequently found in a wide variety of food commodities. Consequently, humans are continuously exposed to OTA, with exposure estimates ranging between 2 and 3 ng/kg body weight (bw) per day and 6–8 ng/kg bw per day for average adult and high consumers, respectively (EFSA, 2006; SCOOP, 2002). Toxicity studies revealed that OTA is nephrotoxic and induced kidney tumors in rodents, raising concern that dietary intake of OTA may present a tumor risk to humans. In particular, high tumor incidences (29 and 72%) were observed in male rats after treatment with OTA at relatively low doses (70 and 210 μg/kg bw, respectively) (NTP, 1989), making OTA one of the most potent renal carcinogens known to date. Moreover, tumors induced by OTA in rats were found to exhibit an unusual, aggressive phenotype and high metastatic potential (Boorman et al., 1992; NTP, 1989). In view of human risk assessment of this important food contaminant, there have been considerable efforts over the past two decades to shed light on the mechanism of renal tumor formation by OTA and above all to establish if OTA acts as a genotoxic carcinogen (Mally and Dekant, 2005), for which by default no tolerable exposure levels can be defined, or by a nongenotoxic (epigenetic, non–DNA reactive), thresholded mode of action (MoA) (Schilter et al., 2005). Although the European Food Safety Authority (EFSA) Scientific Panel on Contaminants in the Food Chain recently applied a threshold approach for OTA risk assessment based on the conclusion that there was insufficient evidence to suggest that OTA is a mutagenic, DNA-reactive carcinogen (EFSA, 2006), the scientific basis for this approach continues to be called into question by some researchers (Mantle et al., 2010) and is likely to remain a matter of debate until the molecular mechanism of OTA carcinogenicity is finally resolved.
Based on cell degeneration accompanied by mitotic abnormalities and prominent nuclear enlargement in kidneys of OTA treated rats, we recently speculated that OTA may interfere with cell division, leading to cell death at mitosis (mitotic catastrophe) or aberrant exit from mitosis without nuclear division (Rached et al., 2007). In addition to these morphological changes, evidence that OTA may cause cell death during mitosis came from the finding that degenerate kidney epithelial cells in OTA-treated rats were frequently positive for phospho-H3 (Ser10), a marker of mitosis (Adler et al., 2009). Moreover, gene expression analyses revealed target organ–specific alterations in the expression of several key regulators of mitosis in response to OTA at doses known to induce renal tumor formation (70 and 210 μg/kg bw) (Adler et al., 2009). These included molecules involved in centrosome function, G2/M progression, chromosome condensation and segregation, spindle checkpoint, and cytokinesis, such as Aurora kinase B, Cdk1cdc2, Cdc20, A-, B-, and E-type cyclins, Bub1b, Plk1, survivin, Espl1 (separase), and Pttg1 (securin). Importantly, many of the genes altered in response to OTA are known to possess oncogenic potential and association with chromosomal instability, aneuploidy, and poor prognosis in tumor patients (Carter et al., 2006), suggesting that overexpression of these mitotic regulators in rat kidney may play a key role and driving force in tumor formation by OTA.
Although there is some evidence to show that OTA may induce apoptosis in vitro, results from several studies suggest that OTA cytotoxicity is primarily linked to growth inhibition (mediated by G2 or M cell cycle arrest) rather than due to direct cytolethality (Dreger et al., 2000; Kamp et al., 2005b; Palma et al., 2007; Rached et al., 2006). In addition, nuclear enlargement and mitotic aberrations were observed in immortalized human kidney epithelial cells (IHKE) exposed to OTA, further supporting mitotic disruption as a critical event in OTA toxicity (Rached et al., 2006).
In this study, we have used live-cell imaging to monitor the effects of OTA on cell division in IHKE cells. These analyses clearly confirm that OTA causes sustained mitotic arrest and exit from mitosis without nuclear or cellular division. We further demonstrate that mitotic chromosomes in OTA-treated cells are characterized by aberrant condensation and sister chromatid separation associated with altered phosphorylation and acetylation of core histones, consistent with the importance of posttranslational histone modifications in determining chromatin structure (Gelato and Fischle, 2008). To understand how OTA mediates these effects, we tested its ability to alter the activity of enzymes involved in the regulation of posttranslational histone modifications. Although we observed loss of histone H3 phosphorylation at threonine 3, OTA showed no significant direct inhibitory effect on Haspin kinase, which regulates recruitment of the chromosomal passenger complex (CPC) to chromosomes via phosphorylation of histone H3 at threonine-3 during mitosis (Higgins, 2010; Kelly et al., 2010; Wang et al., 2010). In contrast, OTA significantly blocked the histone acetyltransferase (HAT) activity of a nuclear cell extract in a concentration-dependent manner (IC50 of 24.5μM), suggesting that OTA may act as an inhibitor of this class of enzymes, which regulate acetylation of histones and nonhistone proteins at lysine residues. Overall, results from this study clearly demonstrate that OTA interferes with the mitotic machinery and suggest HATs as a primary cellular target of OTA.
MATERIALS AND METHODS
Antibodies and reagents.
Primary antibodies used were phospho-histone H3 (Ser10) (polyclonal rabbit, 1:1000; Santa Cruz Biotechnology, Inc., Heidelberg, Germany), phospho-histone H2A/H4 (polyclonal rabbit, 1:1000; Active Motif, Inc., Rixensart, Belgium), acetyl-histone H3 (Lys 9) (polyclonal rabbit, 1:1000; Cell Signaling, Danvers), acetyl-histone H3 (Lys18) (polyclonal rabbit, 1:1000; Cell Signaling), acetyl-histone H4 (Lys16) (polyclonal rabbit, 1:1000; Active Motif, Inc.), anti-phospho-threonine 3 (polyclonal rabbit; Cell Signaling), anti-histone H3 (polyclonal rabbit; Cell Signaling), anti-histone H4 (polyclonal rabbit, 1:1000; Active Motif, Inc.), rabbit anti-histone H2A (polyclonal rabbit, 1:1000; Active Motif, Inc.), and anti-acetyl-lysine (polyclonal rabbit; Cell Signaling). The secondary antibody was a goat horseradish (HRP)-conjugated goat anti-rabbit IgG (Santa Cruz Biotechnology, Inc.; 1:2500–5000). OTA, ochratoxin B (OTB), and anacardic acid (AA) were purchased from Santa Cruz Biotechnology, Inc. Unless otherwise indicated, all other chemicals were from Sigma-Aldrich GmbH (Munich, Germany).
Cell culture.
IHKE cells (originally obtained from S. Mollerup, National Institute of Occupational Health, Norway) were cultured under standard cell culture conditions (37°C, 5% CO2) in Dulbecco’s modified Eagle’s medium/Ham’s F12 (1:1) supplemented with 10% fetal calf serum (PAA Laboratories GmbH, Pasching, Austria), 2mM glutamine, 15mM HEPES, 5 mg/l transferrin, 10 μg/l epidermal growth factor (human), 0.1μM hydrocortisone, 5 mg/l insulin (bovine), 10 μg/l human epidermal growth factor, and 5 μg/l sodium selenite (Sigma-Aldrich GmbH). For subcultivation, 2 × 105 cells were seeded into 75-cm2 cell culture flasks (CELLSTAR Greiner, Frickenhausen, Germany). For cell treatment, compounds were dissolved in ethanol (Carl Roth GmbH, Karlsruhe, Germany) and added to the culture media.
Live-cell imaging.
For live-cell imaging, IHKE cells were seeded into microslide eight-well chamber slides (ibidi GmbH, Munich, Germany) at a density of 10,000 cells per well. Cells were allowed to grow for 24 h before treatment with OTA and OTB at concentrations of 0, 1, 10, 25, and 50μM. After incubation for 2 h, cells were mounted on a Nikon Eclipse Ti-PFS inverted microscope equipped with a Retiga 4000DC camera (QImaging, Surrey, BC, Canada) and Lumen 200 PRO fluorescence illumination system (Prior Scientific, Jena, Germany) and an incubation chamber for temperature, humidity, and CO2 control (Okolab, Ottaviano, Italy). Cells were imaged overnight at 37°C and 5% CO2 by time-lapse microscopy with differential interference contrast (DIC). Images were acquired at ×20 magnification at 15-min intervals for 15 h and were collated into film sequences. Qualitative and quantitative image analysis was performed using Image J (http://rsbweb.nih.gov/ij/). For each individual film sequence, the total number of cells, the number of cells entering mitosis, the time of entry into and exit from mitosis, the duration of mitosis, and the frequency of mitotic aberrations were recorded. Data are presented as mean ± SD of ≥ 3 independent experiments. Statistical analysis was performed by ANOVA + Dunnett multiple comparison test. In a separate series of experiments, fluorescence microscopy of living cells stained with Hoechst 33345 was used to visualize DNA during mitosis. Cells were pretreated with 1 μg/μl Hoechst 33345 dissolved in cell culture medium without phenol red for 30 min prior to treatment with OTA/OTB. Cells were monitored by DIC and fluorescence microscopy for 7 h.
Preparation of metaphase spreads.
To prepare metaphase spreads, cells were seeded into petri dishes (58 cm2) at a density of 7.5 × 105 cells and allowed to grow for 24 h before treatment with 0, 10, 25, and 50M OTA for 24 h. Attached cells were harvested by trypsination, pooled with floating cells and subsequently lysed in 1% sodium citrate containing 1mM sodium vanadate, 1mM sodium fluoride and 10 μl/ml protease inhibitor cocktail (Sigma-Aldrich GmbH) at 37°C for 8 min. Nuclei were pelleted by centrifugation, resuspended in a mixture of glacial acetic acid and methanol (1:3), and incubated overnight at 4°C. Nuclei were then washed and fixed onto microscope slides. Samples were stained with Giemsa dye and mounted on glass coverslips with Eukitt embedding medium. Metaphases (150–200 per treatment condition) were analyzed at ×100 magnification using a Leitz Metaphase Finder system. Data are presented as mean ± SD of four independent experiments. Statistical analysis was performed by ANOVA + Dunnett multiple comparison test.
Posttranslational histone modifications.
IHKE cells were seeded into 150-mm cell culture dishes at a density of 1 × 106 cells per plate and were allowed to grow to subconfluency (∼75%) before treatment with OTA for 24 h. Treatment (3 h) with colchicine (50 ng/ml) (Sigma-Aldrich GmbH), the histone deacetylase inhibitor sodium butyrate (NaB, 2mM) and the histone acetylase inhibitor AA (15μM) served as controls. Histone proteins were extracted using the Histone Purification Mini Kit (Active Motif, Inc.) according to the manufacturer’s instructions and quantified using the Bio-Rad DC protein assay (Bio-Rad, Munich, Germany). Equal amounts of isolated histones (3 μg) were loaded onto 16% sodium dodecyl sulfate-polyacrylamide gels, separated by electrophoresis, and transferred onto nitrocellulose membranes at 100 V for 75 min at 4°C. Membranes were blocked in 5% nonfat dry milk in TBST buffer (50mM Tris, 150mM NaCl, 0.1% Tween 20, pH 7.4; Carl Roth GmbH) for 1 h at room temperature (RT) and subsequently incubated with primary antibody (for source and dilution, see “Antibodies and Reagents” section) overnight at 4°C. Detection was performed using horseradish peroxidase–conjugated secondary antibodies and the ECL Western blotting detection system (GE Healthcare, Munich, Germany). All analyses were performed in three independent experiments.
Haspin kinase assay.
Effects of OTA on Haspin kinase activity were assayed in a cell-free enzyme inhibition assay using recombinant Haspin N-terminal myelin basic protein (MBP) fusion protein (MBP-Haspin) and histone H3 peptide as a substrate as previously described (Cuny et al., 2010; Patnaik et al., 2008). Briefly, kinase reactions were performed for 10 min at room temperature in 50mM Tris, pH 7.5, 5mM MgCl2, 1mM dithiothreitol, 0.01% Brij-35. 200μM ATP (near Km), 1nM MBP-Haspin, and 0.1μM biotinylated H3(1–21) peptide (at the Km) in a volume of 10 μl, using Proxiplate 384 Plus white assay plates (PerkinElmer) and terminated with 10 μl 50mM EDTA, 2nM Europium-labeled anti-Histone H3 threonine 3 phosphorylation (H3T3ph) antibody, and 40nM Streptavidin-APC (PerkinElmer). After 2 h at room temperature, time-resolved fluorescence resonance energy transfer (TR-FRET) measurements were made in a PHERAstar HTS microplate reader (BMG Labtech, Offenberg, Germany), and ratios of acceptor fluorescence at 665 nm over donor fluorescence at 620 nm were determined.
HAT assay.
The effects of OTA on HAT activity of nuclear IHKE cell extracts were assayed using the EpiQuik HAT Activity/Inhibition Assay (Epigentek, Brooklyn, NY) according to the manufacturer’s instructions. This colorimetric HAT activity/inhibition assay is based on the acetylation of histone substrates by active HATs and subsequent quantification of acetylated substrates using a high-affinity anti-acetylated histone antibody. Nuclear extracts of untreated IHKE cells were prepared as follows: Cell cultures were washed twice with ice-cold PBS and lysed in 10mM HEPES, pH 7.5, 150mM NaCl, 0.6% Nonidet P40, 1mM EDTA, 10 μl/ml protease inhibitor cocktail (Sigma-Aldrich GmbH), 1mM sodium vanadate and 1mM sodium fluoride (Th. Geyer GmbH, Renningen, Germany). The lysates were passed five times through a 26-gauge needle. After incubation for 20 min on ice, nuclei were pelleted by centrifugation at 4°C and 1500 revolutions per minute (rpm) for 15 min and homogenized in nuclear extraction buffer (20mM HEPES, pH 7.9, 420mM NaCl, 1.5mM MgCl2, 0.2mM EDTA, 25% glycerin, 5mM dithiothreitol, 10 μl/ml protease inhibitor cocktail, 1mM sodium vanadate, and 1mM sodium fluoride. Protein concentrations were determined using the Bio-Rad DC protein assay (Bio-Rad). For the determination of inhibitory effects of OTA on HAT activity, 6 μg of proteins (nuclear extract) were used per sample. OTA was applied at concentrations from 1 to 100μM. AA (15μM), a known HAT inhibitor of p300 (IC50 = 5μM) and p300/CREB (cAMP-response element binding)–binding protein-associated factor (P/CAF) (IC50 = 8.5μM) served as positive control. Data shown represent mean ± SD of three independent experiments carried out in triplicate.
Acetylation of nonhistone proteins.
IHKE cells were treated as described above (see “Posttranslational Histone Modifications” section). To obtain nuclear and cytosolic fractions, cultures were washed twice with ice-cold PBS and lysed in 10mM HEPES, pH 7.5, 150mM NaCl, 0.6% Nonidet P40, 1mM EDTA, 1 μl/ml protease inhibitor cocktail (Sigma-Aldrich GmbH), 1mM sodium vanadate, and 1mM sodium flouride (Th. Geyer GmbH). The lysates were passed five times through a 26-gauge needle and incubated for 20 min on ice. Nuclei were pelleted by centrifugation at 4°C and 1500 rpm for 15 min and homogenized in nuclear extraction buffer (20mM HEPES, pH 7.9, 420mM NaCl, 1.5 MgCl2, 0.2mM EDTA, 25% glycerol, 5mM dithiothreitol, 1 μl/ml protease inhibitor cocktail, 1mM sodium vanadate, and 1mM sodium fluoride). Protein concentrations in the supernatant (cytosol) and nuclear fraction were determined using the Bio-Rad DC protein assay (Bio-Rad). Equal amounts of protein (50 μg) were separated by SDS-polyacrylamide gel electrophoresis (PAGE) and transferred onto nitrocellulose membranes. Membranes were blocked in 5% nonfat, dry milk in TBST buffer (50mM Tris, 150mM NaCl, 0.1% Tween 20, pH 7.4; Carl Roth GmbH) for 1 h at RT and subsequently incubated with rabbit anti-acetyl-lysine (Cell Signaling, 1:500) (overnight at 4°C) and secondary horseradish peroxidase-conjugated goat anti-rabbit IgG in 5% nonfat, dry milk in TBST (1 h at RT). Membranes were developed using the ECL Western blotting detection system (GE Healthcare) and exposed to Hyperfilm (GE Healthcare).
RESULTS
Live-Cell Imaging Confirms Disruption of Mitosis as a Key Event in OTA Toxicity
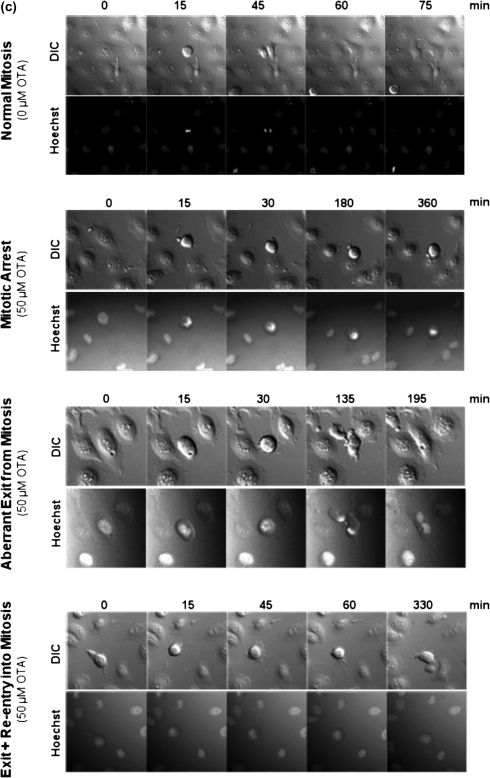
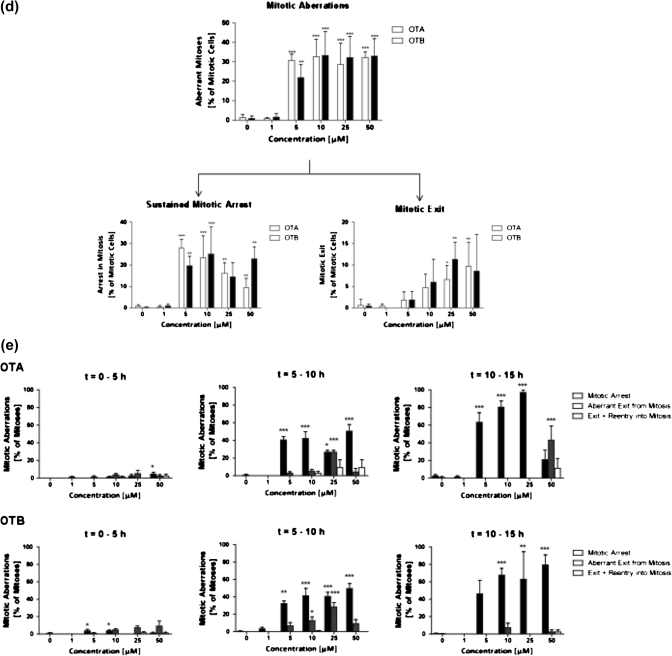
In this study, we have used live-cell imaging to confirm previous findings which suggest that OTA may interfere with cell division. IHKE cells cultured in eight-well microscopy chamber slides were treated with OTA or its structural analogue OTB at 0, 1, 5, 10, 25, and 50μM and were monitored by DIC microscopy overnight. Images were acquired at 15-min intervals for 15 h. In separate experiments, fluorescence microscopy of living cells stained with Hoechst 33345 was combined with DIC to visualize DNA during mitosis. Image analysis clearly demonstrated that OTA perturbs normal mitotic progression. Although cells entering mitosis within the first few hours after addition of OTA remained unaffected and completed a normal mitosis within a time frame comparable to untreated cells, continuing exposure to OTA at concentrations ≥ 5μM resulted in failure of cells to divide (Fig. 1). At these later time points, the percentage of cells that completed normal mitosis dropped to < 15% and these changes were accompanied by a significant increase in the time cells spent in mitosis. Consequently, total cell number, which continued to rise 1.8-fold in controls and cells treated with 1μM OTA, increased by a factor of only 1.2 in the presence of OTA at concentrations ≥ 5μM (Fig. 1). Morphological changes indicative of cell death were not evident throughout the observation period, further supporting a growth-inhibitory rather than cytolethal effect. Mitotic aberrations in cells exposed to OTA were characterized by sustained mitotic arrest or exit from mitosis without nuclear or cellular division (mitotic slippage) (Fig. 1). In addition, a subset of cells which rapidly slipped from mitosis appeared to repeatedly enter (and exit) mitosis as suggested by repetitive cycles of mitotic round-up and reattachment. Maximum responses were reached at 5μM and increasing concentrations did not further increase the total incidence of mitotic aberrations. However, qualitative differences were observed in that sustained mitotic block prevailed at lower concentrations (i.e., 5 and 10μM OTA), whereas the percentage of cells which exited mitosis appeared to increase with higher concentrations (i.e., 25 and 50μM OTA) (Fig. 1). Representative film sequences showing mitotic perturbations by OTA are provided online as Supplementary data.
FIG. 1.
(a) Cell growth of IHKE cells in the presence of OTA and OTB as assessed by image analysis of movies created by live-cell imaging, demonstrating a growth inhibitory effect of OTA and OTB at concentrations ≥ 5μM. (b) In line with the cytostatic effect, prolongation of mitosis is observed in IHKE cells treated with OTA and OTB at concentrations ≥ 5μM. (c) DIC and fluorescence microscopy film sequences demonstrating mitotic aberrations in IHKE cells treated with OTA. In the absence of OTA, normal mitosis of IHKE cells, that is, mitotic roundup, chromatid condensation, chromosome segregation, and cytokinesis, is completed within 75–100 min. In contrast, sustained block of mitosis, aberrant exit from mitosis without cell division, and repeated exits/entries into mitosis are evident in OTA-treated cells. Full film sequences are provided online (Supplementary data). (d) Overall frequency of mitotic aberrations in OTA- and OTB-treated IHKE cells and frequency of mitotic arrest and mitotic exit as the most prominent aberrations. (e) Time-related incidence of mitotic aberrations in OTA- and OTB-treated IHKE cells. Continued on subsequent pages.
Although it has previously been speculated that the presence of the chlorine atom in para position to the phenolic group may represent a structural requirement for OTA toxicity (Malaveille et al., 1994; Gillman et al., 1999), the same mitotic aberrations as induced by OTA were observed following treatment of IHKE cells with OTB, a structural OTA analogue which lacks the chlorine atom. As with OTA, OTB concentrations ≥ 5μM resulted in inhibition of cell growth due to sustained block and aberrant exit from mitosis, although the disruption pattern appeared somewhat different after prolonged treatment at 50μM (Fig. 1). In contrast, 1μM OTB (and OTA) showed no effects, indicating a thresholded dose-response.
OTA Causes Aberrant Chromosome Condensation and Loss of Sister Chromatid Cohesion
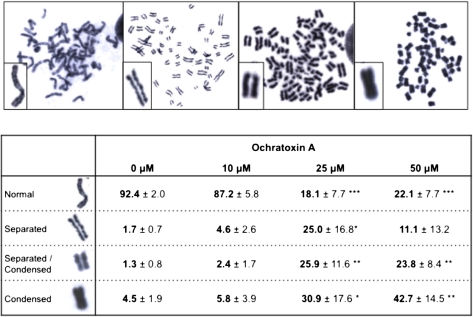
Previous cytogenetic analyses revealed a significant increase in the frequencies of both endoreduplication (two successive rounds of DNA replication without intervening mitosis) and polyploidy in human lymphocytes and V79 Chinese hamster fibroblasts treated with very high concentrations (up to 1mM) of OTA for 3 h (Mosesso et al., 2008). We therefore aimed to determine if the mitotic aberrations observed after continuous exposure to low OTA concentrations, which are more relevant to in vivo conditions of OTA carcinogenicity in rats, are also associated with endoreduplication. In contrast to the previous experiments at higher concentrations (Mosesso et al., 2008), endoreduplication was not evident in metaphase spreads prepared from IHKE cells treated with OTA at concentrations up to 50μM for 24 h. However, overcondensed mitotic chromosomes, sometimes with abnormally separated sister chromatids, were frequently observed (Fig. 2). Loss of centromeric cohesion was evident by clear separation of sister chromatids despite close physical proximity. Based on these observations, mitotic cells were categorized and quantified as follows: (1) normal chromosomes with sister chromatids connected at the centromeres, (2) prematurely separated sister chromatids, (3) prematurely separated, overcondensed chromosomes, and (4) overcondensed chromosomes with sister chromatids connected at the centromeres (Fig. 2). Interestingly, statistically significant increases in cells containing aberrant mitotic chromosomes, that is, overcondensation with or without loss of cohesion, were observed at OTA concentrations ≥ 25μM, that is, at concentrations associated with a higher tendency to exit mitosis, but not at 10μM. Loss of sister chromatid cohesion and overcondensation was also observed in V79 cells, although statistically significant changes were evident only at 50μM, consistent with the somewhat lower toxicity of OTA in this cell line as compared with IHKE cells (data not shown).
FIG. 2.
Metaphase spreads of IHKE treated with OTA, demonstrating overcondensation and aberrant separation of sister chromatids. Table: Frequency of OTA-induced metaphase aberrations in IHKE cells treated with OTA for 24 h (data are shown as % of total metaphases). Data are presented as mean ± SD of four independent experiments. (Statistical analysis was performed by ANOVA + Dunnett post hoc test. Statistically significant changes are indicated by *p < 0.05, **p < 0.01, ***p < 0.001.)
OTA Alters Posttranslational Histone Phosphorylation and Acetylation
Because it is well known that posttranslational histone modifications play an important role in defining chromatin structure and function (Gelato and Fischle, 2008; Ito, 2007), we speculated that OTA-mediated alterations in chromosome condensation and separation may be associated with modifications of core histones. We applied a panel of antibodies which specifically recognize certain phosphorylation and acetylation marks on histones to analyze posttranslational histone modifications in IHKE cells treated with OTA. Phosphorylation of histone H3 serine 10 (H3S10ph) and to a lesser extent histone H4 serine 1 (H4S1ph) was increased at 10 and 25μM OTA as compared with controls, whereas the level of histone H2A phosphorylation at serine 1 (H2AS1ph) remained unchanged (Fig. 3). In contrast, histone H3T3ph, which appears to be required for chromatid cohesion and normal chromosome alignment (Dai et al., 2006), was completely lost in the presence of OTA at concentrations ≥ 10μM (Figure 3). In line with the known association between hypoacetylation of core histones and compacted chromatin conformation (Shogren-Knaak et al., 2006), acetylation of lysine residues on histones H3 and H4, that is, H3K9ac, H3K18ac, and H4K16ac, was consistently reduced in a concentration-dependent manner in IHKE cells treated with OTA (Fig. 3). As with the structural changes, a similar pattern of histone modifications was seen in V79 cells treated with OTA (data not shown).
FIG. 3.
Western blot analysis of posttranslational histone modifications in IHKE cells exposed to OTA for 24 h, demonstrating loss of H3T3 phosphorylation and hypoacetylation of core histones. Sodium butyrate (NaB), a known histone deacetylase inhibitor, and colchicine were used as controls.
OTA Inhibits Histone Acetyltransferase Activity but Has No Direct Effect on Haspin Kinase
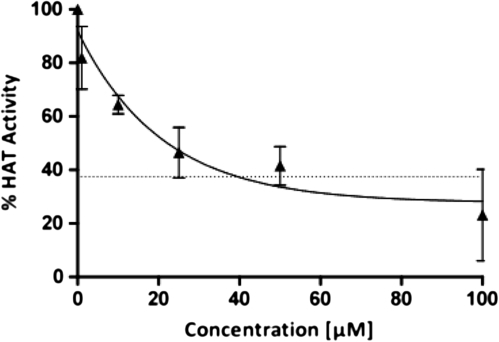
Dai et al. (2005) recently discovered that H3T3ph is mediated by the kinase Haspin. Thus, we were interested to understand if loss of H3T3ph induced by OTA may be the result of direct inhibition of Haspin kinase. However, OTA (and OTB) did not inhibit Haspin kinase activity in a TR-FRET kinase assay until concentrations of almost 1mM were reached, in contrast to the Haspin inhibitor LDN-192960 and the broad-spectrum kinase inhibitor staurosporine, which were used as positive controls (Fig. 4) (Cuny et al., 2010; Patnaik et al., 2008). This suggests that OTA blocks Haspin-mediated H3T3ph by an indirect mechanism in cells. In contrast, OTA significantly inhibited total HAT activity of a nuclear IHKE cell extract in a concentration-dependent manner (IC50 of 24.5μM) (Fig. 5).
FIG. 4.
TR-FRET Haspin kinase assay demonstrating lack of effects of OTA and OTB on Haspin kinase activity.
FIG. 5.
Cell-free HAT activity assay using nuclear extracts of IHKE cells and isolated histones as substrates. A clear concentration-dependent decrease in total HAT activity of the nuclear extract is seen in the presence of OTA. The degree of inhibition by OTA was similar to 15μM AA (indicated by the dashed line), a known inhibitor of HATs, which was used as positive control.
OTA Blocks Posttranslational Acetylation of Nonhistone Proteins
In addition to catalyzing histone acetylation, it is now known that HATs are also involved in the posttranslational regulation of nonhistone proteins (Spange et al., 2009). To obtain further evidence for inhibition of HAT activity as a critical event in OTA toxicity, we were therefore interested to determine if OTA also alters the acetylation pattern of nonhistone proteins. Cytosolic and nuclear protein extracts prepared from OTA-treated IHKE cells were separated by SDS-PAGE and probed with a polyclonal anti-acetyl-lysine antibody. As shown in Fig. 6, OTA blocked overall lysine acetylation of nuclear and cytosolic proteins extracted from OTA-treated IHKE cells in a concentration-dependent manner. Similarly, a trend toward decreasing levels of lysine acetylation was observed in proteins isolated from kidneys of rats treated with OTA at 0, 21, 70, and 210 μg/kg bw for 90 days (data not shown).
FIG. 6.
Western blot analysis of global changes in protein lysine acetylation in response to OTA. A clear dose-related decrease in the overall level of protein acetylation is seen in both cytoplasmic and nuclear fractions of cells treated with OTA. In contrast, no clear changes are observed with sodium butyrate (NaB) and AA, known inhibitors of HDACs and HATs, respectively.
DISCUSSION
Using live-cell imaging, we present unambiguous evidence that OTA acts by interfering with the mitotic machinery, leading to sustained mitotic arrest or exit from mitosis without cell division. Furthermore, we show that disruption of mitosis by OTA is accompanied by aberrant chromosome condensation and segregation and altered posttranslational modification of histones and nonhistone proteins. Finally, our results for the first time suggest HATs as primary cellular targets of OTA.
HATs are a diverse group of more than 30 enzymes, which form multiple subunit complexes and catalyze acetylation of lysine residues on the N-terminus of histones and nonhistone proteins (Choudhary et al., 2009; Spange et al., 2009). HAT complexes participate in a number of key nuclear processes including chromosome decondensation, DNA replication, DNA damage repair, and gene-specific transcription regulation. Although their role in cancer is still under debate, some studies suggest that specific HATs act as tumor suppressors (Gayther et al., 2000; Iyer et al., 2004; Kung et al., 2000; Miller and Rubinstein, 1995). Importantly, recent work demonstrating that depletion of representative HATs (i.e., p300, CBP, and P/CAF) using short hairpin RNAs (shRNAs) results in chromosome misalignment, aberrant delay in metaphase-to-anaphase transition, and subsequent mitotic catastrophe (Ha et al., 2009) provides strong evidence for a direct link between loss of HAT activity and mitotic disruption. Although we do not yet understand how HAT inhibition causes mitotic aberrations, protein acetylation/deacetylation by HATs and histone deacetylases (HDACs) has emerged as a major posttranslational modification involved in the regulation of fundamental cellular processes from chromatin dynamics to cell cycle control, suggesting that an imbalance in the activity of HATs and HDACs may have profound consequences on cell fate.
Considering the importance of posttranslational histone acetylation in determining chromatin architecture (Shogren-Knaak et al., 2006), it is possible that the mitotic delay in response to OTA occurs as a consequence of altered histone acetylation patterns and associated hypercondensation of mitotic chromosomes. With regard to the cohesion defects observed in OTA-treated cells, a direct role of HATs in regulating chromosome cohesion has not yet been reported. However, it is known that histone hyperacetylation in mitosis via inhibition of HDACs impairs sister chromatid resolution, providing evidence that the acetylation status of core histones may be important for timely removal of cohesion during mitosis (Cimini et al., 2003). Interestingly, however, several nonhistone proteins that participate in the control of sister chromatid cohesion also appear to be regulated by acetylation (Choudhary et al., 2009). This includes PDS5A and PDS5B (sister chromatid cohesion protein PDS5 homologs A and B) and the structural maintenance of chromosomes proteins Smc1A, Smc2, Smc3, and Smc4, which are part of the cohesion complex (Choudhary et al., 2009). How acetylation affects sister chromatid cohesion is largely unknown. However, Beckouet et al. (2010) found that establishment of cohesion during S phase requires acetylation of the cohesin subunit Smc3 (Beckouet et al., 2010). Although it remains to be established if Smc3 acetylation is affected by OTA, this suggests that cohesion defects in IHKE cells treated with OTA may in part result from reduced de novo Smc3 acetylation via inhibition of lysine acetyl transferases during earlier stages of the cell cycle. Indeed, the finding that cells completed a normal mitosis within the first few hours after OTA addition might suggest that OTA acts outside mitosis (e.g., in G1 or S phase) by altering transcription or posttranslational acetylation of mitotic regulators rather than by directly altering mitotic chromosome structure.
Accumulating evidence suggests that HATs play a direct role in the regulation of mitosis by mechanisms that are independent of their effects on chromatin architecture. CBP and p300, for instance, have been shown to associate with the anaphase-promoting complex/cyclosome (APC/C), which coordinates progression through mitosis by targeting substrates for ubiquitin-proteasome–mediated degradation (Turnell et al., 2005). Small interfering RNA (siRNA)-mediated knockdown of CBP but not p300 led to a marked reduction in APC/C E3 ubiquitin ligase activity, increased the levels of known APC/C substrates such as cyclin B1 and Plk-1, and blocked progression through mitosis (Turnell et al., 2005). These data point to a role of CBP in regulating APC/C function. Indeed, anaphase-promoting complex subunits 7, 8, and 10 (APC7, APC8, APC10) have recently been identified as nonhistone targets for acetylation (Choudhary et al., 2009). Moreover, modification of cell cycle proteins by acetylation can affect their stability by promoting their proteasomal degradation. Acetylation of cyclin A, for instance, was recently shown to occur simultaneously to ubiquitination at mitosis and appears to be required for cyclin A degradation and mitotic progression (Mateo et al., 2009). Importantly, global analysis of lysine acetylation by high-resolution mass spectrometry identified 243 acetylation sites on 132 proteins involved in cell cycle control, including several cyclins (e.g., Cyclin B1, Cyclin-T1) and cyclin-dependent kinases (e.g., CDK1cdc2, CDK2, CDK5, CDK6) (Choudhary et al., 2009), highlighting the potential importance of lysine acetylation in regulating cell division. In addition to the apparent role in controlling protein stability, acetylation has been shown to occur within the kinase and ATP-binding domain of CDK1cdc2 and other cell cycle kinases (Choudhary et al., 2009), suggesting that acetylation plays a direct role in regulating kinase activity and thus cell cycle progression. Thus, multiple mechanisms involving aberrant acetylation of histone and nonhistone proteins may contribute to OTA-mediated disruption of mitosis (Table 1).
TABLE 1.
Summary of Effects of OTA Consistent with Altered Acetylation of Histone (A) and Nonhistone Proteins (B)
| Proteins regulated by acetylation | Effect of acetylation | Reported effects of OTA consistent with hypoacetylation |
| A. Histones | Open chromatin structure | Hypercondensation (this study) |
| Gene transcription ↑ | Predominantly downregulation of gene expression (Arbillaga et al., 2008; Marin-Kuan et al., 2006; Luhe et al., 2003) | |
| B. Functional categories of nonhistone proteins | ||
| Cell cycle | ||
| Cyclin A | Proteasomal degradation, cell cycle progression (Mateo et al., 2009) | Arrest in mitosis (this study) |
| Cdk1cdc2 | Kinase activity ↑, cell cycle progression (Choudhary et al., 2009) | Arrest in mitosis (this study) |
| Smc3 | Sister chromatid cohesion (Beckouet et al., 2010) | Loss of sister chromatid cohesion (Mosesso et al., 2008; this study) |
| Transcription factors | ||
| Nrf-2 | Promoter-specific DNA binding, transcriptional activity ↑ (Sun et al., 2009) | Inhibition of Nrf-2–dependent gene expression (Marin-Kuan et al., 2006) |
| HNF4 | Nuclear localization and retention, HNF-4 DNA-binding activity, transcriptional activity ↑ (Soutoglou et al., 2000) | Inhibition of HNF4-dependent gene expression (Marin-Kuan et al., 2006) |
| Histone deacetylases | Enzyme activity ↓ (Spange et al., 2009) | Enzyme activity ↑ (Marin-Kuan et al., 2007) |
| DNA damage repair | ? | DNA strand breaks (e.g., Mally et al., 2005; Kamp et al., 2005) |
Interestingly, several features of OTA toxicity such as increased DNA strand breaks and inhibition of Nrf2-mediated antioxidant defense may also be explained by altered acetylation (Table 1). In addition to molecules involved in chromatin remodeling and cell cycle control, a large number of proteins with cellular functions in DNA damage repair are now known to be targets for acetylation (Choudhary et al., 2009). These include p53 and core proteins of its regulatory network as well as members of excision repair (base excision and nucleotide excision) and double-strand break repair pathways (nonhomologous end-joining and homologous recombination). How acetylation affects the activity of these DNA repair pathways remains to be elucidated. However, given that DNA replication/repair also requires access of polymerases/repair enzymes to DNA which is regulated by histone acetylation, it is intriguing to speculate that histone hypoacetylation and altered acetylation of DNA repair proteins might impair DNA repair and thus contribute to the persistence of (oxidative) DNA lesions reported to occur in cells and animals exposed to OTA (Kamp et al., 2005a, 2005b; Lebrun and Follmann, 2002; Mally et al., 2005b). Similarly, inhibition of HNF4alpha and Nrf2-dependent gene expression by OTA (Cavin et al., 2007; Marin-Kuan et al., 2006) may be brought about by reduced acetylation. Accumulating evidence shows that acetylation and deacetylation dynamically regulate the activity of a range of transcription factors at multiple levels including protein-protein interactions, protein stability, nuclear translocation, and DNA-binding affinity (Glozak et al., 2005; Spange et al., 2009). With regard to Nrf2, it was recently shown that p300/CBP–mediated acetylation of Nrf2 enhances promoter-specific DNA binding of Nrf2 and positively regulates its transcriptional activity, whereas lysine-to-arginine mutations at the acetylation sites compromise Nrf2 activity (Sun et al., 2009). Similarly, acetylation of HNF4 has been shown to be critical for HNF4-mediated transcriptional activation through retaining HNF4 in the nucleus and increasing HNF4 DNA binding (Soutoglou et al., 2000). Considering the relationship between acetylation and HNF4α transcriptional activity, Marin-Kuan et al. speculated that OTA may modulate HDACs. Indeed, a small but statistically significant increase in HDAC activity was found in nuclear extracts of kidneys of rats fed diets containing OTA for 7 and 21 days (Marin-Kuan et al., 2007), providing further evidence that OTA interferes with posttranslational protein acetylation. Although it is possible that OTA may exhibit a direct effect on HDACs, it is interesting to note that the activity of HDACs is also known to be negatively regulated by acetylation (Spange et al., 2009). Thus, reduced HDAC acetylation through inhibition of HAT enzymes by OTA may indirectly promote HDAC activity.
Collectively, these data illustrate that posttranslational acetylation of nonhistone proteins through OTA-mediated inhibition of HATs may be responsible for multiple aspects of OTA toxicity from mitotic disruption to altered transcription and stress/DNA damage response (Table 1). Global analysis of acetylation changes in response to OTA and identification of the specific HAT enzymes targeted by OTA will provide critical insight into the molecular mechanism of OTA toxicity and carcinogenicity and contribute to a more fundamental understanding as to how lysine acetylation participates in the regulation of diverse cellular functions.
The mechanistic link between inhibition of HATs and H3T3 hypophosphorylation still remains to be established. It is known that there is significant cross talk between posttranslational histone phosphorylation, acetylation, and methylation, and different histone modifications can mutually influence each other. Depletion of HDAC3 causes a decline in H3T3ph, perhaps through effects on H3K4 acetylation (Eot-Houllier et al., 2008); therefore, it is plausible that loss of H3K4 acetylation also influences H3T3ph. However, many other changes in chromatin structure brought about by hypoacetylation could also prohibit access of Haspin to H3T3, leading to inhibition of H3 phosphorylation at this site. Irrespective of the upstream events, the finding that OTA blocks H3T3 phosphorylation is significant as histone H3 phosphorylated at threonine 3 has recently been shown to be directly recognized by survivin, a member of the CPC (Kelly et al., 2010; Wang et al., 2010). This binding appears to be required for the recruitment of the CPC to chromosomes and subsequent activation of Aurora B, a serine/threonine kinase, and part of the CPC with distinct functions in chromosome condensation, segregation, and cytokinesis through regulation of microtubule kinetochore associations (Kelly et al., 2010; Wang et al., 2010). Loss of H3T3 phosphorylation through depletion of Haspin kinase delocalizes Aurora B from mitotic centromeres, leading to defects in Aurora B–dependent processes and delayed mitotic progression (Dai et al., 2005; Wang et al., 2010).
With regard to toxicological risk assessment, it should be pointed out that the shapes of the dose-response curves in this study varied depending on each specific end point studied. Thus, OTA-mediated inhibition of HAT activity followed a continuous dose-response with a slight reduction in HAT activity even at 1μM. Similarly, acetylation of histones and nonhistone targets was reduced in a concentration-dependent manner. Importantly, however, it appears that the mitotic perturbations induced by OTA follow an “all-or-nothing” response, occurring only when a certain degree of HAT inhibition is exceeded. This has important implications for OTA risk assessment as these clearly thresholded mitotic aberrations might be seen as most critical for OTA carcinogenicity. At the same time, the apparent differences in the dose-response might explain why some features of OTA toxicity also occur in organs or at doses not associated with OTA carcinogenicity, such as DNA strand breaks, which were also reported to occur in the nontarget organ liver and at doses below the threshold of nephrotoxicity (Kamp et al., 2005a; Mally et al., 2005b). In contrast, inhibition of Nrf2 response and mitotic abnormalities appear to be target organ specific and to occur only at doses known to cause tumor formation after chronic treatment (Adler et al., 2009; Cavin et al., 2007; Marin-Kuan et al., 2006). In view of extrapolation to in vivo, it should be emphasized that the concentrations applied in this study are in a similar range as plasma concentrations found in rats treated with OTA under conditions of carcinogenicity (7.4μM at 210 μg/kg bw for 90 days vs. < 1μM at 21 μg/kg bw) (Rached et al., 2007), further highlighting the relevance of our in vitro findings for renal tumor formation by OTA.
Finally, the finding that OTB produces the same mitotic perturbations as OTA with similar potency clearly deserves some attention. It has long been discussed that the chorine atom in para position to the phenolic group may represent a structural requirement for OTA toxicity. Some authors speculated that radical abstraction of the chlorine atom may facilitate DNA adduct formation (Gillman et al., 1999; Malaveille et al., 1994), and indeed, in vitro experiments demonstrate radical attack of the OTA phenoxyl radical generated by photoactivation at the C8 position of 2′-deoxyguanosine (Dai et al., 2003). Under physiological conditions, however, such reaction is unlikely to occur and no evidence for the formation of OTA-derived adducts has been obtained by liquid chromatography/tandem mass spectrometry or accelerator mass spectrometry (Delatour et al., 2008; Mally et al., 2004). In contrast, there is evidence to suggest that the lower toxicity of OTB in vivo may be the result of a more rapid elimination due to the lower lipophilicity and more extensive biotransformation (Li et al., 1997; Mally et al., 2005a). In line with this, our data clearly demonstrate that OTB is equally toxic in vitro, indicating that the chlorine atom is not required for OTA toxicity.
In conclusion, based on results from this and previous studies, we propose an MoA for OTA-mediated renal tumor formation in rats involving the following key events:
Inhibition of HAT activity, leading to hypoacetylation and altered function of histones and nonhistone proteins.
Sustained block in mitosis and aberrant exit from mitosis as a consequence of altered protein acetylation, resulting in both cell death and generation of polyploid cells. Although we did not observe mitotic catastrophe by live-cell imaging within the time frame of our study, there is evidence from both in vitro and in vivo studies to show that cell death at or after failed mitosis is initiated to eliminate at least a proportion of these genetically unstable cells (Rached et al., 2007, 2006; Schwerdt et al., 1999). Combined, single-cell degeneration and failure of cells to divide in the presence of OTA creates a net loss of cells within the renal proximal tubule epithelium.
Stimulation of cell proliferation and survival, including upregulation of mitosis-related genes with known oncogenic potential, potentially as a compensatory mechanism for impaired functioning of the mitotic machinery, leading to evasion of cell death and increasing polyploidy, genomic instability, and acquisition of a malignant phenotype.
SUPPLEMENTARY DATA
Supplementary data are available online at http://toxsci.oxfordjournals.org/.
FUNDING
Deutsche Forschungsgemeinschaft (MA 3323/2); the Society for Mycotoxin Research; the National Institutes of Health (R01 GM074210 to J.M.G.H).
Supplementary Material
Acknowledgments
The authors would like to thank Ursula Tatsch, Michaela Bektheshi, and Elisabeth Rüb-Spiegel for excellent technical assistance.
References
- Adler M, Muller K, Rached E, Dekant W, Mally A. Modulation of key regulators of mitosis linked to chromosomal instability is an early event in ochratoxin A carcinogenicity. Carcinogenesis. 2009;30:711–719. doi: 10.1093/carcin/bgp049. [DOI] [PubMed] [Google Scholar]
- Arbillaga L, Vettorazzi A, Gil AG, Joost HM, van Delft JH, Garcia-Jalon JA, Lopez de Cerain A. Gene expression changes induced by ochratoxin A in renal and hepatic tissues of male F344 rat after oral repeated administration. Toxicol. Appl. Pharmacol. 2008;230:197–207. doi: 10.1016/j.taap.2008.02.018. [DOI] [PubMed] [Google Scholar]
- Beckouet F, Hu B, Roig MB, Sutani T, Komata M, Uluocak P, Katis VL, Shirahige K, Nasmyth K. An Smc3 acetylation cycle is essential for establishment of sister chromatid cohesion. Mol. Cell. 2010;39:689–699. doi: 10.1016/j.molcel.2010.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boorman GA, McDonald MR, Imoto S, Persing R. Renal lesions induced by ochratoxin A exposure in the F344 rat. Toxicol. Pathol. 1992;20:236–245. doi: 10.1177/019262339202000210. [DOI] [PubMed] [Google Scholar]
- Carter SL, Eklund AC, Kohane IS, Harris LN, Szallasi Z. A signature of chromosomal instability inferred from gene expression profiles predicts clinical outcome in multiple human cancers. Nat. Genet. 2006;38:1043–1048. doi: 10.1038/ng1861. [DOI] [PubMed] [Google Scholar]
- Cavin C, Delatour T, Marin-Kuan M, Holzhauser D, Higgins L, Bezencon C, Guignard G, Junod S, Richoz-Payot J, Gremaud E, et al. Reduction in antioxidant defenses may contribute to ochratoxin A toxicity and carcinogenicity. Toxicol. Sci. 2007;96:30–39. doi: 10.1093/toxsci/kfl169. [DOI] [PubMed] [Google Scholar]
- Choudhary C, Kumar C, Gnad F, Nielsen ML, Rehman M, Walther TC, Olsen JV, Mann M. Lysine acetylation targets protein complexes and co-regulates major cellular functions. Science. 2009;325:834–840. doi: 10.1126/science.1175371. [DOI] [PubMed] [Google Scholar]
- Cimini D, Mattiuzzo M, Torosantucci L, Degrassi F. Histone hyperacetylation in mitosis prevents sister chromatid separation and produces chromosome segregation defects. Mol. Biol. Cell. 2003;14:3821–3833. doi: 10.1091/mbc.E03-01-0860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuny GD, Robin M, Ulyanova NP, Patnaik D, Pique V, Casano G, Liu JF, Lin X, Xian J, Glicksman MA, et al. Structure-activity relationship study of acridine analogs as Haspin and DYRK2 kinase inhibitors. Bioorg. Med. Chem. Lett. 2010;20:3491–3494. [PMC free article] [PubMed] [Google Scholar]
- Dai J, Sullivan BA, Higgins JM. Regulation of mitotic chromosome cohesion by Haspin and Aurora B. Dev. Cell. 2006;11:741–750. doi: 10.1016/j.devcel.2006.09.018. [DOI] [PubMed] [Google Scholar]
- Dai J, Sultan S, Taylor SS, Higgins JM. The kinase Haspin is required for mitotic histone H3 Thr 3 phosphorylation and normal metaphase chromosome alignment. Genes Dev. 2005;19:472–488. doi: 10.1101/gad.1267105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai J, Wright MW, Manderville RA. Ochratoxin A forms a carbon-bonded c8-deoxyguanosine nucleoside adduct: implications for c8 reactivity by a phenolic radical. J. Am. Chem. Soc. 2003;125:3716–3717. doi: 10.1021/ja034221r. [DOI] [PubMed] [Google Scholar]
- Delatour T, Mally A, Richoz J, Ozden S, Dekant W, Ihmels H, Otto D, Gasparutto D, Marin-Kuan M, Schilter B, et al. Absence of 2′-deoxyguanosine-carbon 8-bound ochratoxin A adduct in rat kidney DNA monitored by isotope dilution LC-MS/MS. Mol. Nutr. Food Res. 2008;52:472–482. doi: 10.1002/mnfr.200700276. [DOI] [PubMed] [Google Scholar]
- Dreger S, ÓBrien E, Stack M, Dietrich D. Antiproliverative effects and cell-cycle specific effects of ochratoxin A in LLC-PK1, NRK-52E and porcine primary proximal kidney cells. The Toxicologist. 2000;54:70. [Google Scholar]
- The European Commission, Directorate-General Health and Consumer Protection. (2002) Reports on Tasks for Scientific Cooperation (SCOOP): Assessment of Dietary Intake of Ochratoxin A by the Population of EU Member States. http://ec.europa.eu/food/fs/scoop/3.2.7_en.pdf. Accessed May 23, 2011. [Google Scholar]
- European Food Safety Authority (EFSA) Opinion of the scientific panel on contaminants in the food chain on a request from the commission related to ochratoxin A in food, EFSA-Q-2005-154. The EFSA Journal. 2006;365:1–56. [Google Scholar]
- Eot-Houllier G, Fulcrand G, Watanabe Y, Magnaghi-Jaulin L, Jaulin C. Histone deacetylase 3 is required for centromeric H3K4 deacetylation and sister chromatid cohesion. Genes Dev. 2008;22:2639–2644. doi: 10.1101/gad.484108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gayther SA, Batley SJ, Linger L, Bannister A, Thorpe K, Chin SF, Daigo Y, Russell P, Wilson A, Sowter HM, Delhanty JD, Ponder BA, Kouzarides T, Caldas C. Mutations truncating the EP300 acetylase in human cancers. Nat. Genet. 2000;24:300–303. doi: 10.1038/73536. [DOI] [PubMed] [Google Scholar]
- Gelato KA, Fischle W. Role of histone modifications in defining chromatin structure and function. Biol. Chem. 2008;389:353–363. doi: 10.1515/BC.2008.048. [DOI] [PubMed] [Google Scholar]
- Gillman IG, Clark TN, Manderville RA. Oxidation of ochratoxin A by an Fe-porphyrin system: model for enzymatic activation and DNA cleavage. Chem. Res. Toxicol. 1999;12:1066–1076. doi: 10.1021/tx9901074. [DOI] [PubMed] [Google Scholar]
- Glozak MA, Sengupta N, Zhang X, Seto E. Acetylation and deacetylation of non-histone proteins. Gene. 2005;363:15–23. doi: 10.1016/j.gene.2005.09.010. [DOI] [PubMed] [Google Scholar]
- Ha GH, Kim HS, Lee CG, Park HY, Kim EJ, Shin HJ, Lee JC, Lee KW, Lee CW. Mitotic catastrophe is the predominant response to histone acetyltransferase depletion. Cell Death Differ. 2009;16:483–497. doi: 10.1038/cdd.2008.182. [DOI] [PubMed] [Google Scholar]
- Higgins JM. Haspin: a newly discovered regulator of mitotic chromosome behavior. Chromosoma. 2010;119:137–147. doi: 10.1007/s00412-009-0250-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito T. Role of histone modification in chromatin dynamics. J. Biochem. 2007;141:609–614. doi: 10.1093/jb/mvm091. [DOI] [PubMed] [Google Scholar]
- Iyer NG, Ozdag H, Caldas C. p300/CBP and cancer. Oncogene. 2004;23:4225–4231. doi: 10.1038/sj.onc.1207118. [DOI] [PubMed] [Google Scholar]
- Kamp HG, Eisenbrand G, Janzowski C, Kiossev J, Latendresse JR, Schlatter J, Turesky RJ. Ochratoxin A induces oxidative DNA damage in liver and kidney after oral dosing to rats. Mol. Nutr. Food Res. 2005a;49:1160–1167. doi: 10.1002/mnfr.200500124. [DOI] [PubMed] [Google Scholar]
- Kamp HG, Eisenbrand G, Schlatter J, Wurth K, Janzowski C. Ochratoxin A: induction of (oxidative) DNA damage, cytotoxicity and apoptosis in mammalian cell lines and primary cells. Toxicology. 2005b;206:413–425. doi: 10.1016/j.tox.2004.08.004. [DOI] [PubMed] [Google Scholar]
- Kelly AE, Ghenoiu C, Xue JZ, Zierhut C, Kimura H, Funabiki H. Survivin reads phosphorylated histone H3 threonine 3 to activate the mitotic kinase Aurora B. Science. 2010;330:235–239. doi: 10.1126/science.1189505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kung AL, Rebel VI, Bronson RT, Ch'ng LE, Sieff CA, Livingston DM, Yao TP. Gene dose-dependent control of hematopoiesis and hematologic tumor suppression by CBP. Genes Dev. 2000;14:272–277. [PMC free article] [PubMed] [Google Scholar]
- Lebrun S, Follmann W. Detection of ochratoxin A-induced DNA damage in MDCK cells by alkaline single cell gel electrophoresis (comet assay) Arch. Toxicol. 2002;75:734–741. doi: 10.1007/s00204-001-0291-9. [DOI] [PubMed] [Google Scholar]
- Li S, Marquardt RR, Frohlich AA, Vitti TG, Crow G. Pharmacokinetics of ochratoxin A and its metabolites in rats. Toxicol. Appl. Pharmacol. 1997;145:82–90. doi: 10.1006/taap.1997.8155. [DOI] [PubMed] [Google Scholar]
- Luhe A, Hildebrand H, Bach U, Dingermann T, Ahr HJ. A new approach to studying ochratoxin A (OTA)-induced nephrotoxicity: expression profiling in vivo and in vitro employing cDNA microarrays. Toxicol. Sci. 2003;73:315–328. doi: 10.1093/toxsci/kfg073. [DOI] [PubMed] [Google Scholar]
- Malaveille C, Brun G, Bartsch H. Genotoxicity of ochratoxin A and structurally related compounds in Escherichia coli strains: studies on their mode of action. IARC Sci. Publ. 1991;115:261–266. [PubMed] [Google Scholar]
- Malaveille C, Brun G, Bartsch H. Structure-activity studies in E. coli strains on ochratoxin A (OTA) and its analogues implicate a genotoxic free radical and a cytotoxic thiol derivative as reactive metabolites. Mutat. Res. 1994;307:141–147. doi: 10.1016/0027-5107(94)90286-0. [DOI] [PubMed] [Google Scholar]
- Mally A, Dekant W. DNA adduct formation by ochratoxin A: review of the available evidence. Food Addit. Contam. 2005;22(Suppl. 1):65–74. doi: 10.1080/02652030500317544. [DOI] [PubMed] [Google Scholar]
- Mally A, Keim-Heusler H, Amberg A, Kurz M, Zepnik H, Mantle P, Volkel W, Hard GC, Dekant W. Biotransformation and nephrotoxicity of ochratoxin B in rats. Toxicol. Appl. Pharmacol. 2005a;206:43–53. doi: 10.1016/j.taap.2004.11.007. [DOI] [PubMed] [Google Scholar]
- Mally A, Pepe G, Ravoori S, Fiore M, Gupta RC, Dekant W, Mosesso P. Ochratoxin A causes DNA damage and cytogenetic effects but no DNA adducts in rats. Chem. Res. Toxicol. 2005b;18:1253–1261. doi: 10.1021/tx049650x. [DOI] [PubMed] [Google Scholar]
- Mally A, Zepnik H, Wanek P, Eder E, Dingley K, Ihmels H, Völkel W, Dekant W. Ochratoxin A: lack of formation of covalent DNA adducts. Chem. Res. Toxicol. 2004;17:234–242. doi: 10.1021/tx034188m. [DOI] [PubMed] [Google Scholar]
- Mantle PG, Faucet-Marquis V, Manderville RA, Squillaci B, Pfohl-Leszkowicz A. Structures of covalent adducts between DNA and ochratoxin a: a new factor in debate about genotoxicity and human risk assessment. Chem. Res. Toxicol. 2010;23:89–98. doi: 10.1021/tx900295a. [DOI] [PubMed] [Google Scholar]
- Marin-Kuan M, Nestler S, Verguet C, Bezencon C, Piguet D, Delatour T, Mantle P, Cavin C, Schilter B. MAPK-ERK activation in kidney of male rats chronically fed ochratoxin A at a dose causing a significant incidence of renal carcinoma. Toxicol. Appl. Pharmacol. 2007;224:174–181. doi: 10.1016/j.taap.2007.06.014. [DOI] [PubMed] [Google Scholar]
- Marin-Kuan M, Nestler S, Verguet C, Bezencon C, Piguet D, Mansourian R, Holzwarth J, Grigorov M, Delatour T, Mantle P, et al. A toxicogenomics approach to identify new plausible epigenetic mechanisms of ochratoxin A carcinogenicity in rat. Toxicol. Sci. 2006;89:120–134. doi: 10.1093/toxsci/kfj017. [DOI] [PubMed] [Google Scholar]
- Mateo F, Vidal-Laliena M, Canela N, Busino L, Martinez-Balbas MA, Pagano M, Agell N, Bachs O. Degradation of cyclin A is regulated by acetylation. Oncogene. 2009;28:2654–2666. doi: 10.1038/onc.2009.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller RW, Rubinstein JH. Tumors in Rubinstein-Taybi syndrome. Am. J. Med. Genet. 1995;56:112–115. doi: 10.1002/ajmg.1320560125. [DOI] [PubMed] [Google Scholar]
- Mosesso P, Cinelli S, Pinero J, Bellacima R, Pepe G. In vitro cytogenetic results supporting a DNA nonreactive mechanism for ochratoxin A, potentially relevant for its carcinogenicity. Chem. Res. Toxicol. 2008;21:1235–1243. doi: 10.1021/tx800029f. [DOI] [PubMed] [Google Scholar]
- National Toxicology Program. Toxicology and carcinogenesis studies of ochratoxin A (CAS No. 303-47-9) in F344/N rats (Gavage studies) Natl. Toxicol. Program Tech. Rep. Ser. 1989;358:1–142. [PubMed] [Google Scholar]
- Palma N, Cinelli S, Sapora O, Wilson SH, Dogliotti E. Ochratoxin A-induced mutagenesis in mammalian cells is consistent with the production of oxidative stress. Chem. Res. Toxicol. 2007;20:1031–1037. doi: 10.1021/tx700027j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patnaik D, Jun X, Glicksman MA, Cuny GD, Stein RL, Higgins JM. Identification of small molecule inhibitors of the mitotic kinase Haspin by high-throughput screening using a homogeneous time-resolved fluorescence resonance energy transfer assay. J. Biomol. Screen. 2008;13:1025–1034. doi: 10.1177/1087057108326081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rached E, Hard GC, Blumbach K, Weber K, Draheim R, Lutz WK, Ozden S, Steger U, Dekant W, Mally A. Ochratoxin a: 13-week oral toxicity and cell proliferation in male F344/N rats. Toxicol. Sci. 2007;97:288–298. doi: 10.1093/toxsci/kfm042. [DOI] [PubMed] [Google Scholar]
- Rached E, Pfeiffer E, Dekant W, Mally A. Ochratoxin A: apoptosis and aberrant exit from mitosis due to perturbation of microtubule dynamics? Toxicol. Sci. 2006;92:78–86. doi: 10.1093/toxsci/kfj213. [DOI] [PubMed] [Google Scholar]
- Schilter B, Marin-Kuan M, Delatour T, Nestler S, Mantle P, Cavin C. Ochratoxin A: potential epigenetic mechanisms of toxicity and carcinogenicity. Food Addit. Contam. 2005;22(Suppl. 1):88–93. doi: 10.1080/02652030500309319. [DOI] [PubMed] [Google Scholar]
- Schwerdt G, Freudinger R, Mildenberger S, Silbernagl S, Gekle M. The nephrotoxin ochratoxin A induces apoptosis in cultured human proximal tubule cells. Cell Biol. Toxicol. 1999;15:405–415. doi: 10.1023/a:1007662101880. [DOI] [PubMed] [Google Scholar]
- Shogren-Knaak M, Ishii H, Sun JM, Pazin MJ, Davie JR, Peterson CL. Histone H4-K16 acetylation controls chromatin structure and protein interactions. Science. 2006;311:844–847. doi: 10.1126/science.1124000. [DOI] [PubMed] [Google Scholar]
- Soutoglou E, Katrakili N, Talianidis I. Acetylation regulates transcription factor activity at multiple levels. Mol. Cell. 2000;5:745–751. doi: 10.1016/s1097-2765(00)80253-1. [DOI] [PubMed] [Google Scholar]
- Spange S, Wagner T, Heinzel T, Kramer OH. Acetylation of non-histone proteins modulates cellular signalling at multiple levels. Int. J. Biochem. Cell Biol. 2009;41:185–198. doi: 10.1016/j.biocel.2008.08.027. [DOI] [PubMed] [Google Scholar]
- Sun Z, Chin YE, Zhang DD. Acetylation of Nrf2 by p300/CBP augments promoter-specific DNA binding of Nrf2 during the antioxidant response. Mol. Cell Biol. 2009;29:2658–2672. doi: 10.1128/MCB.01639-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reports on Tasks for Scientific Cooperation (SCOOP): The European Commission, Directorate-General Health and Consumer Protection. (2002) Assessment of Dietary Intake of Ochratoxin A by the Population of EU Member States. Available at: http://ec.europa.eu/food/fs/scoop/3.2.7_en.pdf. Accessed May 23, 2011. [Google Scholar]
- Turnell AS, Stewart GS, Grand RJ, Rookes SM, Martin A, Yamano H, Elledge SJ, Gallimore PH. The APC/C and CBP/p300 cooperate to regulate transcription and cell-cycle progression. Nature. 2005;438:690–695. doi: 10.1038/nature04151. [DOI] [PubMed] [Google Scholar]
- Wang F, Dai J, Daum JR, Niedzialkowska E, Banerjee B, Stukenberg PT, Gorbsky GJ, Higgins JM. Histone H3 Thr-3 phosphorylation by Haspin positions Aurora B at centromeres in mitosis. Science. 2010;330:231–235. doi: 10.1126/science.1189435. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.