Abstract
The densin C-terminal domain can target Ca2+/calmodulin-dependent protein kinase IIα (CaMKIIα) in cells. Although the C-terminal domain selectively binds CaMKIIα in vitro, full-length densin associates with CaMKIIα or CaMKIIβ in brain extracts and in transfected HEK293 cells. This interaction requires a second central CaMKII binding site, the densin-IN domain, and an “open” activated CaMKII conformation caused by Ca2+/calmodulin binding, autophosphorylation at Thr-286/287, or mutation of Thr-286/287 to Asp. Mutations in the densin-IN domain (L815E) or in the CaMKIIα/β catalytic domain (I205/206K) disrupt the interaction. The amino acid sequence of the densin-IN domain is similar to the CaMKII inhibitor protein, CaMKIIN, and a CaMKIIN peptide competitively blocks CaMKII binding to densin. CaMKII is inhibited by both CaMKIIN and the densin-IN domain, but the inhibition by densin is substrate-selective. Phosphorylation of a model peptide substrate, syntide-2, or of Ser-831 in AMPA receptor GluA1 subunits is fully inhibited by densin. However, CaMKII phosphorylation of Ser-1303 in NMDA receptor GluN2B subunits is not effectively inhibited by densin in vitro or in intact cells. Thus, densin can target multiple CaMKII isoforms to differentially modulate phosphorylation of physiologically relevant downstream targets.
Keywords: Calcium Calmodulin-dependent Protein Kinase (CaMK); Glutamate Receptors Ionotropic (AMPA, NMDA); Protein Phosphorylation; Protein Targeting; Protein-Protein Interactions; Synapses
Introduction
Activation of N-methyl d-aspartate receptors (NMDARs)5 and/or voltage-gated calcium channels induces postsynaptic calcium signals that vary in frequency, duration, and amplitude. Ca2+/calmodulin-dependent protein kinase II (CaMKII) decodes these signals to elicit multiple cellular responses, especially during learning and memory. Precise targeting of CaMKII to specific subcellular compartments containing upstream activators, other modulators, and/or specific substrates is thought to modulate the specificity and efficiency of CaMKII actions (1, 2). Neuronal postsynaptic densities contain several CaMKII-associated proteins (CaMKAPs), such as NMDAR GluN2B subunits, α-actinin and densin, that presumably collaborate to dynamically control CaMKII targeting to, and perhaps within, the postsynaptic density during synaptic activation. Whereas CaMKII binding to GluN2B is required for some forms of LTP (3, 4), specific roles of other CaMKAPs are less well understood.
Densin was identified as a neuronal postsynaptic density-enriched protein that can bind CaMKII (5–7). Like other LAP protein family members, densin contains leucine-rich repeats (LRRs) at the N terminus and a C-terminal PDZ domain (5). The LRR domain was originally proposed to be extracellular, but more recent studies indicate that densin is entirely intracellular and associates with plasma membranes at least in part via the LRR domain (8–10). Several postsynaptic proteins can interact with the densin PDZ domain (e.g. α-actinin, L-type calcium channel CaV1.3 α1 subunit) (7, 11, 12) or other C-terminal domains (e.g. CaMKIIα, SHANK, δ-catenin) (6, 8, 9, 13). These data suggest that densin is an archetypical scaffolding protein that assembles multiprotein complexes to enhance postsynaptic signaling specificity and efficiency at the plasma membrane. Consistent with this model, densin is required for CaMKIIα to elicit a novel form of CaV1.3 L-type calcium channel facilitation (12).
CaMKII genes (α, β, γ, δ) are differentially expressed in most if not all mammalian tissues and share 80–90% amino acid sequence identity in their catalytic and regulatory domains (14). Consequently, their intrinsic catalytic and regulatory properties are very similar, with the most notable difference being ≈10-fold variations in the affinity for Ca2+/calmodulin (15). However, they are more divergent in C-terminal association domains that are responsible for assembly of dodecameric CaMKII holoenzymes. CaMKII isoforms also may be differentially targeted in cells, for example to the F-actin cytoskeleton or to the nucleus (16–18). Initial studies found that the C-terminal association domain of CaMKIIα, but not CaMKIIβ, interacts with a CaMKII binding site immediately preceding the C-terminal PDZ domain of densin (6, 7), suggesting that densin is a CaMKIIα-specific CaMKAP. Indeed, this C-terminal CaMKII binding site is sufficient to associate with and target CaMKIIα in intact cells (6, 9). However, densin variants are expressed in embryos and in the early postnatal period before expression of CaMKIIα (9). Moreover, densin-dependent facilitation of CaV1.3 L-type calcium channels by CaMKIIα is ablated by deletion of a large intracellular domain in densin (Δ483–1377) even though this protein retained the LRR, C-terminal CaMKIIα binding, and PDZ domains. Surprisingly, this internal deletion also reduced the association of densin with CaMKIIα (12).
Here we show that densin associates with both CaMKIIα and CaMKIIβ in the brain and in transfected HEK293 cells. These interactions involve a second central/internal CaMKII binding site, the densin-IN domain, that displays 50% sequence similarity to the CaMKII inhibitor protein, CaMKIIN. The densin-IN domain interacts with catalytic domains of either isoform, but the densin C-terminal domain interacts efficiently only with CaMKIIα. Notably, we show that the densin-IN domain differentially modulates CaMKII phosphorylation of AMPAR GluA1 subunits and NMDAR GluN2B subunits, suggesting a unique mechanism for fine-tuning CaMKII activity toward different substrates.
EXPERIMENTAL PROCEDURES
Purification of GST Fusion Proteins
GST fusion proteins were expressed in Escherichia coli (BL21-DE3 Gold) and purified using glutathione-agarose (Sigma) as described previously (6). GST-T482 was generated by insertion of cDNA encoding the T482 densin splice variant (9) into a pGEX-4T vector and contains residues 1–482 of regular densin sequence plus 13 novel C-terminal residues due to a frameshift resulting from alternative splicing. GST-D-CTA contains residues 1247–1542(Δ1292–1337), as described (6, 11). For protein interaction studies, GST-N2B contains residues 1260–1339 with Ser-1303 mutated to Ala (11, 19), whereas for phosphorylation studies (Fig. 8B) GST-N2Bs contains residues 1260–1309 with the wild type sequence. GST-GluA1 contains residues 816–889 as described previously (20). Point mutations in GST-densin fusion proteins were created using QuikChange site-directed mutagenesis (Stratagene, Santa Clara, CA). Purified proteins were quantified using Bradford reagent (Bio-Rad).
FIGURE 8.
Densin inhibits phosphorylation of GluA1, but not GluN2B, in intact cells. Triton soluble fractions (Input) of HEK293 cells expressing GluA1 or FLAG-GluN2B with or without CaMKIIα and/or densin were immunoprecipitated (IP) (see “Experimental Procedures”). A, inputs and GluA1 immune complexes were immunoblotted for CaMKII, densin, total GluA1, or phospho-Ser-831 GluA1 (P-Ser-831), as indicated. The graph summarizes the quantification of Ser-831 phosphorylation normalized to total GluA1 from 5 similar experiments. *, p < 0.05 compared with GluA1 alone. **, p < 0.01 compared with GluA1+CAMKII and to GluA1+CAMKII+L815E-densin. B, inputs or FLAG-GluN2B complexes were immunoblotted for CaMKII, densin, total GluN2B, or phospho-Ser-1303 GluN2B (P-Ser1303) as indicated. The graph summarizes the quantification of Ser-1303 phosphorylation normalized to total GluN2B from 4 similar experiments. ***, p < 0.0001 compared with GluN2B alone.
Autophosphorylation of CaMKII Isoforms and Truncation Mutants
Murine CaMKIIα, Xenopus CaMKIIβ, porcine CaMKIIγB, and rat CaMKIIδ2 holoenzymes as well as monomeric CaMKIIα-(1–420) and CaMKIIα-(1–380) truncation mutants were purified from baculovirus-infected Sf9 cells and selectively autophosphorylated at Thr-286/287 essentially as described (11, 19).
Synthetic Peptides
Peptides were custom-synthesized by Macromolecular Resources (University of Colorado, Fort Collins, CO) or by Global Peptide Services (Fort Collins, CO). N-tide (KRPPKLGQIGRSKRVVIEDDRIDDVLK) is a peptide analog of residues 43–69 of the CaMKIIN inhibitor protein. N2B-tide (AQKKNRNKLRRQHAYDTFVD) is an analog of GluN2B residues 1290–1309, with Ala in place of Ser-1303. Syntide-2 is PLARTLSVAGLPGKK. Autocamtide-2 is KKALRRQETVDAL.
GST Co-sedimentation Assays
Unless indicated otherwise, purified GST fusion proteins or GST alone (≈250 nm full-length protein), purified CaMKII isoforms, or truncation mutants (≈250 nm subunit concentration preincubated to autophosphorylate Thr-286/287 or non-phosphorylated, as indicated) and glutathione-agarose (Sigma; 25 μl packed resin) were incubated for 2 h at 4 °C in a final volume of 0.5 ml in PD buffer (50 mm Tris-HCl, pH 7.5, 200 mm NaCl, 0.5% (v/v) Triton X-100). Peptides were tested as competitors at a final concentration of 50 μm. After centrifugation, beads were washed at least four times in PD buffer. Proteins were then eluted from the beads, resolved by SDS-PAGE, and either stained with Coomassie Blue or transferred to nitrocellulose membrane for staining with Ponceau-S followed by immunoblotting.
CaMKII Inhibition Assay
GST-D-IN, GST-D-IN(L815E), N-tide, or a GST control was diluted into ice-cold 50 mm HEPES, pH 7.5, 1 mg/ml bovine serum albumin, 10 mm magnesium acetate, 0.5 mm CaCl2, 1 μm CaM, 1 mm DTT, and 0.4 mm [γ-32P]ATP (≈500 cpm/pmol) containing the indicated substrate: syntide-2 (20 μm), GST-GluA1 (2 μm), or GST-N2Bs (2 μm). Reactions were initiated by the addition of CaMKII (0.75 nm for syntide-2) or autophosphorylated CaMKII (10 nm for GST protein phosphorylation) and incubated at 30 °C for 10 min. Aliquots were spotted on phosphocellulose paper (Whatman P81), washed with water, rinsed with ethanol, and dried for scintillation counting in non-aqueous scintillant. Remaining GST-GluA1 and GST-N2Bs reactions were quenched with SDS, resolved by SDS-PAGE, and stained with Coomassie Blue before autoradiography.
Mammalian Expression Vectors
The expression vector for densin-FLA (densin-Δ1292–1337) with a C-terminal GFP tag was described previously (9). An expression vector for GFP-tagged densin-FLC (densin-Δ1292–1337/Δ1378–1451) was generated similarly in the pEGFP vector (Clontech, BD Biosciences).
The construct to express a constitutively active GFP-CaMKIIα-(1–290) was a generous gift from Dr. Nikolai Otmakhov (Brandeis). The untagged CaMKIIα was described previously (9). GFP-CaMKIIβ (WT and T287D) vectors were a generous gift from Dr. Lori Redmond (Medical College of Georgia) (21). The untagged CaMKIIβ construct was generated by PCR amplification of the entire coding sequence from a GFP-CaMKIIβ WT template and inserted into pcDNA3.1 at EcoRI/XhoI sites. CaMKIIα and CaMKIIβ were also inserted into the mCherry-C3 vector at EcoR1/BamHI or XhoI/EcoRI sites, respectively.
Densin and CaMKII point mutants were created by QuikChange Mutagenesis (Stratagene, Santa Clara, CA) and verified by sequencing. Plasmid DNA was purified using Maxi-Prep kits (Invitrogen).
Cell Culture and Transfection
HEK293 cells were maintained in modified Eagle's medium containing 10% fetal bovine serum (Invitrogen), penicillin-streptomycin (Sigma), and 2 mm glutamate (Sigma) at 37 °C in 5% CO2. Cells were transfected using FuGENE (Roche Applied Science) or PolyJet (SignaGen Laboratories, Gaithersburg, MD) (3 μl/μg of DNA).
HEK293 Cell Lysis, Immunoprecipitation and Pulldown
For immunoprecipitation, HEK293 cells (48 h after transfection) were rinsed with cold PBS twice and lysed on ice with 1.6 ml per 10 cm plate of Buffer A (2 mm Tris-HCl, pH 7.5, 1% (v/v) Triton X-100, 0.1 mm PMSF, 1 mm benzamidine, 5 mg/liter leupeptin, 20 mg/liter soybean trypsin inhibitor). After sonication, lysates were incubated at 4 °C for 1 h and then centrifuged for 10 min at 10,000 × g. NaCl (120 mm final concentration) was added to the supernatants, and equal aliquots were incubated overnight at 4 °C with Densin Ab450 or goat IgG (10 μg each). After the addition of GammaBind Plus-Sepharose (Amersham Biosciences) (30 μl, 1:1 slurry) and continued incubation for 2 h at 4 °C, samples were centrifuged and washed ≥4 times with 1 ml of Buffer B (50 mm Tris-HCl, 150 mm NaCl, 1% (v/v) Triton X-100). The Sepharose was transferred to new microcentrifuge tubes during the first wash. Immune complexes were solubilized in SDS-PAGE sample buffer for immunoblot analysis.
HEK293 cells expressing CaMKII isoforms alone (48 h after transfection) were processed for GST co-sedimentation assays essentially as for immunoprecipitations. The supernatants were incubated for 2 h at 4 °C with 30 μg of GST proteins and glutathione-agarose (50 μl of 1:1 slurry). Agarose was collected and washed at least 4 times with 1 ml of Buffer B, transferring to a new tube at the 2nd wash. Complexes were resolved by SDS-PAGE and transferred to nitrocellulose membranes. CaMKII co-sedimented on the beads was detected by Ponceau-S staining or by immunoblotting.
Phosphorylation of Glutamate Receptor Subunits in HEK293 Cells
Lysates of HEK293 cells expressing GluA1 or FLAG-GluN2B with or without WT CaMKII and/or densin-FLA (WT or L815E) were prepared 24 h after transfection. Cells expressing GluN2B were treated before lysis with 2 mm EGTA in PBS for 5 min and then with 2 mm CaCl2 in PBS for 5 min, all at room temperature. The lysis buffer (Buffer A) also contained 50 mm NaF, 200 μm Na3VO4, and 1 μm microcystin-LR. Immune complexes were isolated from Triton-soluble fractions using either GluA1 (Santa Cruz Biotechnology, Santa Cruz, CA) with a mouse IgG (Jackson ImmunoResearch, West Grove, PA) (2 μg each) control or anti-FLAG antibody-conjugated agarose beads (Sigma) (40 μl) as described above.
Preparation of Mouse Brain Extracts for Immunoprecipitation
Forebrains from WT and CaMKIIα knock-out littermates (generous gift from Dr. Y. Elgersma, Rotterdam) were homogenized in low ionic strength Buffer C (2 mm Tris-HCl, pH 7.5, 0.5% (v/v) Triton X-100, 2 mm EDTA, 2 mm EGTA, 1 mm DTT, 0.2 mm PMSF, 1 mm benzamidine, 10 μg/ml leupeptin, 10 μm pepstatin, and 1 μm microcystin-LR) in a Teflon-glass Wheaton tissue grinder with motorized plunger and incubated at 4 °C for 30–60 min. Total protein concentrations were adjusted to 2.4 mg/ml, and samples were centrifuged at 9000 × g for 10 min at 4 °C. Supernatants were mixed with mouse CaMKIIα (12 μg) or CaMKIIβ (2 μg) antibody or an IgG1 control and incubated at 4 °C for 1 h. After the addition of GammaBind Plus-Sepharose (Amersham Biosciences) (30 μl of 1:1 slurry), incubations were continued overnight at 4 °C. Beads were collected by microcentrifugation and washed ≥3 times with 1 ml of Buffer C containing 50 mm Tris-HCl, pH 7.5, and 150 mm NaCl, transferring samples to a new tube during the second wash. Immune complexes were solubilized in SDS-PAGE sample buffer for immunoblotting.
Immunoblotting
SDS-polyacrylamide gels were transferred to nylon-backed nitrocellulose membranes in 10 mm CAPS buffer. After blocking in TTBS (50 mm Tris-HCl, pH 7.5, 0.1% (v/v) Tween 20, 150 mm NaCl) containing 5% Carnation nonfat milk, membranes were incubated for either 2 h at room temperature or overnight at 4 °C with primary antibodies diluted in TTBS with 5% milk. For densin, antibody R-300 was from Santa Cruz Biotechnology, and Ab450 and Ab650 were described previously (9). Antibodies for total and phospho-Ser-831 GluA1 were from Abcam (Cambridge, MA) and PhosphoSolutions (Aurora, CO), respectively. Antibodies to total and phospho-Ser-1303 GluN2B antibody were from BD Biosciences and Millipore (Billerica, MA), respectively. Mouse monoclonal antibodies to CaMKIIα (Affinity BioReagents, Golden, CO), CaMKIIβ (Invitrogen), GFP (Santa Cruz Biotechnology), and RFP (AbCam, Cambridge, MA) were also used. Membranes were washed 5 times in TTBS and incubated for 1 h at room temperature with secondary antibodies conjugated to either alkaline phosphatase (Jackson ImmunoResearch), horseradish peroxidase (Promega or Santa Cruz Biotechnology), or infrared dyes (LiCor Biosciences, Lincoln, NE) diluted in TTBS with 5% milk. After extensive washing, secondary antibodies were detected colorimetrically with nitro blue tetrazolium/5-bromo-4-chloro-3-indolyl phosphate by enhanced chemiluminescence (PerkinElmer Life Sciences), or using an Odyssey system (LiCor Biosciences), respectively.
Quantification and Statistics
CaMKII associated with GST proteins was quantified using ImageJ from digital scans of Coomassie-stained gels, Ponceau-S-stained membranes, immunoblotted membranes, or x-ray films. Binding of purified CaMKII was normalized to recovered GST fusion protein, and background binding to GST alone was subtracted. For pulldown of CaMKII from HEK293 cells, CaMKII were normalized to both the amount of recovered GST proteins and CaMKII levels in input. Data are reported as the mean ± S.E. from the indicated number of observations and were analyzed by one-way or two-way analysis of variance with post-hoc Tukey or Dunnett's tests. CaMKII inhibition curves (plotted as a % of the “no inhibitor” control) were fitted to sigmoidal dose-response models using GraphPad Prism, constraining top and bottom values to 100 and ≥0, respectively.
RESULTS
Densin Associates with Either CaMKIIα or CaMKIIβ
Initially, we examined interactions of recombinant purified CaMKII isoforms with GST-D-CTA, a fusion protein containing the densin C-terminal domain of the major adult (A) splice variant (amino acid residues 1247–1542(Δ1292–1337); termed GST-densin-A in Strack et al. (6)) (Fig. 1A). A GST fusion protein containing residues 1260–1339 of the NMDA receptor GluN2B subunit (with a S1303A mutation) (GST-N2B) was included as a positive control. CaMKII isoforms were first autophosphorylated at Thr-286 in α or Thr-287 in β/γ/δ within the regulatory domain, generating Ca2+-independent (autonomously active) forms. GST-N2B bound similar amounts of all four CaMKII isoforms, as detected by immunoblot or by protein staining, but GST-D-CTA bound at least 10-fold more CaMKIIα compared with other isoforms (Fig. 1B and data not shown). We also compared interactions of GST-D-CTA and GST-N2B with two purified monomeric C-terminal truncation mutants of CaMKIIα containing amino acid residues 1–420 or 1–380 that were also first autophosphorylated to generate the Ca2+-independent form. Under these conditions, GST-N2B bound comparable amounts of all three forms of autophosphorylated CaMKIIα, but GST-D-CTA bound ≥10-fold more wild type CaMKIIα holoenzyme than either monomeric form (Fig. 1C). Taken together, these data show that the densin C-terminal domain interacts highly selectively with the intact CaMKIIα holoenzyme association domain, consistent with previous yeast two-hybrid data (7).
FIGURE 1.
The densin C-terminal domain binds selectively to CaMKIIα. A, domain structure of densin and the GST-D-CTA fusion protein. TM, originally proposed putative transmembrane domain; CaMKIIBD, known C-terminal CaMKIIα binding domain. B, CaMKII isoforms were pre-autophosphorylated at Thr-286/7 and then incubated with GST alone, GST-D-CTA, or GST-N2B. Left, the protein-stained membrane shows the GST protein preparations used (top: an asterisk on the left marks the full-length GST-D-CTA protein) and a representative immunoblot for CaMKII bound to the GST proteins (bottom). Right, quantification of CaMKII isoform binding to GST-D-CTA, expressed as a percentage of the binding of CaMKIIα (mean ± S.E., n = 3. *, p < 0.001). Binding of CaMKIIδ2 could not be accurately quantified because it co-migrates with GST-D-CTA. C, WT and monomeric CaMKIIα mutants were Thr-286-autophosphorylated, incubated with the indicated GST proteins, and then analyzed as in panel B. Binding of truncation mutants was expressed as a percentage of WT CaMKIIα binding (mean ± S.E., n = 3. *, p < 0.001).
CaMKIIα co-immunoprecipitates with full-length densin-A (densin-FLA) or the C terminus of the A variant from extracts of co-transfected HEK293 cells (9). Surprisingly, in heterologous cells a previously described perinatal densin-FLC variant lacking the C-terminal CaMKII binding domain (6) (Fig. 2A) binds only slightly reduced amounts of CaMKIIα compared with densin-FLA (Fig. 2B). Moreover, similar amounts of CaMKIIβ were readily detected in densin immune complexes isolated from lysates of HEK293 cells co-expressing CaMKIIβ with either densin-FLA or densin-FLC (Fig. 2C). In addition, CaMKIIδ isoforms associated with densin-FLA in HEK293 cell lysates (data not shown). Thus, despite the CaMKII isoform selectivity of the densin C-terminal domain in vitro (Fig. 1B), multiple isoforms associate with full-length densin proteins independently of the C-terminal CaMKII binding domain.
FIGURE 2.
Full-length densin associates with both CaMKIIα and CaMKIIβ. A, comparison of the major adult densin splice variant (densin-FLA) with a natural splice variant lacking the C-terminal CaMKII binding domain (densin-FLC). B, lysates from HEK293 cells expressing CaMKIIα and either GFP-tagged densin-FLA or densin-FLC were immunoprecipitated using densin Ab450 or control goat IgG. Extracts (Input) and immune complexes were immunoblotted for densin and CaMKIIα. C, lysates from HEK293 cells expressing CaMKIIβ and either GFP-tagged densin-FLA or densin-FLC were analyzed as in panel B. D, forebrain extracts from WT or CaMKIIα-KO mice were immunoprecipitated using mouse antibodies to CaMKIIα or CaMKIIβ or control IgG. Extracts and immune complexes were immunoblotted for densin and both CaMKII isoforms.
To investigate the CaMKII isoform selectivity of densin in the brain, mouse forebrain extracts were immunoprecipitated using CaMKIIα- or CaMKIIβ-selective monoclonal antibodies. Each antibody isolated complexes containing both CaMKII isoforms from WT mice (Fig. 2D), as expected due to co-assembly of mixed isoform holoenzymes. In addition, densin was readily detected in both immune complexes. Parallel experiments using forebrain extracts from CaMKIIα knock-out (KO) mice showed that CaMKIIα antibodies failed to immunoprecipitate CaMKIIβ or densin. CaMKIIα also was not detected in CaMKIIβ immune complexes from CaMKIIα-KO mice, demonstrating the specificity of the antibodies in immunoprecipitation assays. However, despite loss of the major CaMKIIα isoform, total levels of densin in CaMKIIβ immune complexes from CaMKIIα-KO mice were only modestly reduced compared with the levels in CaMKIIβ immune complexes from wild type tissue (Fig. 2D). These data indicate that densin associates with CaMKIIβ in brain.
Identification of a Second CaMKII Binding Domain in Densin
Association of multiple CaMKII isoforms with full-length densin variants in HEK293 cells and in the brain, independent of the known C-terminal CaMKII binding domain, suggests that densin contains an additional CaMKII binding domain. Because there are no detectable interactions of CaMKIIα with the N-terminal LRR domain of densin (densin-T482 splice variant) (9), CaMKII may also interact somewhere between the LRR and C-terminal domains. Consistent with this hypothesis, deletion of a large central domain (residues 483–1377) in a naturally occurring densin splice variant substantially reduced the binding of CaMKIIα even though this variant retains the C-terminal CaMKIIα binding domain (12). Therefore, we expressed a family of GST fusion proteins spanning the large central domain of densin in bacteria (Fig. 3A). GST-D-CTA and GST-T482 were included as positive and negative control proteins, respectively, allowing us to compare binding of purified CaMKIIα to all domains in densin. A GST fusion protein containing residues 708–962 of densin interacted strongly with Thr-286-autophosphorylated CaMKIIα in a glutathione-agarose cosedimentation assay, stronger than the binding to GST-D-CTA. Overexposure of the immunoblots revealed weaker interaction of CaMKIIα with a GST protein containing residues 963–1290 of densin but not with residues 482–707 or GST-T482 (Fig. 3B). Two further rounds of truncations within the 708–962 region created progressively smaller fragments, ultimately identifying residues 793–824 as a novel INternal CaMKII binding domain that we termed the densin-IN domain (Fig. 3, C and D).
FIGURE 3.
Identification of a novel CaMKII binding domain in densin. A, summary of truncation mutants created to identify the densin-IN domain. Letters B–D in parentheses indicate GST-densin protein families used in panels B–D; major CaMKII binding fragments are highlighted with thicker lines. Amino acid residues included in each fusion protein are below. B–D, GST or GST fusion proteins were incubated with Thr-286-autophosphorylated CaMKIIα, and proteins isolated using glutathione-agarose were analyzed by protein stain and/or CaMKII immunoblot as indicated. Full-length GST fusion proteins are indicated by asterisks located immediately to their left in each panel. B, first round analysis is shown. CaMKIIα binds to residues 708–962 >GST-D-CTA > residues 963–1290 but not to residues 482–707 or the N-terminal T482 (LRR) domain. C, second round analysis of densin-708–962 region is shown. CaMKIIα binds to residues 708–824 but not to residues 825–915 or 916–962. D, third round analysis of the densin-708–824 region is shown. CaMKIIα binds comparably to residues 761–824 and 793–824, with no detectable interaction with other fragments. Quantification (n ≥ 3) shows that CaMKII binding to densin-708–824 is fully recapitulated in the densin-793–824 fragment, hereafter referred to as the densin-IN domain.
CaMKII Activation Is Required for Interaction with the Densin-IN Domain
The densin-CTA domain interacts with the association domain of CaMKIIα, at least partially independent of kinase activation (6, 7). However, CaMKII activation is required for interactions with GluN2B and some other CaMKAPs. Therefore, we examined the effects of Ca2+/calmodulin, Mg2+, adenine nucleotides, and Thr-286 autophosphorylation on binding of purified CaMKIIα to GST-D-IN, which contains residues 793–824 of densin. Binding of inactive, non-phosphorylated CaMKII to GST-D-IN was not detected, but Ca2+/calmodulin partially stimulated the interaction (Fig. 4A). Further addition of Mg2+ alone had minimal effect, but the addition of Mg2+ with AMP-PNP (a non-hydrolyzable ATP analog) enhanced CaMKIIα binding. Thr-286 autophosphorylation of CaMKIIα was sufficient for strong interaction with GST-D-IN even when divalent cations from the autophosphorylation reaction were chelated with excess EDTA (Fig. 4A). The effects of Ca2+/calmodulin, Mg2+, AMP-PNP, and Thr-286 autophosphorylation on CaMKIIα interaction with the densin-IN domain were broadly similar to their previously reported effects on CaMKIIα binding to GST-N2B (22), but parallel analyses revealed significant quantitative differences (Fig. 4A). When normalized to the binding of Thr-286-autophosphorylated CaMKIIα to GST-N2B, Ca2+/calmodulin (in the absence or presence of Mg2+) supported ≈40% of binding to GST-D-IN but only ≈10% of binding to GST-N2B. In addition, binding of Thr-286 autophosphorylated CaMKIIα to GST-N2B, but not GST-D-IN, was modestly potentiated by Ca2+/calmodulin and nucleotide. Taken together, these data indicate that CaMKII interactions with densin and GluN2B are similar, yet subtly different.
FIGURE 4.
CaMKII activation is required for binding to the densin-IN domain. A, GST-D-IN or GST-N2B were incubated with non-phosphorylated or Thr-286-autophosphorylated CaMKIIα and the indicated additional reagents: 2 mm EDTA, 2 μm calmodulin (CaM), 2 mm calcium chloride (Ca2+),5 mm magnesium acetate (Mg2+), 0.1 mm AMP-PNP. Complexes were isolated using glutathione-agarose and then washed using buffers containing the respective concentrations of EDTA or divalent cations. Binding (mean ± S.E., n = 3) was expressed as a percentage of Thr-286-autophosphorylated CaMKIIα binding to GST-N2B in the presence of excess EDTA (condition g) (*, p < 0.05; **, p < 0.01; ***, p < 0.001; as indicated). B, CaMKIIα or CaMKIIβ were incubated with GST-D-IN or GST-N2B after autophosphorylation at Thr-286/287 and in the presence of excess EDTA (left panel) or without autophosphorylation and in the presence of calcium chloride (2 mm) and calmodulin (0.1 or 5 μm: L or H, respectively) (right panel). Complexes were isolated using glutathione-agarose. Aliquots of inputs, supernatants, and pellets were analyzed on SDS-polyacrylamide gels and stained for protein in parallel. Data are representative of two similar experiments.
We then compared the effects of Ca2+/calmodulin or autophosphorylation on binding of purified CaMKIIα or CaMKIIβ isoforms to GST-D-IN. GST-D-IN and GST-N2B interacted similarly with Thr-286/287-autophosphorylated CaMKIIα and CaMKIIβ (Fig. 4B, left). Effects of low (sub-saturating) and high (saturating) concentrations of calmodulin on binding of non-phosphorylated CaMKII isoforms with GST-D-IN and GST-N2B were also evaluated in the same experiment. As noted above, Ca2+/calmodulin supports more robust binding of CaMKIIα to GST-D-IN than to GST-N2B. However, saturating Ca2+/calmodulin concentrations stimulated comparable binding of CaMKIIβ to GST-D-IN or GST-N2B (Fig. 4B, right), and weak interaction of CaMKIIβ was detected in the presence of subsaturating Ca2+/calmodulin concentrations. Thus, although GST-D-IN binds comparable amounts of the autophosphorylated isoforms, Ca2+/calmodulin supports somewhat more robust interactions of CaMKIIβ than of CaMKIIα.
The Densin-IN Domain Interacts with the T Site in the CaMKII Catalytic Domain
Similarities in requirements for CaMKII activation to support binding to the densin-IN domain and GluN2B suggest a role for the CaMKII catalytic domain in the interactions (23–25). Indeed, GST-D-IN, but not GST alone or GST-D-CTA, interacts with a monomeric constitutively active catalytic domain fragment of CaMKIIα, CaMKIIα-(1–290), expressed as a GFP fusion protein in HEK293 cells (Fig. 5A). Thus, although an intact association domain of CaMKIIα is required for interaction with the densin-CTA domain, a monomeric catalytic domain fragment can bind to the densin-IN domain.
FIGURE 5.
Densin-IN binding to the CaMKII catalytic domain is blocked by CaMKIIN-tide. A, extracts of HEK293 cells expressing GFP-CaMKIIα-(1–290) were incubated with GST, GST-D-IN, or GST-D-CTA. Aliquots of the extract (Input) and complexes isolated using glutathione-agarose were analyzed by GFP immunoblot and by protein staining to detect GST fusion proteins. B, Thr-286-autophosphorylated CaMKIIα was incubated with GST or GST-D-IN (equimolar) in the absence or presence of the indicated peptides (50 μm). Complexes isolated using glutathione-agarose were analyzed by protein staining of nitrocellulose membranes. CaMKII binding (mean ± S.E., n ≥ 3) was expressed as a percentage of binding to GST-D-IN in absence of peptides (*, p < 0.05 versus control). C, extracts of HEK293 cells expressing mCherry-CaMKIIβ(T287D) or mCherry-CaMKIIβ(T287D) with additional I206K or D239R mutations where indicated were incubated with GST-D-IN or GST-N2B. Aliquots of inputs and of complexes isolated using glutathione-agarose were immunoblotted for mCherry using RFP antibodies. Binding of catalytic domain mutants was expressed as a percentage of binding of control mCherry-CaMKIIβ(T287D) (mean ± S.E., n = 3). *, p < 0.05; ***, p < 0.001 compared with control; †, p < 0.001 compared with GST-D-IN.
To further characterize the role of the catalytic domain of CaMKII in binding to the densin-IN domain, several synthetic peptides known to interact with the CaMKII catalytic domain were tested as potential competitors. Peptide concentrations were ≥5-fold higher than their known affinities for CaMKIIα and at an ≈200-fold molar excess over the concentration of GST-D-IN. Syntide-2, a model substrate peptide based on glycogen synthase, had no effect. In contrast, binding was modestly but significantly reduced by ≈25% in the presence of autocamtide-2, a substrate peptide based on the Thr-286 autophosphorylation site in CaMKIIα, or N2B-tide, a homologous peptide based on the Ser-1303 phosphorylation site in GluN2B (Fig. 5B). Moreover, N-tide, an inhibitory peptide based on the naturally occurring CaMKIIN inhibitor protein (26) almost completely blocked binding to GST-D-IN. These data suggest that the densin-IN domain interacts with a region of the CaMKIIα catalytic domain that overlaps the CaMKIIN binding site, perhaps also in proximity to binding sites for autocamtide-2 and GluN2B.
A region of the CaMKIIα catalytic domain, termed the T-site by Bayer, Schulman, and co-workers, appears to be critical for binding to CaMKIIN, GluN2B, and the CaMKII autoinhibitory domain (23, 24, 27, 28). The role of the T-site in interactions of constitutively active and mCherry-tagged T287D-CaMKIIβ mutant with the densin-IN domain was investigated by mutating Ile-206 and Asp-239 to Lys and Arg, respectively. The I206K mutation reduced binding to GST-D-IN by >95%, but the D239R mutation had a much more modest effect (reduced by only ≈20%) (Fig. 5C). However, both the I206K and D239R mutations essentially abolished binding of T287D-CaMKIIβ to GST-N2B under these conditions (Fig. 5C). An I205K mutation of CaMKIIα also decreased interactions with both GST-N2B and GST-D-IN but had no effect on binding to GST-D-CTA (data not shown). Thus, although the Ile to Lys mutants suggest that both densin-IN and GluN2B interact at the T-site of CaMKIIα or CaMKIIβ, the Asp to Arg mutant indicates that interactions with the catalytic domain are somewhat distinct, supporting the interpretation of studies comparing the regulation of CaMKII binding and peptide competition studies (see above).
The Densin-IN Domain Is Homologous to CaMKIIN
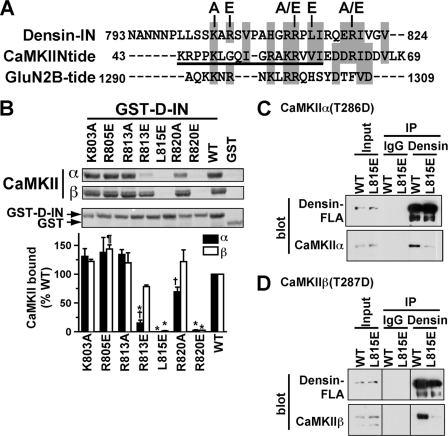
The amino acid sequence of the C-terminal region of the densin-IN domain is similar to that of central/C-terminal regions of N-tide, with 11 identical or similar residues (Fig. 6A; see the color version in supplemental Fig. 1). Possible roles of selected conserved charged or hydrophobic residues in densin were probed by site-directed mutagenesis. Replacement of Leu-815 or Arg-820 with an acidic Glu in GST-D-IN reduced the binding of autophosphorylated CaMKIIα or CaMKIIβ by >95% (Fig. 6, B and C). However, binding was modestly enhanced ≈40% by an R805E mutation. Mutations that only removed basic charge (K803A, R813A, or R820A) had no statistically significant effect on interactions with either isoform in this set of three experiments. However, there was a significant difference in the relative effects of R820A and R813E mutations on binding of CaMKII isoforms (p < 0.05). Binding of CaMKIIα to the R813E mutant was significantly reduced by ≈85%, but the modest ≈20% reduction in binding of CaMKIIβ to the R813E mutant was not statistically significant (Fig. 6B). R820A mutation had no significant effect on binding of CaMKIIβ (Fig. 6B), but combining data from Fig. 6B with data from additional experiments comparing the effects of all mutations on CaMKIIα binding alone showed that R820A mutation significantly, if modestly, reduced binding of CaMKIIα to 74 ± 4% of the WT (n = 8; p < 0.05. Dunnett's) (not shown). Notably, L815E mutation of GST-D-IN also strongly disrupted binding of both mCherry-tagged CaMKIIα(T286D) and CaMKIIβ(T287D) (expressed in HEK293 cells) (data not shown). Taken together, these data suggest an important role for hydrophobic interactions between the CaMKII catalytic domain and residues surrounding Leu-815 in the densin-IN domain that are conserved in CaMKIIN.
FIGURE 6.
L815E mutation of the densin IN domain disrupts CaMKII binding. A, amino acid sequences of the densin-IN domain, the CaMKIIN inhibitory domain, and the CaMKII binding domain of GluN2B are aligned with identical or similar residues in gray boxes. The underlined region of CaMKIIN-tide was resolved in an x-ray crystal structure of the CaMKII-CaMKIIN-tide complex (44). Densin-IN domain mutants characterized in panel B are indicated above. A color version of this panel is available online as supplemental Fig. 1. B, Thr-286/287-autophosphorylated CaMKIIα or CaMKIIβ were incubated with GST-D-IN (WT or mutated) or GST. Complexes isolated on glutathione-agarose were analyzed by protein staining of nitrocellulose membranes. Binding of CaMKII isoforms to mutated GST-D-INs was expressed as a percentage of the binding to WT (mean ± S.E., n = 3). †, p < 0.05 compared with binding of CaMKIIβ (2-way analysis of variance with Bonferroni post-test). *, p < 0.001 compared with WT (1-way analysis of variance with Dunnett's multiple comparison test). ¶, p < 0.05 compared with WT (1-way analysis of variance with Dunnett's multiple comparison test). C, extracts of HEK293 cells expressing mCherry-CaMKIIα-T286D and GFP-tagged densin-FLA (WT or L815E) were incubated with densin Ab450 or control goat IgG. Inputs and immune complexes were immunoblotted for densin and CaMKII. IP, immunoprecipitated. D, lysates from HEK293 cells co-expressing mCherry-CaMKIIβ-T287D with GFP-tagged densin-FLA (WT or L815E) were analyzed as in panel C. Samples were analyzed in parallel with those in panel C but were loaded in a different order to avoid confusion during analysis. The Input, IgG, and Densin lane pairs were each flipped horizontally during preparation of the figure so that all lanes are in the same order as in panel C; lines on the blots indicate these discontinuities.
The Densin-IN Domain Is Critical for CaMKIIα/β Binding to Full-length Densin in Cells
To investigate the importance of the densin-IN domain in the context of full-length densin, the L815E mutation was introduced into densin-FLA. Extracts of HEK293 cells co-expressing GFP-tagged WT or L815E densin-FLA with either CaMKIIα or CaMKIIβ (mCherry-tagged constitutively active T286/7D mutants) were immunoprecipitated with a densin antibody. Both CaMKII isoforms were readily detected in WT densin-FLA samples, and the L815E mutation strongly reduced binding of both isoforms by >90% (Fig. 6, C and D). These data show that the densin-IN domain is critical for interactions with both CaMKII isoforms in cells.
The Densin-IN Domain Is a Substrate-dependent CaMKII Inhibitor
Because CaMKIIN inhibits CaMKII (26), we hypothesized that densin would also inhibit CaMKII. Indeed, GST-D-IN potently and completely inhibited the phosphorylation of syntide-2, a model peptide substrate, by purified CaMKIIα or CaMKIIβ with comparable potencies (EC50 values 49 ± 12 nm, n = 2 or 56 ± 13 nm, n = 3, respectively) similar to those of N-tide (a peptide analog of CaMKIIN) in parallel studies (EC50 values 87 ± 10 nm, n = 2 or 67 ± 24 nm n = 3, respectively) (data not shown). We then tested the effect of densin-IN on CaMKIIα phosphorylation of physiologically relevant synaptic substrates. Phosphorylation of GST-GluA1, containing residues 816–889 of GluA1 including the Ser-831 phosphorylation site (20), was potently and >90% inhibited by both GST-D-IN and N-tide (Fig. 7A) (EC50 values 100 ± 23 and 72 ± 22 nm, respectively; n = 4 similar experiments). Moreover, inhibition by GST-D-IN was completely prevented by the L815E mutation, which disrupts CaMKII binding. On the other hand, phosphorylation of GST-N2Bs, containing residues 1260–1309 of GluN2B including the Ser-1303 phosphorylation site (19, 29), was only modestly inhibited by even micromolar concentrations of GST-D-IN but was completely inhibited by N-tide (Fig. 7B), albeit with somewhat reduced potency (EC50 380 ± 42 nm, n = 3). The extrapolated CaMKII activity at saturating concentrations of GST-D-IN was 79 ± 5% (n = 3) with an estimated half-maximal effective concentration of 571 ± 268 nm, not significantly different from the potency of N-tide toward GluN2B phosphorylation (the low efficacy of inhibition presumably accounts for the increased variability in the estimated half-maximal effective concentration). Thus, the efficacy of densin-IN, but not CaMKIIN, as a CaMKII inhibitor appears to be highly substrate-dependent.
FIGURE 7.
Densin IN potently inhibits CaMKII phosphorylation of GluA1 but not GluN2B. A, GST-GluA1 was incubated with Thr-286-autophosphorylated CaMKIIα, [γ-32P]ATP, and the indicated concentrations of GST-D-IN (WT or L815E), N-tide or GST alone. Top, reaction aliquots were analyzed by SDS-PAGE followed by Coomassie Blue staining and autoradiography. Bottom, quantitative analysis using phosphocellulose papers (see “Experimental Procedures”); the mean of duplicates is plotted. Data are representative of four independent experiments. B, GST-N2Bs was incubated with Thr-286-autophosphorylated CaMKIIα and the indicated concentrations of GST-D-IN (WT or L815E), N-tide, or GST alone and analyzed as described in panel A.
Selective Inhibition of Glutamate Receptor Phosphorylation by Densin in Intact Cells
To begin to explore the effects of the major adult splice variant of densin on CaMKII activity in cells, we compared phosphorylation of full-length GluA1 and GluN2B by CaMKII in transfected HEK293 cells in the absence and presence of densin-FLA. Co-expression of CaMKIIα significantly enhanced the phosphorylation of GluA1 at Ser-831. Densin-FLA significantly reduced the effect of overexpressed CaMKIIα on Ser-831 phosphorylation, but L815E mutation in the densin-IN domain essentially abrogated this effect (Fig. 8A). In contrast, densin-FLA had no effect on CaMKII-stimulated phosphorylation of Ser-1303 in GluN2B (Fig. 8B). Moreover, densin-FLA had no effect on the level of Thr-286 autophosphorylation of CaMKIIα or on the amount of CaMKIIα that co-precipitated with the FLAG-GluN2B (data not shown). These data show that densin-FLA can selectively inhibit CaMKII activity toward some, but not all, substrates in intact cells.
DISCUSSION
Precise physiological control of cell signaling depends on scaffolding and/or anchoring proteins (30). For example, conserved amphipathic α-helical domains in a large family of protein kinase A anchoring proteins competitively interact with type II regulatory subunits to target PKA subpopulations to complexes that also contain diverse upstream regulators and downstream targets (31). Similarly, PP1 targeting subunits containing RVXF sequence motifs compete for a conserved hydrophobic groove on the surface of PP1 catalytic subunits to target phosphatase activity (32). However, CaMKAPs are mechanistically more diverse. NMDAR GluN2B subunits and voltage-gated calcium channel β2a/β1b subunits contain CaMKII binding domains that share sequence similarity with the autoinhibitory/regulatory domain of CaMKII. These domains compete for binding to the CaMKII catalytic domain, and each contains a key phosphorylation site that weakens the interaction (GluN2B, Ser-1303; β2a/β1b, Thr-498; CaMKIIα/β, Thr-286/287) (19, 24, 33, 34). SAP97 also binds to the CaMKII catalytic domain (35) even though there is no obvious sequence similarity with GluN2B, β2a/β1b, or CaMKIIα/β. In contrast, α-actinin appears to bind the calmodulin binding domain of CaMKII (11). Moreover, a C-terminal domain in densin was shown to interact with the association domain of CaMKIIα, but not CaMKIIβ (6, 7), indicating that densin is a CaMKIIα-selective CaMKAP. These diverse interaction mechanisms allow dodecameric CaMKII holoenzymes to form a scaffold for assembly of multiprotein complexes (11). Indeed, CaMKII can have synaptic roles that require scaffolding functions that are independent of ongoing kinase activity (but may be modulated by autophosphorylation) (36–38). The present studies provide new insights into the complexity and function of protein-protein interaction mechanisms employed by CaMKII.
Our data show that rather than being CaMKIIα-specific, densin can target multiple CaMKII isoforms in vivo (Fig. 2). CaMKIIα is first expressed at about postnatal day 5 in rodents and in adulthood is highly expressed in many, but not all, neurons in the forebrain as well as in cerebellar Purkinje neurons. In contrast, CaMKIIβ splice variants are broadly expressed in many neuronal subtypes throughout development.6 The ratio of CaMKII isoform expression is thought to dictate the subunit composition of CaMKII holoenzymes. Long term changes in synaptic activity antagonistically regulate CaMKIIα and CaMKIIβ expression levels (39). Indeed, local CaMKIIα protein synthesis in neuronal dendrites is controlled by synaptic activity and is necessary for normal synaptic plasticity, learning, and memory (40, 41). Thus, neurons likely contain distinct mixtures of CaMKII holoenzymes (CaMKIIβ-specific, CaMKIIα-specific, or with varying isoform ratios), depending on developmental stage and specific cell type. For example, α:β isoform ratios in CaMKII holoenzymes purified from adult forebrain and cerebellum are ≈4:1 and ≈1:4, respectively (42, 43). Thus, investigations of physiological roles for densin in modulating CaMKII signaling need to be expanded to include neurons and other cell types that do not express CaMKIIα.
In addition to showing that the major adult densin-FLA variant can associate with CaMKIIβ, deletion of the known C-terminal CaMKII binding domain (in the naturally occurring densin-FLC variant) has only a modest effect on the binding of either CaMKII isoform (Fig. 2, B and C). These observations were explained by identification of a second internal CaMKII binding domain in densin (residues 793–824) that appears to interact with the catalytic domain of both CaMKII isoforms, but only after kinase activation (Figs. 3, 4, and 5). Although CaMKIIα holoenzymes can also interact with the densin-CTA domain (at least partially independent of activation), mutagenesis studies suggest that the densin-IN domain plays a dominant role in binding to both CaMKIIα and CaMKIIβ in heterologous cells (Fig. 6, C and D). Numerous densin mRNA splice variants are differentially expressed across brain development (6, 9). Several splice variants expressed during embryonic or early postnatal development lack the densin-IN and/or -CTA domains, presumably indicating that densin can have diverse roles, in some cases independent of CaMKII. For example, although densin-FLC may be expressed only in the perinatal period (6), before CaMKIIα expression, the IN domain may allow densin-FLC to modulate CaMKIIβ. In addition, we previously showed that a natural densin splice variant lacking a large central region containing the densin-IN domain is unable to support CaMKIIα-dependent facilitation of CaV1.3 L-type calcium channels (12). A deeper understanding of the expression of densin splice variants in different cell types during development is required to fully appreciate the implications of these findings for CaMKII isoform regulation.
Activation of CaMKII isoforms is required for interactions with both the densin-IN domain and with GluN2B, in contrast to CaMKIIα binding to the densin-CTA domain. Subtle differences in requirements for CaMKIIα/β binding to densin-IN and GluN2B (Figs. 4 and 6B) may relate to the fact that CaMKIIβ has an ≈10-fold higher affinity for Ca2+/calmodulin binding than does CaMKIIα (15). These differences may be relevant to understanding the role of densin in targeting the two CaMKII isoforms in intact cells. In addition, they suggest subtle differences in the mechanisms by which GluN2B and densin-IN bind to CaMKII isoforms (see below).
The densin-IN domain has significant amino acid sequence similarity with the core inhibitory domain of CaMKIIN, a naturally occurring CaMKII inhibitor protein (26). N-tide, a peptide analog containing residues 43–69 of CaMKIIN (26) (termed CN27 by Vest et al. (27)), competes with densin-IN for binding CaMKII in vitro. In a crystal structure of CaMKII bound to an N-tide variant (44), amino acid side chains in the N-terminal and central regions of N-tide (underlined in Fig. 6A and in supplemental Fig. 1; corresponding to the CN17a peptide described in Vest et al. (27)) interact with a series of binding pockets on the catalytic domain that can also be partially occupied by the regulatory domain in autoinhibited CaMKII (45, 46) and that have been collectively termed the T-site (27). Consistent with this structure, potent inhibition by N-tide is highly dependent on N-terminal Lys-43—Arg–Pro residues (27); however, these residues are not conserved in densin-IN (Leu-799–Leu–Ser). Interestingly, N-tide residues beyond amino acid 59 are not resolved in the crystal structure, suggesting that residues 60–63 are not tightly associated with the catalytic domain even though functional studies show that residues 59–63 (IEDDR) are important for potent inhibition (27). Although central and C-terminal regions of N-tide are most similar to the densin-IN domain (residues 803–824), the densin-IN domain contains two Pro residues (808 and 814) that are not present in N-tide (Fig. 6A and supplemental Fig. 1), presumably constraining the conformations that can be adopted when interacting with CaMKII. In addition, N-tide contains five acidic amino acids (red in supplemental Fig. 1), with only one conserved in the densin-IN domain. We identified two basic residues (Arg-813, Arg-820; blue in supplemental Fig. 1) and an intervening hydrophobic residue (Leu-815; green in supplemental Fig. 1) in the densin-IN domain that are important for binding (Fig. 6B) and inhibition (Fig. 7) of CaMKII. These residues are conserved as Arg-56, Val-58, and Arg-63 in N-tide, and truncation mutagenesis studies suggested these residues are important for potent inhibition by N-tide (27). Removal of these basic charges from densin-IN (R813A or R820A mutation) had only a modest effect, but insertion of acidic residues (L815E or R820E mutation) almost completely abrogated binding of CaMKIIα and CaMKIIβ to GST-D-IN. Surprisingly, the R813E and R820A mutations selectively reduced binding of CaMKIIα but not CaMKIIβ for reasons that are not clear (Fig. 6B). In addition, we found that the L815E mutation severely disrupted the association of CaMKIIα or CaMKIIβ with full-length densin in HEK293 cells (Fig. 6, D and E). Taken together, these data suggest that binding of CaMKII isoforms to the densin-IN domain is primarily driven by hydrophobic interactions, in part involving Leu-815.
Although CaMKIIN competes with GluN2B or densin-IN for binding to the CaMKII catalytic domain (Fig. 5B and Refs. 19, 23, and 27), a GluN2B peptide is a poor competitor for densin-IN binding to CaMKII (Fig. 5B). Ca2+/calmodulin also has quantitatively distinct effects on the interactions with GluN2B and densin-IN (see above). Differences in interaction mechanisms are further illuminated by mutations in the CaMKII catalytic domain. I206K mutation in the T site of CaMKIIβ prevented GluN2B binding, consistent with prior studies of CaMKIIα (23), and interactions of the densin-IN domain with CaMKIIα and CaMKIIβ were prevented by the corresponding I205K and I206K mutations, respectively. Thus, Ile-205/206 in the CaMKIIα/β catalytic domains appears to be involved in interactions with densin-IN, CaMKIIN, and GluN2B. In contrast, mutation of Asp-239 to Arg in CaMKIIβ binding disrupted the binding to GluN2B, but this mutation had only a modest effect on binding to the densin-IN domain (Fig. 5C). Taken together, our data indicate that although interactions of the densin-IN domain and N-tide with CaMKII are broadly similar, there are clear differences.
These findings may be reconciled if the “T-site” is considered as an extended/linked series of potential interaction site that can be differentially targeted by the CaMKII regulatory domain or by CaMKII binding domains in CaMKAPs (e.g. CaMKIIN, densin-IN, GluN2B, voltage-gated calcium channel β subunits, SAP97, and potentially other proteins) such that some interactions are strongly competitive (e.g. CaMKIIN competes with densin-IN, GluN2B, β subunits, or SAP97), whereas others are not (e.g. GluN2B does not compete very well with densin-IN or SAP97). Additional mutagenesis and structural studies will be required to fully understand these differences.
CaMKIIN completely and potently inhibits CaMKII activity toward all substrates tested (Fig. 7) (26, 27). The densin-IN domain also potently and essentially completely inhibited CaMKII phosphorylation of either syntide-2, a model peptide substrate, or of the GluA1 AMPA receptor subunit at Ser-831, with potencies similar to those of N-tide (EC50 values 50–100 nm) (Fig. 7A). However, densin-IN was an ineffective inhibitor of CaMKII activity toward Ser-1303 in GluN2B (Fig. 7B), correlating with the weak competition by N2B-tide for CaMKII binding to densin-IN (Fig. 5B). Studies in HEK293 cells confirmed that full-length densin-FLA effectively inhibits phosphorylation of GluA1, but not GluN2B, by CaMKII and that the inhibition of GluA1 phosphorylation required a functional densin-IN domain (Fig. 8). Thus, although CaMKIIN competes with GluN2B for stable interaction with the CaMKII catalytic domain and blocks GluN2B phosphorylation, it appears that the densin-IN domain does not prevent efficient GluN2B phosphorylation.
In summary, the present findings substantially expand our understanding of densin. Rather than functioning as a CaMKIIα-selective targeting protein, our data show that densin can target multiple CaMKII isoforms. Binding of densin appears to have a novel modulatory role; that is, to interfere with phosphorylation of GluA1-AMPARs and favor phosphorylation of GluN2B-NMDARs. Thus, densin may direct CaMKII actions toward discrete subsets of potential downstream targets in dendritic spines in response to synaptic activity, potentially modulating key mechanisms underlying synaptic plasticity. The roles of densin in neurons clearly warrant further investigation. However, it is worth noting that the design and interpretation of such studies needs to consider the diversity of CaMKAPs likely to be present at individual synapses and their overlapping interaction mechanisms as well as the likely presence of multiple densin splice variants in some systems.
Supplementary Material
This work was supported, in whole or in part, by National Institutes of Health Grants R01-MH063232 (to R. J. C.), F32-MH068129 (to A. J. R.), and T32-MH065215 (to A. J. B.). This work was also supported by American Heart Association Predoctoral Fellowship 0815090E (to N. J.-S.) and a UNCF-Merck postdoctoral fellowship (to A. J. B.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Fig. 1.
For purposes of clarity, possible contributions of the CaMKIIγ and -δ isoforms are not further discussed here. Although the brain expresses both of these isoforms, their cellular distribution is less well understood.
- NMDAR
- N-methyl d-aspartate receptor
- AMPAR
- α-amino-3-hydroxyl-5-methyl-4-isoxazole-propionic acid receptor
- CaMKAP
- CaMKII anchoring protein
- CaMKII
- Ca2+/calmodulin-dependent protein kinase II
- LRR
- leucine-rich repeat
- CAPS
- N-cyclohexyl-3-aminopropanesulfonic acid
- AMP-PNP
- adenosine 5′-(β,γ-imino)triphosphate.
REFERENCES
- 1. Colbran R. J. (2004) Biochem. J. 378, 1–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Schulman H. (2004) J. Neurosci. 24, 8399–8403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Barria A., Malinow R. (2005) Neuron 48, 289–301 [DOI] [PubMed] [Google Scholar]
- 4. Zhou Y., Takahashi E., Li W., Halt A., Wiltgen B., Ehninger D., Li G. D., Hell J. W., Kennedy M. B., Silva A. J. (2007) J. Neurosci. 27, 13843–13853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Apperson M. L., Moon I. S., Kennedy M. B. (1996) J. Neurosci. 16, 6839–6852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Strack S., Robison A. J., Bass M. A., Colbran R. J. (2000) J. Biol. Chem. 275, 25061–25064 [DOI] [PubMed] [Google Scholar]
- 7. Walikonis R. S., Oguni A., Khorosheva E. M., Jeng C. J., Asuncion F. J., Kennedy M. B. (2001) J. Neurosci. 21, 423–433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Izawa I., Nishizawa M., Ohtakara K., Inagaki M. (2002) J. Biol. Chem. 277, 5345–5350 [DOI] [PubMed] [Google Scholar]
- 9. Jiao Y., Robison A. J., Bass M. A., Colbran R. J. (2008) J. Neurochem. 105, 1746–1760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Thalhammer A., Trinidad J. C., Burlingame A. L., Schoepfer R. (2009) J. Neurochem. 109, 297–302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Robison A. J., Bass M. A., Jiao Y., MacMillan L. B., Carmody L. C., Bartlett R. K., Colbran R. J. (2005) J. Biol. Chem. 280, 35329–35336 [DOI] [PubMed] [Google Scholar]
- 12. Jenkins M. A., Christel C. J., Jiao Y., Abiria S., Kim K. Y., Usachev Y. M., Obermair G. J., Colbran R. J., Lee A. (2010) J. Neurosci. 30, 5125–5135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Quitsch A., Berhörster K., Liew C. W., Richter D., Kreienkamp H. J. (2005) J. Neurosci. 25, 479–487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Tobimatsu T., Fujisawa H. (1989) J. Biol. Chem. 264, 17907–17912 [PubMed] [Google Scholar]
- 15. Brocke L., Chiang L. W., Wagner P. D., Schulman H. (1999) J. Biol. Chem. 274, 22713–22722 [DOI] [PubMed] [Google Scholar]
- 16. Brocke L., Srinivasan M., Schulman H. (1995) J. Neurosci. 15, 6797–6808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Shen K., Teruel M. N., Subramanian K., Meyer T. (1998) Neuron 21, 593–606 [DOI] [PubMed] [Google Scholar]
- 18. Srinivasan M., Edman C. F., Schulman H. (1994) J. Cell Biol. 126, 839–852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Strack S., McNeill R. B., Colbran R. J. (2000) J. Biol. Chem. 275, 23798–23806 [DOI] [PubMed] [Google Scholar]
- 20. Barria A., Derkach V., Soderling T. (1997) J. Biol. Chem. 272, 32727–32730 [DOI] [PubMed] [Google Scholar]
- 21. Lin Y. C., Redmond L. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 15791–15796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Robison A. J., Bartlett R. K., Bass M. A., Colbran R. J. (2005) J. Biol. Chem. 280, 39316–39323 [DOI] [PubMed] [Google Scholar]
- 23. Bayer K. U., LeBel E., McDonald G. L., O'Leary H., Schulman H., De Koninck P. (2006) J. Neurosci. 26, 1164–1174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Bayer K. U., De Koninck P., Leonard A. S., Hell J. W., Schulman H. (2001) Nature 411, 801–805 [DOI] [PubMed] [Google Scholar]
- 25. Strack S., Colbran R. J. (1998) J. Biol. Chem. 273, 20689–20692 [DOI] [PubMed] [Google Scholar]
- 26. Chang B. H., Mukherji S., Soderling T. R. (1998) Proc. Natl. Acad. Sci. U.S.A. 95, 10890–10895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Vest R. S., Davies K. D., O'Leary H., Port J. D., Bayer K. U. (2007) Mol. Biol. Cell 18, 5024–5033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Yang E., Schulman H. (1999) J. Biol. Chem. 274, 26199–26208 [DOI] [PubMed] [Google Scholar]
- 29. Omkumar R. V., Kiely M. J., Rosenstein A. J., Min K. T., Kennedy M. B. (1996) J. Biol. Chem. 271, 31670–31678 [DOI] [PubMed] [Google Scholar]
- 30. Pawson C. T., Scott J. D. (2010) Nat. Struct. Mol. Biol. 17, 653–658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Wong W., Scott J. D. (2004) Nat. Rev. Mol. Cell Biol. 5, 959–970 [DOI] [PubMed] [Google Scholar]
- 32. Bollen M., Peti W., Ragusa M. J., Beullens M. (2010) Trends Biochem. Sci. 35, 450–458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Grueter C. E., Abiria S. A., Dzhura I., Wu Y., Ham A. J., Mohler P. J., Anderson M. E., Colbran R. J. (2006) Mol. Cell 23, 641–650 [DOI] [PubMed] [Google Scholar]
- 34. Grueter C. E., Abiria S. A., Wu Y., Anderson M. E., Colbran R. J. (2008) Biochemistry 47, 1760–1767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Nikandrova Y. A., Jiao Y., Baucum A. J., Tavalin S. J., Colbran R. J. (2010) J. Biol. Chem. 285, 923–934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Bingol B., Wang C. F., Arnott D., Cheng D., Peng J., Sheng M. (2010) Cell 140, 567–578 [DOI] [PubMed] [Google Scholar]
- 37. Pi H. J., Otmakhov N., El Gaamouch F., Lemelin D., De Koninck P., Lisman J. (2010) Proc. Natl. Acad. Sci. U.S.A. 107, 14437–14442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Hojjati M. R., van Woerden G. M., Tyler W. J., Giese K. P., Silva A. J., Pozzo-Miller L., Elgersma Y. (2007) Nat. Neurosci. 10, 1125–1127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Thiagarajan T. C., Piedras-Renteria E. S., Tsien R. W. (2002) Neuron 36, 1103–1114 [DOI] [PubMed] [Google Scholar]
- 40. Aakalu G., Smith W. B., Nguyen N., Jiang C., Schuman E. M. (2001) Neuron 30, 489–502 [DOI] [PubMed] [Google Scholar]
- 41. Ouyang Y., Rosenstein A., Kreiman G., Schuman E. M., Kennedy M. B. (1999) J. Neurosci. 19, 7823–7833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. McGuinness T. L., Lai Y., Greengard P. (1985) J. Biol. Chem. 260, 1696–1704 [PubMed] [Google Scholar]
- 43. Miller S. G., Kennedy M. B. (1985) J. Biol. Chem. 260, 9039–9046 [PubMed] [Google Scholar]
- 44. Chao L. H., Pellicena P., Deindl S., Barclay L. A., Schulman H., Kuriyan J. (2010) Nat. Struct. Mol. Biol. 17, 264–272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Rellos P., Pike A. C., Niesen F. H., Salah E., Lee W. H., von Delft F., Knapp S. (2010) PLoS Biol. 8, e1000426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Rosenberg O. S., Deindl S., Sung R. J., Nairn A. C., Kuriyan J. (2005) Cell 123, 849–860 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.