Abstract
Occludin is a transmembrane tight junction (TJ) protein that plays an important role in TJ assembly and regulation of the epithelial barrier function, but the mechanisms underlying its post-transcriptional regulation are unknown. The RNA-binding protein HuR modulates the stability and translation of many target mRNAs. Here, we investigated the role of HuR in the regulation of occludin expression and therefore in the intestinal epithelial barrier function. HuR bound the 3′-untranslated region of the occludin mRNA and enhanced occludin translation. HuR association with the occludin mRNA depended on Chk2-dependent HuR phosphorylation. Reduced HuR phosphorylation by Chk2 silencing or by reduction of Chk2 through polyamine depletion decreased HuR-binding to the occludin mRNA and repressed occludin translation, whereas Chk2 overexpression enhanced (HuR/occludin mRNA) association and stimulated occludin expression. In mice exposed to septic stress induced by cecal ligation and puncture, Chk2 levels in the intestinal mucosa decreased, associated with an inhibition of occludin expression and gut barrier dysfunction. These results indicate that HuR regulates occludin mRNA translation through Chk2-dependent HuR phosphorylation and that this influence is crucial for maintenance of the epithelial barrier integrity in the intestinal tract.
INTRODUCTION
Epithelial cells line the intestinal mucosa and establish an important barrier that protects the subepithelial tissue against a wide array of noxious substances in the lumen (1–3). Disruption of this barrier occurs commonly in various pathological conditions, leading to the translocation of luminal toxic substances and bacteria to the bloodstream (3). The integrity and normal function of the epithelial barrier depend on specialized structures composing different intercellular junctions including tight junctions (TJs) and adherens junctions (AJs) (4,5). The TJs located at the apical region of the epithelial lateral membrane provide the barrier that is selectively permeable to certain hydrophilic molecules, ions and nutrients (2,6), whereas AJs mediate strong cell-to-cell adhesions between adjacent epithelial cells and regulate TJ assembly and function (7,8). TJs primarily consist of transmembrane proteins such as occludin, tricellulin and one or more members of the claudin family; these proteins also associate with a cytosolic plaque of proteins such as zonula occludens (ZO)-1 and ZO-2 that link tightly to the cortical cytoskeleton (1,2,9). Despite considerable efforts from many laboratories, the precise mechanisms underlying TJ assembly and the coordination of signals and barrier activity in response to stress are still poorly understood.
To date, four classes of TJ transmembrane proteins and >30 TJ membrane-associated proteins have been identified in mammalian epithelial and endothelial cells (2,3); the major transmembrane and cytosolic TJ proteins expressed in the intestinal epithelium include occludin, claudins, ZO-1 and ZO-2 (1,6). Occludin is an integral membrane protein that is specifically localized at tight junction complexes and is required for normal TJ physiology (10,11). Occludin has a tetraspanning membrane topology with two extracellular loops and three cytoplasmic domains, among which the extracellular loops are important for occludin localization (12,13). Occludin is important for normal function of the gut barrier (12,14) and is also involved in the regulation of other cellular functions such as directional migration of epithelial cells (15). Ectopic expression of wild-type occludin increases the number of TJ strands and promotes the epithelial barrier function (16), while inhibition of occludin function by overexpressing the dominant negative occludin mutant disrupts TJ structures and results in dysfunction of the epithelial barrier (17). The levels of TJs including occludin change dynamically and are tightly regulated by numerous factors (18–20), but the exact mechanism underlying the regulation of TJ expression remains to be fully elucidated.
Although gene expression is critically regulated at the transcriptional level, the essential contribution of post-transcriptional events, particularly altered mRNA turnover and translation, in the control of gene expression programs in the intestinal mucosa is increasingly recognized (21–24). The post-transcriptional fate of a given mRNA is primarily controlled by the interaction of specific mRNA sequences (cis-elements) with specific trans-acting factors such as RNA-binding proteins (RBPs) (25–28) and microRNAs (29,30). The most common cis-elements responsible for rapid regulation of mRNA decay and translation in mammalian cells are U- and AU-rich elements located in the 3′-untranslated regions (3′-UTRs) of many mRNAs (31). Ribonucleoprotein (RNP) associations either increase or decrease mRNA stability and/or translation depending on the particular mRNA sequence, the cellular growth conditions and the stimulus type (25,32,33). Among the RBPs that regulate specific subsets of mRNAs are several that modulate mRNA turnover (HuR, NF90, AUF1, BRF1, TTP, KSRP) and several that modulate translation (HuR, TIAR, NF90, TIA-1), collectively known as translation and turnover-regulatory (TTR)-RBPs (34,35). In cells responding to stressful environmental conditions, TTR-RBPs bind to the specific sequences in the 3′-UTRs of collections of target mRNAs and govern their turnover and translation rates (36–39).
HuR is one of the best-studied RBPs in the intestinal mucosa (20–23,40). It is characterized by the presence of two N-terminal RNA recognition motifs (RRMs) with high affinity for specific U- and AU-rich elements, and a C-terminal RRM that recognizes the poly(A) tail (41). The function of HuR is tightly regulated by altering its subcellular distribution, by its phosphorylation status, and by the presence of other factors, such as microRNAs, interacting with the target mRNA (29,42,43). The checkpoint kinase Chk2 was recently shown to phosphorylate HuR and thereby modulate the affinity of HuR for its target transcripts after exposure to oxidative stress and polyamine depletion (22,42). In addition, HuR phosphorylation by protein kinase Cα elevates its cytoplasmic abundance (38), whereas cyclin-dependent kinase-1-mediated HuR phosphorylation prevents the cytoplasmic accumulation of HuR (44). In this study, we sought to directly investigate the role of HuR in the regulation of occludin expression and therefore in the intestinal epithelial barrier function in vitro as well as in vivo. The data presented here indicate that HuR directly interacts with and enhances occludin mRNA translation through Chk2-dependent HuR phosphorylation, thus promoting the epithelial barrier function. Conversely, reduced HuR association with occludin mRNA following inhibition of Chk2 or exposure to septic stress inhibits occludin expression and results in barrier dysfunction.
MATERIALS AND METHODS
Chemicals and supplies
Tissue culture medium and dialyzed fetal bovine serum were from Invitrogen (Carlsbad, CA, USA) and biochemicals were from Sigma (St Louis, MO, USA). The antibodies recognizing HuR, occludin, Chk2 and β-actin were obtained from Santa Cruz Biotechnology (Santa Cruz, CA) and BD Bioscience, and the antibody against all phosphorylated proteins (Cat No. 61-8300) was from Zymed Laboratories (South San Francisco, CA, USA). The secondary antibody conjugated to horseradish peroxidase was purchased from Sigma. The 12-mm Transwell filters were obtained from Costar Corp; L-[35S]methionine, L-[35S]cysteine, and [14C]mannitol were obtained from NEN Radiopharmaceutical. DFMO (α-difluoromethylornithine) was from GENZYME (Cambridge, MA, USA).
Cell culture and animals
Stable Cdx2-transfected intestinal epithelial cells (IEC-Cdx2L1) were developed from IEC-6 cells (45), derived from normal rat intestinal crypt cells (46), and maintained as described previously (20). Before experiments, IEC-Cdx2L1 cells were grown in DMEM containing 4 mM IPTG for 16 days to induce cell differentiation as described earlier (20,47). The Caco-2 cells (a human colon carcinoma cell line) were cultured similarly to the IEC-Cdx2L1 cells.
A/J mice (male, 6–9 weeks old) were purchased from The Jackson Laboratory and housed in specific pathogen-free animal facility at the Baltimore VA Medical Center. All animal experiments were conducted in accordance with NIH guidelines and were approved by the Institutional Animal Care and Use Committee of University Maryland School of Medicine and Baltimore VA hospital.
Plasmid construction
The Chk2 expression vector was described previously (42). The vectors expressing wild-type HuR-TAP fusion proteins or point-mutated HuR-TAP fusion proteins were generated by site-directed mutagenesis as described (22,42). The chimeric firefly luciferase reporter construct containing the occludin 3′-UTR (http://www.ncbi.nlm.nih.gov/gene/83497) was generated as described (19,21). The full-length 3′-UTR of rat occludin mRNA was amplified and subcloned into the pGL3-Luc plasmid (Promega) at the Xba1 site to generate the chimeric pGL3-Luc-Occludin-3′-UTR (Luc-Occl-3′-UTR) reporter construct. The sequence and orientation of the luciferase reporter were verified by DNA sequencing and enzyme digestion. Transient transfections were performed using the Lipofectamine Reagent following recommendations by the manufacturer (Invitrogen). The luciferase reporter constructs were transfected into cells along with phRL-null, a Renilla luciferase control reporter vector from Promega, to monitor transfection efficiencies as described (22). Luciferase activity was measured using the Dual Luciferase Assay System. To measure translational changes (translational efficiency), the Firefly to Renilla luciferase ratio was further normalized with RNA levels.
RNA interference
HuR and Chk2 were silenced by transfection with specific small interfering RNA (siRNA) as described (22,27). The siRNAs specifically targeting HuR mRNA (siHuR) or Chk2 mRNA (siChk2) and control-siRNA (C-siRNA) were purchased from Dharmacon (Supplementary Table S1). For each 60-mm cell culture dish, 15 µl of the 20 µM stock duplex siHuR, siChk2 or C-siRNA was used. Forty-eight hours after transfection using LipofectAMINE, cells were harvested for analysis.
Western blot analysis
Whole-cell lysates were prepared using 2% SDS, sonicated and centrifuged at 4°C for 15 min. The supernatants were boiled for 5 min and size-fractionated by SDS–PAGE. After transferring proteins onto nitrocellulose filters, the blots were incubated with primary antibodies recognizing occludin, HuR and Chk2 proteins; following incubations with secondary antibodies, immunocomplexes were developed by using chemiluminescence.
RT–PCR and real-time quantitative PCR analysis
Total RNA was isolated by using RNeasy mini kit and used in reverse transcription and PCR amplification reactions as described (43). PCR primers for detecting occludin mRNA were TTGGGACAGAGGCTATGG (sense) and ACCCACTCTTCAACATTGGG (antisense). The levels of glyceraldehyde-3-phosphate dehydrogenase (GAPDH) PCR product were assessed to monitor the evenness in RNA input in RT–PCR samples. Real-time quantitative PCR (qPCR) analysis was performed using 7500-Fast Real-Time PCR Systems with specific primers, probes and software (Applied Biosystems, Foster City, CA, USA).
Measurement of newly translated protein and polysome analysis
New synthesis of occludin protein was measured by L-[35S]methionine and L-[35S]cysteine incorporation assays as described (22,44). Cells were incubated with 1 mCi (1 Ci = 37 GBq) L-[35S]methionine and L-[35S]cysteine per 60-mm plate for 30 min, where upon cells were lysed using RIPA buffer. Immunoprecipitations were carried out for 1 h at 4°C using either an antibody recognizing occludin or IgG1 (BD Pharmingen). Following extensive washes in TNN buffer [50 mM Tris–HCl (pH 7.5), 250 mM NaCl, 5 mM EDTA, 0.5% NP-40], the immunoprecipitated material was resolved by 10% SDS–PAGE, transferred onto PVDF filters, and visualized with a PhosphorImager (Molecular Dynamics).
Polysome analysis was performed as described (48). Briefly, cells were incubated for 15 min in 0.1 mg/ml cycloheximide and then lifted by scraping in 1 ml PEB lysis buffer (0.3 M NaCl, 15 mM MgCl2, 15 mM Tris–HCl, pH 7.6, 1% Triton X-100, 1 mg/ml heparin, and 0.1 mg/ml cycloheximide) and lysed on ice for 10 min. Nuclei were pelleted (10 000 g, 10 min) and the resulting supernatant was fractionated through a 10–50% linear sucrose gradient to fractionate cytoplasmic components according to their molecular weights. The eluted fractions were prepared with a fraction collector (Brandel) and their quality was monitored at 254 nm using a UV-6 detector (ISCO). After the RNA in each fraction was extracted with 8 M guanidine–HCl, the levels of each individual mRNA were quantified by qPCR analysis in each of the fractions, and their abundance represented as a percent of the total mRNA in the gradient.
Biotin pull-down assays
The synthesis of biotinylated transcripts and analysis of RBPs bound to biotinylated RNA were done as described (23). Complementary DNA from IEC-Cdx2L1 cells was used as a template for PCR amplification of the coding region (CR) and 3′-UTR of occludin. The 5′ primers contained the T7 RNA polymerase promoter sequence (T7)CCAAGCTTCTAATACGACTCAC TATAGGGAGA. To prepare the occludin CR template (spanning position 149–1721), oligonucleotides (T7)CCATGTCTGTGAGGCCTTTT and CTCTGGGGAGAACTGCAGAC were used. To prepare the occludin 3′-UTR template (spanning position 1720–4148), oligonucleotides (T7)CAAGAGGATGGTGGGAGACT and ATATAAAGCACATACAACAA AACGTCA were used. All sequences of oligonucleotides for preparation of various short RNA probes for mapping the occludin 3′-UTR were described in Supplementary Table S2. PCR-amplified products were used as templates to transcribe biotinylated RNAs by using T7 RNA polymerase in the presence of biotin-cytidine 5′-triphosphate as described (49). Biotinylated transcripts (6 µg) were incubated with 120 µg of cytoplasmic lysates for 30 min at room temperature. Complexes were isolated with paramagnetic streptavidin-conjugated Dynabeads (Dynal, Oslo, Norway) and analyzed by western blot analysis.
RNP IP assays
To assess the association of endogenous HuR with endogenous occludin mRNA, immunoprecipitation (IP) of ribonucleoprotein (RNP) complexes were performed as described (50). Twenty million cells were collected per sample, and lysates were used for IP for 4 h at room temperature in the presence of excess (30 µg) IP antibody (IgG, or anti-HuR). RNA in IP materials was used in RT followed by PCR and qPCR analysis to detect the presence of occludin and GAPDH mRNAs.
Paracellular tracer flux assay
Flux assays were performed on the 12-mm Transwell as described in previously (51). 14C-mannitol (MW: 184), a membrane-impermeable molecule, served as the paracellular tracer and was added to a final concentration of 3.6 nM to the apical bathing wells that contained 0.5 ml of medium. The basal bathing well had no added tracers and contained 1.5 ml of the same flux assay medium as in the apical compartment. All flux assays were performed at 37°C, and the basal medium was collected 2 h after addition of 14C-mannitol for a Beckman liquid scintillation counter. The results were expressed as percentage of total count values of each tracer.
Surgical procedures
Cecal ligation and puncture (CLP) was induced as described previously (52). Mice were anesthetized by Nembutal (5.5 mg/100-g wt, i.p.), and a midline abdominal incision was performed. The distal portion of the cecum (1 cm) was ligated with 5-0 silk suture. The ligated cecum was then punctured with a 25-gauge needle and slightly compressed with an applicator until a small amount of stool appeared. In sham-operated animals, the cecum was manipulated but without ligation and puncture, and was placed back in the peritoneum. The incision was closed using a 2-layer procedure: 5-0 silk suture on the muscle layer and the skin, respectively. Mice received 1 ml of saline i.p. for fluid resuscitation at the time of closure and 0.1 mg/100-g wt buprenex s.c. 4 times at 12-h intervals to minimize distress.
Intestinal permeability in vivo
Intestinal permeability was determined by examining the appearance in blood of FITC-dextran administered by gavage as described (53). Briefly, mice were gavaged with FITC-dextran at a dose of 60 mg/100-g wt 4 h before harvest. Blood sample was collected by cardiac puncture. The serum concentration of the FITC-dextran was determined using a fluorescence plate reader with an excitation wavelength at 490 nm and an emission wavelength of 530 nm.
Statistics
Values are means ± SEM (standard error of the mean) from 3 to 6 samples. Immunoblotting results were repeated three times. The significance of the difference between means was determined by analysis of variance. The level of significance was determined using Duncan's multiple range test (54).
RESULTS
Occludin mRNA is a direct target of HuR
There are several computationally predicted hits of the HuR motif in the occludin 3′-UTR (55) (Figure 1A), suggesting the involvement of HuR in the post-transcriptional regulation of occludin. To test this possibility, we first examined whether HuR directly interacts with the occludin mRNA by performing RNP–IP assays using anti-HuR antibody under conditions that preserved RNP integrity (50). As shown in Figure 1B and C, the occludin PCR products were highly enriched in HuR samples compared with control IgG samples. The enrichment of c-Myc PCR product was also examined and served as a positive control, since c-Myc mRNA is a target of HuR (22), while the amplification of GAPDH PCR products, found in all samples as low-level contaminating housekeeping transcripts (not HuR targets), served to monitor the evenness of sample input, as reported previously (27,48). In addition, HuR was also found to interact with mRNAs encoding other intercellular junction proteins such as ZO-1, claudin-1, E-cadherin and β-catenin, but it did not bind to mRNAs encoding ZO-2 and JAM-1 (Supplementary Figure S1). (HuR/occludin mRNA) associations were further tested by using biotinylated transcripts which spanned occludin CR and 3′-UTR. Following incubation with cytoplasmic lysates, the interaction between the biotinylated occludin transcripts and HuR was examined by biotin pulldown followed by western blot analysis (23). The occludin 3′-UTR transcripts readily associated with HuR (Figure 1D), but HuR did not interact with the occludin CR. The RBP TIAR also formed complexes with the occludin 3′-UTR but not with its CR. Neither the occludin 3′-UTR nor its CR interacted with β-actin, included here as a negative control.
Figure 1.
HuR binds the occludin mRNA. (A) Schematic representative of the occludin mRNA and the predicted hits of the HuR signature motif in its 3′-UTR. (B) Association of endogenous HuR with endogenous occludin mRNA. After IP of RNA-protein complexes from cell lysates using either anti-HuR antibody (Ab) or control IgG1, RNA was isolated and used in RT reactions. RT–PCR products of occludin and c-Myc were visualized in ethidium bromide-stained agarose gels; low-level amplification of GAPDH (housekeeping mRNA, which is not HuR targets) served as negative controls. (C) Fold differences in occludin transcript abundance in HuR IP compared with IgG IP in cells described in panel (B), as measured by RT–qPCR analysis. Values were means ± SEM from triplicate samples. (D) Representative HuR and TIAR immunoblots using the pull-down materials by biotinylated transcripts of the occludin CR or 3′-UTR. Cytoplasmic lysates were incubated with 6 µg of biotinylated occludin CR or 3′-UTR, and the resulting RNP complexes were pulled down by using streptavidin-coated beads. The presence of HuR or TIAR in the pull-down material was assayed by western blotting. β-actin in the pull-down material was also examined and served as a negative control. (E) HuR-binding to different fractions of 3′-UTR of the occludin mRNA. Top panel, schematic representation of the occludin 3′-UTR biothinylated transcripts. After incubation of cytoplasmic lysates with the full-length (FL) or various fractions (F) of the occludin 3′-UTR, the resulting RNP complexes were pulled down, and the abundance of HuR and β-actin proteins in the pull-down material was examined. (F) Mapping the HuR-binding sites in F3 of the occludin 3′-UTR. Top panel, schematic representation of the biotinylated transcripts of F3 used in this study.
To define the specific HuR-binding regions in the occludin 3′-UTR, various partial biotinylated transcripts spanning the occludin 3′-UTR (spanning positions 1721–4141) were prepared as shown (Figure 1E, schematic). HuR was found to bind predominantly fragment F3 (spanning positions 2791–3425). In contrast, there was no detectable binding of HuR to fragments F2 and F4 and only marginal binding to fragment F1. To further narrow down the HuR-binding sites in the F3 of the occludin 3′-UTR, different sub-fragments (S) of F3 were prepared (Figure 1F, schematic). HuR only formed complexes with sub-fragment S4 (spanning 3247–3390) of the occludin 3′-UTR, the transcript which contained two hits of the HuR signature motif. These results indicate that HuR interacts with the occludin mRNA via specific RNA segments within its 3′-UTR.
HuR interaction with occludin mRNA enhances occludin translation via its 3′-UTR
To examine the functional consequences of HuR interactions with occludin mRNA, we silenced HuR expression by transfection with siRNA targeting the HuR mRNA (siHuR). These specific siHuR nucleotides were designed to reduce HuR mRNA and to have a unique combination of specificity, efficacy and low toxicity (22,42). The levels of HuR protein were decreased by ~85% in cells transfected with siHuR for 48 or 72 h (Figure 2A); this reduction was specific, as the levels of other RBPs such as TIAR were not affected in HuR-silenced populations (Figure 2A). Importantly, HuR silencing reduced occludin protein (Figure 2A, middle) but it failed to alter the levels of total occludin mRNA (Figure 2B).
Figure 2.
Changes in occludin translation after HuR silencing. (A) Representative immunoblots of HuR and occludin proteins. After cells were transfected with either siRNA targeting the HuR mRNA coding region (siHuR) or control siRNA (C-siRNA) for 48 or 72 h, whole-cell lysates were harvested for western blot analysis. (B) Levels of occludin mRNA in cells treated as described in panel (A). Total RNA was harvested, and the levels of occludin mRNA were measured by RT–qPCR analysis. Data were normalized to GAPDH mRNA levels, and values are means ± SEM of data from triplicate experiments. (C) Newly translated occludin protein. Occludin translation was measured by incubating cells with L-[35S]methionine and L-[35S]cysteine for 30 min, followed by immunoprecipitation using an anti-occludin antibody, resolving the immunoprecipitated samples by SDS–PAGE, transferring the samples, and visualization of signals by using a PhosphorImager. (D) Polysomal profiles from cells 48 h after transfection with C-siRNA (a) or siHuR (b). Nuclei were pelleted, and the resulting supernatants were fractionated through a 10–50% linear sucrose gradient. (E) Distributions of occludin (left) and GAPDH (right) mRNAs in each gradient fraction prepared from cells described in panel (D). Total RNA was isolated from the different fractions, and the levels of occludin and GAPDH mRNAs were measured by RT–qPCR analysis and plotted as a percentage of the total occludin or GAPDH mRNA levels in that sample. NB, not bound to polysomes; NT, not translated; LMW, low-molecular-weight polysomes; HMW, high-molecular-weight polysomes. Three independent experiments were performed and showed similar results. (F) Changes in occludin translation efficiency as measured by occludin 3′-UTR-luciferase reporter assays. Left, schematic of plasmids: (a), control (pGL3-Luc); (b), chimeric firefly luciferase-occludin 3′-UTR (Luc-Occl-3′-UTR). Right, levels of occludin translation. The Luc-Occl-3′-UTR or pGL3-Luc (negative control) was cotransfected with a Renilla luciferase reporter. Luciferase values were normalized to the mRNA levels to obtain translation efficiencies and expressed as means ± SEM of data from three separate experiments. *P < 0.05 compared with cells transfected with C-siRNA. (G) Changes in paracellular permeability as measured by using the membrane-impermeable trace molecule 14C-mannitol. Values are means ± SEM of data from six samples. *P < 0.05 compared with cells transfected with C-siRNA.
To ascertain if the reduction in occludin expression in HuR-silenced populations was due to repression of occludin translation, we examined the changes in the rate of new occludin synthesis after HuR silencing. Cells were incubated in the presence of L-[35S]methionine and L-[35S]cysteine for 30 min, whereupon newly translated occludin was visualized by IP. As shown in Figure 2C, newly synthesized occludin was markedly lower in HuR-silenced cells as compared with what was observed in cells transfected with C-siRNA. HuR silencing did not appear to affect global protein translation because it did not influence nascent GAPDH synthesis. To further define the role of HuR in the regulation of occludin mRNA translation, we examined the relative distributions of occludin mRNA on individual fractions from polyribosome gradients after HuR silencing as described previously (19,48). In this study, mRNAs in fractions 1–4 were not considered to be translated, because they were not associated with components of the translation machinery or cosedimented with ribosome subunits (monosomes); and transcripts in fractions 5–7 were bound to single ribosomes or formed polysomes of low molecular weight and they were considered to be translated at low-to-moderate levels. Fractions 8–10 comprised the mRNAs that were associated with polysomes of high molecular weight and they were thus considered to be actively translated. Although there were no significant changes in global polysomal profiles after HuR silencing (Figure 2D), the abundance of occludin mRNA in fractions 8 and 9 of the polysomal profiles decreased in HuR-silenced populations (Figure 2E, left). This redistribution of occludin mRNA in polyribosomes after HuR silencing was specific, as the housekeeping GAPDH mRNA distributed similarly in the two groups (Figure 2E, right).
To examine whether the translational effect of HuR was exerted through specific occludin sequences, the firefly luciferase reporter gene construct pLuc-Occl-3′-UTR (Figure 2F, left) was generated. To distinguish translational output from changes in mRNA levels, luciferase activity was normalized to luciferase-reporter mRNA levels to assess the translation efficiency (29,37). HuR silencing decreased occludin 3′-UTR-luciferase reporter gene activity (Figure 2F, right), indicating that HuR regulates occludin mRNA translation through the occludin 3′-UTR ARE. In contrast, no changes in luciferase activity were seen in response to HuR silencing when testing a negative control pGL3-Luc vector containing no the occludin 3′-UTR ARE (Figure 2F, left). Importantly, decreased levels of occludin by HuR silencing also disrupted the intestinal epithelial barrier function as indicated by an increase in the levels of paracellular flux of 14C-mannitol (Figure 2G). These results indicate that HuR silencing represses occludin translation via its 3′-UTR, thus leading to epithelial barrier dysfunction in vitro.
The results presented in Figure 3 further showed that ectopic HuR overexpression increased occludin translation and enhanced the epithelial barrier function. A vector expressing wild-type HuR-TAP fusion protein (HuR-TAP) was generated as described (42). Overexpression of HuR-TAP increased the levels of exogenous HuR-TAP proteins and occludin without affecting the levels of endogenous HuR (Figure 3A, middle). Increased HuR induced occludin by enhancing its translation, because HuR overexpression increased the occludin 3′-UTR-luciferase reporter gene activity (Figure 3B) but did not induce the levels of total occludin mRNA (Figure 3C). In contrast, neither HuR protein levels nor occludin expression were altered by transfection with the control vector. Consistent with these observations, increased levels of occludin by HuR overexpression also enhanced the epithelial barrier function as shown by a decrease in the levels of paracellular flux of 14C-mannitol (Figure 3D). These results indicate that increasing HuR levels stimulates occludin translation and improves epithelial barrier function.
Figure 3.
Changes in occludin translation after ectopic HuR overexpression. (A) Representative immunoblots of HuR-TAP fusion protein (HuR-TAP), endogenous HuR and occludin proteins. Cells were transfected with the vector expressing wild-type HuR-TAP or control emptying vector; protein levels were measured by western immunoblotting analysis 48 h after the transfection using specific antibody against TAP, HuR or occludin. (B) Changes in occludin translation efficiency as measured by using pGL3-Luc-Occl-3′-UTR reporter assays in cells described in panel (A). Twenty-four hours after cells were transfected with the Luc-Occl-3′-UTR or pGL3-Luc, the levels of luciferase activity were examined and normalized to the mRNA levels to obtain translation efficiencies. Values were expressed as means ± SEM of data from three separate experiments. *P < 0.05 compared with controls and cells transfected with emptying vector. (C) Levels of occludin mRNA as measured by RT–qPCR analysis in cells described in panel (A). The data were normalized to GAPDH mRNA levels, and shown as the means ± SEM of data from triplicate experiments. (D) Changes in paracellular permeability as measured by the membrane-impermeable trace molecule 14C-mannitol flux assays. Values are means ± SEM of data from six samples. *P < 0.05 compared with controls and cells transfected with the emptying vector alone.
Chk2-dependent HuR phosphorylation modulates HuR-binding to occludin mRNA
Recently, Chk2 was shown to physically interact with and modulate HuR function by regulating its phosphorylation (22,42). To determine the implication of Chk2-dependent HuR phosphorylation in the control of occludin mRNA translation, we examined changes in the levels of phosphorylated HuR (p-HuR) and its binding affinity for occludin mRNA after Chk2 silencing. Inhibition of Chk2 by transfection with specific siRNA targeting the coding region of Chk2 mRNA (siChk2) decreased p-HuR levels (Figure 4A), although it did not alter whole-cell HuR content. The inhibition of HuR phosphorylation by Chk2 silencing not only reduced (HuR/occludin mRNA) association (Figure 4B and C) but also repressed occludin mRNA translation, as indicated by a decrease in levels of occludin ARE luciferase reporter gene activity [Figure 4D (a)]. In contrast, Chk2 silencing failed to alter the levels of total occludin mRNA [Figure 4D (b)]. Consistently, occludin protein levels declined in Chk2-silenced populations (Figure 4E); the epithelial barrier function was also disrupted, as indicated by an increase in the levels of paracellular flux of 14C-mannitol (Figure 4F). These results indicate that Chk2 was essential for HuR-binding to the occludin mRNA and played an important role in the regulation of occludin mRNA translation.
Figure 4.
Chk2-dependent HuR phosphorylation regulates HuR-binding to occludin mRNA and alters its translation. (A) Representative immunoblots of Chk2, total HuR (T-HuR) and phosphorylated HuR (p-HuR) after Chk2 silencing. Cells were transfected with either siRNA targeting the Chk2 mRNA coding region (siChk2) or control siRNA (C-siRNA) for 48 h, and whole-cell lysates were harvested for western blot analysis to examine the levels of Chk2 and HuR, and loading control β-actin. To assess the levels of p-HuR, cell lysates were subjected to IP using an anti-HuR antibody, and the precipitates were analyzed by western blotting with the antibody against phosphorylated proteins (pProteins) or anti-HuR antibody. (B) Changes in HuR-binding to occludin mRNA as detected by biotin pull down assays: (a) binding to 3′-UTR; and (b) binding to coding region (CR). (C) Association of endogenous HuR with endogenous occludin mRNA in cells described in (A). Whole-cell lysates were used for IP in the presence of anti-HuR antibody or nonspecific IgG; and the levels of occludin and GAPDH mRNAs in the IP material were examined by RT–qPCR analysis. Values are the means ± SEM of data from three samples. *P < 0.05 compared with cells transfected with the C-siRNA. (D) Changes in occludin translation efficiency (a) and occludin mRNA levels (b) as measured by using Luc-Occl-3′-UTR reporter assays and RT–qPCR analysis in cells described in (A). Data were expressed as means ± SEM of data from three separate experiments. *P < 0.05 compared with cells transfected with Con-siRNA. (E) Levels of occludin protein in Chk2-silenced cells. Data are representative from three independent experiments showing similar results. (F) Changes in paracellular permeability. Values are means ± SEM of data from six samples. *P < 0.05 compared with controls and cells transfected with C-siRNA.
Based on the analysis of the occludin 3′-UTR (Figure 1E and F), HuR predominantly binds occludin transcript F3. To determine if Chk2-dependent HuR phosphorylation regulates occludin translation by interacting specifically with this fragment, we generated reporter constructs that expressed chimeric RNA containing the luciferase and partial transcripts spanning the occludin 3′-UTR, as shown in Figure 5A and B (top panels). The results presented in Figure 5A show that decreased HuR/occludin mRNA interaction by Chk2 silencing repressed translation efficiency, as indicated by a decrease in the levels of reporter gene activity when cells were transfected with pLF3 (containing the HuR-binding site). In contrast, basal levels of reporter gene activity of the constructs pLF1, pLF2 or pLF4 (without HuR-binding sites) were very low and they were also unaffected by Chk2 silencing. Analysis of additional reporter constructs containing sub-fragments of F3 revealed that Chk2 silencing decreased the levels of reporter gene activity when cells were transfected with pLS4 (containing HuR-binding site) but it failed to reduce the reporter activity of pLS1, pLS2, pLS3 and pLS5 in which HuR-binding sites were deleted (Figure 5B). To further define the role of the S4 region in regulating occludin mRNA translation, we generated a reporter construct in which the S4 was deleted from the occludin 3′-UTR (pL-FL-MS4; Figure 5C schematic). Deletion of S4 from the occludin 3′-UTR abrogated the Chk2- and HuR-dependent regulation, since silencing HuR or Chk2 failed to alter the translation efficiency of reporter pL-FL-MS4. Consistent with this observation, deletion mutation of the S4 fragment from the occludin 3′-UTR also disrupted HuR association with the occludin 3′-UTR as measured by RNP/IP–qPCR analysis (Figure 5D). These data strongly suggest that the HuR-binding region located at the F3-S4 fragment is primarily responsible for the regulatory processes mediated by HuR.
Figure 5.
Impact of deleting the HuR-interacting RNA from the occludin 3′-UTR on HuR-mediated occludin translation. (A and B) Changes in the levels of occludin 3′-UTR luciferase reporter activity in Chk2-silenced cells. Top panels: schematic of firefly luciferase (FL) reporter constructs containing different fragments of the occludin 3′-UTR. Twenty-four h after the cells were transfected with either siRNA targeting the Chk2 mRNA coding region (siChk2) or control siRNA (C-siRNA), the cells were further transfected with each of various occludin 3′-UTR luciferase reporter constructs and a Renilla luciferase control reporter. The levels of firefly and Renilla luciferase activities were assayed 24 h later. The results were normalized to the Renilla luciferase activity and are shown as the means ± SEM of data from three separate experiments. *P < 0.05 compared with cells transfected with C-siRNA. (C) Activity of luciferase reporters containing the occludin 3′-UTR with the S4 mutation after silencing HuR or Chk2. Cells were initially transfected with either siHuR, siChk2 or C-siRNA and 24 h later they were further transfected with pL-FL or pL-FL-MS4 together with the Renilla luciferase reporter. *P < 0.05 compared with cells transfected with C-siRNA. (D) Changes in HuR association with the occludin 3′-UTR after deleting the S4 sequence. Twenty-four hours after cells were transfected with pL-FL or pL-FL-MS4, whole-cell lysates were used for IP in the presence of anti-HuR antibody (left) or non-specific IgG1 (right). RNA in the IP material was used in RT–PCR reactions to detect the presence of occludin 3′-UTR (S3–S5), and the resulting PCR product (402 bp) were visualized in agarose gels. Three separate experiments were performed that showed similar results.
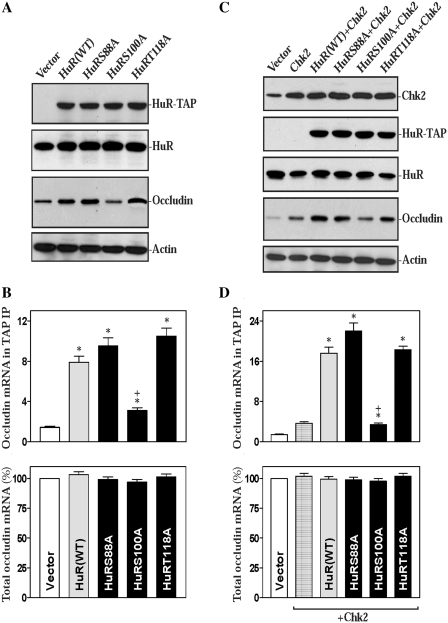
Given the fact that Chk2 phosphorylates HuR at residues S88, S100 and T118 and that each individual phosphorylation site by Chk2 plays a distinct role in regulating HuR- binding to different target mRNAs (22,42), we further tested HuR mutants with alanine substitutions at each of the Chk2 phosphorylation sites. As shown in Figure 6A and B, point mutation at Chk2 phosphorylation sites modulated HuR-binding to the occludin mRNA and altered occludin expression. Point mutations at each individual Chk2 phosphorylation site in HuR affected its binding to the occludin mRNA differently (Figure 6B, top), but they did not affect the levels of total occludin mRNA (Figure 6B, bottom). The S100A mutation decreased HuR association with the occludin mRNA and reduced occludin protein level when compared with those obtained from cells expressing HuR(WT)-TAP. In contrast, S88A and T118A mutations did not affect their binding to the occludin mRNA, as the levels of the binding complexes and occludin protein in cells expressing HuR(S88A)-TAP or HuR(T118A)-TAP were similar to those observed in cells expressing HuR(WT)-TAP. We also examined the effect of coexpressing Chk2 and different HuR-TAP mutants on HuR association with the occludin mRNA and found that point mutation at Chk2 phosphorylation sites in HuR also altered Chk2-induced HuR-binding to the occludin mRNA, in turn affecting occludin translation. The results presented in Figure 6C show that Chk2 overexpression alone slightly increased occludin protein expression level compared with those observed in cells transfected with the control vector, but coexpression of Chk2 with HuR(WT)-TAP, HuR(S88A)-TAP or HuR(T118A)-TAP remarkably increased the levels of occludin protein. On the other hand, coexpression of Chk2 and HuR(S100A)-TAP did not increase occludin translation, because the levels of occludin protein were similar in the Chk2 alone and Chk2 + HuR(S100A) groups. When the association of HuR with the occludin mRNA was examined, the level of the binding complexes increased marginally in cells transfected with the Chk2 alone but was remarkably elevated in cells coexpressing Chk2 and HuR(WT)-TAP, HuR(S88A)-TAP or HuR(T118A)-TAP, but not HuR(S100A)-TAP (Figure 6D, top). In addition, coexpression of Chk2 with either HuR(WT)-TAP or different HuR-TAP mutants failed to alter total occludin mRNA levels (Figure 6D, bottom). Taken together, these findings indicate that phosphorylation at residue S100 by Chk2 enhances HuR-binding to the occludin mRNA, while phosphorylation at S88 and T118 reduces this interaction; these modifications in turn regulate occludin translation.
Figure 6.
Ectopic overexpression of HuR carrying point mutations modulates occludin expression by altering (HuR/occludin mRNA) association. (A) Representative immunoblots of HuR-TAP fusion proteins (HuR-TAP), endogenous HuR, and occludin proteins. Cells were transfected with the vector expressing wild-type (WT) HuR-TAP or mutated HuR-TAP fusion proteins, and the protein levels were measured by western immunoblotting analysis 48 h after the transfection using specific antibody against TAP, HuR or occludin. (B) Changes in levels of the occludin mRNA in TAP IP materials (top) and total occludin mRNA (bottom) in cells described in panel (A). Total occludin mRNA levels were measured by RT–qPCR analysis, while binding of chimeric HuR-TAP proteins to occludin mRNA was examined by performing TAP IP followed by RT–qPCR analysis. Values are means ± SEM of data from three separate experiments. *, +P < 0.05 compared with controls (vector) and cells transfected with the HuR(WT)-TAP, respectively. (C) Changes in levels of occludin protein after overexpression of Chk2 and chimeric HuR-TAP proteins. Cells were cotransfected with the Chk2 expression vector (Chk2) and vectors expressing HuR (WT)-TAP or mutated HuR-TAP fusion proteins. The levels of Chk2, HuR-TAP, endogenous HuR and occludin proteins were measured 48 h after the transfection. (D) Changes in levels of the occludin mRNA in TAP IP materials (top) and total occludin mRNA (bottom) in cells described in panel (C) as measured by RT–qPCR analysis. *P < 0.05 compared with cells transfected with the emptying vector; +P < 0.05 compared with cells cotransfected with Chk2 and HuR(WT)-TAP.
Inhibition of Chk2 by polyamine depletion reduces (HuR/occludin mRNA) complexes and represses occludin translation
Polyamines (spermidine, spermine and their precursor putrescine) are ubiquitous polycationic molecules in eukaryotic cells and are implicated in many aspects of biological functions including the control of the gut barrier (40,55). In this study, we examined if polyamines regulate occludin translation by altering Chk2-dependent HuR phosphorylation. Consistent with our previous studies (56,57), inhibition of ODC (a key enzyme for polyamine biosynthesis) by treatment with DFMO for 4 days completely depleted the cellular polyamines putrescine and spermidine and significantly decreased spermine level (Supplementary Figure S2). As shown in Figure 7A, polyamine depletion by DFMO inhibited Chk2 expression and decreased the levels of p-HuR, although it did not change whole-cell HuR levels. Polyamine depletion also inhibited HuR association with the occludin mRNA (Figure 7B and C); this reduction was mediated through occludin 3′-UTR. The decreased levels of (HuR/occludin mRNA) complexes in polyamine-deficient cells were accompanied by reductions of occludin translation (Figure 7D) and occludin protein abundance (Figure 7E). Furthermore, the reduced occludin levels in cells with decreased p-HuR through polyamine depletion were also associated with the epithelial barrier dysfunction as indicated by an increase in levels of paracellular flux of [14C]mannitol (Figure 7F). In the presence of DFMO, exogenous putrescine not only prevented reduced levels of Chk2 and (HuR/occludin mRNA) complexes but also returned occludin translation to near-normal levels. Consistently, the epithelial barrier function also returned to normal when putrescine was given together with DFMO. In addition, there were no significant differences in the levels of total occludin mRNA between control cells and cells exposed to DFMO alone or DFMO plus putrescine (Supplementary Figure S3). These results indicate that polyamine depletion represses occludin translation by decreasing (HuR/occludin mRNA) association as a result of a reduction in Chk2-dependent HuR phosphorylation.
Figure 7.
Polyamine depletion represses occludin translation by decreasing Chk2-dependent HuR phosphorylation. (A) Representative immunoblots of Chk2, total HuR (T-HuR), and phosphorylated HuR (p-HuR) in polyamine-deficient cells. After cells were grown in control media and in media containing DFMO (5 mM) or DFMO plus putrescine (Put; 10 µM) for 4 days, whole cell lysates were harvested. The levels of Chk2, T-HuR and p-HuR were measured by western blot analysis, while the levels of p-HuR were examined by immunoprecipitation (IP) assays. (B) Changes in HuR-binding to occludin mRNA as detected by biotin pull down assays in cells described in panel (A): (a) binding to 3′-UTR; and (b) binding to coding region (CR). (C) Association of endogenous HuR with endogenous occludin mRNA as measured by RNP–IP/RT–qPCR assays. Values are means ± SEM of data from six samples. *P < 0.05 compared with controls and cells treated with DFMO plus Put. (D) Changes in occludin translation efficiency as measured by Luc-Occl-3′-UTR reporter assays in cells described in (A). Data were expressed as means ± SEM of data from three samples. *P < 0.05 compared with controls and cells exposed to DFMO plus Put. (E) Representative immunoblots of occludin protein. (F) Changes in paracellular permeability in cells described in panel (A). Values are means ± SEM of data from six samples. *P < 0.05 compared with controls and cells exposed to DFMO plus Put.
Septic stress decreases Chk2 and reduces HuR/occludin mRNA association in vivo
To assess the physiologic role of Chk2/HuR in the regulation of occludin expression in vivo and their involvement in pathogenesis of the gut barrier dysfunction after surgical stress, we investigated if intestinal Chk2 expression and HuR/occludin mRNA interactions were changed in sepsis using the cecal ligation and puncture (CLP) model (52). As shown in Figure 8A, mucosal Chk2 and occludin expression levels in the small intestine decreased markedly after sublethal CLP, although there were no changes in total HuR levels in CLP-mice. The maximal decrease in the levels of Chk2 and occludin proteins occurred between 24 and 48 h after CLP, whereas expression of Chk2 and occludin began to gradually recover at 72 h and almost returned to normal level at 120 h after CLP. To examine changes in cellular distribution of Chk2 and microscopic injury of the small intestine after CLP, we found that Chk2 immunostaining was predominantly located at the villous area of the mucosa and its intensity decreased dramatically after CLP (Figure 8B). Focal villous necrosis in the intestine was observed at 24 and 48 h after CLP; the damage was repaired within the next 48 h. Sham operation did not affect expression of Chk2, occludin levels or gut epithelial integrity. On the other hand, CLP stress also resulted in gut barrier dysfunction as indicated by an increase in intestinal mucosal permeability to FITC-dextran (Figure 8C), in which the maximal increase in gut permeability occurred at 24 and 48 h after the stress. Consistent with the recovery of Chk2 and occludin expression, gut barrier function partially recovered at 72 and 96 h after CLP and restored to near-normal level at 120 h thereafter. To determine the mechanism underlying the repression of occludin expression induced by CLP, we examined the changes in levels of total occludin mRNA and its interaction with HuR. Exposure to CLP stress decreased the association of occludin mRNA with HuR as measured by RNP–IP/qPCR analysis (Figure 8D), but it failed to affect total occludin mRNA levels (Figure 8E). The maximal reduction in the levels of (HuR/occludin mRNA) complexes occurred at 24 and 48 h after CLP and recovered gradually thereafter. The levels of (HuR/occludin mRNA) complexes at 120 h after CLP were similar to those observed in control interventions. These findings suggest that CLP-induced repression of occludin expression occurs at the post-transcriptional level and that decreased HuR association with the occludin mRNA following Chk2 reduction plays a role in pathogenesis of the gut barrier dysfunction in response to CLP stress.
Figure 8.
Reduction in Chk2 associates with inhibition of the occludin expression and gut barrier dysfunction in mice exposed to CLP. (A) Representative immunoblots of Chk2, occludin and HuR proteins in the small intestinal mucosa after CLP. The mucosal tissues were harvested at different times as indicated after CLP surgery; and changes in the levels of Chk2, occludin and HuR proteins were measured by western immunoblotting analysis. The results in each panel were from separate experiments. (B) Immunohistochemical staining of Chk2 in the small intestinal mucosa in control (a) and mice exposed to CLP for 24 h (b). c, crypt; v, villous; arrow, Chk2 immunostaining. (C) Changes in gut permeability as measured by FITC-dextran tracer flux assays in animals described in panel (A). FITC-dextran was given orally, and blood samples were collected 4 h thereafter for measurement. Values are means ± SEM of data from five samples. *,+P < 0.05 compared with controls (0 h) and animals exposed to CLP for 24 h, respectively. (D) Association of HuR with occludin mRNA in the small intestinal mucosa as measured by RNP–IP/RT–qPCR analysis. Values are means ± SEM of data from five samples. *,+P < 0.05 compared with controls (0 h) and animals exposed to CLP for 24 h, respectively. (E) The levels of total occludin mRNA in the intestinal mucosa as measured by RT–qPCR analysis in mice exposed to CLP. Values are means ± SEM of data from five samples.
Polyamine depletion represses occludin expression and delays gut barrier recovery
Given the fact that decreasing cellular polyamines by DFMO inhibited Chk2 and dissociated (HuR/occludin mRNA) complexes in cultured IECs (Figure 7), we examined the effect of Chk2 inhibition by polyamine depletion on HuR/occludin mRNA interaction in the intestinal mucosa after CLP. In this study, half of the mice were given DFMO, 50 mg/100-g body weight, i.p. immediately after CLP, and then 2% DFMO was given in the drinking water throughout the entire period of experiment. Our previous studies show that this dose of DFMO completely prevents increases in ODC and decreases tissue polyamine content after the stress (58,59). As shown in Figure 9A and B, the DFMO-decreased levels of cellular polyamines did not additionally reduce the levels of (HuR/occludin mRNA) complexes and occludin protein at 24 h after CLP. The normal recovery of HuR-binding to the occludin mRNA and occludin expression that occurred by 72 and 96 h after CLP, however, was prevented by polyamine depletion. In the animals treated with DFMO, there was no significant increase in the levels of (HuR/occludin mRNA) complexes and occludin protein at 72 and 96 h after CLP. In contrast, DFMO treatment did not affect total occludin mRNA levels in CLP-mice. Consistently, polyamine depletion also delayed recovery of the gut barrier function at 72 and 96 h after CLP, although it failed to additionally increase gut permeability at 24 h in CLP-mice (Figure 9C). These results suggest that recovery of HuR/occludin mRNA association via Chk2 and the ensuing occludin upregulation are involved in the reestablishment of gut barrier function after septic stress.
Figure 9.
Polyamine depletion reduces (HuR/occludin mRNA) association and delays the recovery of occludin expression and gut barrier function in CLP-mice. (A) Changes in the levels of occludin mRNA in HuR IP materials (top) and total occludin mRNA (bottom). After CLP surgery, mice were injected i.p. with saline or DFMO at the dose of 5 mg/10-g body wt, followed by 2% DFMO in their drinking water throughout the experiment. Binding of HuR to occludin mRNA was examined by RNP-IP/RT–qPCR analysis, while total occludin mRNA levels were measured by RT–qPCR analysis. Values are means ± SEM of data from five samples. *P < 0.05 compared with CLP-mice treated with saline. (B) Representative immunoblots of occludin protein in the small intestinal mucosa from mice described in panel (A). (C) Changes in gut permeability as measured by FITC-dextran tracer flux assays. Values are means ± SEM of data from five samples. *P < 0.05 compared with CLP-mice treated with saline.
DISCUSSION
Although the importance of occludin in the assembly and regulation of TJs is well documented (1–3,9–11,14), little is known about the post-transcriptional control of occludin expression. In the present study, we uncover a novel function of the RBP HuR in the regulation of occludin mRNA translation and therefore in the intestinal barrier function. We found that HuR bound the occludin mRNA by directly interacting with its 3′-UTR and promoted occludin translation in vitro as well as in vivo. Experiments aimed at characterizing the molecular aspects of this process suggested that HuR associates with the occludin mRNA through Chk2-dependent HuR phosphorylation, likely at residue S100. Reduced HuR phosphorylation by Chk2 silencing or inhibition of Chk2 activity via polyamine depletion decreased the affinity of HuR for the occludin mRNA and repressed occludin translation, whereas Chk2 overexpression enhanced HuR association with the occludin mRNA and stimulated its expression. Our results also suggest that Chk2-dependent HuR phosphorylation is a critical factor for maintenance of the gut barrier integrity during septic stress in mice.
The results reported herein provide a comprehensive view of HuR interaction with the occludin mRNA and its regulation. HuR associated with the occludin 3′-UTR at a segment spanning position 3164–3306, which contains two predicted hits of the HuR signature motif (Figure 1), but it did not interact with the occludin CR. We did not identify the specific occludin 3′-UTR nucleotides with which HuR interacts, since those experiments would require more specialized biochemical, crystallographic and molecular methods than those used here. Our results further indicate that HuR association with the occludin mRNA enhances occludin translation without affecting total occludin mRNA levels. The occludin mRNA translation was decreased in HuR-silenced cells, but it was increased in cells overexpressing HuR. These findings are consistent with observations from others who demonstrated that HuR binds to the specific signature motifs in its target mRNAs and plays a general role as an enhancer of mRNA translation, as described for transcripts such as p53 (49), c-Myc (22), thrombospondin-1 (60), prothymosin-α (61), cytochrome c (48), MKP-1 (62), HIF-1α (63) and MEK1 (21) mRNAs. In addition, HuR also stabilizes SIRT1 (42), XIAP (27), VEGF (64), nucleophosmin (43) and ATF-2 (23) mRNAs, in turn inducing protein expression levels.
An important finding from the present study is that Chk2-dependent HuR phosphorylation is essential for HuR-binding to the occludin mRNA and thus plays a role in maintenance of normal epithelial barrier function. In Chk2-silenced cells, the levels of phosphorylated HuR decreased, (HuR/occludin mRNA) complexes dissociated and occludin mRNA translation declined, leading to barrier dysfunction (Figure 4). HuR is ubiquitously expressed and an RNA signature motif recognized by HuR is widely found in numerous mRNAs (26), suggesting that HuR can bind to a broad range of mRNAs and can influence their post-transcriptional fates. Increasing evidence indicates that the ability of HuR to bind a given mRNA and the specificity of HuR function are tightly regulated by numerous factors including HuR phosphorylation and binding of other RBPs and microRNAs (22,29,30,42). HuR was reported to be phosphorylated by Chk2 kinase in vivo as well as in vitro and this post-translational modification altered HuR's association with given target transcripts (22,42). HuR association with the SIRT1 and c-Myc mRNAs depends upon its phosphorylation by Chk2 (22,42), but HuR-binding to the mRNAs encoding p53, nucleophosmin, ATF-2 and XIAP appears to be independent of HuR phosphorylation (23,27,43). HuR is also shown to recruit the microRNA let-7/RISC complex to c-Myc 3′-UTR and repress c-Myc translation in HeLa cells (30), whereas it enhances c-Myc translation in intestinal epithelial cells that lack microRNA let-7 (22).
Studies using various HuR mutations revealed that HuR phosphorylation at residue S100 by Chk2 is crucial for the modulation of (HuR/occludin mRNA) association, although the exact role of HuR phosphorylation at the putative Chk2 target residues is not yet understood in full detail. The association of HuR(WT) with occludin mRNA was disrupted by the S100A mutation, but not by the S88A or T118A mutations (Figure 6). Consistent with changes in the pattern of (HuR/occludin mRNA) interactions, cells overexpressing HuR(S100A) showed decreased occludin protein levels relative to cells expressing HuR(WT), HuR(S88A) or HuR(T118A). Our recent study also shows that compared with HuR(WT), HuR(S100A) showed reduced binding to the c-Myc mRNA, while HuR(S88A) showed enhanced binding; HuR(T118A) showed the same binding as HuR(WT) (22). These observations, together with our previous studies (22,42), support the notion that HuR mutants lacking a given Chk2 phosphorylation site display distinct binding affinities for different target mRNAs.
Our results also indicate that Chk2-dependent HuR phosphorylation is regulated by cellular polyamines. Maintenance of intestinal mucosal epithelial integrity requires polyamines and their intracellular levels are tightly controlled under biological conditions (40,56–59,65). The regulation of cellular polyamines has been recognized for many years as a central convergence point for the multiple signaling pathways driving various epithelial cell functions (65). We recently demonstrated that decreasing cellular polyamines inhibited occludin expression and also decreased the levels of phosphorylated HuR (20,22). Although the mechanisms by which polyamines regulate occludin expression remain to be elucidated in full, our current studies show that polyamine depletion by DFMO decreased (HuR/occludin mRNA) complexes and repressed occludin mRNA translation (Figure 7). Exogenous polyamine putrescine when given together with DFMO prevented Chk2 inhibition and restored HuR phosphorylation and its binding affinity for the occludin mRNA, thereby promoting occludin translation and the epithelial barrier function. Consistent with our current findings, polyamines are also shown to increase HuR-binding to the c-Myc mRNA through enhancement of Chk2-dependent HuR phosphorylation (22).
Another major finding of the present study is that Chk2-dependent HuR phosphorylation is implicated in the maintenance and re-establishment of the gut barrier integrity under critical surgical stress. CLP stress decreased Chk2 levels, reduced (HuR/occludin mRNA) complexes, and inhibited occludin abundance, thus contributing to the pathogenesis of gut barrier dysfunction (Figure 8). In contrast, induced Chk2-dependent HuR phosphorylation at later phase after CLP appears to be crucial for the recovery of occludin expression and the gut barrier function, because Chk2 inhibition by polyamine depletion prevented the induction in occludin expression and delayed the re-establishment of barrier function at 72 and 96 h in septic mice with administration with DFMO (Figure 9). Although there are potential concerns about the specificity of DFMO, Chk2 inhibition, and the involvement of other TJ proteins or factors in this process, to our knowledge, these results are the first to show an association between the expression of occludin via Chk2-dependent HuR phosphorylation and the maintenance of gut barrier integrity under pathological conditions. In summary, together our current findings indicate that Chk2-dependent HuR phosphorylation regulates occludin mRNA translation and helps to maintain and re-establish epithelial barrier function in the intestinal tract in response to stress.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Merit Review Grants (to J.-Y.W. and J.N.R.) from US Department of Veterans Affairs and by National Institutes of Health (DK57819, DK61972, DK68491 to J.-Y.W.). National Institute on Aging-Intramural Research Program, National Institutes of Health (to M.G.). Funding for open access charge: National Institute of Diabetes and Digestive and Kidney Diseases.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
J.-Y.W. is a Senior Research Career Scientist, Medical Research Service, US Department of Veterans Affairs.
REFERENCES
- 1.Turner JR. Intestinal mucosal barrier function in health and disease. Nat. Rev. Immunol. 2009;9:799–809. doi: 10.1038/nri2653. [DOI] [PubMed] [Google Scholar]
- 2.Schulzke JD, Fromm M. Tight junctions: molecular structure meets function. Ann. NY Acad. Sci. 2009;1165:1–6. doi: 10.1111/j.1749-6632.2009.04925.x. [DOI] [PubMed] [Google Scholar]
- 3.Förster C. Tight junctions and the modulation of barrier function in disease. Histochem. Cell Biol. 2008;130:55–70. doi: 10.1007/s00418-008-0424-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Funke L, Dakoji S, Bredt DS. Membrane-associated guanylate kinases regulate adhesion and plasticity at cell junctions. Annu. Rev. Biochem. 2005;74:219–245. doi: 10.1146/annurev.biochem.74.082803.133339. [DOI] [PubMed] [Google Scholar]
- 5.Nejsum LN, Nelson WJ. A molecular mechanism directly linking E-cadherin adhesion to initiation of epithelial cell surface polarity. J. Cell Biol. 2007;178:323–335. doi: 10.1083/jcb.200705094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schneeberger EE, Lynch RD. The tight junction: a multifunctional complex. Am. J. Physiol. Cell Physiol. 2004;286:1213–1228. doi: 10.1152/ajpcell.00558.2003. [DOI] [PubMed] [Google Scholar]
- 7.Liu L, Guo X, Rao JN, Zou T, Xiao L, Yu T, Timmons JA, Turner DJ, Wang JY. Polyamines regulate E-cadherin transcription through c-Myc modulating intestinal epithelial barrier function. Am. J. Physiol. Cell Physiol. 2009;296:C801–C810. doi: 10.1152/ajpcell.00620.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Guo X, Rao JN, Liu L, Zou T, Turner DJ, Bass BL, Wang JY. Regulation of adherens junctions and epithelial paracellular permeability: a novel function for polyamines. Am. J. Physiol. Cell Physiol. 2003;285:C1174–C1187. doi: 10.1152/ajpcell.00015.2003. [DOI] [PubMed] [Google Scholar]
- 9.Raleigh DR, Marchiando AM, Zhang Y, Shen L, Sasaki H, Wang Y, Long M, Turner JR. Tight junction-associated MARVEL proteins marveld3, tricellulin, and occludin have distinct but overlapping functions. Mol. Biol. Cell. 2010;21:1200–1213. doi: 10.1091/mbc.E09-08-0734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Suzuki T, Elias BC, Seth A, Shen L, Turner JR, Giorgianni F, Desiderio D, Guntaka R, Rao R. PKCη regulates occludin phosphorylation and epithelial tight junction integrity. Proc. Natl Acad. Sci. USA. 2009;106:61–66. doi: 10.1073/pnas.0802741106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Marchiando AM, Shen L, Graham WV, Weber CR, Schwarz BT, Austin JR, 2nd, Raleigh DR, Guan Y, Watson AJ, Montrose MH, et al. Caveolin-1-dependent occludin endocytosis is required for TNF-induced tight junction regulation in vivo. J. Cell Biol. 2010;189:111–126. doi: 10.1083/jcb.200902153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen Y, Merzdorf C, Paul DL, Goodenough DA. COOH terminus of occludin is required for tight junction barrier function in early Xenopus embryos. J. Cell Biol. 1997;138:891–899. doi: 10.1083/jcb.138.4.891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lacaz-Vieira F, Jaeger MM, Farshori P, Kachar B. Small synthetic peptides homologous to segments of the first external loop of occludin impair tight junction resealing. J. Membr. Biol. 1999;168:289–297. doi: 10.1007/s002329900518. [DOI] [PubMed] [Google Scholar]
- 14.Rao R. Occludin phosphorylation in regulation of epithelial tight junctions. Ann. NY Acad. Sci. 2009;1165:62–68. doi: 10.1111/j.1749-6632.2009.04054.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Du D, Xu F, Yu L, Zhang C, Lu X, Yuan H, Huang Q, Zhang F, Bao H, Jia L, et al. The tight junction protein, occludin, regulates the directional migration of epithelial cells. Dev. Cell. 2010;18:52–63. doi: 10.1016/j.devcel.2009.12.008. [DOI] [PubMed] [Google Scholar]
- 16.McCarthy KM, Skare IB, Stankewich MC, Furuse M, Tsukita S, Rogers RA, Lynch RD, Schneeberger EE. Occludin is a functional component of the tight junction. J. Cell Sci. 1996;109:2287–2298. doi: 10.1242/jcs.109.9.2287. [DOI] [PubMed] [Google Scholar]
- 17.Bamforth SD, Kniesel U, Wolburg H, Engelhardt B, Risau W. A dominant mutant of occludin disrupts tight junction structure and function. J. Cell Sci. 1999;112:1879–1888. doi: 10.1242/jcs.112.12.1879. [DOI] [PubMed] [Google Scholar]
- 18.Krishnan M, Singh AB, Smith JJ, Sharma A, Chen X, Eschrich S, Yeatman TJ, Beauchamp RD, Dhawan P. HDAC inhibitors regulate claudin-1 expression in colon cancer cells through modulation of mRNA stability. Oncogene. 2010;29:305–312. doi: 10.1038/onc.2009.324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen J, Xiao L, Rao JN, Zou T, Liu L, Bellavance E, Gorospe M, Wang JY. JunD represses transcription and translation of the tight junction protein zona occludens-1 modulating intestinal epithelial barrier function. Mol. Biol. Cell. 2008;19:3701–3712. doi: 10.1091/mbc.E08-02-0175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Guo X, Rao JN, Liu L, Zou T, Keledjian KM, Boneva D, Marasa BS, Wang JY. Polyamines are necessary for synthesis and stability of occludin protein in intestinal epithelial cells. Am. J. Physiol. Gastrointest. Liver Physiol. 2005;288:G1159–G1169. doi: 10.1152/ajpgi.00407.2004. [DOI] [PubMed] [Google Scholar]
- 21.Wang PY, Rao JN, Zou T, Liu L, Xiao L, Yu TX, Turner DJ, Gorospe M, Wang JY. Post-transcriptional regulation of MEK-1 by polyamines through the RNA-binding protein HuR modulating intestinal epithelial apoptosis. Biochem. J. 2010;426:293–306. doi: 10.1042/BJ20091459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu L, Rao JN, Zou T, Xiao L, Wang PY, Turner DJ, Gorospe M, Wang JY. Polyamines regulate c-Myc translation through Chk2-dependent HuR phosphorylation. Mol. Biol. Cell. 2009;20:4885–4898. doi: 10.1091/mbc.E09-07-0550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Xiao L, Rao JN, Zou T, Liu L, Marasa BS, Chen J, Turner DJ, Zhou H, Gorospe M, Wang JY. Polyamines regulate the stability of activating transcription factor-2 mRNA through RNA-binding protein HuR in intestinal epithelial cells. Mol. Biol. Cell. 2007;18:4579–4590. doi: 10.1091/mbc.E07-07-0675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li L, Liu L, Rao JN, Esmaili A, Strauch ED, Bass BL, Wang JY. JunD stabilization results in inhibition of normal intestinal epithelial cell growth through p21 after polyamine depletion. Gastroenterology. 2002;123:764–779. doi: 10.1053/gast.2002.35386. [DOI] [PubMed] [Google Scholar]
- 25.Keene JD. RNA regulons: coordination of post-transcriptional events. Nat. Rev. Genet. 2007;8:533–543. doi: 10.1038/nrg2111. [DOI] [PubMed] [Google Scholar]
- 26.Abdelmohsen K, Kuwano Y, Kim HH, Gorospe M. Posttranscriptional gene regulation by RNA-binding proteins during oxidative stress: implications for cellular senescence. Biol. Chem. 2008;389:243–255. doi: 10.1515/BC.2008.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang X, Zou T, Rao JN, Liu L, Xiao L, Wang PY, Cui YH, Gorospe M, Wang JY. Stabilization of XIAP mRNA through the RNA binding protein HuR regulated by cellular polyamines. Nucleic Acids Res. 2009;37:7623–7637. doi: 10.1093/nar/gkp755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yeh CH, Hung LY, Hsu C, Le SY, Lee PT, Liao WL, Lin YT, Chang WC, Tseng JT. RNA-binding protein HuR interacts with thrombomodulin 5′ untranslated region and represses internal ribosome entry site-mediated translation under IL-3 treatment. Mol. Biol. Cell. 2008;19:3812–3822. doi: 10.1091/mbc.E07-09-0962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bhattacharyya SN, Habermacher R, Martine U, Closs EI, Filipowicz W. Relief of microRNA-mediated translational repression in human cells subjected to stress. Cell. 2006;125:1111–1124. doi: 10.1016/j.cell.2006.04.031. [DOI] [PubMed] [Google Scholar]
- 30.Kim HH, Kuwano Y, Srikantan S, Lee EK, Martindale JL, Gorospe M. HuR recruits let-7/RISC to repress c-Myc expression. Genes Dev. 2009;23:1743–1748. doi: 10.1101/gad.1812509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chen CY, Shyu AB. AU-rich elements: characterization and importance in mRNA degradation. Trends Biochem. Sci. 1995;20:465–470. doi: 10.1016/s0968-0004(00)89102-1. [DOI] [PubMed] [Google Scholar]
- 32.Wilusz CJ, Wormington M, Peltz SW. The cap-to-tail guide to mRNA turnover. Nat. Rev. Mol. Cell Biol. 2001;2:237–246. doi: 10.1038/35067025. [DOI] [PubMed] [Google Scholar]
- 33.Wilusz CJ, Wiluszm J. Bringing the role of mRNA decay in the control of gene expression into focus. Trends Genet. 2004;20:491–497. doi: 10.1016/j.tig.2004.07.011. [DOI] [PubMed] [Google Scholar]
- 34.Kedersha N, Stoecklin G, Ayodele M, Yacono P, Lykke-Andersen J, Fitzler MJ, Scheuner D, Kaufman RJ, Golan DE, Anderson P. Stress granules and processing bodies are dynamically linked sites of mRNP remodeling. J. Cell Biol. 2005;169:871–884. doi: 10.1083/jcb.200502088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gherzi R, Lee KY, Briata P, Wegmuller D, Moroni C, Karin M, Chen CY. A KH domain RNA binding protein, KSRP, promotes ARE-directed mRNA turnover by recruiting the degradation machinery. Mol. Cell. 2004;14:571–583. doi: 10.1016/j.molcel.2004.05.002. [DOI] [PubMed] [Google Scholar]
- 36.Pan YX, Chen H, Kilberg MS. Interaction of RNA-binding proteins HuR and AUF1 with the human ATF3 mRNA 3'-untranslated region regulates its amino acid limitation-induced stabilization. J. Biol. Chem. 2005;280:34609–34616. doi: 10.1074/jbc.M507802200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liao B, Hu Y, Brewer G. Competitive binding of AUF1 and TIAR to MYC mRNA controls its translation. Nat. Struct. Mol. Biol. 2007;14:511–518. doi: 10.1038/nsmb1249. [DOI] [PubMed] [Google Scholar]
- 38.Doller A, Huwiler A, Müller R, Radeke HH, Pfeilschifter J, Eberhardt W. Protein kinase C alpha-dependent phosphorylation of the mRNA-stabilizing factor HuR: implications for posttranscriptional regulation of cyclooxygenase-2. Mol. Biol. Cell. 2007;18:2137–2148. doi: 10.1091/mbc.E06-09-0850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gorospe M. HuR in the mammalian genotoxic response: post-transcriptional multitasking. Cell Cycle. 2003;2:412–414. [PubMed] [Google Scholar]
- 40.Wang JY. Polyamines and mRNA stability in regulation of intestinal mucosal growth. Amino Acids. 2007;33:241–252. doi: 10.1007/s00726-007-0518-z. [DOI] [PubMed] [Google Scholar]
- 41.Fan XC, Steitz JA. Overexpression of HuR, a nuclear-cytoplasmic shuttling protein, increases the in vivo stability of ARE-containing mRNAs. EMBO J. 1998;17:3448–3460. doi: 10.1093/emboj/17.12.3448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Abdelmohsen K, Pullmann R, Jr, Lal A, Kim HH, Galban S, Yang X, Blethrow JD, Walker M, Shubert J, Gillespie DA, et al. Phosphorylation of HuR by Chk2 regulates SIRT1 expression. Mol. Cell. 2007;25:543–557. doi: 10.1016/j.molcel.2007.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zou T, Mazan-Mamczarz K, Rao JN, Liu L, Marasa BS, Zhang AH, Xiao L, Pullmann R, Gorospe M, Wang JY. Polyamine depletion increases cytoplasmic levels of RNA-binding protein HuR leading to stabilization of nucleophosmin and p53 mRNAs. J. Biol. Chem. 2006;281:19387–19394. doi: 10.1074/jbc.M602344200. [DOI] [PubMed] [Google Scholar]
- 44.Kim HH. Nuclear HuR accumulation through phosphorylation by Cdk1. Genes Dev. 2008;22:1804–1815. doi: 10.1101/gad.1645808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Suh E, Traber PG. An intestine-specific homeobox gene regulates proliferation and differentiation. Mol. Cell Biol. 1996;16:619–625. doi: 10.1128/mcb.16.2.619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Quaroni A, Wands J, Trelstad RL, Isselbacher KJ. Epithelioid cell cultures from rat small intestine. Characterization by morphologic and immunologic criteria. J. Cell Biol. 1979;80:248–265. doi: 10.1083/jcb.80.2.248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rao JN, Liu SV, Zou T, Liu L, Xiao L, Zhang X, Bellavance E, Yuan JX, Wang JY. Rac1 promotes intestinal epithelial restitution by increasing Ca2+ influx through interaction with phospholipase C-γ1 after wounding. Am. J. Physiol. Cell Physiol. 2008;295:C1499–C1509. doi: 10.1152/ajpcell.00232.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kawai T, Lal A, Yang X, Galban S, Mazan-Mamczarz K, Gorospe M. Translational control of cytochrome c by RNA-binding proteins TIA-1 and HuR. Mol. Cell. Biol. 2006;26:3295–3307. doi: 10.1128/MCB.26.8.3295-3307.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mazan-Mamczarz K, Galban S, de Silanes IL, Martindale JL, Atasoy U, Keene JD, Gorospe M. RNA-binding protein HuR enhances p53 translation in response to ultraviolet light irradiation. Proc. Natl Acad. Sci. USA. 2003;100:8354–8359. doi: 10.1073/pnas.1432104100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.López de Silanes I, Lal A, Gorospe M. HuR: post-transcriptional paths to malignancy. RNA Biol. 2004;1:135–137. doi: 10.4161/rna.2.1.1552. [DOI] [PubMed] [Google Scholar]
- 51.Guo X, Rao JN, Liu L, Zou TT, Turner DJ, Bas BL, Wang JY. Regulation of adherens junctions and epithelial paracellular permeability: a novel function for polyamines. Am. J. Physiol. Cell Physiol. 2003;285:C1174–C1187. doi: 10.1152/ajpcell.00015.2003. [DOI] [PubMed] [Google Scholar]
- 52.Hubbard WJ, Choudhry M, Schwacha MG, Kerby JD, Rue LW, 3rd, Bland KI, Chaudry IH. Cecal ligation and puncture. Shock. 2005;24:S52–S57. doi: 10.1097/01.shk.0000191414.94461.7e. [DOI] [PubMed] [Google Scholar]
- 53.Napolitano LM, Koruda MJ, Meyer AA, Baker CC. The impact of femur fracture with associated soft tissue injury on immune function and intestinal permeability. Shock. 1996;5:202–207. doi: 10.1097/00024382-199603000-00006. [DOI] [PubMed] [Google Scholar]
- 54.Harter JL. Critical values for Duncan's new multiple range test. Biometrics. 1960;16:671–685. [Google Scholar]
- 55.López de Silanes I, Zhan M, Lal A, Yang X, Gorospe M. Identification of a target RNA motif for RNA-binding protein HuR. Proc. Natl Acad. Sci. USA. 2004;101:2987–2992. doi: 10.1073/pnas.0306453101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Patel AR, Wang JY. Polyamine depletion is associated with an increase in JunD/AP-1 activity in small intestinal crypt cells. Am. J. Physiol. Gastrointest. Liver Physiol. 1999;276:G441–G450. doi: 10.1152/ajpgi.1999.276.2.G441. [DOI] [PubMed] [Google Scholar]
- 57.Zou T, Liu L, Rao JN, Marasa BS, Chen J, Xiao L, Zhou H, Gorospe M, Wang JY. Polyamines modulate the subcellular localization of RNA-binding protein HuR through AMP-activated protein kinase-regulated phosphorylation and acetylation of importin α1. Biochem. J. 2008;409:389–398. doi: 10.1042/BJ20070860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wang JY, Johnson LR. Polyamines and ornithine decarboxylase during repair of duodenal mucosa after stress in rats. Gastroenterology. 1991;100:333–343. doi: 10.1016/0016-5085(91)90200-5. [DOI] [PubMed] [Google Scholar]
- 59.Li L, Rao JN, Guo X, Liu L, Santora R, Bass BL, Wang JY. Polyamine depletion stabilizes p53 resulting in inhibition of normal intestinal epithelial cell proliferation. Am. J. Physiol. Cell Physiol. 2001;281:C941–C953. doi: 10.1152/ajpcell.2001.281.3.C941. [DOI] [PubMed] [Google Scholar]
- 60.Mazan-Mamczarz K, Hagner PR, Corl S, Srikantan S, Wood WH, Becker KG, Gorospe M, Keene JD, Levenson AS, Gartenhaus RB. Post-transcriptional gene regulation by HuR promotes a more tumorigenic phenotype. Oncogene. 2008;27:6151–6163. doi: 10.1038/onc.2008.215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Galbán S, Kuwano Y, Pullmann R, Jr, Martindale JL, Kim HH, Lal A, Abdelmohsen K, Yang X, Dang Y, Liu JO, et al. RNA-binding proteins HuR and PTB promote the translation of hypoxia-inducible factor 1alpha. Mol. Cell. Biol. 2008;28:93–107. doi: 10.1128/MCB.00973-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kuwano Y, Kim HH, Abdelmohsen K, Pullmann R, Jr, Martindale JL, Yang X, Gorospe M. MKP-1 mRNA stabilization and translational control by RNA-binding proteins HuR and NF90. Mol. Cell. Biol. 2008;28:4562–4575. doi: 10.1128/MCB.00165-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lal A, Kawai T, Yang X, Mazan-Mamczarz K, Gorospe M. Antiapoptotic function of RNA-binding protein HuR effected through prothymosin-α. EMBO J. 2005;24:1852–1862. doi: 10.1038/sj.emboj.7600661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Levy NS, Chung S, Furneaux H, Levy AP. Hypoxic stabilization of vascular endothelial growth factor mRNA by the RNA-binding protein HuR. J. Biol. Chem. 1998;273:6417–6423. doi: 10.1074/jbc.273.11.6417. [DOI] [PubMed] [Google Scholar]
- 65.Casero RA, Jr, Marton LJ. Targeting polyamine metabolism and function in cancer and other hyperproliferative diseases. Nat. Rev. Drug Discov. 2007;6:373–390. doi: 10.1038/nrd2243. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.