A perspective on the article by Damgaard and Lykke-Andersen (published in the October 1, 2011, issue) dealing with the regulation of 5′TOP mRNA translation by stress granule proteins under starvation conditions.
Keywords: 5′TOP, TIA-1, TIAR, translation regulation
Abstract
Under conditions of limited nutrients, eukaryotic cells reprogram protein expression in a way that slows growth but enhances survival. Recent data implicate stress granules, discrete cytoplasmic foci into which untranslated mRNPs are assembled during stress, in this process. In the October 1, 2011, issue of Genes & Development, Damgaard and Lykke-Andersen (p. 2057–2068) provide mechanistic insights into the regulation of a specific subset of mRNAs bearing 5′-terminal oligopyrimidine tracts (5′TOPs) by the structurally related stress granule proteins TIA-1 and TIAR.
Cells employ both general and transcript-specific post-transcriptional control mechanisms to reprogram protein expression in response to stress (for review, see Holcik and Sonenberg 2005; Spriggs et al. 2010). Reduced expression of growth-associated proteins is needed to conserve limited resources when nutrients are scarce. At the same time, increased expression of proteins that repair stress-induced damage (e.g., heat-shock proteins and DNA damage-responsive factors) or promote a transition to a quiescent state facilitates cell survival. This transition is initiated by differentially modulating the translation of specific mRNAs, allowing rapid response to altered environmental conditions.
Global translation is largely regulated by the activity and/or availability of two key translation initiation factors: eIF4E and eIF2 (Ma and Blenis 2009; Sonenberg and Hinnebusch 2009). Under conditions favorable to growth, the mRNA cap-binding protein eIF4E binds to the 5′ cap structure of mRNA and interacts with the scaffolding protein eIF4G to promote 40S docking and consequent translation of the mRNA. In nutritionally stressed cells, eIF4E-binding proteins (4E-BPs) disrupt eIF4E–eIF4G interactions, which reduces translation initiation. The assembly of translation-promoting (eIF4E–eIF4G) and translation inhibitory (eIF4E–4E-BP) complexes is regulated by the master kinase mTORC1 (mammalian target of rapamycin complex 1) in an energy- and nutrition-dependent manner (Ma and Blenis 2009). The initiation factor eIF2α/β/γ-dependent recruitment of tRNAiMet to the ribosome is regulated by phosphorylation of eIF2α and mediated by a family of stress-activated serine/threonine kinases. Phosphorylation of eIF2α depletes the levels of eIF2–GTP–tRNAiMet ternary complexes to inhibit translation initiation. One of the eIF2α kinases, GCN2 (general control nonderepressible 2), is specifically activated in response to amino acid starvation (Dong et al. 2000; Qiu et al. 2001). Thus, activation of both mTORC1 and GCN2 conspires to dampen general protein synthesis in cells deprived of amino acids. Damgaard and Lykke-Andersen (2011) show for the first time how cross-talk between these kinases regulates the translation of a subset of housekeeping mRNAs marked by 5′-terminal oligopyrimidine tracts (5′TOPs).
Who's on TOP?
5′TOP mRNAs are defined by a m7G cap structure followed by a 4- to 15-nucleotide CU-rich element in the context of a relatively short and unstructured 5′ untranslated region (UTR) (Avni et al. 1994; Davuluri et al. 2000; Hamilton et al. 2006; Iadevaia et al. 2008). This motif is found in transcripts encoding components of the translational machinery, including ribosomal proteins and translation factors (Hamilton et al. 2006). In response to nutrient deprivation, the translation of 5′TOP transcripts is selectively repressed, an adaptive response that prevents the energy-intensive assembly of surplus ribosomes. Although 5′TOP mRNAs are considered to be a subset of housekeeping mRNAs, the proportion of 5′TOP transcripts engaged in protein synthesis (∼70%) is significantly lower than that of other housekeeping transcripts (∼90%) (Meyuhas et al. 1996). The selective repression of 5′TOP mRNA translation is even more pronounced in response to amino acid starvation, suggesting that a specific regulatory circuit selectively inhibits 5′TOP mRNA translation.
Numerous studies have identified signaling pathways and trans-acting factors that inhibit the translation of 5′TOP mRNAs. Mitogenic stimulation of quiescent cells or reintroduction of amino acids following starvation activates translation of 5′TOP mRNAs in an mTOR-, PI3-kinase (PI3K)-, and S6 kinase (S6K)-dependent manner (Hamilton et al. 2006). The relative contribution of these signaling pathways and their molecular links to the 5′TOP motif are not well understood and data are often contradictory. Initially, a positive correlation between translational activation of 5′TOP mRNAs and mitogen-induced phosphorylation of ribosomal protein S6 (RPS6) by the closely related kinases S6K1 and S6K2 was noted (Jefferies et al. 1994; Jefferies and Thomas 1994). In addition, pharmacological inhibition of S6K activities by the TOR inhibitor rapamycin (Jefferies et al. 1994) or by overexpression of dominant-negative mutants of S6K1 led to the reduced recruitment of 5′TOP transcripts to polysomes (Jefferies et al. 1997). These data suggested a model wherein phosphorylation of RPS6 directly regulates translation of 5′TOP mRNAs. Further investigations challenged this idea, as translation of 5′TOP mRNAs in double-knockout S6K1/S6K2 embryonic stem cells remains responsive to mitogens, although in a rapamycin-sensitive manner (Pende et al. 2004). Recent data suggest that both mTOR and PI3K signaling pathways (which are closely integrated with each other) regulate translation of 5′TOP transcripts (Tang et al. 2001; Stolovich et al. 2002), although whether mitogenic/hormonal and nutritional stimuli use different signaling routes and/or signaling kinetics for this regulation remains to be clarified.
Beyond regulation by signaling pathways, existence of a “titratable” factor regulating translation of 5′TOP transcripts has been postulated, based on in vitro competition studies using synthetic RNA oligos resembling the 5′TOP motif (Biberman and Meyuhas 1999). The RNA-binding proteins La (Pellizzoni et al. 1997, 1998; Cardinali et al. 2003), AUF1 (Kakegawa et al. 2007), and ZNF9 (Pellizzoni et al. 1998; Huichalaf et al. 2009), have been proposed as candidates that selectively regulate translation of 5′TOP transcripts.
La protein is an abundant nucleocytoplasmic shuttling protein that interacts with different classes of RNA such as tRNAs, snoRNAs, and 5S rRNA and is implicated in their maturation, stabilization, and nucleocytoplasmic transport (for review, see Wolin and Cedervall 2002). Intriguingly, La protein also binds selected 5′TOP mRNAs, implying a common function in the coordinated biogenesis of the translational machinery (Pellizzoni et al. 1996; Cardinali et al. 2003). In line with this idea, Lhp1p, a yeast homolog of La protein, also associates with mRNAs encoding ribosomal proteins, although canonical 5′TOP motifs have not been found in yeast (Inada and Guthrie 2004). Functional consequences of La protein association with 5′TOP mRNAs are not well characterized and data are even radically contradictory: While some reports suggest that La protein stimulates translation of 5′TOP mRNAs (Crosio et al. 2000), others indicate an inhibitory effect (Zhu et al. 2001). AUF1, the protein that promotes decay of selected short-lived mRNAs bearing AU-rich elements in their 3′ UTRs (Brewer 1991), also binds mRNA encoding ribosomal protein L32 in a rapamycin-sensitive manner (Kakegawa et al. 2007). Similarly, ZNF9 (also known as CNBP) binds 5′ UTRs of 5′TOP mRNAs encoding the ribosomal protein RPL17, elongation factors eEF1A and eEF2, and poly(A)-binding protein (PABP) (Pellizzoni et al. 1997, 1998; Huichalaf et al. 2009). Notably, ZNF9 levels are reduced in patients suffering from myotonic dystrophy 2 (DM2), a multisystemic musculoskeletal disease, leading to reduced association of ZNF9 with these mRNAs and observed reduction of protein expression (Huichalaf et al. 2009).
Although biochemical data indicate that all of these factors bind 5′ UTRs of selected 5′TOP transcripts, direct association of these proteins with 5′TOP motifs has not been demonstrated. More importantly, functional data proving their roles in the regulation of 5′TOP mRNA translation are lacking, as are the molecular details of their recruitment to these transcripts under nutrient starvation/mitogenic stimulation.
In contrast to putative negative regulators, the microRNA miR-10a may work as a positive regulator of 5′TOP mRNA translation (Orom et al. 2008). Interestingly, miR-10a binds immediately downstream from the regulatory 5′TOP motif and enhances translation of 5′TOP mRNAs. Moreover, miR-10a overexpression alleviates amino acid starvation-induced translational repression of 5′TOP mRNAs (Orom et al. 2008). These leads notwithstanding, the mechanism that mediates selective translational inhibition of 5′TOP transcripts in response to nutritional stress remains elusive. Other data suggest that specific 5′TOP mRNAs are regulated in a tissue- and cell type-specific manner. For example, eEF2 mRNA confers growth-dependent regulation in cells of hematopoietic, but not in cells of nonhematopoietic, origins, while translation of β1-tubulin mRNA, which possesses all classical features of TOP mRNAs, is absolutely refractory to growth arrest in the same cells (Avni et al. 1997). Further mutational analysis of cis-elements within these mRNAs suggests that downstream sequences proximal to the TOP element are critical in the differential response of 5′TOP mRNAs to growth arrest (Avni et al. 1997). Whether cell type-specific heterogeneity of the 5′TOP mRNA regulation reflects cell type-specific abundance of distinct regulatory trans-factors binding to selective regulatory sequences within 5′TOP mRNAs remains to be determined.
TIA proteins: it's all about ‘U'
Damgaard and Lykke-Andersen (2011) show that the related RNA-binding proteins TIA-1 and TIAR are required for amino acid starvation-induced repression of 5′TOP mRNA translation. TIA-1 and TIAR are involved in multiple aspects of cellular metabolism, including mRNA splicing, mRNA translation and decay, cell survival, and the stress response. The proteins share a very similar domain architecture, consisting of three RNA recognition motifs (RRM1–3) and a C-terminal glutamine/asparagine (Q/N)-rich prion-related domain (PRD). The RRMs (RRM1–3) of TIA-1 and TIAR share 79%, 89%, and 91% amino acid identity, respectively (Dember et al. 1996). In contrast, their PRDs are only 51% identical (Kawakami et al. 1992; Dember et al. 1996). Both proteins predominantly use RRM2 to selectively bind RNAs containing U-rich motifs. Similar U-rich stretches are components of adenine/uridine-rich (ARE) regulatory motifs found in the 3′ UTRs of a subset of transcripts that are subject to post-transcriptional control (for review, see Anderson 2008). Many of these mRNAs encode immune, proliferative, or stress-responsive proteins whose translation is selectively repressed by TIA-1 and TIAR (Gueydan et al. 1999; Lopez de Silanes et al. 2005). The localization, stability, and translatability of these transcripts are subject to very complex regulation involving interactions with several different RNA-binding proteins. Studies of mice lacking TIA-1 or TIAR reveal that these proteins dampen the translation of ARE-containing transcripts without affecting mRNA levels by holding them in a translationally repressed state (Piecyk et al. 2000; Mazan-Mamczarz et al. 2006).
Biochemical analysis revealed that ARE-bearing mRNAs are not the sole targets of TIA-1 and TIAR. SELEX analysis and gene array studies have identified cytosine/uridine-rich (C-rich) elements and a hairpin consensus motif that are targeted by TIA-1/TIAR (Lopez de Silanes et al. 2005; Kim et al. 2007). Features common to all three TIA-1/TIAR-binding motifs are pyrimidine-rich content, location within the 3′ UTRs of targeted transcripts, and repressive effects on translation when bound by TIA-1/TIAR.
TIA-1/TIAR act as general translational repressors in stressed cells (for review, see Anderson and Kedersha 2002). Stress-induced phosphorylation of eIF2α results in the assembly of noncanonical, translationally stalled 48S* preinitiation complexes that lack eIF2 and eIF5 and are assembled into stress granules (SGs) by TIA-1/TIAR binding and prion domain-mediated aggregation (Kedersha et al. 2002). Transiently overexpressed TIA-1/TIAR can nucleate SG assembly while repressing the translation of generic reporters (Gilks et al. 2004), but effects are blocked in mutant cells expressing only nonphosphorylatable eIF2α. SG competence is restored when phosphomimetic eIF2α is cotransfected into the mutant cells along with TIA-1 (McEwen et al. 2005). Recruitment of mRNA transcripts into SGs is a selective process that depends on mRNA structure, cytoplasmic levels of specific SG-associated mRNA-binding proteins, and the nature/severity of the stress. The RNA and protein composition of SGs is dynamic and is in equilibrium with translating polysomes (Kedersha et al. 2000), providing necessary plasticity for stress adaptation by preferential inhibition or activation of translation of selected mRNAs.
Several observations suggest redundant functions for the TIA proteins. Both TIA-1 and TIAR bind to U-rich RNA motifs, promote the nucleation of SGs, function as general translational repressors, and have similar tissue distributions, subcellular localizations, and nucleocytoplasmic shuttling properties. Macrophages obtained from mice lacking tiar or tia-1 genes similarly overexpress proinflammatory proteins such as TNFα, interleukin-6, interleukin-1β, and cyclooxygenase-2. As a consequence of this cytokine overexpression, mutant mice lacking TIA-1 or TIAR have a hyperinflammatory phenotype (for review, see Anderson 2008). Mice lacking both of these proteins die before embryonic day 7, suggesting that at least one of these proteins must be present for normal embryonic development (Piecyk et al. 2000).
Genetic studies also demonstrate both shared and unique characteristics of these closely related proteins. Targeted disruption of TIA-1 or TIAR results in embryonic lethality, but the penetrance of the lethality varies between these proteins. The rate of embryonic lethality is close to 100% in TIAR knockout mice, but is <50% in TIA-1 knockout mice. However, TIA-1 nullizygotes are fully fertile, whereas mice lacking the tiar gene are sterile (Beck et al. 1998) due to a specific requirement for TIAR in primordial germ cell development and survival (Beck et al. 1998). Moreover, overexpression of TIAR impairs embryonic development of mice at late preimplantation and post-implantation stages (Kharraz et al. 2010), suggesting that balanced TIAR expression is absolutely required for early embryogenesis. The molecular mechanisms by which TIAR regulates these developmental processes are largely unknown.
TIA s(TOP)s translation
Using an RNA immunoprecipitation assay, Damgaard and Lykke-Andersen (2011) identified 5′TOP mRNAs as novel targets of TIA-1/TIAR regulation. TIA-1/TIAR-binding sites on 5′TOP mRNAs were mapped to the cap-proximal region containing pyrimidine-rich sequences, revealing that TIA-1/TIAR RNA-binding motifs are not restricted to the 3′ UTRs of target mRNAs. Consistent with a role for TIA-1/TIAR as stress-regulated translational repressors, amino acid starvation stabilizes TIA-1/TIAR binding to 5′TOP mRNAs as it inhibits their translation. Several observations suggest that these proteins inhibit translation initiation. Upon amino acid starvation, 5′TOP mRNAs are released from polysomes in a TIA-1/TIAR-dependent manner and are assembled into SGs, sites where translationally repressed transcripts accumulate. Codepletion and complementation studies show that TIA-1 and TIAR inhibit translation of 5′TOP mRNAs in a functionally redundant manner that requires the RRMs, but not the PRDs. Additionally, mRNAs with TIA-1/TIAR-binding sites in the 3′ UTR are refractory to such regulation, suggesting that proximity of the TIA-1/TIAR-binding site to the m7G cap is required for starvation-mediated repression of 5′TOP mRNA translation. Since other studies have shown that miR-10a binds the region adjacent to the 5′TOP motif and activates translation of 5′TOP transcripts (Orom et al. 2008), TIA-1/TIAR and miR-10a may work in a mutually exclusive and stress-regulated manner. Whether TIAR/TIA-1 proteins and miR-10a-binding sites overlap and compete for binding to 5′TOP mRNAs remains to be determined.
This study also reveals that TIA-1/TIAR-dependent regulation of 5′TOP mRNA translation requires both activation by GCN2 kinase and inactivation of mTOR signaling (Fig. 1). GCN2 activation causes phosphorylation of eIF2α and consequently depletes levels of translationally competent eIF2–GTP–tRNAiMet ternary complexes. Since phospho-eIF2α is the primary physiological trigger of SG formation, GCN2 activation appears to collaborate with TIA-1/TIAR to selectively assemble translationally stalled 5′TOP-containing mRNPs into SGs. Similarly, inactivation of mTOR promotes the assembly of eIF4E-BP complexes at the cap structure of 5′TOP mRNAs, adjacent to the TIA-1/TIAR-binding region. Since both increased phospho-eIF2α levels and inhibition of cap-dependent translation cause global translation repression, GCN2 activation and mTOR inactivation alone do not explain the selective repression of 5′TOP mRNA translation induced by amino acid starvation; TIA-1/TIAR appear to be required for this selective effect. How these signaling pathways modulate the activity of TIA-1/TIAR and perhaps other trans-factors that repress 5′TOP mRNA translation is unclear, but either kinase might phosphorylate TIA-1/TIAR proteins to modulate their activities and/or localization. Similarly, questions as to whether the proximity of the TIA-1/TIAR-binding site to the cap structure in 5′TOP mRNAs is absolutely required for this regulation and whether TIA-1/TIAR interact with early initiation factors are yet to be answered.
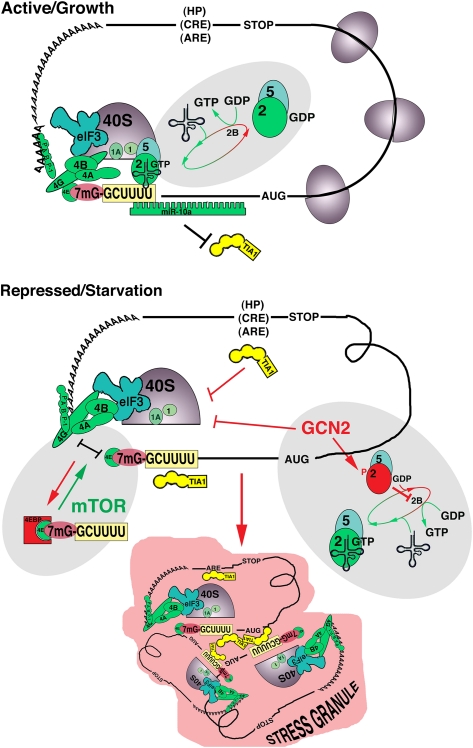
Figure 1.
Translation of TOP mRNAs under conditions of active growth versus starvation. (Active/Growth) Under nutrient-rich conditions, translation of TOP mRNAs is initiated when the cap-binding eIF4F complex (eIF4E/G/A) binds to the m7G cap and recruits the activated 43S complex (40S subunit, eIF1, eIF1A, eIF3, and eIF2–GTP–tRNAiMet/eIF5) to assemble the 48S preinitiation complex. (Top shaded area) The assembly of the eIF2–GTP–tRNAiMet ternary complex associated with eIF5 is a key regulatory event requiring the eIF2B-mediated activation of eIF2 by GDP–GTP exchange and recruitment of tRNAiMet. Translation is enhanced by miR-10a binding to sequences downstream from the 5′TOP motif, which may prevent the inhibitory effect of TIA-1/R binding to the TOP motif. (Repressed/Starvation) Under amino acid starvation conditions, Damgaard and Lykke-Andersen (2011) show that TIA-1/R is recruited to the 5′TOP motif to prevent the assembly of the canonical 48S preinitiation complex. (Shaded area at right) TIA-1/R:5′TOP binding appears to require the GCN2-mediated phosphorylation of eIF2α, which prevents GDP/GTP exchange and subsequent charging of the eIF2:eIF5 complex with tRNAiMet. Formation of noncanonical 48S complexes lacking the charged ternary complex (48S*) may facilitate TIA-1/R binding to the 5′TOP motif and thereby repress initiation. In non-TOP mRNAs, TIA-1/R can bind to U-rich elements in the 3′ UTR (hairpin [HP], C-rich element [CRE]; and adenine/uridine-rich element [ARE]) to inhibit translation initiation by an uncharacterized mechanism. Damgaard and Lykke-Andersen (2011) also implicate mTOR inactivation in the TIA-1/R-mediated repression of TOP mRNA translation. (Gray area at left) A possible mechanism is shown wherein mTOR inactivation results in dephosphorylation of 4EBP and consequent disruption of eIF4G:eIF4E interactions. TIA-1/R-inactivated 48S* complexes are assembled into stress granules (SGs) for mRNA triage and possible cross-talk with other signaling pathways.
Perspectives
Selective mRNA translation mediates both metabolic and energetic transitions accompanying cell growth, stress, and death. Earlier models postulated that stress activates signaling pathways that inhibit bulk mRNA translation, causing “global” translational repression concurrent with the assembly of translationally repressed mRNAs into SGs. At the same time, a minor mRNA fraction escapes translational repression to synthesize proteins necessary for stress adaptation, using noncanonical translation initiation mechanisms (Holcik and Sonenberg 2005; Spriggs et al. 2010) and/or by cotranslational mRNA localization to protected regions of the cytoplasm such as the ER (Unsworth et al. 2010). More recent data suggest that mechanisms of stress-induced translational repression or activation are not this simple and implicate trans-factors such as RNA-binding proteins and microRNAs in these processes. The selective regulation of 5′TOP mRNA translation during amino acid starvation demonstrates how specific trans-acting factors selectively repress a specific subset of mRNAs. Additional complex systems, in which other mRNA families are coordinately regulated in response to specific stimuli (“mRNA regulons”), likely remain to be discovered. Cancer cells often exhibit uncoupling of translational inhibition in response to nutrient deprivation and hypoxia; thus, elucidation of the multifaceted signaling networks that regulate functionally linked mRNA subsets should further our understanding of gene expression both at the bench and in the clinic.
Footnotes
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.17838411.
References
- Anderson P 2008. Post-transcriptional control of cytokine production. Nat Immunol 9: 353–359 [DOI] [PubMed] [Google Scholar]
- Anderson P, Kedersha N 2002. Stressful initiations. J Cell Sci 115: 3227–3234 [DOI] [PubMed] [Google Scholar]
- Avni D, Shama S, Loreni F, Meyuhas O 1994. Vertebrate mRNAs with a 5′-terminal pyrimidine tract are candidates for translational repression in quiescent cells: characterization of the translational cis-regulatory element. Mol Cell Biol 14: 3822–3833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avni D, Biberman Y, Meyuhas O 1997. The 5′ terminal oligopyrimidine tract confers translational control on TOP mRNAs in a cell type- and sequence context-dependent manner. Nucleic Acids Res 25: 995–1001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beck AR, Miller IJ, Anderson P, Streuli M 1998. RNA-binding protein TIAR is essential for primordial germ cell development. Proc Natl Acad Sci 95: 2331–2336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biberman Y, Meyuhas O 1999. TOP mRNAs are translationally inhibited by a titratable repressor in both wheat germ extract and reticulocyte lysate. FEBS Lett 456: 357–360 [DOI] [PubMed] [Google Scholar]
- Brewer G 1991. An A+U-rich element RNA-binding factor regulates c-myc mRNA stability in vitro. Mol Cell Biol 11: 2460–2466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardinali B, Carissimi C, Gravina P, Pierandrei-Amaldi P 2003. La protein is associated with terminal oligopyrimidine mRNAs in actively translating polysomes. J Biol Chem 278: 35145–35151 [DOI] [PubMed] [Google Scholar]
- Crosio C, Boyl PP, Loreni F, Pierandrei-Amaldi P, Amaldi F 2000. La protein has a positive effect on the translation of TOP mRNAs in vivo. Nucleic Acids Res 28: 2927–2934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damgaard CK, Lykke-Andersen J 2011. Translational coregulation of 5′TOP mRNAs by TIA-1 and TIAR. Genes Dev 25: 2057–2068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davuluri RV, Suzuki Y, Sugano S, Zhang MQ 2000. CART classification of human 5′ UTR sequences. Genome Res 10: 1807–1816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dember LM, Kim ND, Liu KQ, Anderson P 1996. Individual RNA recognition motifs of TIA-1 and TIAR have different RNA binding specificities. J Biol Chem 271: 2783–2788 [DOI] [PubMed] [Google Scholar]
- Dong J, Qiu H, Garcia-Barrio M, Anderson J, Hinnebusch AG 2000. Uncharged tRNA activates GCN2 by displacing the protein kinase moiety from a bipartite tRNA-binding domain. Mol Cell 6: 269–279 [DOI] [PubMed] [Google Scholar]
- Gilks N, Kedersha N, Ayodele M, Shen L, Stoecklin G, Dember LM, Anderson P 2004. Stress granule assembly is mediated by prion-like aggregation of TIA-1. Mol Biol Cell 15: 5383–5398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gueydan C, Droogmans L, Chalon P, Huez G, Caput D, Kruys V 1999. Identification of TIAR as a protein binding to the translational regulatory AU-rich element of tumor necrosis factor α mRNA. J Biol Chem 274: 2322–2326 [DOI] [PubMed] [Google Scholar]
- Hamilton TL, Stoneley M, Spriggs KA, Bushell M 2006. TOPs and their regulation. Biochem Soc Trans 34: 12–16 [DOI] [PubMed] [Google Scholar]
- Holcik M, Sonenberg N 2005. Translational control in stress and apoptosis. Nat Rev Mol Cell Biol 6: 318–327 [DOI] [PubMed] [Google Scholar]
- Huichalaf C, Schoser B, Schneider-Gold C, Jin B, Sarkar P, Timchenko L 2009. Reduction of the rate of protein translation in patients with myotonic dystrophy 2. J Neurosci 29: 9042–9049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iadevaia V, Caldarola S, Tino E, Amaldi F, Loreni F 2008. All translation elongation factors and the e, f, and h subunits of translation initiation factor 3 are encoded by 5′-terminal oligopyrimidine (TOP) mRNAs. RNA 14: 1730–1736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inada M, Guthrie C 2004. Identification of Lhp1p-associated RNAs by microarray analysis in Saccharomyces cerevisiae reveals association with coding and noncoding RNAs. Proc Natl Acad Sci 101: 434–439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jefferies HB, Thomas G 1994. Elongation factor-1α mRNA is selectively translated following mitogenic stimulation. J Biol Chem 269: 4367–4372 [PubMed] [Google Scholar]
- Jefferies HB, Reinhard C, Kozma SC, Thomas G 1994. Rapamycin selectively represses translation of the ‘polypyrimidine tract' mRNA family. Proc Natl Acad Sci 91: 4441–4445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jefferies HB, Fumagalli S, Dennis PB, Reinhard C, Pearson RB, Thomas G 1997. Rapamycin suppresses 5′TOP mRNA translation through inhibition of p70s6k. EMBO J 16: 3693–3704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kakegawa T, Ohuchi N, Hayakawa A, Hirata S, Matsuda M, Kogure K, Kobayashi H, Inoue A, Kaspar RL 2007. Identification of AUF1 as a rapamycin-responsive binding protein to the 5′-terminal oligopyrimidine element of mRNAs. Arch Biochem Biophys 465: 274–281 [DOI] [PubMed] [Google Scholar]
- Kawakami A, Tian Q, Duan X, Streuli M, Schlossman SF, Anderson P 1992. Identification and functional characterization of a TIA-1-related nucleolysin. Proc Natl Acad Sci 89: 8681–8685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kedersha N, Cho MR, Li W, Yacono PW, Chen S, Gilks N, Golan DE, Anderson P 2000. Dynamic shuttling of TIA-1 accompanies the recruitment of mRNA to mammalian stress granules. J Cell Biol 151: 1257–1268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kedersha N, Chen S, Gilks N, Li W, Miller IJ, Stahl J, Anderson P 2002. Evidence that ternary complex (eIF2–GTP–tRNAiMet)-deficient preinitiation complexes are core constituents of mammalian stress granules. Mol Biol Cell 13: 195–210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kharraz Y, Salmand PA, Camus A, Auriol J, Gueydan C, Kruys V, Morello D 2010. Impaired embryonic development in mice overexpressing the RNA-binding protein TIAR. PLoS ONE 5: e11352 doi: 10.1371/journal.pone.0011352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim HS, Kuwano Y, Zhan M, Pullmann R Jr, Mazan-Mamczarz K, Li H, Kedersha N, Anderson P, Wilce MC, Gorospe M, et al. 2007. Elucidation of a C-rich signature motif in target mRNAs of RNA-binding protein TIAR. Mol Cell Biol 27: 6806–6817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez de Silanes I, Galban S, Martindale JL, Yang X, Mazan-Mamczarz K, Indig FE, Falco G, Zhan M, Gorospe M 2005. Identification and functional outcome of mRNAs associated with RNA-binding protein TIA-1. Mol Cell Biol 25: 9520–9531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma XM, Blenis J 2009. Molecular mechanisms of mTOR-mediated translational control. Nat Rev Mol Cell Biol 10: 307–318 [DOI] [PubMed] [Google Scholar]
- Mazan-Mamczarz K, Lal A, Martindale JL, Kawai T, Gorospe M 2006. Translational repression by RNA-binding protein TIAR. Mol Cell Biol 26: 2716–2727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McEwen E, Kedersha N, Song B, Scheuner D, Gilks N, Han A, Chen JJ, Anderson P, Kaufman RJ 2005. Heme-regulated inhibitor kinase-mediated phosphorylation of eukaryotic translation initiation factor 2 inhibits translation, induces stress granule formation, and mediates survival upon arsenite exposure. J Biol Chem 280: 16925–16933 [DOI] [PubMed] [Google Scholar]
- Meyuhas O, Avni D, Shama S 1996. Translational control of ribosomal protein mRNAs in eukaryotes. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- Orom UA, Nielsen FC, Lund AH 2008. MicroRNA-10a binds the 5′UTR of ribosomal protein mRNAs and enhances their translation. Mol Cell 30: 460–471 [DOI] [PubMed] [Google Scholar]
- Pellizzoni L, Cardinali B, Lin-Marq N, Mercanti D, Pierandrei-Amaldi P 1996. A Xenopus laevis homologue of the La autoantigen binds the pyrimidine tract of the 5′ UTR of ribosomal protein mRNAs in vitro: implication of a protein factor in complex formation. J Mol Biol 259: 904–915 [DOI] [PubMed] [Google Scholar]
- Pellizzoni L, Lotti F, Maras B, Pierandrei-Amaldi P 1997. Cellular nucleic acid binding protein binds a conserved region of the 5′ UTR of Xenopus laevis ribosomal protein mRNAs. J Mol Biol 267: 264–275 [DOI] [PubMed] [Google Scholar]
- Pellizzoni L, Lotti F, Rutjes SA, Pierandrei-Amaldi P 1998. Involvement of the Xenopus laevis Ro60 autoantigen in the alternative interaction of La and CNBP proteins with the 5′UTR of L4 ribosomal protein mRNA. J Mol Biol 281: 593–608 [DOI] [PubMed] [Google Scholar]
- Pende M, Um SH, Mieulet V, Sticker M, Goss VL, Mestan J, Mueller M, Fumagalli S, Kozma SC, Thomas G 2004. S6K1−/−/S6K2−/− mice exhibit perinatal lethality and rapamycin-sensitive 5′-terminal oligopyrimidine mRNA translation and reveal a mitogen-activated protein kinase-dependent S6 kinase pathway. Mol Cell Biol 24: 3112–3124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piecyk M, Wax S, Beck AR, Kedersha N, Gupta M, Maritim B, Chen S, Gueydan C, Kruys V, Streuli M, et al. 2000. TIA-1 is a translational silencer that selectively regulates the expression of TNF-α. EMBO J 19: 4154–4163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu H, Dong J, Hu C, Francklyn CS, Hinnebusch AG 2001. The tRNA-binding moiety in GCN2 contains a dimerization domain that interacts with the kinase domain and is required for tRNA binding and kinase activation. EMBO J 20: 1425–1438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonenberg N, Hinnebusch AG 2009. Regulation of translation initiation in eukaryotes: mechanisms and biological targets. Cell 136: 731–745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spriggs KA, Bushell M, Willis AE 2010. Translational regulation of gene expression during conditions of cell stress. Mol Cell 40: 228–237 [DOI] [PubMed] [Google Scholar]
- Stolovich M, Tang H, Hornstein E, Levy G, Cohen R, Bae SS, Birnbaum MJ, Meyuhas O 2002. Transduction of growth or mitogenic signals into translational activation of TOP mRNAs is fully reliant on the phosphatidylinositol 3-kinase-mediated pathway but requires neither S6K1 nor rpS6 phosphorylation. Mol Cell Biol 22: 8101–8113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang H, Hornstein E, Stolovich M, Levy G, Livingstone M, Templeton D, Avruch J, Meyuhas O 2001. Amino acid-induced translation of TOP mRNAs is fully dependent on phosphatidylinositol 3-kinase-mediated signaling, is partially inhibited by rapamycin, and is independent of S6K1 and rpS6 phosphorylation. Mol Cell Biol 21: 8671–8683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unsworth H, Raguz S, Edwards HJ, Higgins CF, Yague E 2010. mRNA escape from stress granule sequestration is dictated by localization to the endoplasmic reticulum. FASEB J 24: 3370–3380 [DOI] [PubMed] [Google Scholar]
- Wolin SL, Cedervall T 2002. The La protein. Annu Rev Biochem 71: 375–403 [DOI] [PubMed] [Google Scholar]
- Zhu J, Hayakawa A, Kakegawa T, Kaspar RL 2001Binding of the La autoantigen to the 5′ untranslated region of a chimeric human translation elongation factor 1A reporter mRNA inhibits translation in vitro. Biochim Biophys Acta 1521: 19–29 [DOI] [PubMed] [Google Scholar]