Abstract
Human cytomegalovirus (HCMV) contributes its own set of microRNAs (miRNAs) during lytic infection of cells, likely fine-tuning conditions important for viral replication. To enhance our understanding of this component of the HCMV-host transcriptome, we have conducted deep-sequencing analysis of small RNAs (smRNA-seq) from infected human fibroblast cells. We found that HCMV-encoded miRNAs accumulate to ∼20% of the total smRNA population at late stages of infection, and our analysis led to improvements in viral miRNA annotations and identification of two novel HCMV miRNAs, miR-US22 and miR-US33as. Both of these miRNAs were capable of functionally repressing synthetic targets in transient transfection experiments. Additionally, through cross-linking and immunoprecipitation (CLIP) of Argonaute (Ago)-bound RNAs from infected cells, followed by high-throughput sequencing, we have obtained direct evidence for incorporation of all HCMV miRNAs into the endogenous host silencing machinery. Surprisingly, three HCMV miRNA precursors exhibited differential incorporation of their mature miRNA arms between Ago2 and Ago1 complexes. Host miRNA abundances were also affected by HCMV infection, with significant upregulation observed for an miRNA cluster containing miR-96, miR-182, and miR-183. In addition to miRNAs, we also identified novel forms of virus-derived smRNAs, revealing greater complexity within the smRNA population during HCMV infection.
INTRODUCTION
Human cytomegalovirus (HCMV), a member of the herpesvirus family, is prevalent in the majority of the global population and the main viral cause of birth defects (19). HCMV is capable of infecting a wide variety of human cell types in vivo and is known to establish a latent form of infection that persists throughout the life of the host. Immunocompromised patients are especially susceptible to problems associated with infection, further motivating HCMV studies.
Expression of virus-encoded microRNAs (miRNAs) likely contributes to the molecular transformation necessary for productive infection in human cells. Since the initial discovery of viral miRNAs in 2004 (24), several viruses have been documented to express their own sets of miRNAs (4, 5, 7, 11, 12, 23). These small RNAs (smRNAs) are highly attractive from a viral standpoint in that they require minimal space within the genome and potentially offer a robust mechanism for specific targeting of host defense genes. HCMV is known to encode at least 11 miRNA precursors that are expressed and processed into mature miRNAs during infection (12).
High-throughput sequencing approaches have been instrumental in recent viral miRNA studies, acquiring a level of resolution that was previously unachievable through cloning efforts (17, 27, 36). Deep-sequencing approaches have not been extended to smRNA analysis of HCMV-infected cells, and previous microarray-based studies were limited in assessing only host miRNA levels (30, 37). Thus, we set out to conduct a more comprehensive analysis by deep sequencing all smRNAs (smRNA-seq) from HCMV Towne-infected human foreskin fibroblast (HFF) cells. We have fully characterized both the viral and human miRNA fractions, leading us to identify novel HCMV miRNA precursors and differentially regulated human miRNAs, including the miR-182/-96/-183 cluster, which was not previously reported. Additionally, the unbiased sequencing refined the 5′ ends of three HCMV mature miRNAs, with miR-US4 differing the most from its annotated boundaries. To further characterize the functional attributes of smRNAs, we performed high-throughput sequencing of Argonaute (Ago)-associated RNAs from infected HFFs to obtain direct evidence that HCMV-encoded miRNAs are incorporated into endogenous cellular silencing complexes. Finally, we also observed production of non-miRNA smRNAs from discrete regions of the HCMV genome during infection, including areas that encode long noncoding RNA (lncRNA) transcripts. Our results revealed a greater diversity within the smRNA component of HCMV-host interactions than was previously appreciated.
MATERIALS AND METHODS
HCMV infections and culture conditions.
Primary human foreskin fibroblasts (HFFs) were cultured as previously described (29) and maintained within passages 16 to 20. At the time of infection, G0-synchronized cells were trypsinized, resuspended in the presence of virus, and plated at near confluence. Unless otherwise stated, all infections were performed with the wild-type Towne strain at a multiplicity of infection (MOI) of 3 PFU per cell. For mock infections, tissue culture supernatant (spent medium from 3 days of culture) containing 1% dimethyl sulfoxide (DMSO) was used in place of virus stock. At different times postinfection, cells were washed with 1× phosphate-buffered saline (PBS) before being harvested. 293T cells (ATCC) were cultured in high-glucose Dulbecco's modified Eagle's medium (DMEM), supplemented with 10% heat-inactivated fetal bovine serum (FBS), 100 units/ml penicillin, 100 μg/ml streptomycin, and 2 mM l-glutamine (Invitrogen).
Library preparation for smRNA-seq and CLIP-seq experiments.
For smRNA-seq, total RNA from HCMV-infected and mock-treated HFFs was isolated using TRIzol reagent (Invitrogen) and subsequently treated with Turbo DNase (Ambion), all according to instructions from the manufacturers. Libraries were then generated from 18- to 35-nucleotide (nt) cDNAs with the Illumina Small RNA Sample Prep, version 1.5, kit, using 5 μg of DNase-treated total RNA. CLIP-seq (cross-linking and immunoprecipitation [CLIP] followed by high-throughput sequencing) libraries were prepared as described previously (40, 43). Four 15-cm dishes (approximately 2 × 107 cells) of either infected or mock-treated cells were used for each condition. Cells were subjected to 400 mJ of UV irradiation, in 1× PBS on ice, using a Stratalinker machine (Stratagene, Agilent Technologies). Following cross-linking, cells were manually scraped off the dishes, pelleted, and snap frozen. Cell pellets were only minimally stored at −80°C prior to proceeding with radioimmunoprecipitation assay (RIPA)-mediated lysis and the remainder of the protocol. Wako anti-Ago1 (2A7) and Millipore anti-Ago2 (07-590) antibodies were used for immunoprecipitation steps. Illumina Genome Analyzer IIx sequencing for all smRNA-seq and CLIP-seq libraries was performed with 36 cycles.
Northern analysis of miRNAs.
Total RNA was isolated using TRIzol reagent (Invitrogen) according to instructions from the manufacturer up until the addition of isopropanol. Samples were precipitated overnight at −80°C and centrifuged the following day at 14,000 rpm for 30 min at 4°C, followed by one ethanol wash and a final 15-min spin. RNA isolated in this manner was subjected to polyacrylamide Northern blot analysis (10 μg per lane) as described previously (22). Modifications to the protocol included gel transfer to GeneScreen Plus membranes (PerkinElmer) and additional posthybridization washes. StarFire probes were used for miRNA detection (Integrated DNA Technologies), and hybridizations were performed for 16 h at 50°C in 50-ml conical tubes. After two 30-min washes in nonstringent buffer (3× SSC [1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate], 5% SDS, 0.025 M sodium phosphate, 10× Denhardt's solution) at 50°C, membranes were moved to new conical tubes and washed twice for 30 min in 2× SSC–1% SDS at room temperature. Membranes were washed briefly once more in a tray containing 2× SSC–1% SDS at 42° and subjected to one final wash in 1× SSC–1% SDS, rotating in a conical tube at 42°C for 30 min. Exposure to film was performed for 5 h to overnight between two intensifying screens at −80°C. All membranes were reprobed with 5′ end-labeled traditional oligonucleotides for the U6 snRNA to determine loading efficiencies. Probe sequences used are listed in Table 1.
Table 1.
Probe sequences used for Northern blot analysis
| smRNA name | Probe sequence (source) |
|---|---|
| HCMV miR-US22-5p | ACGCCCGCGGACACACGCTGAAACA (StarFire) |
| HCMV miR-US33as-5p | CACCGTCACGGTCCGAGCACATCC (StarFire) |
| hsa-miR-182-5p | AGTGTGAGTTCTACCATTGCCAAA (StarFire) |
| U6 snRNA | GCGTGTCATCCTTGCGCAGGGGCC |
miRNA transfection experiments and luciferase assays.
At 24 h prior to miRNA transfection, 293T cells were replated in medium containing 5% FBS and 2 mM l-glutamine (no antibiotics) at a density of 5 × 106 cells per six-well dish. HCMV miRNA precursors, with approximately 100 bases of flanking viral genomic context, were PCR amplified from HCMV Towne genomic DNA and cloned into the XhoI and BamHI restriction sites of pcDNA3.1 (Invitrogen). A total of 200 ng of plasmid and 5 μl of Lipofectamine 2000 were diluted in Opti-MEM (Invitrogen) and added to cells according to the manufacturer's instructions. No medium changes were performed, and total RNA was harvested for Northern blot analysis after 48 h. For luciferase activity measurements, 293T cells were seeded as above at a density of 4 × 106 cells per 24-well dish. Each well was transfected with 2 μl of Lipofectamine 2000, 750 ng of miRNA expression plasmid, and 50 ng of either unmodified psiCHECK2 vector (Promega) or psiCHECK2 containing a synthetic target site. Target sites were generated by annealing complementary oligonucleotides that were compatible with the XhoI and NotI restriction sites located downstream of Renilla luciferase. Activity measurements were obtained at 36 h posttransfection using a Dual-Luciferase Assay System from Promega. This assay was performed according to the manufacturer's instructions except cells were lysed in 200 μl of passive lysis buffer and further diluted 1:20 prior to acquiring measurements. All primers used for generating the constructs described above are listed in Table 2.
Table 2.
Primers used for construction of miRNA expression and luciferase reporter constructs
| miRNA name or target site | Forward primer | Reverse primer |
|---|---|---|
| HCMV miR-US22 | CGCTCGAGCTGTGCAGAATCATAAGTTTATGATG | GCGGATCCGAAAACGAGGACGACACGA |
| HCMV miR-US33as | CTCGAGCGACCATTTCCGTGCGATTAGC | GGATCCAATTGACGGTCCACTGAGGTG |
| miR-US22-5p-full | TCGAGCCCCGCGGACACACGCTGAAACAGC | GGCCGCTGTTTCAGCGTGTGTCCGCGGGGC |
| miR-US22-5p-seed | TCGAGCCCGGGAATTCGTGAAACTCTAGAGC | GGCCGCTCTAGAGTTTCACGAATTCCCGGGC |
| miR-US33as-5p-full | TCGAGCCGTCACGGTCCGAGCACATCCAGC | GGCCGCTGGATGTGCTCGGACCGTGACGGC |
| miR-US33as-5p-seed | TCGAGCCCGGGAATTCGACATCCTCTAGAGC | GGCCGCTCTAGAGGATGTCGAATTCCCGGGC |
Computational processing of smRNA-seq and CLIP-seq data.
First, 3′ adapter sequences were trimmed from sequencing reads. For bar-coded libraries, which included the smRNA-seq and Ago2 CLIP-seq data sets, only reads harboring fully intact 5′ barcode sequences were considered in the analysis. Filtered data sets were then mapped against an index composed of both the human (University of California, Santa Cruz [UCSC] hg19 [http://genome.ucsc.edu]) and HCMV Towne (GenBank accession number FJ616285.1) genomes, using Bowtie software (version 0.12.7; the following parameters were used: -k [valid alignments per read], 1; -m [number of possible alignments], 10; -l [seed length], 25; --best [optimal alignments]). smRNA annotations were obtained from miRBase, version 16.0 (http://www.mirbase.org), and the UCSC Table Browser. For rRNAs, hg18-based coordinates were converted to hg19 using the UCSC LiftOver utility (http://genome.ucsc.edu/cgi-bin/hgLiftOver). Custom Perl scripts were used to overlap mapped smRNA-seq coordinates with these annotations.
miRNA expression analysis.
Abundance levels for human and HCMV miRNAs were determined by summing the number of smRNA-seq reads that pattern matched to mature miRNA sequences or mapped to mature miRNA coordinates (allowing up to two mismatches). Normalized values were obtained by dividing each miRNA count by the total number of human and viral miRNA reads in the data set and then multiplying the fraction by 105. Ago-CLIP miRNA levels were calculated using the same metric. To identify human miRNAs that significantly changed in abundance during infection, HCMV miRNA counts were removed from the data sets, and infected libraries were compared to mock-infected libraries from the same time point. Normalized human miRNA counts were binned by expression level into two groups. Within the low and high expression groups, Z-scores were calculated for each miRNA, and values obtained that were less than −1.96 or greater than 1.96 were deemed statistically significant (25). miRNA Z-scores were required to meet this threshold in both infected versus mock or mock versus infected comparisons.
RNA-seq library preparation and HCMV transcriptome analysis.
Strand-specific RNA-seq libraries were prepared as described previously (21). Ten micrograms of total RNA, isolated with TRIzol (Invitrogen) according to the manufacturer's instructions, was treated with Turbo DNase (Ambion) prior to library preparation. Illumina sequencing was performed with 76 cycles. As above, sequencing reads were adapter trimmed and mapped to the human and HCMV genomes using Bowtie (parameters, -m 25 -k 1 -l 28 --best). HCMV-mapped RNA-seq reads were visualized using R software (version 2.10.1).
Sequencing data accession number.
Data sets from the smRNA-seq and CLIP-seq experiments have been deposited in the GEO database under accession number GSE33584.
RESULTS
Global statistics of short cDNA libraries in HCMV-infected cells.
We prepared short cDNA libraries corresponding to 18- to 35-nt RNAs from infected and mock-treated HFFs at 24 and 72 h postinfection (hpi). High-throughput sequencing of the four smRNA libraries using the Illumina Genome Analyzer IIx platform generated 2.5 to 4.5 million 36-nt reads per condition. An average of 84% of smRNA-seq reads were mapped uniquely to the human and viral genomes, allowing for two base mismatches. Greater than 95% of mapped reads were classified into different categories of annotated noncoding RNAs (ncRNAs). As expected, the majority of smRNA-seq reads mapped to human and virus-encoded miRNAs, as well as snoRNAs, another class of ncRNAs (Fig. 1A).
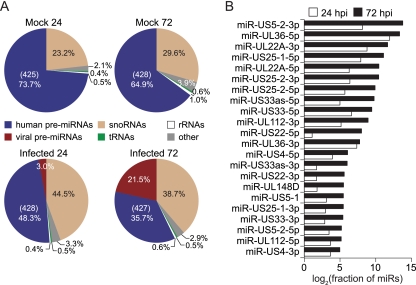
Fig 1.
Genomic analysis of smRNAs during HCMV infection of HFFs. (A) Distribution of smRNA-seq reads across categories of annotated smRNAs (pre-miRNAs, snoRNAs, tRNAs, and rRNAs) in HCMV-infected (Towne strain) or mock-treated HFFs at 24 and 72 hpi. Reads that did not match current annotations were classified as “other.” For human miRNA fractions, values listed in parentheses indicate the total number of unique pre-miRNAs detected. (B) Expression levels of HCMV mature miRNAs during infection, measured by smRNA-seq. Total read counts for each viral miRNA, including reads that exactly matched the mature miRNA sequence and those that were uniquely mapped to its genomic coordinates, were normalized by the total number of miRNA reads (human and viral) in the data set.
To validate that the cDNA libraries accurately recapitulated virus-encoded miRNA expression, the number of reads corresponding to HCMV miRNAs, as a fraction of all smRNA-seq reads, was measured at the two time points. HCMV miRNAs increased in overall abundance more than 7-fold over the course of infection, accounting for over 30% of the total (human and HCMV) miRNAs sequenced at 72 hpi. Consistent with previous observations regarding cytomegalovirus miRNA expression kinetics (9, 12), all HCMV miRNAs were found to individually accumulate from 24 to 72 hpi (Fig. 1B). In fact, at 72 hpi particular HCMV miRNAs reached levels comparable to the most highly expressed human miRNAs in HFFs, including miR-21 and let-7f.
Of the 11 annotated HCMV precursor miRNAs (pre-miRs), 10 were well supported by the smRNA-seq data at both time points, with 416 reads corresponding to mature miR-US4-3p and 184,877 reads corresponding to miR-US5-2-3p at 72 hpi, the lowest and highest expressed viral miRNAs at this time point (5p and 3p follow the miRNA name to designate arms processed from a common pre-miR hairpin structure). The annotated HCMV pre-miR-UL70 was not supported by smRNA-seq. Except for miR-UL148 and miR-US5-1, all HCMV miRNA names have been assigned a 5p or 3p designation in this study to avoid confusion with miR/miR* (where miR* indicates a passenger strand) nomenclature. Despite the substantial presence of viral miRNAs in the infected samples, miRNAs as a fraction of the total smRNA pool decreased upon infection, while the proportion of snoRNAs relative to other smRNAs increased, as has been observed to some extent in previous studies (24). Nevertheless, similar numbers of individual human miRNAs were detected across all conditions (Fig. 1A).
Identification of novel viral miRNAs.
Since smRNA-seq reads were also detected outside annotated HCMV miRNA regions, we subjected the data sets to the MIResque miRNA prediction algorithm (G. W. Yeo, unpublished method) to search for novel HCMV-encoded miRNAs. Two candidate pre-miRs were further inspected, having satisfied established guidelines for miRNA annotation (1) and rules for Drosha processing of primary miRNA transcripts (14, 41). One candidate region, which is in antisense orientation to the protein-coding gene US22 and thus named pre-miR-US22, was found to contain a significant level of smRNA-seq coverage (3,410 reads for miR-US22-5p at 72 hpi) and featured a distribution of sequence alignments that mapped to a predicted stem-loop hairpin structure characteristic of a pre-miR (Fig. 2A). As an independent method of validation, miR-US22-5p was detected through Northern blot analysis by 48 hpi (Fig. 2C).
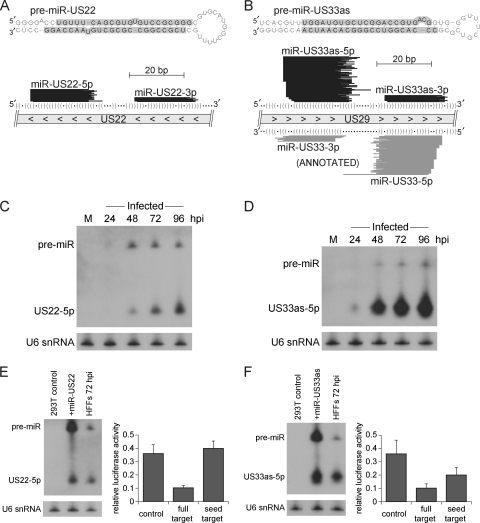
Fig 2.
Identification of novel HCMV miRNA precursors miR-US22 and miR-US33as. (A and B) Uniquely mapped smRNA-seq reads align to predicted RNA stem-loop structures characteristic of a pre-miR hairpin. RNA stem-loop structures of pre-miR-US22 (A) and pre-miR-US33as (B) were generated using mFold software (http://mfold.rna.albany.edu). Highlighted arms indicate where the majority of smRNA-seq reads aligned at 72 hpi (percentages are listed in Table 3). Genomic locations of the pre-miRs are depicted below the stem-loop structures (direction of transcription is indicated by arrowheads). Only nonredundant sequences represented by at least five smRNA-seq reads from the 72-hpi data set are depicted. The predicted RNA structures of the precursors are illustrated by parentheses indicating bases that are paired and dots that correspond to unpaired bases. (C and D) Northern blot analysis of miR-US22-5p (C) and miR-US33as-5p (D) in infected HFFs. Mock-treated cells were harvested at 72 h for this analysis. The U6 snRNA serves as a loading control. (E) Processing of HCMV miR-US22 in 293T cells and repression of a synthetic target. An miR-US22 expression construct or empty vector was transfected into 293T cells, and Northern blot analysis was performed to compare miRNA levels in these samples to those of HFFs at 72 hpi. A luciferase assay was used to demonstrate repression of a perfectly complementary target sequence by miR-US22-5p (P value of <0.01). 293T cells were transfected with miR-US22 expression vector and a construct containing a luciferase reporter with either no targeting site (control), a target consisting of the perfectly complementary target of miR-US22-5p (full target), or a canonical seed match target corresponding to only positions 2 to 7 of miR-US22-5p (seed target). Luciferase measurements shown are averages of three independent experiments. Within each experiment two independent reporter plasmid preparations were used to assess each type of target site. Error bars represent standard deviation, and significance values were calculated using an unpaired, two-tailed Student's t test. (F) Analysis of miR-US33as-5p in transfected 293T cells, performed as described above for panel E (P value of <0.01 for repressed full and seed versions of synthetic targets).
Substantial coverage by smRNA-seq was also detected in antisense orientation to the region encoding HCMV pre-miR-US33 (Fig. 2B). This candidate, pre-miR-US33as, is in the sense orientation to the US29 locus and again harbored a distribution of reads characteristic of a pre-miR stem-loop. While the existence of this miRNA was suggested before through computational prediction efforts and termed miR-US29-1 (12), no further experimental evidence was previously obtained. miR-US33as-5p was also validated by Northern analysis and could be detected by 24 hpi (Fig. 2D).
Characterization of miR-US22 and miR-US33as outside infection.
Studies from Grey et al. have demonstrated that HCMV miRNAs such as miR-UL112 can be transiently expressed in noninfected cells (13). Placement of the miR-UL112 precursor and flanking viral genomic context within an expression construct allowed for efficient processing in the absence of viral factors, which was evident by Northern analysis of transfected cells. We followed a similar approach for miR-US22-5p, yet the precursor context of this miRNA did not permit accumulation in transfected 293T cells beyond the moderate levels present in HFFs at 72 hpi (Fig. 2E). Nevertheless, significant repression of a fully complementary target site was observed within the context of a luciferase assay, but miR-US22 was not able to regulate a target site bearing its 5p miRNA seed match in our system (Fig. 2E). In the case of miR-US33as-5p, higher levels of this miRNA could be obtained in transfected 293T cells, and repression was observed for both fully complementary and seed match target sites (Fig. 2F). For comparison purposes, we have also performed these experiments for the highly expressed miR-US25-1 and obtained similar results for its 5p mature form (data not shown).
Refinement of HCMV miRNA annotations.
miRNA-mRNA interactions occur primarily through partial sequence complementarity between the miRNA seed region (nt 2 to 7 from the 5′ end) and a targeted mRNA. Consequently, accurate definition of the 5′ termini of miRNA sequences is important in the elucidation of HCMV miRNA functions. We determined the most abundant sequences associated with each miRNA at both 24 and 72 hpi, and, based on analysis of the 72-hpi data set, 7 of the 17 existing HCMV mature miRNA annotations can be revised (Table 3). Notably, the sequence obtained for miR-US4-5p differs from the annotated version by 5 bp at the 5′ end, complicating the interpretation of the seed-mediated targeting described in the recent work of Kim and colleagues (16). In fact, no reads obtained from our smRNA-seq experiments matched the annotated miR-US4 mature sequence. The miR-US4 precursor and upstream genomic sequence are completely conserved between the Towne, AD169, and Merlin strains of HCMV. Interestingly, five miRNAs (miR-US25-1-5p, miR-US33-5p, miR-US5-2-3p, miR-US22-5p, and miR-US33as-3p) differed in their most abundant forms between 24 and 72 hpi (Table 3). Globally, HCMV miRNAs exhibited heterogeneous 5′ and 3′ ends in the smRNA-seq data, as shown in the depiction of read alignments across pre-miR-US22 and pre-miR-US33as/miR-US33 in Fig. 2A and B. We have also observed comparable levels of end heterogeneity for human miRNAs.
Table 3.
Comparison of smRNA-seq results with miRBase, version 16.0, HCMV miRNA annotationsa
| Annotation and name | Most abundant sequences at 24 hpi |
Most abundant sequences at 72 hpi |
||||
|---|---|---|---|---|---|---|
| Sequence | No. of reads | % of total | Sequenceb | No. of reads | % of total | |
| Annotated in miRBase |
||||||
| miR-UL112-3p | AAGTGACGGTGAGATCCAGGCT | 452 | 78.6 | AAGTGACGGTGAGATCCAGGCT | 3,319 | 67.4 |
| AAGTGACGGTGAGATCCAGGCTT | 26 | 4.5 | AAGTGACGGTGAGATCCAGGCTA | 215 | 4.4 | |
| miR-UL148D | TCGTCCTCCCCTTCTTCACCGT | 24 | 47.1 | TCGTCCTCCCCTTCTTCACCGT | 317 | 61.1 |
| TCGTCCTCCCCTTCTTCACCG | 17 | 33.3 | TCGTCCTCCCCTTCTTCACCG | 116 | 22.4 | |
| TCGTCCTCCCCTTCTTCACCG* | ||||||
| miR-UL22A-5p | CTAACTAGCCTTCCCGTGAGA | 631 | 46.0 | TAACTAGCCTTCCCGTGAGA | 9,656 | 55.7 |
| TAACTAGCCTTCCCGTGAGA | 619 | 45.1 | CTAACTAGCCTTCCCGTGAGA | 6,069 | 35.0 | |
| miR-UL22A-3p | TCACCAGAATGCTAGTTTGTAG | 5,652 | 73.7 | TCACCAGAATGCTAGTTTGTAG | 33,536 | 83.7 |
| TCACCAGAATGCTAGTTTGTAGT | 1,301 | 17.0 | TCACCAGAATGCTAGTTTGTAGA | 3,178 | 7.9 | |
| miR-UL36-5p | TCGTTGAAGACACCTGGAAAGA | 63,877 | 97.3 | TCGTTGAAGACACCTGGAAAGA | 115,685 | 96.0 |
| TCGTTGAAGACACCTGGAAAGAA | 543 | 0.8 | TCGTTGAAGACACCTGGAAAGAA | 2,010 | 1.7 | |
| miR-UL36-3p | TTTCCAGGTGTTTTCAACGTG | 926 | 52.6 | TTTCCAGGTGTTTTCAACGTG | 1,068 | 56.3 |
| TTTCCAGGTGTTTTCAACGTGT | 317 | 18.0 | TTTCCAGGTGTTTTCAACGTGT | 359 | 18.9 | |
| TTTCCAGGTGTTTTCAACGTGC* | ||||||
| miR-US25-1-5p | AACCGCTCAGTGGCTCGGACCG | 1,667 | 38.7 | AACCGCTCAGTGGCTCGGACC | 11,057 | 41.3 |
| AACCGCTCAGTGGCTCGGACC | 879 | 20.4 | AACCGCTCAGTGGCTCGGACCG | 7,415 | 27.7 | |
| miR-US25-1-3p | GTCCGAACGCTAGGTCGGTTCT | 71 | 93.4 | GTCCGAACGCTAGGTCGGTTCT | 168 | 94.4 |
| GTCCGAACGCTAGGTCGGTTCTAA | 2 | 2.6 | GTCCGAACGCTAGGTCGGTTCTA | 2 | 1.1 | |
| TCCGAACGCTAGGTCGGTTCTC* | ||||||
| miR-US25-2-3p | ATCCACTTGGAGAGCTCCCGCGGT | 1,344 | 93.0 | ATCCACTTGGAGAGCTCCCGCGGT | 13,512 | 86.7 |
| ATCCACTTGGAGAGCTCCCGCGGTT | 31 | 2.1 | ATCCACTTGGAGAGCTCCCGCGGTA | 729 | 4.7 | |
| ATCCACTTGGAGAGCTCCCGCGG* | ||||||
| miR-US25-2-5p | AGCGGTCTGTTCAGGTGGATGA | 802 | 87.9 | AGCGGTCTGTTCAGGTGGATGA | 11,185 | 94.6 |
| TAGCGGTCTGTTCAGGTGGATGA | 50 | 5.5 | TAGCGGTCTGTTCAGGTGGATGA | 168 | 1.4 | |
| miR-US33-3p | TCACGGTCCGAGCACATCCAA | 74 | 64.3 | TCACGGTCCGAGCACATCCAA | 316 | 69.9 |
| TCACGGTCCGAGCACATCCA | 17 | 14.8 | TCACGGTCCGAGCACATCCA | 71 | 15.7 | |
| TCACGGTCCGAGCACATCCA* | ||||||
| miR-US33-5p | ATTGTGCCCGGACCGTGGGCGC | 554 | 42.6 | GATTGTGCCCGGACCGTGGGCG | 2,327 | 37.2 |
| GATTGTGCCCGGACCGTGGGCG | 311 | 23.9 | ATTGTGCCCGGACCGTGGGCGC | 1,706 | 27.3 | |
| miR-US4-5p | TGGACGTGCAGGGGGATGTCTG | 162 | 68.9 | TGGACGTGCAGGGGGATGTCTG | 439 | 56.9 |
| TGGACGTGCAGGGGGATGTCTGT | 29 | 12.3 | TGGACGTGCAGGGGGATGTCTGT | 80 | 10.4 | |
| CGACATGGACGTGCAGGGGGAT* | ||||||
| miR-US5-1 | TGACAAGCCTGACGAGAGCGT | 73 | 66.4 | TGACAAGCCTGACGAGAGCGT | 420 | 78.2 |
| TGACAAGCCTGACGAGAGCGTT | 33 | 30.0 | TGACAAGCCTGACGAGAGCGTT | 53 | 9.9 | |
| miR-US5-2-3p | TTATGATAGGTGTGACGATGTC | 3,296 | 68.1 | TATGATAGGTGTGACGATGTCT | 85,959 | 53.1 |
| TATGATAGGTGTGACGATGTCT | 1,107 | 22.9 | TTATGATAGGTGTGACGATGTC | 64,215 | 39.7 | |
| TTATGATAGGTGTGACGATGTC* | ||||||
| Not annotated in miRBase |
||||||
| miR-UL112-5p | CCTCCGGATCACATGGTTACTCAG | 107 | 49.5 | CCTCCGGATCACATGGTTACTCA | 151 | 39.2 |
| CCTCCGGATCACATGGTTACTCA | 82 | 38.0 | CCTCCGGATCACATGGTTACTCAG | 150 | 39.0 | |
| miR-US4-3p | TGACAGCCCGCTACACCTCTCT | 60 | 44.8 | TGACAGCCCGCTACACCTCTCT | 102 | 38.5 |
| TGACAGCCCGCTACACCTCTCTG | 46 | 34.3 | TGACAGCCCGCTACACCTCTCTG | 84 | 31.7 | |
| miR-US5-2-5p | CTTTCGCCACACCTATCCTGAAAG | 56 | 28.6 | CTTTCGCCACACCTATCCTGAAAG | 125 | 26.8 |
| GCTTTCGCCACACCTATCCTGA | 39 | 19.9 | CTTTCGCCACACCTATCCTGAAAGT | 78 | 16.7 | |
| miR-US22-5p | TGTTTCAGCGTGTGTCCGCGGGC | 15 | 48.4 | TGTTTCAGCGTGTGTCCGCGGG | 1,362 | 46.3 |
| TGTTTCAGCGTGTGTCCGCGGG | 14 | 45.2 | TGTTTCAGCGTGTGTCCGCGGGC | 836 | 28.4 | |
| miR-US22-3p | TCGCCGGCCGCGCTGTAACCAGG | 38 | 88.4 | TCGCCGGCCGCGCTGTAACCAGG | 389 | 87.8 |
| TCGCCGGCCGCGCTGTAACCAGGT | 4 | 9.3 | TCGCCGGCCGCGCTGTAACCAGGT | 25 | 5.6 | |
| miR-US33as-5p | TGGATGTGCTCGGACCGTGACG | 94 | 17.7 | TGGATGTGCTCGGACCGTGACG | 2,547 | 26.8 |
| GGATGTGCTCGGACCGTGACG | 91 | 17.1 | GGATGTGCTCGGACCGTGACGGTGT | 1,472 | 15.5 | |
| miR-US33as-3p | CCCACGGTCCGGGCACAATCAA | 11 | 18.6 | CCCACGGTCCGGGCACAATCA | 318 | 42.9 |
| CCCACGGTCCGGGCACAATCAAT | 11 | 18.6 | CCCACGGTCCGGGCACAATCAAC | 139 | 18.7 | |
Only the top two most abundant mapped sequences associated with each HCMV mature miRNA are listed. Total read counts for each sequence and percentages (out of the total reads aligned to the given miRNA) are indicated.
miRBase sequences (*) are listed only for those miRNAs that differed from annotations based on their most abundant form sequenced at 72 hpi.
In the process of defining mature arms within each miRNA precursor, we found sufficient evidence for only a single mature miRNA expressed from miR-UL148D and miR-US5-1, while all other pre-miRs generated two individual mature miRNA species (Table 3). In total, 22 mature miRNAs were identified during HCMV infection, originating from 12 HCMV miRNA precursors, including the novel miR-US22 and miR-US33as (Fig. 1B).
Association of HCMV miRNAs with Argonaute proteins.
Recent reports have identified that miRNAs encoded by other herpesviruses associate with host silencing complexes (8, 27). To confirm whether HCMV miRNAs are indeed incorporated into these complexes, we performed CLIP-seq (cross-linking and immunoprecipitation followed with high-throughput sequencing) experiments against Ago proteins for mock-infected and infected HFFs at 24 and 72 hpi (Fig. 3A). This technique has been shown to offer insight into Ago-mediated miRNA-mRNA interactions (6, 43). We determined that all HCMV miRNAs detected by smRNA-seq, including the novel miRNAs described above, could be found in CLIP-seq libraries generated from infected cells, using antibodies against either Ago1 or Ago2. Therefore, we conclude that all HCMV miRNAs are capable of functional activity, given that they associate with endogenous Ago proteins during productive infection.
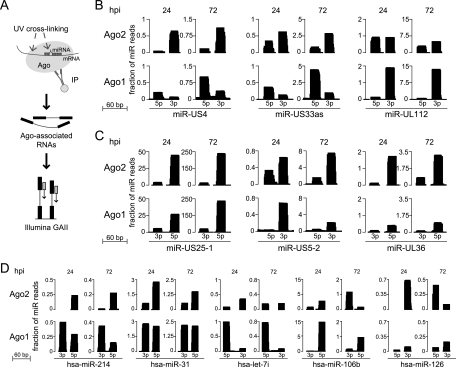
Fig 3.
Functional association of HCMV miRNAs with endogenous Argonaute proteins. (A) Experimental strategy for generation of Ago CLIP-seq libraries. Mock-infected and infected cells were UV cross-linked at 24 and 72 hpi. Immunoprecipitated (IP) RNA associated with Ago complexes was further processed and subjected to Illumina sequencing. (B) Differential incorporation of particular HCMV miRNAs between Ago2 and Ago1 complexes, as measured by the ratio of 5p over 3p mature miRNA arm incorporation between Ago2 and Ago1 complexes. Statistical significance was assessed by chi-square analysis (P value threshold of 6 × 10−5). Read count heights at each position represent the total number of overlapping CLIP-seq reads, normalized by the total number of miRNA reads in the data set. (C) Nondifferential efficiencies of miRNA association with Ago2 and Ago1 demonstrated by other HCMV miRNAs. Three representative examples are shown. (D) Human miRNAs that exhibit differential 5p-3p miRNA arm incorporation into Ago2 and Ago1 complexes. CLIP-seq read counts were used from infected data sets (24 and 72 hpi) for this assessment. Similar differential incorporation levels of these miRNAs could be observed in Ago2-Ago1 comparisons for the mock data sets.
From analysis of the Ago1 and Ago2 CLIP-seq data sets, Ago-associated miRNA expression values (each miRNA abundance measured as a fraction of the total miRNA reads) correlated with abundance levels derived from smRNA-seq. Additionally, positive correlation was observed between Ago2- and Ago1-associated miRNA abundance levels under mock-infected and infected conditions alike (correlation coefficients for pairwise comparisons ranged from 0.77 to 0.81). To further investigate if this correlation was similarly true for both the 5p and 3p mature arms of pre-miRNAs, we used 5p over 3p enrichment as a metric to compare Ago2 versus Ago1 incorporation. At a P value threshold of 6 × 10−5, three HCMV miRNAs (miR-US4, miR-US33as, and miR-UL112) were found to exhibit significantly different miRNA arm incorporation levels at one or both time points of infection (Fig. 3B), while others did not demonstrate this characteristic (three examples are shown in Fig. 3C). Using the same significance threshold, only five human pre-miRNAs of the 150 expressed during infection (considering only those annotated to express two mature miRNAs) were found to display variable incorporation of their 5p and 3p arms into Ago2 and Ago1 complexes (Fig. 3D). These included hsa-miR-214, hsa-miR-31, hsa-let-7i, hsa-miR-106b, and hsa-miR-126.
Altered levels of specific human miRNAs during HCMV infection.
In order to explore the impact of viral infection on the host cell smRNA profile, we focused on identifying human miRNAs that were differentially expressed by comparing mock-infected to virus-infected cells at both 24 and 72 hpi (Fig. 4A). To avoid detecting differences at the level of transcriptional noise, we required a minimum of 50 reads in at least one of the conditions to consider the miRNA detected; 318 mature miRNAs remained for further analysis. Within each data set, individual miRNA read counts were normalized using the total number of human and viral miRNA reads found in the library, and significantly changing miRNAs were identified as described in Materials and Methods. The large abundance of viral miRNAs at 72 hpi had the potential to complicate the statistics; however, the results did not change when viral miRNA counts were removed in the normalization process. From the perspective of individual miRNA fold changes upon infection, our results largely supported previous studies that utilized microarray technology to detect human miRNAs (30, 37).
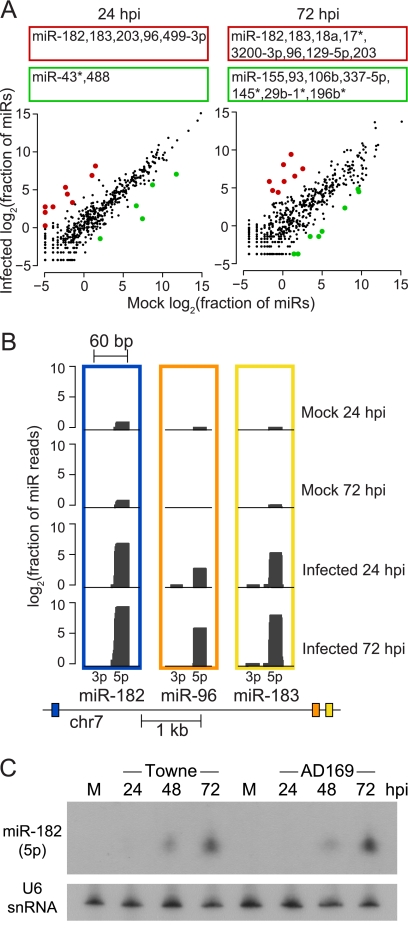
Fig 4.
Effects of HCMV infection on human miRNA levels in HFFs. (A) Scatter plot comparisons of infected and mock-infected human miRNA expression values at 24 and 72 hpi. Highlighted in red and green are significantly upregulated and downregulated miRNAs during infection, respectively (P value of < 0.05). Listed above the scatter plots are the names of the significantly altered miRNAs (any that changed between the two mock infection time points are not listed) (B) Expression of miR-182/-96/-183 during infection. The region of chromosome 7 encoding these miRNAs is depicted below the distribution of smRNA-seq reads that aligned to the 182, 96, and 183 miRNA precursors (transcription is from right to left). Read count heights at each position represent log2-transformed values of the total number of overlapping smRNA-seq reads, normalized by the total number of miRNA reads in the data set. (C) Northern blot analysis of mature miR-182 in both HCMV Towne- and AD169-infected cells. HFFs were either mock-treated or infected with the Towne strain at an MOI of 3 or AD169 at an MOI of 5. The U6 snRNA serves as a loading control.
Intriguingly, among the miRNAs that were most highly upregulated at both stages of infection, miR-96, miR-182, and miR-183 stood out in that they belong to the same miRNA genomic cluster (Fig. 4B). Also of interest, these three miRNAs share nearly the same canonical seed sequences. Since this miRNA cluster was not identified as upregulated in previous studies, we also verified increased expression of miR-182 in HFFs infected with AD169, another laboratory-adapted strain of HCMV (Fig. 4C).
Non-miRNA regions of concentrated HCMV smRNA production.
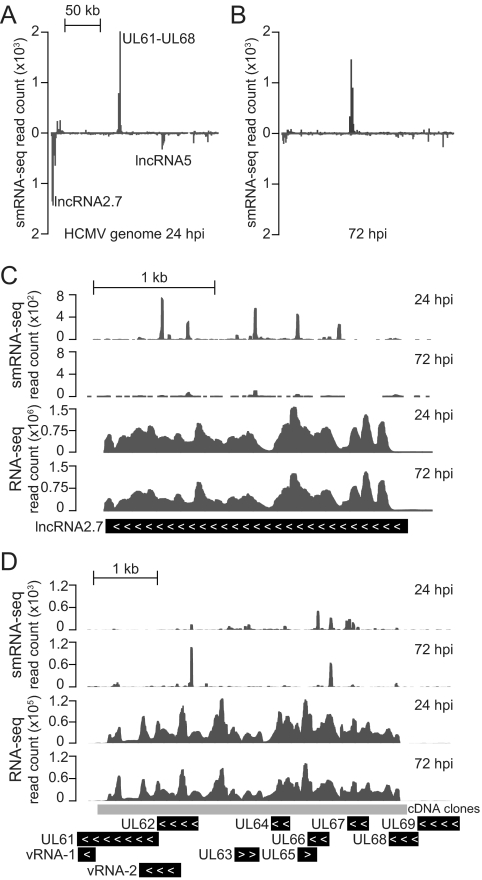
While the majority of smRNA-seq reads aligned to human and viral smRNAs, ∼3% of the mapped reads from each data set remained unclassified (Fig. 1A). From the infected data sets we found that this uncategorized set of smRNAs was concentrated in particular regions (Fig. 5A and B), with the greatest coverage observed across the HCMV long noncoding RNA 2.7 (lncRNA2.7), a large region covering UL61 to UL68 (transcribed in sense orientation to UL63 and UL65), and the lncRNA5 (located between UL105 and UL111). Upon individual inspection, these areas differed from miRNA-encoding regions with respect to their smRNA-seq read distributions and predicted secondary structures.
Fig 5.
Analysis of regions of the HCMV genome enriched for production of non-miRNA smRNAs. (A and B) Distribution of uniquely aligned non-miRNA smRNA-seq reads at 24 and 72 hpi, with peaks representing stacked reads that aligned to the Watson (above) and Crick (below) strands of the HCMV genome. Reads overlapping annotated and predicted miRNA precursor regions were excluded, and the viral genome was interrogated in 1-kb segments for smRNA-seq alignments. (C) smRNAs that are generated in sense orientation to the area encoding lncRNA2.7. (D) Representation of smRNA-seq alignments across a transcriptionally active region that falls within the UL61-UL68 region. Expression data from RNA-seq experiments is shown to illustrate transcription of the regions. Coverage by cDNA clones from Zhang et al. (42) is shown, providing independent support of RNA-seq-detected expression of the RNAs.
In an assessment of the transcriptional activity within these regions, we have used preliminary data from mRNA deep-sequencing (RNA-seq) experiments, which we have conducted to extend our studies beyond the smRNA fraction of the virus and host transcriptomes. The lncRNA2.7 (Fig. 5C) has been previously described as one of the most active transcriptional units in the HCMV genome (33), and our RNA-seq data showed that the region of UL61 to UL68 was also highly transcribed during infection (Fig. 5D). While this correlation with enrichment for non-miRNA smRNAs might suggest that these sequences were simply degradation products, other highly transcribed regions of the viral genome were not correspondingly enriched for production of such smRNA species. Accordingly, at 24 hpi the lncRNA2.7 was the most highly transcribed region of the HCMV genome by far, but the lesser-transcribed region of UL61 to UL68 was more enriched for smRNA-seq alignments.
DISCUSSION
Among the complex network of virus-host interactions present within HCMV-infected human cells, viral miRNAs expand this regulatory framework, capable of modulating both host and virus gene expression. In our smRNA-focused characterization of lytically infected HFFs, we found that a total of 22 HCMV-encoded mature miRNAs are expressed, including two novel precursors, miR-US22 and miR-US33as. Characterization of these miRNAs in transfection experiments revealed that both can be processed by the host cellular machinery and are able to repress synthetic targets. Since miR-US22 lies in antisense orientation to the US22 coding region, we predict that it may directly regulate US22 transcript levels, as was previously shown to be the case for the Epstein-Barr virus (EBV) miR-BART2, which is transcribed antisense to the viral DNA polymerase BALF5 (2). Similarly, the location of the miR-US33as precursor within the putative US29 coding region has interesting implications, given that Drosha processing would likely negatively impact US29 expression levels.
The set of miRNAs described in this study are conserved across different laboratory and clinical isolate strains of HCMV, despite instances of sequence variability across other genomic loci (10, 15). Our smRNA-seq data did not support expression of HCMV miR-UL70, despite the recent quantitative PCR experiments by Stern-Ginossar et al. using two clinical strains (35). Consistent with our smRNA-seq, Pfeffer et al. also failed to detect this particular miRNA in their cloning efforts (23). In the initial northern characterization of miR-UL70, Gray et al. indicated that the probe used for detection sometimes hybridized to an ∼22-nt species in noninfected samples, questioning its true validity as an HCMV-encoded miRNA (12).
Nevertheless, we were able to refine annotations for the other HCMV miRNAs, clarifying that all but two HCMV pre-miRNAs express two individual mature miRNAs. Except in the case of miR-US4, any discrepancies between the results from our smRNA-seq and current annotations involved only a single base difference at either the 5′ or 3′ mature HCMV miRNA boundaries. However, as stated above, sequences obtained for miR-US4-5p were shifted by 5 bp at the 5′ end from previous predictions (12). The recent studies performed by Kim et al. involved overexpression of the annotated sequence for miR-US4-5p and featured a mutant form of miR-US4-5p in which three point mutations were made within the first 7 nt of its annotated sequence (16). This third final change reaches the seed region of the form detected by smRNA-seq, which may have fortuitously reduced miRNA-mRNA targeting. While the precursor sequence of miR-US4 and more than 50 bp of upstream sequence are conserved between Towne, AD169, and several other strains of HCMV, we cannot yet rule out whether strain-specific issues account for the observed difference in mature miRNA sequence.
While it has been suggested that mammalian miRNAs associate at similar frequencies with different Ago proteins in the cell (3), we found, surprisingly, that three of the HCMV miRNA precursors possessed differential incorporation of their individual mature arms into Ago1 and Ago2 complexes. The differential incorporation of the 5p and 3p forms of miR-UL112 at 24 hpi is important to consider, given that studies focused on identification of miR-UL112 targets have primarily followed the 3p form (13, 34). Furthermore, only the 3p form of HCMV miR-UL112 is listed within current miRNA annotations.
Aside from virus-encoded miRNAs, our smRNA-seq efforts also revealed human miRNAs that responded significantly in terms of expression levels during HCMV infection. In this study, we identified a genomic cluster of human miRNAs that was highly upregulated at both 24 and 72 hpi. Consisting of miR-96, miR-182, and miR-183, this miRNA cluster has been suggested to arise from processing of a single independent primary transcript (38, 39). The upregulation of this cluster in multiple forms of cancer and tumorigenesis (31, 32) is of particular interest in the debate surrounding the potential association of HCMV infection and higher risks of tumor formation (18). Furthermore, studies using mice have implicated a potential role for the murine orthologs of these miRNAs in the development of the inner ear (28).
Our study represents the first unbiased genome-wide smRNA sequencing of HCMV-infected cells and enabled us to identify a novel class of HCMV non-miRNA smRNAs. Interestingly, the region of UL61 to UL68, where a substantial number of these smRNAs were produced, overlaps with the virus origin of lytic replication. Two previously characterized short RNA species, viral RNA-1 (vRNA-1) and vRNA-2 (26), also fall within this region (locations shown in Fig. 5D). Further studies would have to be conducted to determine if any functional relationship exists between the smRNAs we have detected and these short RNAs. While the non-miRNA group of smRNAs did not appear to collectively increase in abundance in the way that HCMV-encoded miRNAs accumulate during infection, these elements may also harbor similar regulatory potential. Recent deep-sequencing analysis of the smRNA content of host models infected with different RNA viruses has illustrated that non-miRNA viral smRNAs are found in a wide variety of virus-infected systems (20). It has been suggested that this class of smRNAs may elicit general host RNA interference responses, similar to the antiviral mechanisms known to exist in plants and lower-eukaryotic organisms.
In summary, through a battery of genomic analyses, we report the following findings. First, we have improved viral miRNA annotations and identified new HCMV-encoded miRNAs, bringing the total number of known HCMV miRNA precursors to 12. Beyond computational evidence and Northern analysis of the novel HCMV miR-US22 and miR-US33as, we have further characterized and validated the processing of these miRNA precursors in transiently transfected cells and demonstrated that their mature forms are capable of target repression. In addition to showing that HCMV miRNAs account for one-fifth of the total smRNA population at late stages of infection, we used CLIP-seq technology to demonstrate that all of these miRNAs associate directly with endogenous human Ago proteins. This analysis yielded the surprising result that mature miRNAs from three HCMV pre-miRs are differentially incorporated into Ago1 and Ago2 complexes. With regard to human miRNA levels, we observed significant upregulation of the entire host miR-182/-96/-183 cluster, previously reported to have roles in cancer and implications for hearing loss. Last, we identified novel virus-derived smRNAs emanating from coding and noncoding regions of the HCMV genome, especially across long noncoding RNAs, illustrating that a complex level of regulatory activity likely exists within the viral and host smRNA fractions of the transcriptome.
ACKNOWLEDGMENTS
We thank members of the Yeo and Spector laboratories for critical reading of the manuscript, especially S. Huelga, K. Massirer, and B. Roberts. We thank J. Nathanson and T. Liang for technical assistance with sequencing, N. Patel for comments on the manuscript, and R. Sanders for insight and suggestions during initial stages of this work. We thank S. Hoon, K. W. Kong, and S. B. Brenner for providing sequencing resources at the Molecular Engineering Laboratory at A*STAR, Singapore.
T.J.S. was supported in part by the University of California, San Diego, Genetics Training Program through an institutional training grant from the National Institute of General Medical Sciences, T32 GM008666. This work was partially supported by grants from the U.S. National Institutes of Health (HG004659 and GM084317 to G.W.Y.) and the Stem Cell Program at the University of California, San Diego (G.W.Y.).
Footnotes
Published ahead of print 19 October 2011
REFERENCES
- 1. Ambros V, et al. 2003. A uniform system for microRNA annotation. RNA 9:277–279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Barth S, et al. 2008. Epstein-Barr virus-encoded microRNA miR-BART2 down-regulates the viral DNA polymerase BALF5. Nucleic Acids Res. 36:666–675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Burroughs AM, et al. 2011. Deep-sequencing of human Argonaute-associated small RNAs provides insight into miRNA sorting and reveals Argonaute association with RNA fragments of diverse origin. RNA Biol. 8:158–177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Cai X, et al. 2005. Kaposi's sarcoma-associated herpesvirus expresses an array of viral microRNAs in latently infected cells. Proc. Natl. Acad. Sci. U. S. A. 102:5570–5575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Cai X, et al. 2006. Epstein-Barr virus microRNAs are evolutionarily conserved and differentially expressed. PLoS Pathog. 2:e23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Chi SW, Zang JB, Mele A, Darnell RB. 2009. Argonaute HITS-CLIP decodes microRNA-mRNA interaction maps. Nature 460:479–486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Cui C, et al. 2006. Prediction and identification of herpes simplex virus 1-encoded microRNAs. J. Virol. 80:5499–5508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Dolken L, et al. 2010. Systematic analysis of viral and cellular microRNA targets in cells latently infected with human gamma-herpesviruses by RISC immunoprecipitation assay. Cell Host Microbe 7:324–334 [DOI] [PubMed] [Google Scholar]
- 9. Dolken L, et al. 2007. Mouse cytomegalovirus microRNAs dominate the cellular small RNA profile during lytic infection and show features of posttranscriptional regulation. J. Virol. 81:13771–13782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Dolken L, Pfeffer S, Koszinowski UH. 2009. Cytomegalovirus microRNAs. Virus Genes 38:355–364 [DOI] [PubMed] [Google Scholar]
- 11. Dunn W, et al. 2005. Human cytomegalovirus expresses novel microRNAs during productive viral infection. Cell Microbiol. 7:1684–1695 [DOI] [PubMed] [Google Scholar]
- 12. Grey F, et al. 2005. Identification and characterization of human cytomegalovirus-encoded microRNAs. J. Virol. 79:12095–12099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Grey F, Meyers H, White EA, Spector DH, Nelson J. 2007. A human cytomegalovirus-encoded microRNA regulates expression of multiple viral genes involved in replication. PLoS Pathog. 3:e163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Han J, et al. 2006. Molecular basis for the recognition of primary microRNAs by the Drosha-DGCR8 complex. Cell 125:887–901 [DOI] [PubMed] [Google Scholar]
- 15. Jung GS, et al. 2011. Full genome sequencing and analysis of human cytomegalovirus strain JHC isolated from a Korean patient. Virus Res. 156:113–120 [DOI] [PubMed] [Google Scholar]
- 16. Kim S, et al. 2011. Human cytomegalovirus microRNA miR-US4-1 inhibits CD8+ T cell responses by targeting the aminopeptidase ERAP1. Nat. Immunol. 12:984–991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Meyer C, et al. 2011. Cytomegalovirus microRNA expression is tissue specific and is associated with persistence. J. Virol. 85:378–389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Michaelis M, et al. 2011. Oncomodulation by human cytomegalovirus: novel clinical findings open new roads. Med. Microbiol. Immunol. 200:1–5 [DOI] [PubMed] [Google Scholar]
- 19. Mocarski E, Shenk T, Pass R. 2007. Cytomegaloviruses, p 2702–2772 In Knipe DM, et al. (ed), Fields virology, 5th ed Lippincott Williams & Wilkins, Philadelphia, PA [Google Scholar]
- 20. Parameswaran P, et al. 2010. Six RNA viruses and forty-one hosts: viral small RNAs and modulation of small RNA repertoires in vertebrate and invertebrate systems. PLoS Pathog. 6:e1000764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Parkhomchuk D, et al. 2009. Transcriptome analysis by strand-specific sequencing of complementary DNA. Nucleic Acids Res. 37:e123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Pasquinelli AE, et al. 2003. Expression of the 22 nucleotide let-7 heterochronic RNA throughout the Metazoa: a role in life history evolution? Evol. Dev. 5:372–378 [DOI] [PubMed] [Google Scholar]
- 23. Pfeffer S, et al. 2005. Identification of microRNAs of the herpesvirus family. Nat. Methods 2:269–276 [DOI] [PubMed] [Google Scholar]
- 24. Pfeffer S, et al. 2004. Identification of virus-encoded microRNAs. Science 304:734–736 [DOI] [PubMed] [Google Scholar]
- 25. Polymenidou M, et al. 2011. Long pre-mRNA depletion and RNA missplicing contribute to neuronal vulnerability from loss of TDP-43. Nat. Neurosci. 14:459–468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Prichard MN, et al. 1998. Identification of persistent RNA-DNA hybrid structures within the origin of replication of human cytomegalovirus. J. Virol. 72:6997–7004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Riley KJ, Rabinowitz GS, Steitz JA. 2010. Comprehensive analysis of rhesus lymphocryptovirus microRNA expression. J. Virol. 84:5148–5157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Sacheli R, et al. 2009. Expression patterns of miR-96, miR-182 and miR-183 in the development inner ear. Gene Expr. Patterns 9:364–370 [DOI] [PubMed] [Google Scholar]
- 29. Sanders RL, Del Rosario CJ, White EA, Spector DH. 2008. Internal deletions of IE2 86 and loss of the late IE2 60 and IE2 40 proteins encoded by human cytomegalovirus affect the levels of UL84 protein but not the amount of UL84 mRNA or the loading and distribution of the mRNA on polysomes. J. Virol. 82:11383–11397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Santhakumar D, et al. 2010. Combined agonist-antagonist genome-wide functional screening identifies broadly active antiviral microRNAs. Proc. Natl. Acad. Sci. U. S. A. 107:13830–13835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Sarver AL, et al. 2009. Human colon cancer profiles show differential microRNA expression depending on mismatch repair status and are characteristic of undifferentiated proliferative states. BMC Cancer 9:401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Schaefer A, et al. 2010. Diagnostic and prognostic implications of microRNA profiling in prostate carcinoma. Int. J. Cancer 126:1166–1176 [DOI] [PubMed] [Google Scholar]
- 33. Spector DH. 1996. Activation and regulation of human cytomegalovirus early genes. Intervirology 39:361–377 [DOI] [PubMed] [Google Scholar]
- 34. Stern-Ginossar N, et al. 2007. Host immune system gene targeting by a viral miRNA. Science 317:376–381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Stern-Ginossar N, et al. 2009. Analysis of human cytomegalovirus-encoded microRNA activity during infection. J. Virol. 83:10684–10693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Umbach JL, Cullen BR. 2010. In-depth analysis of Kaposi's sarcoma-associated herpesvirus microRNA expression provides insights into the mammalian microRNA-processing machinery. J. Virol. 84:695–703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Wang FZ, et al. 2008. Human cytomegalovirus infection alters the expression of cellular microRNA species that affect its replication. J. Virol. 82:9065–9074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Weston MD, Pierce ML, Rocha-Sanchez S, Beisel KW, Soukup GA. 2006. MicroRNA gene expression in the mouse inner ear. Brain Res. 1111:95–104 [DOI] [PubMed] [Google Scholar]
- 39. Xu S, Witmer PD, Lumayag S, Kovacs B, Valle D. 2007. MicroRNA (miRNA) transcriptome of mouse retina and identification of a sensory organ-specific miRNA cluster. J. Biol. Chem. 282:25053–25066 [DOI] [PubMed] [Google Scholar]
- 40. Yeo GW, et al. 2009. An RNA code for the FOX2 splicing regulator revealed by mapping RNA-protein interactions in stem cells. Nat. Struct. Mol. Biol. 16:130–137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Zeng Y, Yi R, Cullen BR. 2005. Recognition and cleavage of primary microRNA precursors by the nuclear processing enzyme Drosha. EMBO J. 24:138–148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Zhang G, et al. 2007. Antisense transcription in the human cytomegalovirus transcriptome. J. Virol. 81:11267–11281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Zisoulis DG, et al. 2010. Comprehensive discovery of endogenous Argonaute binding sites in Caenorhabditis elegans. Nat. Struct. Mol. Biol. 17:173–179 [DOI] [PMC free article] [PubMed] [Google Scholar]