Abstract
Total enterococci and vancomycin-resistant enterococci (VRE) were enumerated in samples of effluent (n = 50) and water (n = 167) from a number of sources. VRE were detected in the outflow of a wastewater treatment plant and in a single rural drinking water supply, suggesting potential for transmission to humans through environmental contamination.
TEXT
Enterococci with acquired high-level (≥256 μg/ml) resistance to vancomycin were first reported in Europe in 1988 and have since been reported worldwide with increasing frequency (10). In Ireland, 44% of invasive isolates of Enterococcus faecium were vancomycin resistant in the second quarter of 2011 (http://www.hpsc.ie/hpsc/A-Z/MicrobiologyAntimicrobialResistance/EuropeanAntimicrobialResistanceSurveillanceSystemEARSS/EARSSSurveillanceReports/2011Reports/File,12962,en.pdf). Environmental contamination is a factor in dissemination of vancomycin-resistant enterococci (VRE) in hospitals (13); however, the role of contamination of the general environment is less well explored. It has been reported that enterococci can persist for long periods in the environment (2, 5). This is the first report on the occurrence of VRE in waters and effluents from a number of sources in Ireland.
Fifty samples of effluent were collected between 2006 and 2008: hospital effluent (hospital A [HA] [n = 17] and hospital B [HB] [n = 2]), municipal effluent upstream and downstream from the effluent discharge points of hospital A (USA [n = 5] and DSA [n = 5], respectively) and hospital B (USB [n = 2] and DSB [n = 2], respectively), and final treated effluent from a secondary wastewater treatment process (TE) (n = 13) and, on a single occasion in 2008, from a number of points throughout the wastewater treatment process (n = 4). Seawater (n = 29), river water (n = 28), and lake water (n = 8) samples were collected in 2006 and 2007. Source (S) (n = 17) and distributed (P) (n = 17) water samples were taken from 3 rural group water supplies, K, M, and C, between October 2006 and June 2007.
Total enterococci were enumerated using an Enterolert Quantitray system (Idexx Technopath, Limerick, Ireland) (6), and the proportion of VRE was determined by addition of vancomycin to a final concentration of 256 μg/ml. Effluent from hospital A contained the highest numbers of enterococci (up to 1.1 × 107 most probable number [MPN]/100 ml detected) (Table 1) and percentages of VRE (0.02 to 27%) (Table 1). VRE were also detected in 2 of 5 (40%) samples of municipal effluent downstream of hospital A, in 2 of 13 (15%) samples of final treated effluent, in 1 source water sample from the K supply, and in a single river water sample (Table 1).
Table 1.
Quantitative analysis of effluents and waters for total enterococci and percentages of vancomycin-resistant enterococci (VRE)
| Sample site | Sample size | Total enterococci (MPN/100 ml)b | Percentage of VRE |
|---|---|---|---|
| Hospital effluent | |||
| Hospital A | 17 | NDa to 1.1 × 107 | ND to 27 |
| Hospital B | 2 | 4.7 × 105 to 1.97 × 107 | ND |
| Municipal effluent | |||
| Upstream of hospital A | 5 | 3.1 × 104 to 1.58 × 106 | ND to 2.8 |
| Downstream of hospital A | 5 | 1 × 105 to 2.75 × 106 | ND |
| Upstream of hospital B | 2 | ND | ND |
| Downstream of hospital B | 2 | 4.1 × 105 to 1.45 × 106 | ND |
| Wastewater treatment plant | |||
| Raw intake | 1 | 1.32 × 106 | ND |
| Postreturn | 1 | 1.45 × 105 | ND |
| Primary treated effluent | 1 | 1.83 × 105 | ND |
| Aeration effluent | 1 | 9.8 × 105 | ND |
| Final treated effluent | 13 | 9.7 × 102 to 2.6 × 104 | ND to 2 |
| Rural group water supplies | |||
| K source water | 17 | ND to 7.93 × 101 | ND to 6 |
| K piped water | 17 | ND to 5.16 × 102 | ND |
| C source water | 17 | ND to 5.2 | ND |
| C piped water | 17 | ND to 8.05 | ND |
| M source water | 17 | ND to 7.57 × 101 | ND |
| M piped water | 17 | ND to 2.28 × 101 | ND |
| River water | 28 | ND to 2.42 × 103 | ND to 1.6 |
| Lake water | 8 | ND to 2.61 × 102 | ND |
| Seawater | 29 | ND to 4.2 × 102 | ND |
ND, not detected.
Limit of detection is 1 × 105 MPN/100 ml for untreated effluent, 1 × 102 MPN/100 ml for treated effluent, and 1 × 101 MPN/100 ml for seawater.
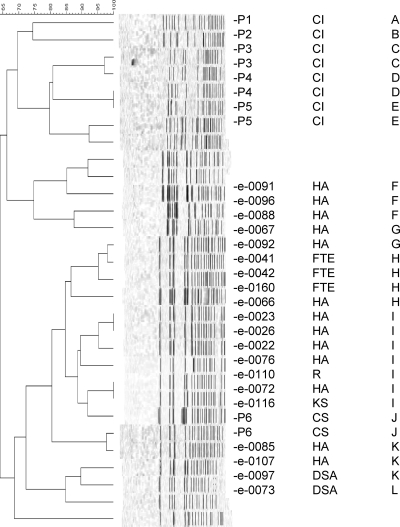
Aliquots (1 ml) were removed from one positive well per tray as previously described (6), leading to the collection of 20 isolates of VRE (HA, n = 13; DSA, n = 2; TE, n = 3; KS, n = 1; urban river water, n = 1). All were identified as Enterococcus faecium and had vancomycin MICs of >256 μg/ml as determined by Etest (bioMérieux Inc., Marcy l'Etoile, France), validating the modification of the Enterolert Quantitray system for detection of VRE. The vanA gene was confirmed in all isolates by PCR (4). Pulsed-field gel electrophoresis (PFGE) analysis using SmaI identified 18 major pulsed-field profiles (PFPs) and 5 major clusters (clusters A to E) containing 2 to 7 isolates based on a PFP similarity of ≥85% (Fig. 1). The 10 bloodstream isolates of VRE included 9 distinct PFPs and were not similar to environmental isolates (Fig. 1).
Fig 1.
Comparison of VRE from environmental and clinical sources. From left to right, the columns show the following: percent homology, PFGE profile, sample identifier, sample type, and cluster. CI and CS, clinical isolates; FTE, final treated effluent; R, river water.
Caplin et al. reported high-level (>128 μg/ml) vancomycin resistance in 4/26 (15%) hospital effluent samples and in 15/21 (71%) untreated municipal effluent samples in England (3). Kuhn et al. reported VRE in 36% of urban wastewater and 16% of hospital effluent samples examined from 4 countries (Sweden, Spain, United Kingdom, and Denmark) (7). Novais et al. reported detection of VRE in 11/14 (79%) wastewater samples downstream from 4 hospitals in Portugal (12). We have evaluated VRE as a percentage of total enterococci and demonstrate that VRE are present in hospital effluent not only frequently but also in high numbers compared with previous findings (4, 10). The influence of hospital effluent on the presence of VRE in municipal effluent downstream from the hospital is also clearly demonstrated.
While VRE appear to be partly removed by wastewater treatment, VRE were detected in two treated effluent samples (2% of enterococci detected). Araújo et al. reported no significant differences in the proportions of VRE between the inflow and the effluent from wastewater treatment plants (1). Vilanova et al. also found that VRE persisted after the treatment process (14).
Lanthier et al. reported that enterococci resistant to ciprofloxacin (2 μg/ml) or vancomycin (16 μg/ml) were uncommon in a river basin in Canada (8). Novais et al. found VRE (all E. faecium of the vanA genotype) in 2/3 samples collected from a river in Portugal (12). Moore et al. reported no VRE in surface and ocean waters from California (11). In these reports, vancomycin at 16 μg/ml was used for screening, compared with 256 μg/ml in this study. The higher concentration used here excludes detection of low-level intrinsic resistance. Our data indicate that VRE are detected infrequently in river water (1/28), lake water (0/8), and seawater (0/29) samples. This is the first report of vanA VRE in a drinking water supply. Although VRE detected in 1 of 51 drinking water sources seems a low frequency, it is worth noting that this represents detection once in a total volume of just 5.1 liters. The situation in Ireland whereby some rural dwellers are served by small water supply systems with limited or inconsistent treatment also pertains to rural areas in a number of other developed countries.
PFGE does not demonstrate a close relationship between the clinical and environmental isolates of VRE. The clinical isolates were bloodstream isolates. Studies have demonstrated that particular clonal groups of VRE, such as clonal complex 17 (CC17), are associated predominantly with infection (9). Therefore, invasive isolates are unlikely to represent the diversity of VRE present in the gastrointestinal tracts of patients in the hospital from which the effluent is sourced, and this may relate to the apparent lack of similarity. Multilocus sequence typing (MLST) or whole-genome analyses may reveal relationships that are less apparent by PFGE, as clonal groups such as CC17 may be quite diverse on PFGE (15).
From an ecological perspective, vanA E. faecium VRE in the aqueous environment represent a disturbance of natural biodiversity, as a relatively novel anthropogenic biological entity related to antimicrobial use. There may be other impacts of microbial biodiversity that are more difficult to detect. From a human health perspective, the discharge of VRE into the environment in hospital effluent is a concern as a potential avenue for spread of VRE into the community; however, it is difficult to assess its significance at this time.
Footnotes
Published ahead of print 18 November 2011
REFERENCES
- 1. Araujo C, et al. 2010. Vancomycin-resistant enterococci from Portuguese wastewater treatment plants. J. Basic Microbiol. 50:605–609 [DOI] [PubMed] [Google Scholar]
- 2. Barcina I, Lebaron P, Vives-Rego J. 1997. Survival of allochthonous bacteria in aquatic systems: a biological approach. FEMS Microbiol. Ecol. 23:1–9 [Google Scholar]
- 3. Caplin JL, Hanlon GW, Taylor HD. 2008. Presence of vancomycin and ampicillin-resistant Enterococcus faecium of epidemic clonal complex-17 in wastewaters from the south coast of England. Environ. Microbiol. 10:885–892 [DOI] [PubMed] [Google Scholar]
- 4. Depardieu F, Perichon B, Courvalin P. 2004. Detection of the van alphabet and identification of enterococci and staphylococci at the species level by multiplex PCR. J. Clin. Microbiol. 42:5857–5860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Duran AE, et al. 2002. Removal and inactivation of indicator bacteriophages in fresh waters. J. Appl. Microbiol. 92:338–347 [DOI] [PubMed] [Google Scholar]
- 6. Galvin S, et al. 2010. Enumeration and characterization of antimicrobial-resistant Escherichia coli bacteria in effluent from municipal, hospital, and secondary treatment facility sources. Appl. Environ. Microbiol. 76:4772–4779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Kuhn I, et al. 2005. Occurrence and relatedness of vancomycin-resistant enterococci in animals, humans, and the environment in different European regions. Appl. Environ. Microbiol. 71:5383–5390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Lanthier M, et al. 2011. Distribution of selected virulence genes and antibiotic resistance in Enterococcus species isolated from the South Nation River drainage basin, Ontario, Canada. J. Appl. Microbiol. 110:407–421 [DOI] [PubMed] [Google Scholar]
- 9. Leavis HL, Bonten MJ, Willems RJ. 2006. Identification of high-risk enterococcal clonal complexes: global dispersion and antibiotic resistance. Curr. Opin. Microbiol. 9:454–460 [DOI] [PubMed] [Google Scholar]
- 10. Leclercq R, Derlot E, Duval J, Courvalin P. 1988. Plasmid-mediated resistance to vancomycin and teicoplanin in Enterococcus faecium. N. Engl. J. Med. 319:157–161 [DOI] [PubMed] [Google Scholar]
- 11. Moore DF, Guzman JA, McGee C. 2008. Species distribution and antimicrobial resistance of enterococci isolated from surface and ocean water. J. Appl. Microbiol. 105:1017–1025 [DOI] [PubMed] [Google Scholar]
- 12. Novais C, Coque TM, Ferreira H, Sousa JC, Peixe L. 2005. Environmental contamination with vancomycin-resistant enterococci from hospital sewage in Portugal. Appl. Environ. Microbiol. 71:3364–3368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ott M, Wirick H. 2008. Vancomycin-resistant enterococci (VRE) and the role of the healthcare worker. Can. Oper. Room Nurs. J. 26:21–24, 26-29, 32 [PubMed] [Google Scholar]
- 14. Vilanova X, Manero A, Cerda-Cuellar M, Blanch AR. 2004. The composition and persistence of faecal coliforms and enterococcal populations in sewage treatment plants. J. Appl. Microbiol. 96:279–288 [DOI] [PubMed] [Google Scholar]
- 15. Werner G, Klare I, Witte W. 2007. The current MLVA typing scheme for Enterococcus faecium is less discriminatory than MLST and PFGE for epidemic-virulent, hospital-adapted clonal types. BMC Microbiol. 7:28. [DOI] [PMC free article] [PubMed] [Google Scholar]