Abstract
Pseudouridine synthase 1 (Pus1p) is an unusual site-specific modification enzyme in that it can modify a number of positions in tRNAs and can recognize several other types of RNA. No consensus recognition sequence or structure has been identified for Pus1p. Human Pus1p was used to determine which structural or sequence elements of human tRNASer are necessary for pseudouridine (Ψ) formation at position 28 in the anticodon stem-loop (ASL). Some point mutations in the ASL stem of tRNASer had significant effects on the levels of modification and compensatory mutation, to reform the base pair, restored a wild-type level of Ψ formation. Deletion analysis showed that the tRNASer TΨC stem-loop was a determinant for modification in the ASL. A mini-substrate composed of the ASL and TΨC stem-loop exhibited significant Ψ formation at position 28 and a number of mutants were tested. Substantial base pairing in the ASL stem (3 out of 5 bp) is required, but the sequence of the TΨC loop is not required for modification. When all nucleotides in the ASL stem other than U28 were changed in a single mutant, but base pairing was retained, a near wild-type level of modification was observed.
INTRODUCTION
Pseudouridine (Ψ) is the most abundant post-transcriptional RNA base modification and the enzymes responsible for this isomerization of uridine and the positions modified in the RNAs, are conserved (1–4). This modification is important in the structure and function of small RNAs (5–11), including small nuclear RNAs (snRNAs), cofactors in pre-mRNA splicing (12–16). There are two pathways for the formation of Ψ in eukaryotes. One pathway is involved in the formation of Ψ in rRNA and snRNA and requires Dyskerin or its homologs (Cbf5p in yeast for example) and RNP cofactors [most often H/ACA small nucleolar ribonucleoprotein particles (snoRNPs)] that enable one enzyme to recognize many different sites for modification on different substrates (17–25). The other pathway for Ψ formation employs site-specific Ψ synthases that require no cofactors to recognize and modify the RNA substrate. A number of enzymes have been identified in this pathway and are grouped in six families that all share a common basic structure (4). It is safe to say the cofactor ‘guided’ pathway has received a great deal of attention because of its similarity to aspects of RNA editing, but the site-specific pseudouridine synthases accomplish the same task, on more substrates, with fewer tools, arguably making them more interesting from an enzymatic perspective.
One of the most studied site-specific enzymes is Pseudouridine synthase 1 (Pus1p) (26–35). Pus1p modifies uridines in several regions of many different tRNAs (27,33,34) and one position in yeast U2 snRNA (36). It is a transcriptional coactivator in nuclear receptor signaling, modifying Steroid Receptor Activator RNA (SRA) (37,38), and when mutated or missing, leads to the disorder Mitochondrial myopathy and sideroblastic anemia with lactic acidemia (MLASA) (39–42).
In vivo and in vitro Pus1p modifies positions 1, 26, 27 and 28 in many tRNAs and positions 34 and 36 in the anticodons of certain intron-containing tRNAs (26–28,33). In addition, yeast Pus1p has been shown to modify positions 65 and 67 in vivo (34) and mouse (mPus1p) and human Pus1p (hPus1p) modify position 30 in tRNAs, whereas yeast Pus1p does not (29,33). The presence of the intron in yeast pre-tRNAIle(UAU) is required for the formation of Ψ at positions 34 and 36 in the anticodon by Pus1p in vitro (26,34). The intron allows for the formation of a stem that contains the uridines to be modified (26,34,43). Residues that are modified in tRNA by Pus1p are part of a base-paired stem, suggesting RNA secondary structure is important for recognition by Pus1p (26,27,33–35). Even with all of the studies on Pus1p, the only sequence consensus (not requirement) that has been proposed for Pus1p is the presence of a pyrimidine nucleotide 3′ of the site of modification (44).
Interestingly, yeast Pus1p has been shown to modify position 44 on yeast U2 snRNA, a position that is considered to be in a non-base paired region of U2 snRNA (36). When mouse Pus1p is expressed in a pus1 deleted yeast strain, the mouse enzyme can also modify yeast U2 snRNA at the same position (33). However, the modification of U2 snRNA in Caenorhabditis elegans does not require Pus1p, since the loss of the pus-1 gene product in worms results in the absence of Ψ at known Pus1p modification sites on tRNA, but has no effect on the modification of U2 snRNA or other snRNAs (28,45).
Crystal structures for Pus1p have not been solved. However, the structure of a member of the same synthase family, bacterial TruA, in the presence of tRNA substrates, has been revealed (46,47). TruA modifies uridines at positions 38–40 in many tRNAs and the equivalent activity in eukaryotes is associated with Pseudouridine synthase 3, another member of the TruA family [Pus3p; (48,49)]. The substrate tRNA binds across a dimer of TruA (46,47). The anticodon stem-loop (ASL) of the tRNA, as well as the dihydrouridine (D) stem and the ‘elbow’ where the D and TΨC loops interact, contact the synthase (47). TruA did not interact with the variable (V) stem-loop or the acceptor stem of the tRNA substrate (47).
In order to begin to understand how Pus1p can modify several positions in tRNAs and also modify other RNAs such as U2 snRNA and SRA, we undertook an investigation of the sequence and structural requirements for hPus1p activity on human tRNASer. The activity of hPus1p with wild-type and point and deletion mutants of the tRNA substrate indicates there are structural requirements in the formation of Ψ at position 28 in tRNASer. A minimum level of base pairing in the ASL (the location of the modification) and the presence of an additional 3′ stem-loop, constitute the minimal substrate for hPus1p. Even when all nucleotides in the ASL stem other than U28 were changed in a single minimal substrate, but base pairing was retained, a near wild-type level of modification was observed.
MATERIALS AND METHODS
Isolation of recombinant hPus1p and site directed mutagenesis of tRNASer
Recombinant hPus1p was expressed and isolated as described (29). Site-directed mutagenesis of human tRNASer (UGA) was performed with a Quikchange II kit (Stratagene), utilizing primers containing the mutations and the pHtS plasmid [human tRNASer(UGA); gift of H. Gross (50)] to create the mutants. The resulting constructs have a T7 RNA polymerase promoter at the 5′-end of the tRNA gene and a BstN1 site at the 3′-end of the gene. Plasmid DNA digested with BstN1 will yield tRNAs with CCA at the 3′-terminus after transcription (29,50). The primers used for mutagenesis are given below with just the forward primers listed; the reverse primers are the inverse complement. The forward primer for:
27C was 5′CGAGTGGTTAAGGCGCTGGACTTGAAATCCA,
27G was 5′CGAGTGGTTAAGGCGGTGGACTTGAAATCCA,
28C was 5′GAGTGGTTAAGGCGACGGACTTGAAATCC used with the 42G mutant to create 28C/42G,
29C was 5′GAGTGGTTAAGGCGATCGACTTGAAATCCATT,
30C was 5′GTGGTTAAGGCGATGCACTTGAAATCCATTG,
31C was 5′GTGGTTAAGGCGATGGCCTTGAAATCCATTGG,
39G was 5′CGATGGACTTGAAAGCCATTGGGGTC,
40G was 5′GATGGACTTGAAATGCATTGGGGTCTC,
41G was 5′GACTTGAAATCGATTGGGGTCTCC, and used in the 29C/41G mutant starting with 29C template,
42C was 5′GGACTTGAAATCCCTTGGGGTCTCC,
42G was 5′GGACTTGAAATCCGTTGGGGTCTCC,
43G was 5′GGACTTGAAATCCAGTGGGGTCTCC, and used in the 27C/43G mutant starting with 27C template and in 27G/43G mutant starting with the 27G template,
30C/40G was 5′GATGCACTTGAAATGCATTGGGGTCTC, starting with the 30G template,
31C/39G was 5′CGATGGCCTTGAAAGCCATTGGGGTC, starting with the 31C template,
Δ D stem-loop was 5′GTAGTCGTGGCGATGGACTTGAAATCC,
Δ D stem-loop with 28C/42G was 5′GTAGTCGTGGCGACGGACTTGAAATCCGTTGGGGTCTCCCC.
RNA synthesis, the tritium-release assay and binding assays
The digested plasmid DNAs were transcribed with T7 RNA polymerase in the presence of 25 µCi [5-3H]-UTP (11.1 or 23.9 Ci/mmol, Moravek) or 25 µCi [5,6-3H]-UTP (38 Ci/mmol, Moravek) with 100 µM cold UTP and 1.0 mM, non-labeled, ATP, GTP and CTP (51). Additional RNA substrates were synthesized using a sense T7 DNA oligonucleotide, an antisense template oligonucleotide and high concentration T7 RNA Polymerase, as described (52,53). The oligodeoxynucleotides used for this type of synthesis are listed below, note that the oligodeoxynucleotides that serve as the template for synthesis are antisense and have antisense T7 primer sequence at the 3′-end.
The T7 primer used was 5′TAATACGACTCACTATAG. The template for the:
ASL stem-loop was 5′AATGGATTTCAAGTCCATCTATAGTGAGTCGTATTA,
ASL & D stem-loops was 5′ATGGATTTCAAGTCCATCGCCTTAACCACTCGGCTATAGTGAGTCGTATTA,
ASL & V stem-loops was 5′GGGGAGACCCCAATGGATTTCAAGTCCATCTATAGTGAGTCGTATTA,
ASL, V, & TΨC stem-loops was 5′GCAGGATTCGAACCTGCGCGGGGAGACCCCAATGGATTTCAAGTCCATCTATAGTGAGTCGTATTA,
ASL, D, & V stem-loops was 5′GGGGAGACCCCAATGGATTTCAAGTCCATCGCCTTAACCACTCGGCTATAGTGAGTCGTATTA,
ASL, D, V, & TΨC stem-loops was 5′GCAGGATTCGAACCTGCGCGGGGAGACCCCAATGGATTTCAAGTCCATCGCCTTAACCACTCGGCTATAGTGAGTCGTATTA,
Δ TΨC stem-loop was 5′TGGCGTAGTCGGCGCGGGGAGACCCCAATGGATTTCAAGTCCATCGCCTTAACCACTCGGCCACGACTACTATAGTGAGTCGTATTA,
Δ V stem-loop was 5′ TGGCGTAGTCGGCAGGATTCGAACCTGCGAATGGATTTCAAGTCCATCGCCTTAACCACTCGGCCACGACTACTATAGTGAGTCGTATTA.
ASL & TΨC stem-loop mini-substrate required the following oligodeoxynucleotide template: 5′GCAGGATTCGAACCTGCGCCAATGGATTTCAAGTCCATCTATAGTGAGTCGTATTA. The small size of this mini-substrate and the fact that the first nucleotide transcribed is a ‘G’, allows for efficient synthesis of RNAs in vitro. The following mutations were incorporated into the oligodeoxynucleotide template to generate variations of the ASL & TΨC stem-loop mini-substrate:
GGACG starting at 61 was 5′CGTCCATTCGAACCTGCGCCAATGGATTTCAAGTCCATCATAGTGAGTCGTATTA,
AAGCUUA in TΨC loop was 5′GCAGGTAAGCTTCCTGCGCCAATGGATTTCAAGTCCATCATAGTGAGTCGTATTA,
28C/42G was 5′GCAGGATTCGAACCTGCGCCAACGGATTTCAAGTCCGTCTATAGTGAGTCGTATTA,
27C/39G was 5′GCAGGATTCGAACCTGCGCCAATGGCTTTCAAGTCCAGCTATAGTGAGTCGTATTA,
27C/42C was 5′GCAGGATTCGAACCTGCGCCAAGGGATTTCAAGTCCAGCTATAGTGAGTCGTATTA,
27C/29C/39G was 5′GCAGGATTCGAACCTGCGCCAATGGCTTTCAAGTCGAGCTATAGTGAGTCGTATTA,
27C/39G/40G was 5′GCAGGATTCGAACCTGCGCCAATGCCTTTCAAGTCCAGCTATAGTGAGTCGTATTA,
27C/29C/42G was 5′GCAGGATTCGAACCTGCGCCAAGGGATTTCAAGTCGAGCTATAGTGAGTCGTATTA,
27C/29C/39G/40G was 5′GCAGGATTCGAACCTGCGCCAATGCCTTTCAAGTCGAGCTATAGTGAGTCGTATTA,
27C/29C/39G/40G/42C was 5′GCAGGATTCGAACCTGCGCCAAGGCCTTTCAAGTCGAGCTATAGTGAGTCGTATTA,
Δ55 & 56 was 5′GCAGGATTCACCTGCGCCAATGGATTTCAAGTCCATCTATAGTGAGTCGTATTA,
Δ55–58 was 5′GCAGGATACCTGCGCCAATGGATTTCAAGTCCATCTATAGTGAGTCGTATTA,
GCAA inserted after 55 was 5′GCAGGATTCGTTGCAACCTGCGCCAATGGATTTCAAGTCCATCTATAGTGAGTCGTATTA,
Y for Y R for R stem mini-substrate mutant was 5′GCAGGATTCGAACCTGCGCCAGCAAGTTTCAAGCTTACCTATAGTGAGTCGTATTA.
Before incubation with hPus1p, the RNA substrates were heated to 78°C for 3 min and allowed to cool slowly to at least 37°C (30–40 min). Substrate reactions with the Ψ synthases were carried out in 50 mM ammonium chloride, 5 mM DTT, 25 mM Tris (pH 7.5) and 1 mM MgCl2. The amount of Ψ synthase used was ~200 ng at 37°C for the times indicated.
The tritium-release assay is described in detail elsewhere (54,55). Briefly, it involves mixing the previously incubated reaction with Norit A charcoal. Norit A charcoal binds all of the 3H counts in the sample except those that have been ‘released’ to water in the process of forming Ψ. The released counts are separated from the charcoal-bound counts using a SPIN-X column (Costar), the filtrate is mixed with scintillation fluid, and the released 3H counts are measured on a scintillation counter.
The equilibrium binding assays were carried out as described (32) using hPus1p concentrations of 0, 50, 100, 200, 400 and 800 nM in 50 µl reactions, with the 3H-labeled RNA substrates at 500 pM. The RNAs were synthesized as described above except that the cold UTP concentration in the synthesis reaction was 250 µM. A slot-blot apparatus (BioRad) was used for filtration of the samples, the resulting filters were air dried, cut into appropriate pieces and counted in scintillation fluid.
RESULTS
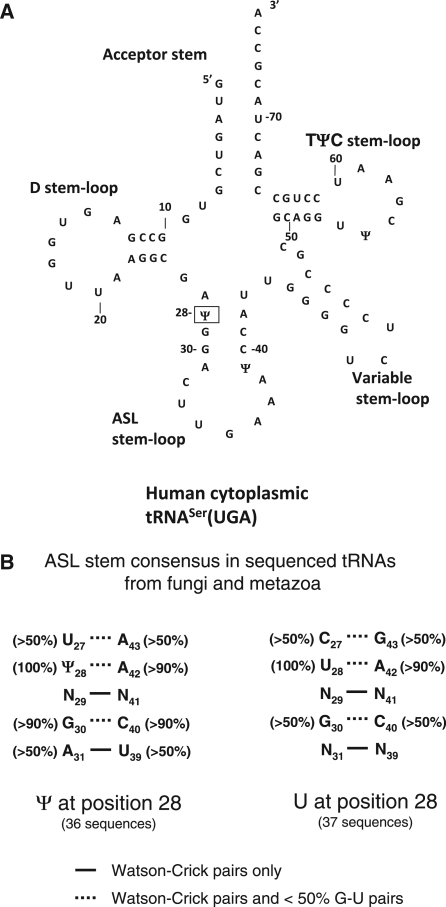
As an initial investigation into the substrate requirements for hPus1p, it was important to choose a known Pus1p substrate tRNA that has only one site modified by the enzyme (29,35), to simplify the analysis of the activity. A tRNA that is typical in structure and has a V stem-loop that was >6 nt, was preferred in order to be able to test the contribution of all structural components to substrate recognition by hPus1p. In addition, a tRNA with traditional Watson-Crick base-pairing in the ASL stem was desired. The tRNA sequence database, tRNAdb (56,57), was consulted to compile consensus sequences for the ASL stem where a Ψ or U is found at position 28 in sequenced tRNAs from the fungi and metazoan group in tRNAdb. At least 36 tRNA sequences were compiled to generate each consensus shown in Figure 1B. Several features are apparent in these two stems. First, with a Ψ at position 28, G30–C40 is the most conserved base pair (>90% conserved) but when a U is found at position 28, this sequence is not as well conserved. Therefore, one might expect that disruption of this base pair or replacement with the inverse (C30–G40) will affect modification. Second, for sequenced tRNAs that have a Ψ at 28, the preferred 27–43 bp is U27–A43, whereas with those tRNAs with a U at 28, the preferred base pair is C27–G43. Third, a A31–U39 base pair is more common in those ASL stems that have a Ψ at position 28 than those with a U at position 28. Finally, the base pairs 29–41 and 31–39, below the 28–42 bp, are Watson-Crick only but the rest of the base pairs in the stem can have G-U pairing in either consensus. In the selection of a tRNA, a good match to this consensus was preferred, and human tRNASer(UGA) meets most of the criteria. The only difference is that this tRNA has an A27–U43 base pair instead of a U–A base pair at that position (Figure 1A).
Figure 1.
Human tRNASer(UGA) sequence and secondary structure and ASL stem consensus sequence. (A) The sequence and proposed secondary structure of Human tRNASer(UGA) (50). The major aspects of the secondary structure are labeled on the diagram and it is numbered without including most of the Variable stem-loop. The single site of Pus1p modification, position 28, is boxed. (B) Consensus sequences for sequenced tRNAs from the fungi and metazoa group in the tRNAdb (57) that have a Ψ or a U at position 28. The figure is presented in the output style from tRNAdb. A solid line between nucleotides indicates Watson-Crick pairs only and a dash line indicates G–U pairs (<50%) are also found at this position.
The point mutations in the ASL of full-length tRNASer are diagrammed in Figure 2H and were obtained by site-directed mutagenesis of the plasmid containing wild-type human tRNASer [pHtS, gift from H. Gross; (50)] followed by in vitro transcription of linear templates (see ‘Materials and Methods’ section). Time course analyses of the in vitro activity of the substrate RNAs and hPus1p are presented in Figure 2 (panels A–G), with the same wild-type tRNASer curve in all panels (Figure 2A–G). The formation of Ψ in this tRNA by hPus1p is reduced to background levels (0.02 mol Ψ/mol RNA) if the U at position 28 is changed to C and A42 is changed to G to preserve base pairing (Table 2, second experiment). This shows that the only U converted to Ψ by hPus1p is the one at position 28. The panels in Figure 2 each contain the wild-type tRNASer curve and either two or three curves generated with tRNASer mutant substrates. This style allows for a simplification of the presentation and for the focus in most panels to be on 1 bp at a time.
Figure 2.
Time course experiments with single and double mutants in the ASL of human tRNASer. (A–G), 3H-labeled RNAs were incubated with hPus1p for the times indicated and the amount of Ψ formed assayed. Each panel has the same time course of wild-type (wt) tRNASer values. Each mutant is plotted separately and standard deviation (SD) bars are displayed for all time points. (H) Diagram of ASL point mutations made in the context of full-length tRNASer that are presented in panels A–G.
Table 2.
Activity of hPus1p on tRNASer(UGA) point and deletion mutants and mini-substrates
| RNA substrate | mole Ψ/mole RNA (SD)a | Percent of wild-type (SD) |
|---|---|---|
| Wild-type human tRNASer | 0.60 (0.08) | 100 (13) |
| Δ TΨC stem-loop (deleted nucleotides 59–73) | 0.09 (0.05) | 15 (8) |
| Δ Variable stem-loop (deleted nucleotides 45–56) | 0.40 (0.08) | 67 (13) |
| Wild-type human tRNASer | 0.43 (0.07) | 100 (16) |
| 28C/42G (no Ψ formation expected) | 0.02 (0) | 5 (0) |
| Δ D stem-loop (deleted nucleotides 11–24) | 0.44 (0.16) | 102 (37) |
| Δ D stem-loop with 28C/42G | 0.01 (0.01) | 2 (2) |
| Wild-type human tRNASer | 0.68 (0.02) | 100 (3) |
| ASL stem loop alone (includes nucleotides 26–43) | 0.02 (0.01) | 3 (1) |
| ASL & D stem-loops (includes nucleotides 10–43) | 0.07 (0.01) | 10 (1) |
| ASL & V stem-loops (includes nucleotides 26–47) | 0.05 (<0.01) | 7 (1) |
| ASL, D, & V stem-loops (includes nucleotides 10–47) | 0.14 (<0.01) | 21 (1) |
| ASL, V, & TΨC stem-loops (includes nucleotides 26–65) | 0.48 (0.04) | 71 (6) |
| ASL, D, V, & TΨC stem-loops (includes nucleotides 7–65) | 0.64 (0.16) | 94 (24) |
aActivities are from 2-h incubations, an average of two assays, with duplicate samples, and reported as mol Ψ/mol RNA and as the percent of the level seen with wild-type tRNASer(UGA) with the standard deviation (SD) also reported as percent of wild-type. These results are grouped as separate experiments with a separate wild-type control for each experiment.
When the A at 27 is changed to C or G (Figure 2A), the levels of Ψ formed are not significantly different from wild-type tRNASer. The curve that results when the U at 43 is changed to G is distinct from the wild-type tRNASer curve but the difference is not statistically significant because of the relatively large error bars (SD).
Mutation of the A at the position across from U28 to either C or G has little effect on time course of Ψ formed (Figure 2B). Even though the A at position 42 is preferred (Figure 1B), the formation of Ψ28 does not require canonical or even U–G base pairing, something unanticipated given the consensus sequence (Figure 1B).
When G29 is changed to C or C41 is changed to G (Figure 2C), there is a significant decrease in Ψ formation for both mutations in the formation of Ψ at 28. However, when the base pair is reformed, but is now C-G in the case of the double mutant 29C/41G, the curve obtained is indistinguishable from that of wild-type tRNASer (Figure 2F). This introduction of a base-paired pyrimidine 3′ to U28 did not enhance the formation of Ψ in the substrate, an expectation if a pyrimidine is preferred at position 29, as suggested in the Introduction.
The consensus sequence shown in Figure 1B revealed the G30–C40 base pair to be highly conserved in the putative Pus1p substrates with a Ψ at position 28. It was expected that disruption of this base pair would affect the formation of Ψ by hPus1p. In fact, the 30C and 40G mutants have significantly lower amounts of Ψ formed than wild-type tRNASer (Figure 2D). If the base pair is reformed but inverted, as with the 30C/40G double mutant, the time course of Ψ formed is the same as that of wild-type tRNASer (Figure 2F). This result argues against the requirement for a G at position 30, even though the Ψ28 consensus suggests it (Figure 1B).
When the base pair at A31–U39 is disrupted by either the 31C or 39G point mutations, the results are interesting. The curve of Ψ formation seen with the 31C substrate is significantly different from wild-type tRNASer (Figure 2E), but with the 39G mutant the curve mimics that seen with wild-type tRNASer (Figure 2E). Reforming the base pair with the 31C/39G double mutant results in a curve that matches that of wild-type tRNASer (Figure 2F).
The only difference between the Ψ28 consensus stem (Figure 1B) and the stem of the tRNASer (Figure 1A) is the 27–43 bp, which is A–U in tRNASer. The Ψ formation curves for the 27C, 27G and 43G mutants were not significantly different from wild-type tRNASer (Figure 2A). This would suggest that the enzyme does not require a base pair at 27–43, and may in fact be indifferent to the presence of a base pair. That statement was supported by the Ψ formation curve seen with the 27G/43G double mutant, which was not significantly different from the curve seen with wild-type tRNASer (Figure 2G), even though there is no canonical base pair formed. The results with the double mutant 27C/43G, which would form a stronger base pair than the wild-type A–U, are also not significantly different from wild-type tRNASer (Figure 2G) even though this mutant now mimics the U28 consensus in Figure 1B.
Equilibrium dissociation constants (Kd) were determined for wild-type tRNASer and the single- and double-mutant substrates found in the panels of Figure 2. The means and standard deviations of two separate Kd determinations are presented in Table 1 and that of wild-type tRNASer (240 nM) agrees with a previous determination of the Kd (250 nM) of hPus1p and yeast pre-tRNAIle (29). All of the mutants have a higher mean Kd than wild-type, with all but 41G and 43G >300 nM. There is not a correlation between the Ψ formation levels and the Kds for the mutants. It is interesting to note that a tRNA that is not a substrate for hPus1p, such as the 28C/42G double mutant, binds to the enzyme with a mean Kd of 332 nM. This was also as seen previously with yeast Pus1p and a non-substrate tRNA (31).
Table 1.
Equilibrium dissociation constants (Kd) of hPus1p with various RNAs
| RNA substrate | Kd Mean (SD)a |
|---|---|
| Wild-type tRNASer | 240 (14) nM |
| Single point mutants in tRNASer | |
| 27C | 328 (46) nM |
| 27G | 370 (14) nM |
| 29C | 408 (11) nM |
| 30C | 335 (120) nM |
| 31C | 358 (53) nM |
| 39G | 380 (28) nM |
| 40G | 305 (49) nM |
| 41G | 282 (18) nM |
| 42C | 360 (57) nM |
| 42G | 338 (46) nM |
| 43G | 248 (32) nM |
| Double mutants in tRNASer | |
| 27C/43G | 302 (39) nM |
| 27G/43G | 375 (7) nM |
| 28C/42G | 332 (18) nM |
| 29C/41G | 328 (74) nM |
| 30G/40C | 345 (21) nM |
| 31C/39G | 315 (92) nM |
aListed are the means and standard deviations (SD) of two separate binding experiments of hPus1p and the substrates listed (see ‘Materials and Methods’ section for details).
In order to identify additional recognition elements of the hPus1p substrate, stem or stem-loop deletions of, and mini-substrates derived from tRNASer, were tested in the Ψ formation assay. These mutant substrates were generated either by site-directed mutagenesis of pHtS and subsequent in vitro transcription or by in vitro transcription using oligodeoxynucleotide templates [see ‘Materials and Methods’ section; (53)]. It is understood that, with the deletion of whole stem-loops or the creation of mini-substrates, the tertiary structure of the entire tRNA is affected, but surprisingly, many of these mutants can function as substrates for hPus1p. The results for 120 min incubations are listed in Table 2 relative to the activity seen with wild-type tRNASer. If the D stem-loop is missing in the substrate, there is no decrease in hPus1p activity at position 28. If the U at position 28 in the ΔD stem-loop mutant is changed to C (and the A at 42 is changed to G) so that Ψ cannot be formed at position 28, there is essentially no Ψ formation (2% of wild-type tRNASer Ψ formation, see Table 2). This result indicates the level of Ψ formation seen with the ΔD stem-loop mutant is not due to Ψ formation at an additional site created by an altered secondary structure in the deletion mutant. If the TΨC stem-loop is missing from the tRNA, there is a significant reduction in the formation of Ψ at U28 (15% of wild-type tRNASer levels, Table 2) but the reduction is not as substantial when the V stem-loop is missing (67% of wild-type, Table 2).
When a mini-substrate of just the ASL stem-loop was incubated with hPus1p, Ψ formation was at background levels for 2 h of incubation (3% of wild-type levels, Table 2). In addition, if either the D stem-loop or the V stem-loop was combined with the ASL stem-loop, there was little increase in the amount of Ψ formed (10% and 7%, respectively, of the Ψ formation seen in 120 min with wild-type tRNASer, Table 2). This implies that adding a single stem-loop to the 5′ or 3′ side of the ASL stem-loop does not constitute an efficient substrate for hPus1p. If both the D and V stem-loops are added to the ASL stem-loop there is a noticeable increase in Ψ formed but it is still low (21%) compared to wild-type tRNASer. When the TΨC stem-loop was combined with the V and ASL stem-loops, there was a substantial level of hPus1p activity (71% of wild-type tRNASer levels of Ψ). With just the acceptor stem missing from tRNASer (ASL, D, V and TΨC stem-loops present), the activity is essentially the same as with wild-type tRNASer (94% of wild-type Ψ formation at 120 min, Table 2).
These results suggest that the secondary structure of the tRNASer is very important for the modification at position 28, and a minimal substrate could be created by simply connecting the ASL and TΨC stem-loop as diagrammed (Figure 3A). The numbering on the diagram is consistent with full-length tRNASer to allow comparisons with the results reported in Figure 2 and Table 2. The level of Ψ formation at position 28 in this minimal substrate (wild-type ASL & TΨC mini-substrate) was ~0.60 mol Ψ/mol RNA after 2 h of incubation, a substantial level of modification (Table 3). A mini-substrate incorporating a 28C/42G double mutation was used in a separate experiment with little or no Ψ formation observed (Table 3), providing evidence that in this mini-substrate, the U at position 28 is the only one being converted to Ψ, as expected.
Figure 3.
ASL & TΨC stem-loop mini-substrate and mutations. (A) Predicted structure of the ASL & TΨC stem loop mini-substrate, retaining the numbering found on the full-length tRNASer and indicating the position of the Ψ formed. The structure for the Y for Y R for R stem mini-substrate mutant and the C28 mutant are also shown. (B) Diagram of the possible base pairing in the ASL stem mutants in the mini-substrate listed in Table 3. The activities in percent of the level observed with wild-type ASL & TΨC stem-loop mini-substrate without the SD are listed above the diagrams. The mutations found in each stem are listed below the diagrams.
Table 3.
Activity of hPus1p on the wild-type ASL & TΨC stem-loop mini-substrate and mutants
| RNA substrate | mole Ψ/mole RNA (SD)a | mole Ψ/mole RNA (SD) | Percent of wild-type (SD) |
|---|---|---|---|
| wild-type ASL & TΨC stem-loop mini-substrate | 0.60 (0.11) | 0.54 (0.16) | 100 (18) (30) |
| 28C/42G (no U at expected site of Ψ formation) | 0.01 (0.01) | 2 (2) | |
| 27C/42C | 0.48 (0.04) | 80 (7) | |
| 27C/39G | 0.24 (0.05) | 40 (8) | |
| 27C/29C/39G | 0.10 (0.03) | 17 (5) | |
| 27C/29C/42C | 0.04 (0.01) | 7 (2) | |
| 27C/39G/40G | <0.01 (0.01) | 2 (2) | |
| 27C/29C/39G/40G | 0.02 (0.03) | 3 (5) | |
| 27C/29C/39G/40G/42C | 0.02 (<0.01) | 3 (2) | |
| Δ55 & 56 | 0.37 (0.10) | 62 (17) | |
| Δ55–58 | 0.17 (0.06) | 28 (10) | |
| GCAA inserted between 55 and 56 | 0.46 (0.04) | 77 (7) | |
| wild-type ASL & TΨC stem-loop mini-substrate | 0.30 (0.04) | 100 (13) | |
| GGACG starting at 61 | 0.22 (0.03) | 73 (10) | |
| AAGCUUA in TΨC loop | 0.34 (0.04) | 113 (13) |
aActivities are from 2-h incubations, an average of two assays with duplicate samples, and reported as mol Ψ/mol RNA and as the percent of the level seen with wild-type ASL & TΨC stem loop mini substrate, with the standard deviation (SD) also reported as percent of wild-type. These results are grouped as separate experiments with separate controls for each experiment. All of the listed mutants are in the context of the ASL & TΨC stem-loop mini-substrate.
Concentrating on the ASL stem, a number of mutant substrates were synthesized that pick up where the previous mutations in the full length tRNA (Figure 2 and Table 2) left off. The ASL stem of tRNASer has five Watson-Crick base pairs and the mini-substrate mutants listed in Table 3 were designed to eliminate two or more of those base pairs in each mutant. The base pairs that may still form in each mutant are diagrammed in Figure 3B and the levels of activity, as a percentage of the activity observed with the wild-type ASL & TΨC mini-substrate, are given in Table 3 and Figure 3B. When there is no base-pairing in the stem there is little formation of Ψ, and the possibility of 1 or even 2 bp in the stem, does not substantially elevate the amount of Ψ formed. However, with the 27 C/42C mutant substrate, which has 3 bp in the stem below the site of modification, 80% of the Ψ level seen with the wild-type ASL & TΨC mini-substrate was observed. The level of Ψ formation by hPus1p on the 27C/39G mutant substrate was only 40%, even though it too has 3 bp in the stem.
With respect to the TΨC stem-loop, if the nucleotides at 55 and 56 are deleted in this ASL & TΨC mini-substrate, there is a decrease in Ψ formation relative to the wild-type ASL & TΨC mini-substrate (62% of wild-type tRNASer, Table 3). If the size of the TΨC loop is reduced further (Δ55–58) the resulting substrate is a poor one, just 28% of wild-type levels of Ψ were observed (Table 3). If 4 nt (GCAA) are inserted into the TΨC loop to make it larger, the substrate still allows for a significant amount of Ψ to be formed (77% of wild-type ASL & TΨC mini-substrate levels). In a separate experiment, if Watson-Crick base-pairing is disrupted in the TΨC stem, with the sequence GGACG starting at nucleotide 61, the level of hPus1p activity was also substantial (73% or wild-type ASL & TΨC mini-substrate levels), suggesting a stem loop is not required. If the sequence of the TΨC loop was replaced with its complement (AAGCUUA), the amount of Ψ formed at position 28 was 113% of wild-type ASL & TΨC mini-substrate levels, suggesting that any loop of at least 5 nt will suffice.
To summarize the results from the mutation and deletion studies, the levels of Ψ formation with the single and double mutants in the context of the full-length tRNASer suggest that the sequence of the ASL stem is not an absolute determinant of hPus1p activity, except of course that there must be a U at position 28 to modify (Tables 2 and 3). The preference for a pyrimidine 3′ to the site of modification (at position 29; see ‘Introduction’ section) was not confirmed since even though the 29 C single mutant showed a decreased amount of Ψ formed and the double mutant 29 C/41 G exhibited an almost identical curve as wild-type tRNASer, suggesting that a pyrimidine is not preferred, only the base pair. The point mutations in full-length tRNASer show that the base pairing below the modified position (U28) affects Ψ formation, whereas the 27 A–U43 base pair is dispensable. In addition, the modified U28 does not need to participate in base pairing. The results with the ASL & TΨC stem loop mini-substrate mutants also showed that the base pairs below U28 were the most critical. The sequence of the stem nucleotides is of less importance than the presence of base pairs, which agrees with the fact that known substrates of Pus1p exhibit no absolute sequence requirement (26,27,33–35). In terms of the requirements for a particular secondary structure of tRNASer, the D-stem loop and acceptor stem are dispensable for Ψ formation at U28. In fact, a single stem loop, 3′ to the ASL, is all that is required for substantial Ψ formation at position 28. The sequence, and possibly the structure of this additional RNA, is not constrained.
Given the above summary of the mutation experiments, a mini-substrate where all of the nucleotides in the ASL stem have been changed, except of course for the U at position 28 which is modified, should be a substrate for hPus1p. A substrate fitting this description (Figure 3A), along with another where the U at position 28 was changed to C, were incubated with hPus1p and assayed for activity. The Y for Y R for R stem mini-substrate mutant has A replaced by G, G replaced by A, C replaced by U, and U replaced by C when compared with the wild-type ASL & TΨC mini-substrate, except at position 28. The 28 C mutant has complete replacement of the stem sequence, and should not be modified by hPus1p. The results with these substrates, as well as with the wild-type ASL & TΨC mini-substrate, are presented in Table 4 and show that this mutant is modified by hPus1p (0.33 mol Ψ/mol RNA) at 80% of wild-type levels for this experiment. In addition, the 28C mutant is not modified (0.01 mol Ψ/mol RNA; 2% of wild-type levels), proving that only the U at position 28 is modified. So, even though all the ASL stem nucleotides in the mutant are different from the wild-type tRNASer ASL stem, and there is a U28–G42 base pair in the mutant, the U at position 28 is converted to Ψ at near wild-type levels.
Table 4.
Activity of hPus1p with wild-type ASL & TΨC stem-loop mini-substrate and stem sequence mini-substrate mutants
| RNA substrate | mole Ψ/mole RNA (SD)a | Percent of wild-type (SD) |
|---|---|---|
| Wild-type ASL & TΨC stem-loop mini-substrate | 0.41 (0.05) | 100 (12) |
| Y for Y R for R stem mini-substrate mutant 27G/29A/30A/31G/39C/40U/41U/42G/43C | 0.33 (0.02) | 80 (5) |
| Y for Y R for R stem mini-substrate 28C mutant 27G/28C/29A/30A/31G/39C/40U/41U/42G/43C | 0.01 (0.01) | 2 (2) |
aActivities are from 2-h incubations, an average of three separate assays, and reported as mol Ψ/mol RNA and as the percent of the level seen with wild-type ASL & TΨC stem loop mini substrate, with the standard deviation (SD) also reported as percent of wild-type. All of the listed mutants are in the context of the ASL & TΨC stem-loop mini-substrate.
DISCUSSION
A number of studies have determined the sites in tRNAs that are modified by Pus1p (26,27,29,33,34). Yeast and mammalian Pus1p are the most extensively studied and in vivo and in vitro, the enzymes are responsible for the formation of Ψ at positions 27 and 28 in many tRNAs, at 34 and 36 in certain intron containing tRNAs, and at position 1 in a few substrates (29,33,34). The yeast enzyme can also modify uridine at positions 26, 65 and 67 in vivo but not in vitro (34). When the mouse enzyme was expressed in a Δpus1 yeast strain, position 26 was not modified, and the modification states of positions 65 and 67 were not determined (33). Sequenced human tRNAs do not have Ψ at positions 26, 65, or 67 (58), so it is unlikely mPus1p or hPus1p exhibit activity at these sites in vivo. The mammalian enzymes do modify position 30 in a few tRNAs but the yeast enzyme lacks this activity (29,33).
For the purposes of the discussion of the current results, the modification at positions 27 and 28 will be considered, since these are formed in vitro in a number of substrates by the mammalian enzymes (27,33,35). Nearly all (17 out of 18) of the sequenced yeast and human tRNAs that have a U at position 27 have converted it to Ψ (58). However, in the six sequenced yeast tRNAs that have a U at position 28, the three that have a 27C–43G base pair lack Ψ at position 28, whereas, the other three have a 27Ψ–43A base pair and a Ψ at position 28 (58). This completely agrees with the two consensus sequences shown in Figure 1B. In the sequenced human tRNAs with a U at 28 (eight total) this correlation does not hold up completely. Three of the tRNAs have a 27Ψ–43A base pair, two have a Ψ at position 28, but one does not. A human tRNA with an 27A–43U base pair does have a Ψ at position 28 (58), suggesting that an U–A or A–U base pair at 27–43 allows for Ψ formation at 28, whereas a 27C–43G base pair is not conducive to Ψ formation at position 28. However, with the four remaining tRNAs, which have a 27C–43G base pair, two have a Ψ at position 28 and two do not (58). The current finding that the loss of base pairing at, or the presence of, a C–G base pair at 27–43 does not significantly affect the formation of Ψ at position 28 in tRNASer, correlates with the data from human tRNAs. Human mitochondrial tRNALys is known to be a substrate of hPus1p in vivo (41) but the sequence of the stem only matches the Ψ28 consensus at the 27–43 and 28–42 bp. Finally, the Y for Y R for R stem mini-substrate mutant is an efficient substrate for hPus1p in vitro, even though the sequence in the stem does not match the consensus at any position except 28 (Figure 3A and Table 4). There must be other determinants allowing for or suppressing Ψ formation in these other tRNAs with a U at 28.
The uridine to be modified does not have to participate in Watson-Crick base-pairing but nucleotides participating in base-pairing 3′ to the modified position appear to be required to constitute an efficient substrate. These results correlate well with the observation that, in vivo, yeast Pus1p formed Ψ at position 26 in yeast tRNATrp(CCA), where the uridine is in the hinge between the D stem-loop and the ASL, and is followed by 5 bp in the ASL stem (33). In addition, both yeast and mouse Pus1p modify the uridine in position 1 on yeast tRNAArg(ACG), with base pairing present in the acceptor stem 3′ to the position of modification (33). And finally, yeast and mammalian Pus1p modifies several uridines in the extensively base-paired ASL of yeast pre-tRNAIle(UAU) in vitro and in vivo (27,29,33).
The lack of a strict substrate sequence requirement for hPus1p can be contrasted with the rigid recognition motif of yeast Pseudouridine synthase 7 (Pus7p), another synthase that can recognize more than one site in tRNA and other types of RNA (44). With Pus7p there is an absolute requirement for a U at −2 and an A at +1 relative to the uridine modified (44). Of the sequenced yeast and human tRNAs, all that have the proscribed Pus7p recognition sequence (17 total) have a Ψ at position 13 or 35 (58). The other site-specific Ψ synthases modify only one site in tRNAs (Pseudouridine synthases 4/10, 6, 8/9) or one small region in many tRNAs (Pus3p) (48,49,59–62). With these latter Ψ synthases the local tRNA structure appears to be a determining factor for substrate recognition when one considers the results of TruA (Pus3p homolog) and TruB (Pseudouridine synthase 4 homolog) structural studies (46,47,63,64).
Given the requirement that the uridine to be modified be in a base-paired stem, why is the uridine at position 44 in yeast U2 snRNA modified by Pus1p (33) when that position is not considered to be in a region that is base paired? Yeast Pus1p can modify U2 snRNA in vitro and in vivo, but mPus1p only modifies U44 on U2 snRNA in vivo, in a Δpus1 strain (33,36). An alternative structure, perhaps in the ribonucleoprotein particle where U44 is base-paired, may form in vivo. It is also possible that another RNA, such as a snoRNA, base pairs in the region of U44 and forms a stem that can be recognized by yeast or mouse Pus1p. It is interesting to note that Ψ formation in U2 snRNA in C. elegans is not dependent on Pus1p activity (28).
The results with mutations in the TΨC stem-loop suggest that there is a minimum size requirement for this loop but the actual sequence, or base pairing in the stem, are not critical. A minimum size requirement may be a reason that the V stem-loop of tRNASer(UGA) does not substitute for the TΨC stem-loop when combined with the ASL. The V stem-loop in tRNASer(UGA) is predicted to have only 3 nt in the loop. The fact that the TΨC stem-loop is separated from the ASL by 4 nt in the mini-substrate and the V stem-loop has no separation, may also be a factor in why the TΨC stem-loop can participate in creating a mini-substrate that is recognized by hPus1p.
The only Ψ synthase in the TruA family that has been crystallized is bacterial TruA (see above) and the structures were obtained in the presence of substrate tRNAs (46,47). TruA is a dimer and makes contact with the ASL (positions 38–40 are modified if a U is present at those sites), the D stem, and the ‘elbow’ where the D and TΨC loops interact. The V loop or stem-loop and the acceptor stem did not interact with TruA (47). In this report the D stem-loop of tRNASer(UGA) was found to be dispensable for hPus1p activity, as was the acceptor stem, but the absence of the V stem-loop from the tRNA does reduce the amount of Ψ formed. In this tRNA substrate, the TΨC stem-loop appears to be the more critical component for recognition by hPus1p. Therefore, even though there is considerable homology between TruA and hPus1p, the substrate contacts with the protein are probably different. Yeast Pus1p has been shown to be a monomer when bound to tRNA (30) and it is reasonable to assume the substrate/enzyme interactions will be fundamentally different between TruA and Pus1p. Crystal structures for any of the Pus1p proteins in the presence of tRNA substrates will go a long way towards reconciling these differences.
With what appear to be relatively lax requirements for substrate recognition in tRNASer, what prevents other uridines, such as those at 39, 43, 44, 63, or 70 in tRNASer (Figure 1A) from being modified by hPus1p in vitro? Modifications in the ASL (26, 27, 28, 30) of all natural substrates of Pus1p are only found on the 5′ side of the stem. This specificity is most likely due to the presentation of this side of the ASL to the enzyme's active site cleft, in much the same way the opposite side of the stem is presented to TruA for modification of positions 38–40 (47). This flipping of the orientation of the substrate in the active site cleft could explain why the TΨC stem-loop is a critical part of the recognition motif for hPus1p rather than the D stem-loop. These other sites that have uridines in a stem are not recognized because they are not presented in a conformation compatible with placing the uridine in the active site.
Additional natural and mini-substrates will need to be studied to refine the parameters of hPus1p substrate recognition to construct a recognition site that can be used to narrow down the possible modification sites in SRA, a relatively large, non-tRNA substrate of Pus1p, where most of the actual Ψ residues have not been identified (37,38), or to identify additional substrates. The known substrates of Pus1p may just be the tip of the iceberg of actual RNA substrates for this enigmatic enzyme.
FUNDING
National Institutes of Health (grant number DK074368-01); South Carolina Honors College at the University of South Carolina (Undergraduate Research Fellowship and Thesis Preparation grants to B.S.S.). Funding for open access charge: university funds.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
The plasmid (pHtS) for the synthesis of human tRNASer(UGA) and the template for mutations of the full-length tRNASer(UGA) was a gift of H. Gross.
REFERENCES
- 1.Auffinger P, Westhof E. In: Modification and Editing of RNA. Grosjean H, Benne R, editors. Washinton, D.C.: ASM Press; 1998. pp. 569–576. [Google Scholar]
- 2.Massenet SA, Mougin A, Branlant C. In: Modification and Editing of RNA. Grosjean H, Benne R, editors. Washington, D.C.: ASM Press; 1998. pp. 201–227. [Google Scholar]
- 3.Ofengand J, Fournier MJ. In: Modification and Editing of RNA. Grosjean H, Benne R, editors. Washington, D.C.: ASM Press; 1998. pp. 229–253. [Google Scholar]
- 4.Hamma T, Ferre-D'Amare AR. Pseudouridine synthases. Chem. Biol. 2006;13:1125–1135. doi: 10.1016/j.chembiol.2006.09.009. [DOI] [PubMed] [Google Scholar]
- 5.Davis DR. Stabilization of RNA stacking by pseudouridine. Nucleic Acids Res. 1995;23:5020–5026. doi: 10.1093/nar/23.24.5020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Davis DR, Veltri CA, Nielsen L. An RNA model system for investigation of pseudouridine stabilization of the codon-anticodon interaction in tRNALys, tRNAHis and tRNATyr. J. Biomol. Struct. Dyn. 1998;15:1121–1132. doi: 10.1080/07391102.1998.10509006. [DOI] [PubMed] [Google Scholar]
- 7.Durant PC, Davis DR. The effect of pseudouridine and pH on the structure and dynamics of the anticodon stem-loop of tRNA(Lys,3) Nucleic Acids Symp. Ser. 1997;36:56–57. [PubMed] [Google Scholar]
- 8.Durant PC, Davis DR. Stabilization of the anticodon stem-loop of tRNALys,3 by an A+-C base- pair and by pseudouridine. J. Mol. Biol. 1999;285:115–131. doi: 10.1006/jmbi.1998.2297. [DOI] [PubMed] [Google Scholar]
- 9.Zerfass K, Beier H. Pseudouridine in the anticodon G psi A of plant cytoplasmic tRNA(Tyr) is required for UAG and UAA suppression in the TMV-specific context. Nucleic Acids Res. 1992;20:5911–5918. doi: 10.1093/nar/20.22.5911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Perret V, Garcia A, Grosjean H, Ebel JP, Florentz C, Giege R. Relaxation of a transfer RNA specificity by removal of modified nucleotides. Nature. 1990;344:787–789. doi: 10.1038/344787a0. [DOI] [PubMed] [Google Scholar]
- 11.Lecointe F, Namy O, Hatin I, Simos G, Rousset J-P, Grosjean H. Lack of pseudouridine 38/39 in the anticodon arm of yeast cytoplasmic tRNA decreases in vivo recoding efficiency. J. Biol. Chem. 2002;277:30445–30453. doi: 10.1074/jbc.M203456200. [DOI] [PubMed] [Google Scholar]
- 12.Segault V, Will CL, Sproat BS, Luhrmann R. In vitro reconstitution of mammalian U2 and U5 snRNPs active in splicing: Sm proteins are functionally interchangeable and are essential for the formation of functional U2 and U5 snRNPs. EMBO J. 1995;14:4010–4021. doi: 10.1002/j.1460-2075.1995.tb00072.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yu YT, Shu MD, Steitz JA. Modifications of U2 snRNA are required for snRNP assembly and pre-mRNA splicing. EMBO J. 1998;17:5783–5795. doi: 10.1093/emboj/17.19.5783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Newby MI, Greenbaum NL. A conserved pseudouridine modification in eukaryotic U2 snRNA induces a change in branch-site architecture. RNA. 2001;7:833–845. doi: 10.1017/s1355838201002308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhao X, Yu Y-T. Pseudouridine in and near the branch site recognition region of U2 snRNA are required for snRNP biogenesis and pre-mRNA splicing in Xenopus oocytes. RNA. 2004;10:681–690. doi: 10.1261/rna.5159504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Donmez G, Hartmuth K, Luhrmann R. Modified nucleotides at the 5' end of human U2 snRNA are required for spliceosomal E-complex formation. RNA. 2004;10:1925–1933. doi: 10.1261/rna.7186504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Heiss NS, Knight SW, Vulliamy TJ, Klauck SM, Wiemann S, Mason PJ, Poustka A, Dokal I. X-linked dyskeratosis congenita is caused by mutations in a highly conserved gene with putative nucleolar functions. Nat. Genet. 1998;19:32–38. doi: 10.1038/ng0598-32. [DOI] [PubMed] [Google Scholar]
- 18.Ni J, Tien AL, Fournier MJ. Small nucleolar RNAs direct site-specific synthesis of pseudouridine in ribosomal RNA. Cell. 1997;89:565–573. doi: 10.1016/s0092-8674(00)80238-x. [DOI] [PubMed] [Google Scholar]
- 19.Ganot P, Bortolin ML, Kiss T. Site-specific pseudouridine formation in preribosomal RNA is guided by small nucleolar RNAs. Cell. 1997;89:799–809. doi: 10.1016/s0092-8674(00)80263-9. [DOI] [PubMed] [Google Scholar]
- 20.Zebarjadian Y, King T, Fournier MJ, Clarke L, Carbon J. Point mutations in yeast CBF5 can abolish in vivo pseudouridylation of rRNA. Mol. Cell Biol. 1999;19:7461–7472. doi: 10.1128/mcb.19.11.7461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kiss T. Small nucleolar RNA-guided post-transcriptional modification of cellular RNAs. EMBO J. 2001;20:3617–3622. doi: 10.1093/emboj/20.14.3617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Darzacg X, Jady BE, Verheggen C, Kiss AM, Bertrand E, Kiss T. Cajal body-specific small nuclear RNAs: a novel class of 2'-O-methylation and pseudouridylation guide RNAs. EMBO J. 2002;21:2746–2756. doi: 10.1093/emboj/21.11.2746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jady BE, Kiss T. A small nucleolar guide RNA functions both in 2'-O-ribose methylation and pseudouridylation of the U5 spliceosomal RNA. EMBO J. 2001;20:541–551. doi: 10.1093/emboj/20.3.541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhao X, Li Z-H, Terns RM, Terns MP, Yu Y-T. An H/ACA guide RNA directs U2 pseudouridylation at two different sites in the branch point recognition region in Xenopus oocytes. RNA. 2002;8:1515–1525. [PMC free article] [PubMed] [Google Scholar]
- 25.Kiss AM, Jady BE, Bertrand E, Kiss T. Human box H/ACA pseudouridylation guide RNA machinery. Mol. Cell. Biol. 2004;24:5797–5807. doi: 10.1128/MCB.24.13.5797-5807.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Simos G, Tekotte H, Grosjean H, Segref A, Sharma K, Tollervey D, Hurt EC. Nuclear pore proteins are involved in the biogenesis of functional tRNA. EMBO J. 1996;15:2270–2284. [PMC free article] [PubMed] [Google Scholar]
- 27.Chen J, Patton JR. Cloning and characterization of a mammalian pseudouridine synthase. RNA. 1999;5:409–419. doi: 10.1017/s1355838299981591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Patton JR, Padgett RW. Caenorhabditis elegans pseudouridine synthase 1 activity in vivo: tRNA is a substrate but not U2 small nuclear RNA. Biochem. J. 2003;372:595–602. doi: 10.1042/BJ20021938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sibert BS, Fischel-Ghodsian N, Patton JR. Partial activity is seen with many substitutions of highly conserved active site residues in human Pseudouridine synthase 1. RNA. 2008;14:1895–1906. doi: 10.1261/rna.984508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Arluison V, Batelier G, Ries-Kautt M, Grosjean H. RNA:pseudouridine synthetase Pus1 from Saccharomyces cerevisiae: oligomerization property and stoichiometry of the complex with yeast tRNA(Phe) Biochimie. 1999;81:751–756. doi: 10.1016/s0300-9084(99)80133-3. [DOI] [PubMed] [Google Scholar]
- 31.Arluison V, Buckle M, Grosjean H. Pseudouridine synthetase Pus1 of Saccharomyces cerevisiae: kinetic characterisation, tRNA structural requirement and real-time analysis of its complex with tRNA. J. Mol. Biol. 1999;289:491–502. doi: 10.1006/jmbi.1999.2789. [DOI] [PubMed] [Google Scholar]
- 32.Arluison V, Hountondji C, Robert B, Grosjean H. Transfer RNA-pseudouridine synthetase Pus1 of Saccharomyces cerevisiae contains one atom of zinc essential for its native conformation and tRNA recognition. Biochemistry. 1998;37:7268–7276. doi: 10.1021/bi972671o. [DOI] [PubMed] [Google Scholar]
- 33.Behm-Ansmant I, Massenet S, Immel F, Patton JR, Motorin Y, Branlant C. A previously unidentified activity of yeast and mouse RNA:pseudouridine synthases 1 (Pus1p) on tRNAs. RNA. 2006;12:1583–1593. doi: 10.1261/rna.100806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Motorin Y, Keith G, Simon C, Foiret D, Simos G, Hurt E, Grosjean H. The yeast tRNA:pseudouridine synthase Pus1p displays a multisite substrate specificity. RNA. 1998;4:856–869. doi: 10.1017/s1355838298980396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chen J, Patton JR. Mouse pseudouridine synthase 1: gene structure and alternative splicing of pre-mRNA. Biochem. J. 2000;352:465–473. [PMC free article] [PubMed] [Google Scholar]
- 36.Massenet S, Motorin Y, Lafontaine DL, Hurt EC, Grosjean H, Branlant C. Pseudouridine mapping in the Saccharomyces cerevisiae spliceosomal U small nuclear RNAs (snRNAs) reveals that pseudouridine synthase pus1p exhibits a dual substrate specificity for U2 snRNA and tRNA. Mol. Cell Biol. 1999;19:2142–2154. doi: 10.1128/mcb.19.3.2142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhao X, Patton JR, Davis SL, Florence B, Ames SJ, Spangaard RA. Regulation of nuclear receptor activity by a pseudouridine synthase through posttranscriptional modification of Steriod Receptor RNA Activator. Mol. Cell. 2004;15:549–558. doi: 10.1016/j.molcel.2004.06.044. [DOI] [PubMed] [Google Scholar]
- 38.Zhao X, Patton JR, Ghosh SK, Fischel-Ghodsian N, Shen L, Spanjaard RA. Pus3p- and Pus1p-dependent pseudouridylation of steroid receptor RNA activator controls a functional switch that regulates nuclear receptor signaling. Mol. Endocrinol. 2007;21:686–699. doi: 10.1210/me.2006-0414. [DOI] [PubMed] [Google Scholar]
- 39.Bykhovskaya Y, Casas K, Mengesha E, Inbal A, Fischel-Ghodsian N. Missense mutation in pseudouridine synthase 1 (PUS1) causes mitochondrial myopathy and sideroblastic anemia (MLASA) Am. J. Hum. Genet. 2004;74:1303–1308. doi: 10.1086/421530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zeharia A, Fischel-Ghodsian N, Casas K, Bykhovskaya Y, Tamari H, Lev D, Mimouni M, Lerman-Sagie T. Mitochondrial myopathy, sideroblastic anemia, and lactic acidosis: an autosomal recessive syndrome in Persian Jews caused by a mutation in the PUS1 gene. J. Child Neurol. 2005;20:449–452. doi: 10.1177/08830738050200051301. [DOI] [PubMed] [Google Scholar]
- 41.Patton JR, Bykhovskaya Y, Mengesha E, Bertolotto C, Fischel-Ghodsian N. Mitochondrial myopathy and sideroblastic anemia (MLASA): missense mutation in the pseudouridine synthase 1 (PUS1) gene is associated with the loss of tRNA pseudouridylation. J. Biol. Chem. 2005;280:19823–19828. doi: 10.1074/jbc.M500216200. [DOI] [PubMed] [Google Scholar]
- 42.Fernandez-Vizarra E, Berardinelli A, Valente L, Tiranti V, Zeviani M. Nonsense mutation in pseudouridylate synthase 1 (PUS1) in two brothers affected by myopathy, lactic acidosis and sideroblastic anaemia (MLASA) J. Med. Genet. 2007;44:173–180. doi: 10.1136/jmg.2006.045252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Grosjean H, Szweykowska-Kulinska Z, Motorin Y, Fasiolo F, Simos G. Intron-dependent enzymatic formation of modified nucleosides in eukaryotic tRNAs: a review. Biochimie. 1997;79:293–302. doi: 10.1016/s0300-9084(97)83517-1. [DOI] [PubMed] [Google Scholar]
- 44.Urban A, Behm-Ansmant I, Branlant C, Motorin Y. RNA sequence and two-dimensional structure features required for efficient substrate modification by the Saccharomyces cerevisiae RNA:{Psi}-synthase Pus7p. J. Biol. Chem. 2009;284:5845–5858. doi: 10.1074/jbc.M807986200. [DOI] [PubMed] [Google Scholar]
- 45.Patton JR, Padgett RW. Pseudouridine modification in Caenorhabditis elegans spliceosomal snRNAs: unique modifications are found in regions involved in snRNA-snRNA interactions. BMC Mol. Biol. 2005;6:20. doi: 10.1186/1471-2199-6-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Foster PG, Huang L, Santi DV, Stroud RM. The structural basis for tRNA recognition and pseudouridine formation by pseudouridine synthase I. Nat. Struct. Biol. 2000;7:23–27. doi: 10.1038/71219. [DOI] [PubMed] [Google Scholar]
- 47.Hur S, Stroud RM. How U38, 39, and 40 of many tRNAs become the targets for pseudouridylation by TruA. Mol. Cell. 2007;26:189–203. doi: 10.1016/j.molcel.2007.02.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lecointe F, Simos G, Sauer A, Hurt EC, Motorin Y, Grosjean H. Characterization of yeast protein Deg1 as pseudouridine synthase (Pus3) catalyzing the formation of psi 38 and psi 39 in tRNA anticodon loop. J. Biol. Chem. 1998;273:1316–1323. doi: 10.1074/jbc.273.3.1316. [DOI] [PubMed] [Google Scholar]
- 49.Chen J, Patton JR. Pseudouridine Synthase 3 from Mouse Modifies the Anticodon Loop of tRNA. Biochemistry. 2000;39:12723–12730. doi: 10.1021/bi001109m. [DOI] [PubMed] [Google Scholar]
- 50.Wu XQ, Gross HJ. The long extra arms of human tRNA((Ser)Sec) and tRNA(Ser) function as major identify elements for serylation in an orientation-dependent, but not sequence-specific manner. Nucleic Acids Res. 1993;21:5589–5594. doi: 10.1093/nar/21.24.5589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Melton DA, Krieg PA, Rebagliati MR, Maniatis T, Zinn K, Green MR. Efficient in vitro synthesis of biologically active RNA and RNA hybridization probes from plasmids containing a bacteriophage SP6 promoter. Nucleic Acids Res. 1984;12:7035–7056. doi: 10.1093/nar/12.18.7035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Milligan JF, Groebe DR, Witherell GW, Uhlenbeck OC. Oligoribonucleotide synthesis using T7 RNA polymerase and synthetic DNA templates. Nucleic Acids Res. 1987;15:8783–8798. doi: 10.1093/nar/15.21.8783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Milligan JF, Uhlenbeck OC. Synthesis of small RNAs using T7 RNA polymerase. Methods Enzymol. 1989;180:51–62. doi: 10.1016/0076-6879(89)80091-6. [DOI] [PubMed] [Google Scholar]
- 54.Cortese R, Kammen HO, Spengler SJ, Ames BN. Biosynthesis of pseudouridine in transfer ribonucleic acid. J. Biol. Chem. 1974;249:1103–1108. [PubMed] [Google Scholar]
- 55.Patton JR. Pseudouridine modification of U5 RNA in ribonucleoprotein particles assembled in vitro. Mol. Cell Biol. 1991;11:5998–6006. doi: 10.1128/mcb.11.12.5998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lowe TM, Eddy SR. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 1997;25:955–964. doi: 10.1093/nar/25.5.955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Juhling F, Morl M, Hartmann RK, Sprinzl M, Stadler PF, Putz J. tRNAdb 2009: compilation of tRNA sequences and tRNA genes. Nucleic Acids Res. 2009;37:D159–D162. doi: 10.1093/nar/gkn772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sprinzl M, Horn C, Brown M, Ioudovitch A, Steinberg S. Compilation of tRNA sequences and sequences of tRNA genes. Nucleic Acids Res. 1998;26:148–153. doi: 10.1093/nar/26.1.148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Becker HF, Motorin Y, Planta RJ, Grosjean H. The yeast gene YNL292w encodes a pseudouridine synthase (Pus4) catalyzing the formation of psi55 in both mitochondrial and cytoplasmic tRNAs. Nucleic Acids Res. 1997;25:4493–4499. doi: 10.1093/nar/25.22.4493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Roovers M, Hale C, Tricot C, Terns MP, Terns RM, Grosjean H, Droogmans L. Formation of the conserved pseudouridine at position 55 in archaeal tRNA. Nucleic Acids Res. 2006;34:4293–4301. doi: 10.1093/nar/gkl530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ansmant I, Motorin Y, Massenet S, Grosjean H, Branlant C. Identification and characterization of the tRNA:Psi 31-synthase (Pus6p) of Saccharomyces cerevisiae. J. Biol. Chem. 2001;276:34934–34940. doi: 10.1074/jbc.M103131200. [DOI] [PubMed] [Google Scholar]
- 62.Behm-Ansmant I, Grosjean H, Massenet S, Motorin Y, Branlant C. Pseudouridylation at position 32 of mitochondrial and cytoplasmic tRNAs requires two distinct enzymes in Saccharomyces cerevisiae. J. Biol. Chem. 2004;279:52998–53006. doi: 10.1074/jbc.M409581200. [DOI] [PubMed] [Google Scholar]
- 63.Hoang C, Ferre-D'Amare AR. Cocrystal structure of a tRNA Psi55 pseudouridine synthase: nucleotide flipping by an RNA-modifying enzyme. Cell. 2001;107:929–939. doi: 10.1016/s0092-8674(01)00618-3. [DOI] [PubMed] [Google Scholar]
- 64.Pan H, Agarwalla S, Moustakas DT, Finer-Moore J, Stroud RM. Structure of tRNA pseudouridine synthase TruB and its RNA complex: RNA recognition through a combination of rigid docking and induced fit. Proc. Natl Acad. Sci. USA. 2003;100:12648–12653. doi: 10.1073/pnas.2135585100. [DOI] [PMC free article] [PubMed] [Google Scholar]