Abstract
The precise nature of antisense transcripts in eukaryotes such as Saccharomyces cerevisiae remains elusive. Here we show that the 3′ regions of genes possess a promoter architecture, including a pre-initiation complex (PIC), which mirrors that at the 5′ region and which is much more pronounced at genes with a defined antisense transcript. Remarkably, for genes with an antisense transcript, average levels of PIC components at the 3′ region are ∼60% of those at the 5′ region. Moreover, at these genes, average levels of nascent antisense transcription are ∼45% of sense transcription. We find that this 3′ promoter architecture persists for highly transcribed antisense transcripts where there are only low levels of transcription in the divergent sense direction, suggesting that the 3′ regions of genes can drive antisense transcription independent of divergent sense transcription. To validate this, we insert short 3′ regions into the middle of other genes and find that they are capable of both initiating antisense transcripts and terminating sense transcripts. Our results suggest that antisense transcription can be regulated independently of divergent sense transcription in a PIC-dependent manner and we propose that regulated production of antisense transcripts represents a fundamental and widespread component of gene regulation.
INTRODUCTION
Genome-wide mapping of RNA transcripts in the budding yeast Saccharomyces cerevisiae has revealed an extensive array of transcripts that do not encode proteins, the product of an apparently pervasive mechanism of transcription in which individual genes can possess multiple, overlapping transcripts, giving rise to a genome that is heavily interleaved (1,2). Amongst these non-coding transcripts are a class that are transcribed from the 3′-ends of protein-coding genes, running antisense to the protein-coding sense transcript and terminating at the 5′-end of the gene (3–7), which have been shown to have a general regulatory effect on sense transcription, increasing gene expression variability (8).
The presence of such antisense transcripts raises the question of how they are initiated. Transcription initiation at the gene promoters requires a complex assembly of proteins collectively termed the ‘pre-initiation complex’ (PIC) upstream of the transcription start site (TSS) and within a nucleosome-depleted region (NDR) that allows access to the underlying DNA. If the initiation of antisense transcripts is driven by a similar mechanism then one would expect to find elements of the transcription initiation machinery within the 3′ regions of genes, and indeed there is evidence to suggest this is true. Components of the PIC, including TBP and TFIIB, have been found within the 3′ regions of certain genes, as have histone modifications thought to be specific to the 5′ region (9,10). Furthermore, NDRs have been reported in the 3′ regions of genes (9,11), although the veracity of these studies is complicated by their use of the enzyme micrococcal nuclease (MNase), given that the 3′ region is very A T rich and MNase has a strong preference for such sequences (12).
It has been suggested that antisense transcripts initiate from the promoters of downstream genes, in the vicinity of the TSS of the downstream gene (13). This has given rise to the idea that a single promoter, termed a ‘bidirectional promoter’, can direct transcription of both transcripts. What is unclear is whether antisense transcription is an inevitable consequence of transcription being intrinsically bidirectional, a possible consequence of transcriptional infidelity and which could perhaps explain why many antisense transcripts are rapidly degraded following transcription (4,14), or whether antisense transcription can be regulated independently of divergent sense transcription, i.e. whether their promoters, though in close proximity, can respond to distinct regulatory inputs. In support of the latter, the levels of sense and antisense transcription from bidirectional promoters have been shown to be only moderately correlated (15). What also remains unclear is whether bidirectional promoters consist of a single PIC, from which RNA polymerase II (Pol II) transcribes in both directions, or whether there are two divergently oriented PICs, as is the case in the model presented by Neil et al. (13).
Here we show that the 3′ regions of genes are marked by strikingly high average levels of PIC components, whose presence is associated with antisense initiation. Furthermore, we find that high levels of antisense transcription are supported by a pronounced promoter architecture comprised of the PIC components Spt15 (TBP) and Sua7 (TFIIB), an NDR and chromatin remodelling factors. We show evidence of two PICs being present at bidirectional promoters, suggesting that sense and antisense transcription from bidirectional promoters may arise from two distinct transcription complexes. Most notably, we find evidence for antisense transcripts that are transcribed to high levels in the absence of even moderate levels of sense transcription, and that this, too, requires a distinct PIC within an NDR. We propose that antisense transcription can be driven by PIC formation in the 3′ regions of genes independently of adjacent, divergent sense transcription and provide experimental validation of this model.
MATERIALS AND METHODS
Classification of genes
ORF start and end sites were obtained from the SGD browser (http://www.yeastgenome.org/). The transcription start and transcript termination sites (TSS and TTS, respectively), as determined by Nagalakshmi et al. (16), were then mapped to the ORFs. The 3′ region of a gene was defined as being delimited by a point 100 bp upstream of a gene's translation stop codon and by the border of the nearest downstream ORF (or 600 bp downstream of the stop codon otherwise). A gene was defined as not having a modestly transcribed gene in the vicinity of its 3′ region if there was no other ORF border (start or stop codon) within 1000 bp of the gene's TSS, belonging to a gene with a transcription value above 80 reads (Supplementary Figures S2 and S5; see below for an explanation of how a gene's transcription value was calculated). Genes labelled as dubious were considered for this purpose, but were otherwise excluded from all analyses. A complete list of genes together with their annotated ends is included in the Supplementary Data (Supplementary Table S2).
Collated transcript map
Transcript coordinates were obtained from Nagalakshmi et al. (16), Xu et al. (17), Yassour et al. (18), Granovskaia et al. (19) and Yassour et al. (20) and mapped to the S. cerevisiae genome. Transcripts were defined as being the same between different sources if both their start and end sites fell within 75 bp of one another and they were of the same orientation. Transcripts whose orientation was unknown were discarded for the purposes of this analysis. Antisense transcripts were defined as those transcripts whose TSS lay within the 3′ region of a gene (as defined above) and which ran antisense to that gene. Such genes were in turn defined as those possessing an antisense transcript. The collated list of transcripts is included in the Supplementary Data (Supplementary Table S1).
Genome-wide chromatin immunoprecipitation data
Genome-wide levels of Spt15, Sua7, Swr1 and Rsc9 were obtained from Venters and Pugh (10). Normalized nucleosome occupancy values were obtained from Kaplan et al. (11) and Fan et al. (21) to obtain the MNase-dependent and -independent occupancy, respectively. The genome-wide data sets were all normalized such that the mean of the data was 0 and the standard deviation was 1. Average levels around either the TSS or the TTS of a particular gene set were calculated by aligning the genes by either the TSS (for the 5′ region) or the TSS (for the 3′ region) and averaging the occupancy at each base. The resultant average occupancy was then smoothed with a 41-bp window. When aligning the protein-coding genes by their TSS or TTS ends the mapped ends determined by Nagalakshmi et al. (16) were used. When aligning the antisense transcripts by their TSS the annotated ends from the collated transcript map described above were used. To determine the levels of Spt15 and Sua7 in the 3′ region as a percentage of those in the 5′ region we divided the difference between the maximum and minimum average level in the 3′ region by the difference between the maximum and minimum average level in the 5′ region.
Calculating A T richness
The A T richness for a specific base was defined as the average frequency of A or T nucleotides within a 21-bp window centred on that base. The average A T richness around the TTS was determined by aligning all the genes in a given set by the TTS of their protein-coding transcript and averaging the A T richness at each base. The resultant average A T richness was then smoothed with a 41-bp window.
Transcription levels and Pol II density
Genome-wide levels of elongating Pol II (nascent transcription) were obtained from Churchman and Weissman (15). To quantify the level of transcription of a given transcript we used a method similar to that described previously (15). In summary, we calculated the sum of the read density in 500-bp windows, starting 50 bp upstream of the transcript start site and ending 700 bp downstream of the start site, or else the end of the transcript if it was shorter than 700 bp. The transcription level was defined as the maximum value obtained in this way. To determine the level of transcription of a gene's antisense strand for those genes without an antisense transcript we used the TTS of its protein-coding sense transcript in place of the TSS. For the purposes of determining if a gene was modestly transcribed (Supplementary Figures S2 and S5), it was necessary to determine the transcription level of all genes, even those without mapped TSSs (including dubious ORFs). For these genes we used a point 100 bp upstream of the start codon to calculate the transcription level. To obtain the average Pol II density across the antisense strand (Figure 1F) we aligned all genes by the TTS of their protein-coding sense transcript, as determined by Nagalakshmi et al. (16), and calculated the average number of reads at each nucleotide relative to the anchor point. The average Pol II density in the 5′ regions of the antisense transcripts was determined in the same manner, save that we also determined the average Pol II density upstream of the TSS and on the opposite strand, in order to assess divergent sense transcription (Figures 4D and 5A).
Figure 1.
Evidence for PIC formation at the 3′ region of protein-coding genes. (A) The average levels of Spt15 and Sua7 at gene 5′ regions, obtained by aligning genes by their transcription start site (TSS). Nucleosome occupancy as determined by Fan et al. (2010) is included for comparative purposes, and is discussed in greater detail in the text. (B) The average levels of Spt15 and Sua7 at the 3′ regions of genes with antisense transcripts, obtained by aligning genes by their transcript termination site (TTS). (C and D) Spt15 and Sua7 levels at the 3′ regions of genes, comparing genes with antisense transcripts to those without. P-values were calculated using the Wilcoxon rank-sum test, comparing the distribution of values at the maximum point of each averaged curve. Note that these relationships remained when those genes whose TTSs were close to other genes were excluded from the analysis (see Supplementary Figure S2). (E) A comparison of the average transcription levels of the sense and antisense strands of protein-coding genes. The antisense strands of genes with antisense transcripts are considered separately from those without. Error bars were calculated by bootstrapping (the standard deviation of the mean of 1000 bootstrap pseudoreplicates). (F) Average levels of Pol II density across the antisense strands of protein-coding genes aligned by their 3′-ends. Shown is the average Pol II density of those genes possessing 3’ initiating antisense transcripts compared with those genes that do not.
Figure 4.
3′ nucleosome occupancy determined using a MNase independent genome-wide assay (Fan et al. 2010). (A) The average occupancy at the 3′ region of those genes possessing antisense transcripts, compared with those genes that do not. P-values were calculated using the Wilcoxon rank-sum test and comparing the distribution of values at the minimum point of each averaged curve. (B) The transcription level of each protein-coding sense transcript plotted against the transcription level of its associated antisense transcript (if it has one). Shown is the Pearson correlation coefficient (−0.1). The Spearman correlation coefficient was −0.06.
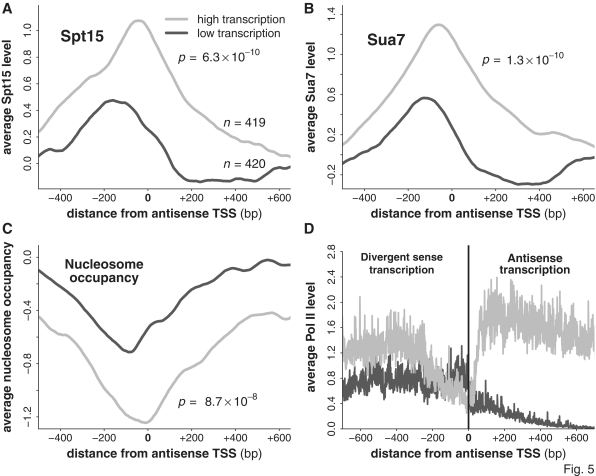
Figure 5.
Antisense initiation is supported by PIC formation upstream of the antisense TSS. (A–C) The average levels of Spt15, Sua7 and nucleosome occupancy around the TSS of both highly and lowly transcribed antisense transcripts. All P-values were calculated using the Wilcoxon rank-sum test, comparing the distribution of values at the maximum point of each averaged curve (the minimum point in the case of nucleosome occupancy). (D) The average level of nascent transcription around the TSS of the antisense transcripts. Shown left of the TSS is the average level of divergent sense transcription while on the right of the TSS is the average level of antisense transcription.
Transcription analysis and selecting for antisense transcripts with low divergent sense transcription
The highly transcribed group of antisense transcripts were defined as the top 20% of the data, based upon the transcription values obtained above. Before defining the lowly transcribed group the bottom 10% of the data was masked from consideration, to remove potential false-positive antisense transcripts for which we might expect no PIC formation to be present. The lowly transcribed group was then defined as the 20% above this. To determine antisense transcripts with low levels of divergent sense transcription we calculated the sum of the Pol II read density in an 800-bp window placed 50 bp upstream of the antisense TSS. From this we determined which antisense transcripts had low divergent sense transcription (the bottom 35% of the data). From this group, we then selected for those antisense transcripts which were not adjacent to the 5′-end of any non-Pol II transcribed elements. This group included rRNAs, tRNAs, snRNAs, snoRNAs, retrotransposons and transposable elements. Their coordinates were obtained from the SGD database. The highly and lowly transcribed groups were selected as described above, save that each group contained 25% of the total antisenses to obtain larger n values.
Transcription factor binding sites
The transcription factor binding site map was downloaded from the Fraenkel lab's website [http://fraenkel.mit.edu/improved_map/, (22)]. A binding site was defined as a ‘true’ binding site based upon the confidence of the chip–chromatin immunoprecipitation (ChIP) data (P < 0.005).
Analysing bidirectional promoters
Classes of bidirectional promoters were selected as described in the ‘Results’ section. The antisense transcripts considered were those defined above. Promoters were only considered bidirectional if both sense and antisense transcripts were transcribed beyond a certain threshold (20 reads).
Yeast strains and experimental methods
Experiments were done in yeast strain BY4741. The ScADH1 and AgTEF 3′ (terminator) regions used here are the sequences present in the vectors used to construct yeast gene deletions (23,24) and were engineered into GAL1 at +757 and into GAL10 at position +1453, after deletion of the region to +2007 to remove the internal bidirectional promoter of GAL10 (25), using modified DNA in which loxP sites (26) were inserted either after the ScADH1 insertion and at the end of the AgTEF terminator or before the AgTEF promoter and before the AgTEF terminator so that expression of Cre leaves just the ScADH1 terminator followed by a loxP site or the AgTEF terminator preceded by a loxP site in the locus. Note the results obtained with the ScADH1 terminator were the same whether the TEFKanMX6TEF cassette was present or not. Strains were grown in YPD and induced at O.D. 600 = 0.5 in YP +2% galactose for up to 3 h, total RNA prepared and separated on formaldehyde gels, blotted and hybridized with strand-specific DNA probes (see Supplementary Data and ‘Materials and Methods’ section for details).
RESULTS
The 3′-ends of genes show a pronounced promoter architecture that mirrors the 5′-end, and which is associated with antisense transcription
We sought to assess whether TBP and TFIIB, components of the PIC associated with transcription initiation, are a feature of the 3′ region of genes, and whether their presence correlates with antisense transcripts. We compiled a genome-wide transcript map from a range of RNA-seq and tiling array experiments performed in S. cerevisiae (16–20) and used this to provide a comprehensive map of antisense transcripts (Supplementary Table S1). This gave us a map consisting of over 20 000 protein-coding and non-coding transcripts, and allowed us to identify those genes which had one or more non-coding antisense transcripts initiating in the vicinity of their 3′ region (Supplementary Table S2). The 3′ region was defined as being delimited by a point 100 bp upstream of a gene's translation stop codon and by the border of the nearest downstream ORF (or 600 bp downstream of the stop codon otherwise). 1896 (36.5%) of genes with mapped 3′ transcript termination sites (TTSs) for their sense transcript were defined as possessing an antisense transcript, where antisense transcript refers to a non-coding transcript (ncRNA) from the compiled map defined above that initiates within the 3′ region of a verified gene and runs antisense to that gene.
To assess the average levels of PIC components Spt15 (TBP) and Sua7 (TFIIB) at the 3′ region, and compare this to their levels in 5′ promoter regions, we used the genome-wide data set from Venters and Pugh (10). When considering the set of genes with antisense transcripts (Supplementary Table S2), we found a substantial peak of both PIC components downstream of the 3′ TTS, which mirrored what was seen upstream of the 5′ TSS (Figure 1A and B). These peaks were substantially more pronounced at genes with an antisense transcript than for those genes without (Figure 1C and D), although interestingly a smaller but similar peak was still observed for the latter group. Other factors associated with transcription, namely Swr1, which deposits the histone variant H2A.Z at gene promoters (27) and Rsc9, a component of the RSC chromatin remodelling complex (28), were found to be significantly enriched within the 3′ regions of genes with antisense transcripts compared to those without (Supplementary Figure S1A and S1B). Our results demonstrate the presence of a promoter architecture at the 3′ region, which is associated with antisense transcripts and which is similar to that at the promoters of protein-coding genes. Next we asked whether the PIC observed at the 3′ region reflects the presence of adjacent protein-coding genes. We excluded from the gene set all protein-coding genes with another gene (in either orientation) within 1000 bp of the TTS which also showed evidence of being moderately transcribed (see ‘Materials and Methods’ section). This left a set of 1097 genes, of which 412 (37.6%, similar to the total gene set) had an antisense transcript and were significantly enriched for Spt15 (TBP) and Sua7 (TFIIB) in their 3′ region relative to the remaining 685 genes (Supplementary Figure S2). Thus, there is an association between PIC components at the 3′ region and antisense transcripts even when those genes with moderately transcribed protein-coding genes downstream are removed from the analysis, suggesting that the formation of a PIC at the 3′ region is not linked to the presence of a promoter of a downstream gene.
Levels of PIC components at the 3′ region reflect levels of nascent antisense transcription
We compared the average peaks of PIC components at the 5′ regions of all protein-coding genes with those at the 3′ regions of genes with antisense transcripts. Surprisingly, the average peaks of Spt15 and Sua7 at the 3′ regions of genes with antisense transcripts were more than half those at the 5′ regions (Spt15 levels were 62% of what they were at the 5′ regions, Sua7 levels were 60%; Figure 1A and B). We reasoned that this might reflect the fact that antisense transcripts are transcribed at higher levels than evident from steady-state levels, which are generally low due to transcript degradation (4,14). Thus, we sought to determine their average levels of transcription. We utilized a genome-wide map of elongating Pol II to obtain the level of nascent sense and antisense transcription and to assign a measure of transcription to each transcript [(15), see ‘Materials and Methods’ section). Remarkably, we found that the average level of antisense transcription, at genes with an antisense transcript, was 45% that of sense transcription at all protein-coding genes (Figure 1E). This is strikingly similar to the difference in average levels of PIC components between the 5′ region of all protein-coding genes and 3′ region of genes with antisense transcripts. We conclude that there is an association between levels of PIC components and levels of transcription for both the protein-coding sense and the non-coding antisense transcripts. In addition, the levels of PIC components at the 5′ and 3′ regions are much more similar than one might expect from steady-state transcript levels.
The small peaks of Spt15 and Sua7 observed for the group of genes without antisense transcripts suggests that many of these genes may support low levels of antisense transcription which were not detected in any of the transcript maps utilized. Indeed, we found evidence for antisense transcription within these genes, although the average level of elongating Pol II was significantly lower than for those genes with defined antisense transcripts initiating from the 3′-end (Figure 1F). An alternative explanation could be that these peaks of Spt15 and Sua7 are a consequence of conditional long-range chromosomal juxtapositions between a promoter and a terminator, also known as gene loops (29), which are Sua7 (TFIIB) dependent (30,31). It is certainly possible that antisense transcription could itself be a consequence of gene looping, and that in presenting a PIC to the 3′ region one also produces an antisense transcript, which could explain why those genes with antisense transcripts have higher levels of PIC components in their 3′ region than those without.
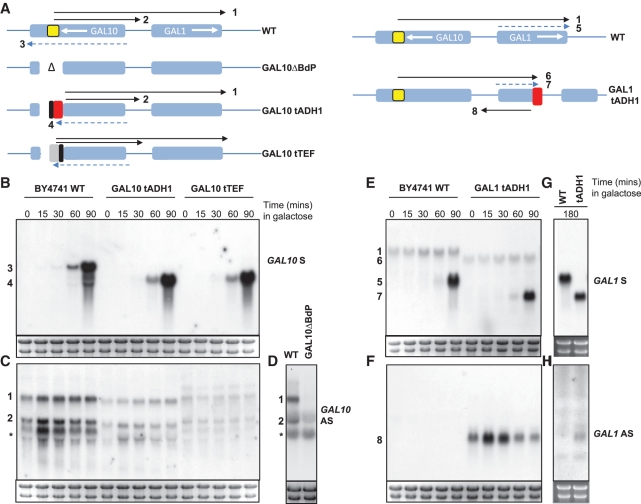
Short 3′ regions placed within GAL1 and GAL10 drive production of antisense transcripts whilst also behaving as terminators
We sought some experimental validation for our hypothesis that sequences at the 3′ regions of genes are capable of driving antisense transcription in addition to their roles in transcription termination and 3′-end processing (Figure 2). So that we could assess functions associated with 3′ regions, we inserted short (∼250 bp) sequences from the 3′ regions of either ScADH1 or AgTEF into GAL10, after deletion of an internal promoter, or into the middle of GAL1 (Figure 2A and D). In all cases, when induced, we observed high levels of truncated protein-coding transcript, implying that the inserted sequences were sufficient to direct transcription processing and termination, as was anticipated (Figure 2B, E and G). Strikingly, the 3′ regions of both ScADH1 and AgTEF1 were also sufficient to drive the production of antisense transcripts, both when the galactose inducible GAL1 and GAL10 promoters were repressed and also during induction (Figure 2C, F and H). These antisense transcripts persist through induction, although the levels of the antisense transcript produced from ScADH1 in GAL1 after 3 h in galactose (steady-state) are barely detectable, a possible consequence of degradation or repression of the antisense promoter due to high levels of sense transcription (Figure 2G and H). Our results demonstrate that both of these short sequences are capable of recapitulating functions associated with both a promoter and a terminator when placed within the middle of a gene, demonstrating that the 3′ regions of genes may often direct antisense transcription in addition to their expected functions in termination of the protein-coding sense transcript. Interestingly, in this system we also found evidence for a gene loop that formed between the promoter of GAL10 and the inserted terminator, and which was present under conditions in which the sense transcript was repressed but the antisense transcript was being transcribed (Supplementary Figure S3). We observed similar behaviour in the unmodified GAL10 gene (Supplementary Figure S4), raising the intriguing possibility that a gene loop can be formed in the absence of sense transcription but the presence of antisense transcription.
Figure 2.
Evidence that 3′ regions can function as both promoters of antisense transcription and as terminators of protein-coding sense transcripts. (A) Schematics showing the derivatives of GAL10 and GAL1. The direction of sense transcription is shown by the white arrows. (B–H) Autoradiographs of northern blots hybridized to strand-specific probes designed to detect the sense (S) or antisense (AS) transcripts at GAL10 (B–D) and GAL1 (E–H) in WT strains (BY4741), or after insertion of the ScADH1 3′ region (red box) into GAL10ΔBdP (A–C) and GAL1 (A and E–H) or the AgTEF 3′ region (grey box) into GAL10ΔBdP (A–C). The black box is the residual loxP sequences used in the construction. Total RNA was prepared from cells cultured in YPD or after induction in galactose for the time shown. GAL10 produces two major antisense transcripts (black arrows) in glucose medium, one extends over the GAL10-1 promoter (2) and the second longer transcript (1) extends sense to the end of GAL1 and persists for 90–100 min after induction but then is no longer detectable. There are other antisense transcripts (asterisks) that do not initiate at the BdP. Loss of the GAL10 internal promoter (yellow box) causes loss of both AS transcripts (GAL10ΔBdP; D) which can be restored by inserting either ScADH1 or AgTEF 3′ regions (C). The 18S and 25S rRNAs are used to estimate loading. For (B–F) exposure times and specific activities of probes are similar, although probe lengths are three times longer for GAL10 compared to GAL1 (see Supplementary Materials and Methods section). G is exposed four times shorter than B–F which reflects accumulation of GAL1 transcript to high levels. H is exposed 10 times longer than B–F to reveal very low levels of GAL1 AS in GAL1tADH1 3 h after induction.
An NDR at the 3′-end of genes supports antisense transcription
We next wished to assess the nucleosome occupancy in the 3′ region of genes, to ask whether a relationship could be identified between occupancy and antisense transcription. The majority of promoters are found in regions depleted of nucleosomes (32), and the extent of this depletion has been related to transcriptional activity (33). We reasoned that genes showing antisense transcription would have a more pronounced 3′ NDR to support PIC formation. To this end, we utilized data from a micrococcal nuclease (MNase)-dependent assay to assess nucleosome occupancy at the 3′-ends of genes (11). Using this map, we found only a very slight difference in the apparent average occupancy levels between those genes with antisense transcripts and those without (Figure 3A). However, we found that the 3′ regions of both sets of genes (with and without antisense transcripts) showed highly similar patterns of average A T richness (Figure 3B), thus the observed similarity may reflect the strong preference of MNase for A T-rich sequences (12), rather than similar occupancy levels.
Figure 3.
3′ nucleosome occupancy determined using a MNase-dependent genome-wide assay (11). (A) The average occupancy at the 3′ region of those genes with antisense transcripts, compared to those without. (B) Comparison of the average A T richness at the 3′ region between those genes with antisense transcripts and those without. Note that the y-axis has been inverted for comparative purposes. All P-values were calculated using the Wilcoxon rank-sum test, comparing the distribution of values at the minimum point of each averaged curve.
To assess whether this was the case, we next utilized a data set obtained in a MNase independent assay of global nucleosome levels (21). Using this map we found that the level of depletion at the 3′ region was substantially more pronounced for those genes with antisense transcripts than for those without (Figure 4A). This relationship remained even when those genes with other moderately transcribed genes within the vicinity of their 3′ region were removed from consideration as before (Supplementary Figure S5). Fan et al. (21) proposed that increased nucleosome depletion at the 3′ region is a consequence of increased Pol II passage from the gene promoter, suggesting that antisense transcription might arise from high levels of sense transcription on the same gene. If antisense transcription levels were a function of sense transcription levels then we might expect the two to be correlated. However, we observed only a very small and negative correlation between the levels of sense and associated antisense transcription on the same gene (Figure 4B, Pearson's correlation coefficient = −0.1). Our results suggest that nucleosome depletion at the 3′ region is associated with antisense initiation, and that this depletion is independent of gene transcription in the sense direction. We cannot rule out the contributions of other transcription-related events (such as termination) toward depletion; however, we propose that the additional depletion observed for those genes with antisense transcripts is a consequence of antisense transcription.
Extensive antisense transcription necessitates a more pronounced promoter architecture
We next wished to assess whether high levels of antisense transcription were supported by a correspondingly pronounced promoter architecture at the 3′ region, and whether the reverse was true for lowly transcribed antisense transcripts. We defined a group of highly transcribed and lowly transcribed antisense transcripts (the top 20% and bottom 20% of the data, after masking the bottom 10% from consideration to remove potentially false-positive antisense transcripts) and determined the average levels of Spt15, Sua7 and nucleosome occupancy at the 5′-end of the antisense transcripts (see ‘Materials and Methods’ section). We identified peaks of Spt15 and Sua7 at the 3′ region that were substantially higher for the highly transcribed antisense transcripts (Figure 5A and B). Nucleosome depletion was also found to be more pronounced for the highly expressed antisense transcripts (Figure 5C). These results support the notion that antisense transcription is driven by PIC formation, in a manner similar to protein-coding sense transcription. In support of this, average levels of Swr1 and Rsc9 were high over the antisense TSS (Supplementary Figure S6). Our results demonstrate that highly transcribed antisense transcripts are driven by similarly high levels of the PIC components Spt15 and Sua7.
Bidirectional promoters show evidence of two distinct PICs
It has been proposed that promoters are inherently bidirectional (13). It would be reasonable to expect that for these highly expressed antisense transcripts one would observe similarly high levels of transcription in the sense direction. Using the two groups of genes with highly or lowly expressed antisense transcripts, we assessed levels of divergent sense transcription. Although the average level of transcription was higher in the sense direction for the highly transcribed group than for the lowly transcribed group, the level was not as high as the average level of antisense transcription itself (Figure 5D). This agrees with the finding by Churchman and Weissman (15) that sense and antisense transcription from a bidirectional promoter is only modestly correlated, and suggests that high levels of antisense transcription can be supported in the absence of high sense transcription, i.e. that inherently bidirectional promoters can be biased in both the sense and antisense direction, and perhaps that the two can be regulated independently of one another.
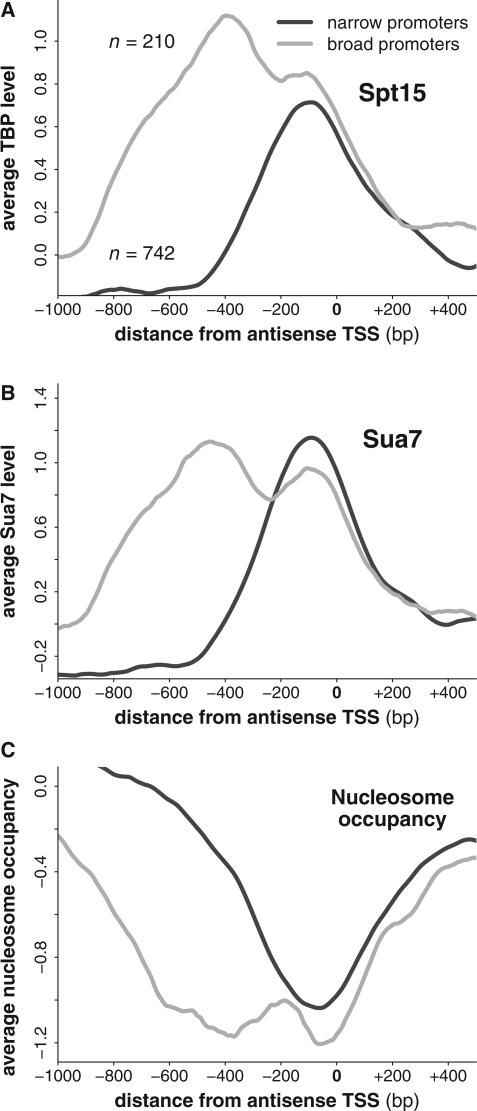
To address this we defined two classes of bidirectional promoter to see if we could find evidence of two distinct PICs at bidirectional promoters. The classes were defined on the basis of how far the TSS of the antisense transcript was from the TSS of the sense transcript. In the first group, termed the narrow promoters, the TSSs were within 300 bp. In the second group, termed the broad promoters, they were within 700 bp but further than 400 bp (with no other transcript TSS between them). The antisense transcripts considered were the same as defined above (i.e. they initiated within the 3′ region of a gene). We assessed the average levels of Spt15, Sua7 and nucleosome occupancy around the TSS of the antisense transcripts as before.
The two classes of bidirectional promoter showed distinct patterns. Overlaying the Spt15 profile of the broad promoters with that of the narrow promoters demonstrated a substantial enrichment of Spt15 upstream of the antisense 5′-end despite similar levels over the 5′-end itself, supporting a model in which two distinct PICs form at bidirectional promoters (Figure 6A). Additionally, two partially overlapping peaks of Sua7 were observed over the broad promoters, whilst only one peak was discernible over the group of narrow promoters (Figure 6B). Of these two peaks, one corresponded to the single peak seen in the narrow promoters, both of which lay over the antisense 5′-end. The second peak was further upstream, potentially representing the PIC driving the divergent sense transcript. Finally, we observed a much broader nucleosome depletion in the broad promoter group, which one would expect if the two PICs were further apart in the broad group than in the narrow group (Figure 6C). This analysis supports a model in which two PIC recruitment sites exist at bidirectional promoters, providing a mechanism to explain how sense and antisense levels could be independently regulated.
Figure 6.
Bidirectional promoters show evidence of possessing two distinct PICs. (A–C) The average levels of Spt15, Sua7 and nucleosome occupancy around the TSS of antisense transcripts with an adjacent and divergent sense transcript. Shown are both the narrow and broad classes of bidirectional promoter.
Extensive antisense transcription can be supported by a promoter architecture in the absence of adjacent divergent sense transcription
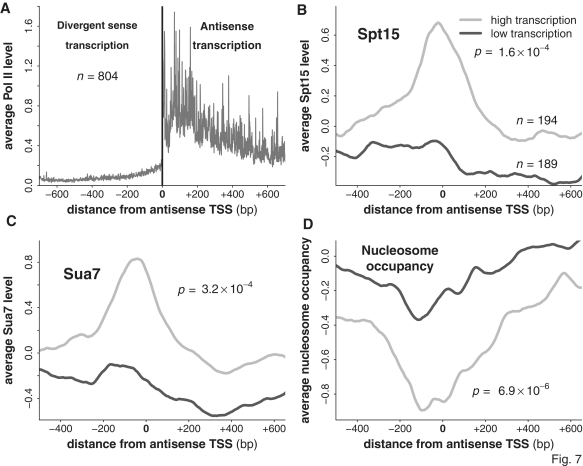
If antisense transcription can be regulated independently of divergent sense direction from the same promoter region, then we might expect to find highly transcribed antisense transcripts supported by a pronounced promoter architecture in the absence of transcription in the sense direction, i.e. a unidirectional promoter driving antisense transcription. To this end, we selected for antisense transcripts (as defined above) with low levels of adjacent divergent sense transcription (see ‘Materials and Methods’ section), and which were not also adjacent to any non-Pol II transcribed elements, including tRNAs and rRNAs. This gave us a set of 804 transcripts, with an average level of nascent transcription that was much higher in the antisense direction than in the divergent sense direction (Figure 7A).
Figure 7.
PIC formation supports high levels of antisense transcription in the absence of high levels of divergent sense transcription. (A) We selected for a group of antisense transcripts with low levels of divergent sense transcription. Shown is the average level of nascent transcription around the TSS of antisense transcripts belonging to this group, with nascent sense transcription to the left of the TSS and nascent antisense transcription to the right. (B–D) The average levels of Spt15, Sua7 and nucleosome occupancy around the TSS of both highly and lowly transcribed antisense transcripts belonging to this group. All P-values were calculated using the Wilcoxon rank-sum test, comparing the distribution of values at the maximum point of each averaged curve (the minimum point in the case of nucleosome occupancy).
We selected both highly and lowly transcribed subgroups (the top 25% and bottom 25% of the data, after masking the bottom 10% from consideration to remove potentially false-positive antisense transcripts) from these antisense transcripts, and compared the average level of Spt15, Sua7 and nucleosome occupancy at their TSSs. We found that the highly expressed antisense transcripts had a more pronounced peak of Spt15 and Sua7 than the lowly expressed antisense transcripts, and were significantly more depleted for nucleosomes (Figure 7B–D). These results demonstrate that high levels of antisense can be supported in a PIC-dependent manner in the absence of adjacent divergent sense transcription, demonstrating that antisense transcription is not a consequence of divergent sense transcription, and that the process of antisense transcription can be regulated independently of divergent sense transcription. In support of this, by using a genome-wide map of over a 100 different transcription factors [(22,34); note that this map only assessed intergenic regions], we found that 40% of those antisense transcripts with low divergent sense transcription and whose TSSs fell within a region covered by the genome-wide map had at least one transcription factor bound within a 300-bp window placed immediately upstream of their TSS (292 out of 738). By comparison, 64% of protein-coding transcripts had at least one bound transcription factor upstream, an increase which may be due to the higher average level of sense transcription compared to antisense transcription (Figure 1E). That these binding sites are present (and occupied) suggests that antisense transcript production could potentially be regulated by transcription factor binding in the absence of divergent sense transcription.
DISCUSSION
Here we have explored the origins of antisense transcription from the 3′ regions of genes. To do this, we compiled two major collections of genome-wide maps. The first was a collated list of transcripts, in which every transcript was assigned a level of nascent transcription using a map of strand-specific elongating Pol II (15–20). The second map comprised genome-wide levels of critical promoter components, chiefly Spt15 and Sua7, as well as the levels of nucleosome occupancy and regulated transcription factors (10,21,22,34). Using these data, we show that the average levels of the PIC components Spt15 and Sua7 are high in the 3′ region of genes with antisense transcripts, and that the extent of nascent antisense transcription at these genes is correspondingly high. Perhaps most strikingly, we find that high levels of antisense transcription can be supported by PIC formation in the absence of adjacent divergent sense transcription, demonstrating that antisense transcription is not dependent upon the formation of transcription complexes driving protein-coding sense transcription. We validate this by creating hybrid genes in which short 3′ regions are inserted into the middle of protein-coding genes, ensuring that there are no promoters in the vicinity, and find that these 3′ regions can reconstitute both promoter and terminator activities, giving rise to antisense transcription whilst redefining the length of the protein-coding sense transcript.
In general, antisense transcripts have much lower steady-state levels than protein-coding sense transcripts, which is likely to reflect their differential processing. At first sight the high levels of PIC components associated with the 3′ regions of genes with antisense transcripts appears to be at odds with the very low steady-state levels of many antisense transcripts. However, antisense transcripts have been shown to be targeted for degradation by a number of different exonucleases (4,5,13,14,17,35). Indeed, over 1000 transcripts antisense to open reading frames are polyadenylated, exported and degraded in the cytoplasm by the 5′ to 3′ exonuclease Kem1 (35). As a consequence, their steady-state levels do not accurately reflect the levels of nascent transcription within the genome and this gives an impression that antisense transcription from the 3′ region of genes is not significant. In support of this, the repressed GAL1 gene produces a Kem1-degraded antisense from its 3′ region and thus is not detectable in steady state [Figure 2E, (35)]. When we consider genes with antisense transcripts, average levels of antisense transcription are 45% of the average levels of protein-coding sense transcription. This value is consistent with a direct correlation between levels of PIC components and levels of nascent transcription and allows us to suggest that in other genomes, levels of PIC components at sites at which transcription is initiated will reflect levels of nascent transcription (though not necessarily steady state transcript levels). That antisense transcription is so abundant genome-wide (Figure 1E) supports a possible regulatory function which, given that antisense transcripts are so often unstable, is likely to be exerted by the act of transcription itself, and not by the resultant transcripts.
Niel et al. (13) proposed a model describing the initiation of bidirectional transcription, in which two independent PICs form within an NDR to direct sense and antisense transcription. This model was based on the observation in TPI1 that the deletion of a TATA box abrogates sense transcription of the protein-coding mRNA whilst enhancing levels of the non-coding antisense transcript. Neil et al. (13) suggest that the PICs at bidirectional promoters compete for transcription factors, such that the resultant distribution of factors determines the relative PIC stability and the extent of sense and antisense transcription. This would imply an inverse correlation between antisense transcription and divergent sense transcription at bidirectional promoters; however, it has also been shown that bidirectional promoters can be coregulated in a positive manner (Xu et al. 2011). Furthermore, Churchman and Weissman (15) have demonstrated that sense and antisense levels from bidirectional promoters are only modestly and positively correlated (Spearman correlation coefficient, r = 0.34). We believe our work sheds light on two important features of antisense transcription from bidirectional promoters. Firstly, that it is a consequence of directed PIC formation, rather than of transcriptional infidelity [Figures 1, 5, 6 and 7; (36)], and secondly that high levels of antisense transcription can be supported by high levels of PIC components (implying a highly stable PIC) in both the presence and absence of high levels of divergent sense transcription (Figures 5 and 7). We therefore propose that bidirectional promoters are in many respects two distinct but closely spaced promoters that can be coregulated in a positive manner, such as at the SUT719/GAL80 promoter (8), an inverse manner or indeed independently from one another.
There is evidence to suggest that antisense transcripts initiating from the 3′ regions of protein-coding genes may regulate sense gene transcription and thus gene expression. Antisense transcripts have been shown to be capable of regulating protein-coding sense transcription both by recruiting histone-modifying enzymes and by transcriptional interference (5,37) and there is evidence that they can both upregulate and downregulate expression (25,38). Furthermore, by assessing the levels of gene expression across a number of different environmental conditions, it has been shown that genes with antisense transcripts show a higher variability of expression, i.e. they are more transcriptionally plastic than genes without (8). Here we find that antisense transcripts are remarkably abundant genome wide. Given that they are so frequently degraded, one might argue that the process of antisense transcription is wasteful. However, if the act of antisense transcription is involved in changing the pattern of gene expression (8), or in expediting the transition in expression in response to changing conditions (25), then it may represent a necessary investment to ensure a rapid and appropriate change in the transcriptional landscape.
If antisense transcription does indeed represent a genome-wide mechanism of gene regulation, then understanding how antisense transcription is in turn regulated becomes crucially important. We show that antisense transcription is directed by PIC formation at the 3′ regions of genes, and that 3′ PIC formation does not necessarily predicate divergent sense transcription. Moreover, we find that DNA bound regulatory transcription factors are frequently present at sites of antisense transcription where there is little divergent sense transcription, supporting a model in which antisense transcription can be regulated independently of divergent sense transcription. It follows that if the promoters of antisense transcripts can be regulated independently of downstream gene promoters, antisense-mediated gene regulation is not merely a function of downstream gene transcription. We suggest that this allows for a more exquisite level of control over the transcriptional landscape of the organism, with regulatory signals able to impact upon both the 5′ and 3′ promoters of a gene, influencing sense transcription and thus gene expression.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Methods, Supplementary Figures 1–6, Supplementary Tables 1 and 2 and Supplementary Reference [39].
FUNDING
The Engineering and Physical Sciences Research Council (to S.C.M.); the Fundação para a Ciência e a Tecnologia (to A.S.B.); Cancer Research UK (to J.A.); Oxford Biodynamics Ltd. (ALRNEI1 to J.M.); Epigenesys Network of Excellence (to J.M.); and the Wellcome Trust (WT089156MA to J.M.). Funding for open access charge: The Wellcome Trust.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors would like to thank Michael Youdell for helpful and informative discussions, and Françoise Howe for proof-reading the manuscript.
REFERENCES
- 1.Kapranov P, Willingham AT, Gingeras TR. Genome-wide transcription and the implications for genomic organization. Nat. Rev. Genet. 2007;8:413–423. doi: 10.1038/nrg2083. [DOI] [PubMed] [Google Scholar]
- 2.Berretta J, Morillon A. Pervasive transcription constitutes a new level of eukaryotic genome regulation. EMBO Rep. 2009;10:973–982. doi: 10.1038/embor.2009.181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hongay CF, Grisafi PL, Galitski T, Fink GR. Antisense transcription controls cell fate in Saccharomyces cerevisiae. Cell. 2006;127:735–745. doi: 10.1016/j.cell.2006.09.038. [DOI] [PubMed] [Google Scholar]
- 4.Perocchi F, Xu Z, Clauder-Münster S, Steinmetz LM. Antisense artifacts in transcriptome microarray experiments are resolved by actinomycin D. Nucleic Acids Res. 2007;35:e128. doi: 10.1093/nar/gkm683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Camblong J, Iglesias N, Fickentscher F, Dieppois G, Stutz F. Antisense RNA stabilization induces transcriptional gene silencing via histone deacetylation in S. cerevisiae. Cell. 2007;131:706–717. doi: 10.1016/j.cell.2007.09.014. [DOI] [PubMed] [Google Scholar]
- 6.Nishizawa M, Komai T, Katou Y, Shirahige K, Ito T, Toh-e A. Nutrient-regulated antisense and intragenic RNAs modulate a signal transduction pathway in yeast. PloS Biol. 2008;6:2817–2830. doi: 10.1371/journal.pbio.0060326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pinskaya M, Gourvennec S, Morillon A. H3 lysine 4 di- and tri-methylation deposited by cryptic transcription attenuates promoter activation. EMBO J. 2009;28:1697–1707. doi: 10.1038/emboj.2009.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Xu Z, Wei W, Gagneur J, Clauder-Münster S, Smolik M, Huber W, Steinmetz LM. Antisense expression increases gene expression variability and locus interdependency. Mol. Syst. Biol. 2011;7:468. doi: 10.1038/msb.2011.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mavrich TN, Ioshikhes IP, Venters BJ, Jiang C, Tomsho LP, Qi J, Schuster SC, Albert I, Pugh BF. A barrier nucleosome model for statistical positioning of nucleosomes throughout the yeast genome. Genome Res. 2008;18:1073–1083. doi: 10.1101/gr.078261.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Venters BJ, Pugh BF. A canonical promoter organization of the transcription machinery and its regulators in the Saccharomyces genome. Genome Res. 2009;19:360–371. doi: 10.1101/gr.084970.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kaplan N, Moore IK, Fondufe-Mittendorf Y, Gossett AJ, Tillo D, Field Y, LeProust EM, Hughes TR, Lieb JD, Widom J, et al. The DNA-encoded nucleosome organization of a eukaryotic genome. Nature. 2009;458:362–366. doi: 10.1038/nature07667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chung HR, Dunkel I, Heise F, Linke C, Krobitsch S, Ehrenhofer-Murray AE, Sperling SR, Vingron M. The effect of micrococcal nuclease digestion on nucleosome positioning data. PloS ONE. 2010;5:e15754. doi: 10.1371/journal.pone.0015754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Neil H, Malabat C, d'Aubenton-Carafa Y, Xu Z, Steinmetz LM, Jacquier A. Widespread bidirectional promoters are the major source of cryptic transcripts in yeast. Nature. 2009;457:1038–1042. doi: 10.1038/nature07747. [DOI] [PubMed] [Google Scholar]
- 14.Wyers F, Rougemaille M, Badis G, Rousselle JC, Dufour ME, Boulay J, Régnault B, Devaux F, Namane A, Séraphin B, et al. Cryptic pol II transcripts are degraded by a nuclear quality control pathway involving a new poly(A) polymerase. Cell. 2005;121:725–737. doi: 10.1016/j.cell.2005.04.030. [DOI] [PubMed] [Google Scholar]
- 15.Churchman LS, Weissman JS. Nascent transcript sequencing visualizes transcription at nucleotide resolution. Nature. 2011;469:368–373. doi: 10.1038/nature09652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nagalakshmi U, Wang Z, Waern K, Shou C, Raha D, Gerstein M, Snyder M. The transcriptional landscape of the yeast genome defined by RNA sequencing. Science. 2008;320:1344–1349. doi: 10.1126/science.1158441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xu Z, Wei W, Gagneur J, Perocchi F, Clauder-Münster S, Camblong J, Guffanti E, Stutz F, Huber W, Steinmetz LM. Bidirectional promoters generate pervasive transcription in yeast. Nature. 2009;457:1033–1037. doi: 10.1038/nature07728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yassour M, Kaplan T, Fraser HB, Levin JZ, Pfiffner J, Adiconis X, Schroth G, Luo S, Khrebtukova I, Gnirke A, et al. Ab initio construction of a eukaryotic transcriptome by massively parallel mRNA sequencing. Proc. Natl Acad. Sci. USA. 2009;106:3264–3269. doi: 10.1073/pnas.0812841106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Granovskaia MV, Jensen LJ, Ritchie ME, Toedling J, Ning Y, Bork P, Huber W, Steinmetz LM. High-resolution transcription atlas of the mitotic cell cycle in budding yeast. Genome Biol. 2010;11:R24. doi: 10.1186/gb-2010-11-3-r24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yassour M, Pfiffner J, Levin JZ, Adiconis X, Gnirke A, Nusbaum C, Thompson DA, Friedman N, Regev A. Strand-specific RNA sequencing reveals extensive regulated long antisense transcripts that are conserved across yeast species. Genome Biol. 2010;11:R87. doi: 10.1186/gb-2010-11-8-r87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fan X, Moqtaderi Z, Jin Y, Zhang Y, Liu XS, Struhl K. Nucleosome depletion at yeast terminators is not intrinsic and can occur by a transcriptional mechanism linked to 3’-end formation. Proc. Natl Acad. Sci. USA. 2010;107:17945–17950. doi: 10.1073/pnas.1012674107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.MacIsaac KD, Wang T, Gordon DB, Gifford DK, Stormo GD, Fraenkel E. An improved map of conserved regulatory sites for Saccharomyces cerevisiae. BMC Bioinform. 2006;7:113. doi: 10.1186/1471-2105-7-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wach A, Brachat A, Pohlmann R, Philippsen P. New heterologous modules for classical or PCR-based gene disruptions in Saccharomyces cerevisiae. Yeast. 1994;10:1793–1808. doi: 10.1002/yea.320101310. [DOI] [PubMed] [Google Scholar]
- 24.Longtine MS, McKenzie A, Demarini DJ, Shah NG, Wach A, Brachat A, Philippsen P, Pringle JR. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast. 1998;14:953–961. doi: 10.1002/(SICI)1097-0061(199807)14:10<953::AID-YEA293>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- 25.Houseley J, Rubbi L, Grunstein M, Tollervey D, Vogelauer M. A ncRNA modulates histone modification and mRNA induction in the yeast GAL gene cluster. Mol. Cell. 2008;32:685–695. doi: 10.1016/j.molcel.2008.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Güldener U, Heck S, Fielder T, Beinhauer J, Hegemann JH. A new efficient gene disruption cassette for repeated use in budding yeast. Nucleic Acids Res. 1996;24:2519–2524. doi: 10.1093/nar/24.13.2519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mizuguchi G, Shen X, Landry J, Wu W, Sen S, Wu C. ATP-driven exchange of histone H2AZ variant catalyzed by SWR1 chromatin remodeling complex. Science. 2004;303:343–348. doi: 10.1126/science.1090701. [DOI] [PubMed] [Google Scholar]
- 28.Cairns BR, Lorch Y, Li Y, Zhang M, Lacomis L, Erdjument-Bromage H, Tempst P, Du J, Laurent B, Kornberg RD. RSC, an essential, abundant chromatin-remodeling complex. Cell. 1996;87:1249–1260. doi: 10.1016/s0092-8674(00)81820-6. [DOI] [PubMed] [Google Scholar]
- 29.O'Sullivan JM, Tan-Wong SMM, Morillon A, Lee B, Coles J, Mellor J, Proudfoot NJ. Gene loops juxtapose promoters and terminators in yeast. Nature Genet. 2004;36:1014–1018. doi: 10.1038/ng1411. [DOI] [PubMed] [Google Scholar]
- 30.Singh BNN, Hampsey M. A transcription-independent role for TFIIB in gene looping. Mol. Cell. 2007;27:806–816. doi: 10.1016/j.molcel.2007.07.013. [DOI] [PubMed] [Google Scholar]
- 31.Medler S, Al Husini N, Raghunayakula S, Mukundan B, Aldea A, Ansari A. Evidence for a complex of transcription factor IIB with poly(A) polymerase and cleavage factor 1 subunits required for gene looping. J. Biol. Chem. 2011;286:33709–33718. doi: 10.1074/jbc.M110.193870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bernstein BE, Liu CLL, Humphrey EL, Perlstein EO, Schreiber SL. Global nucleosome occupancy in yeast. Genome. Biol. 2004;5:R62. doi: 10.1186/gb-2004-5-9-r62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lee CK, Shibata Y, Rao B, Strahl BD, Lieb JD. Evidence for nucleosome depletion at active regulatory regions genome-wide. Nature Genet. 2004;36:900–905. doi: 10.1038/ng1400. [DOI] [PubMed] [Google Scholar]
- 34.Harbison CT, Gordon DB, Lee TI, Rinaldi NJ, MacIsaac KD, Danford TW, Hannett NM, Tagne J, Reynolds DB, Yoo J, et al. Transcriptional regulatory code of a eukaryotic genome. Nature. 2004;431:99–104. doi: 10.1038/nature02800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.van Dijk EL, Chen CL, d'Aubenton-Carafa Y, Gourvennec S, Kwapisz M, Roche V, Bertrand C, Silvain M, Legoix-Ne P, Loeillet S, et al. XUTs are a class of Xrn1-sensitive antisense regulatory non-coding RNA in yeast. Nature. 2011;475:114–117. doi: 10.1038/nature10118. [DOI] [PubMed] [Google Scholar]
- 36.Struhl K. Transcriptional noise and the fidelity of initiation by RNA polymerase II. Nat. Struct. Mol. Biol. 2007;14:103–105. doi: 10.1038/nsmb0207-103. [DOI] [PubMed] [Google Scholar]
- 37.Berretta J, Pinskaya M, Morillon A. A cryptic unstable transcript mediates transcriptional trans-silencing of the Ty1 retrotransposon in S. cerevisiae. Genes. Dev. 2008;22:615–626. doi: 10.1101/gad.458008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Uhler JP, Hertel C, Svejstrup JQ. A role for noncoding transcription in activation of the yeast PHO5 gene. Proc. Natl Acad. Sci. USA. 2007;104:8011–8016. doi: 10.1073/pnas.0702431104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Thoden JB, Holden HM. The molecular architecture of galactose mutarotase/UDP-galactose 4-epimerase from Saccharomyces cerevisiae. J. Biol. Chem. 2005;280:21900–21907. doi: 10.1074/jbc.M502411200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.