Abstract
Background
Men with familial prostate cancer (PCA) and African American men are at risk for developing PCA at younger ages. Genetic markers predicting early-onset PCA may provide clinically useful information to guide screening strategies for high-risk men. We evaluated clinical information from six polymorphisms associated with early-onset PCA in a longitudinal cohort of high-risk men enrolled in PCA early detection with significant African American participation.
Methods
Eligibility criteria include ages 35–69 with a family history of PCA or African American race. Participants undergo screening and biopsy per study criteria. Six markers associated with early-onset PCA (rs2171492 (7q32), rs6983561 (8q24), rs10993994 (10q11), rs4430796 (17q12), rs1799950 (17q21), and rs266849 (19q13)) were genotyped. Cox models were used to evaluate time to PCA diagnosis and PSA prediction for PCA by genotype. Harrell’s concordance index was used to evaluate predictive accuracy for PCA by PSA and genetic markers.
Results
460 participants with complete data and ≥1 follow-up visit were included. 56% were African American. Among African American men, rs6983561 genotype was significantly associated with earlier time to PCA diagnosis (p=0.005) and influenced prediction for PCA by the PSA (p<0.001). When combined with PSA, rs6983561 improved predictive accuracy for PCA compared to PSA alone among African American men (PSA= 0.57 vs. PSA+rs6983561=0.75, p=0.03).
Conclusions
Early-onset marker rs6983561 adds potentially useful clinical information for African American men undergoing PCA risk assessment. Further study is warranted to validate these findings.
Impact
Genetic markers of early-onset PCA have potential to refine and personalize PCA early detection for high-risk men.
Keywords: genetics, population, mass screening, African Americans, prostatic neoplasms
BACKGROUND
Prostate cancer (PCA) is the second-leading cause of cancer-related deaths in men in the United States. (1) Men with a family history of PCA and African American men are at significantly increased risk for the disease (1, 2) and of onset at younger ages.(3, 4) A report from 2009 found that the diagnosis of PCA among men ≤ 55 years has risen from 2.3% between 1988–1991 to 9% between 2000–2003,(5) and these men with young-onset PCA have more threat of morbidity and mortality from PCA especially if they are diagnosed with high-grade or advanced disease. (5) The challenge however, is predicting which high-risk men are at risk of developing PCA at younger ages and appropriately recommending screening to those men while sparing other men unnecessary tests and procedures.
Prostate-specific antigen (PSA)-based methods have been the traditional tests used to screen men for PCA and make risk-management decisions. Among high-risk men undergoing prostate biopsies based on PSA criteria, PCA detection rates range between 10–17%.(3, 4) Therefore, a subset of high-risk men undergo unnecessary prostate biopsies while others have aggressive PCA detected. Genetic markers have the potential to personalize and refine PCA screening and interpretation of PSA if their clinical role is better understood.
Several genetic single nucleotide polymorphisms (SNPs) have been identified from genome-wide association studies (GWAS) to be associated with PCA risk,(6) and several GWAS markers have been evaluated for association to early-onset PCA in substudies.(7–10) A chromosomal region of great interest in PCA genetics research is 8q24.(8, 11–14) One study found that carrying the minor allele of one polymorphism (rs6983561) at chromosomal locus 8q24 was associated with an increased risk for PCA in men < 50 years.(7) Another chromosomal region of interest for potentially harboring PCA susceptibility variants is 17q12.(15) One study found that the A-allele of a polymorphism (rs4430796) at 17q12 increased the risk for PCA in men < 50 years.(16) New genomic regions of interest for PCA risk have continued to be identified. Some of these regions are on chromosomes 3, 6, 7, 10, 11, 19, and X.(9) One polymorphism (rs266849) on chromosome 19 was found to be associated with an increased risk for PCA in men < 60 years.(9) Another study examining genetic variants at 10q11 and Xp11 found that polymorphism rs10993994 was associated with a 2-fold increased risk for PCA in men ≤ 65 years.(10)
A subset of men in families with potential hereditary PCA may be at increased risk for early-onset disease, and one study found a common variant in BRCA1 (rs1799950) was overtransmitted to younger men (age <50 years) affected with PCA from hereditary and early-onset PCA families.(17)
Other polymorphisms that reside in known genes have been characterized for association to early-onset PCA.(18, 19) One study found a polymorphism (rs2171492) in Carboxypeptidase 4 to be associated with risk for aggressive PCA in men < 66 years.(18) Therefore, while markers associated with early-onset PCA from various studies are promising for identifying risk for disease diagnosis at younger ages, further study of the clinical information gained from these and other genetic markers is needed to help guide patients and physicians in making individualized screening recommendations.
Here we evaluate the potential clinical information gained from six genetic markers associated with early-onset PCA among high-risk men undergoing PCA screening.(7–10, 17, 18) These markers were chosen for study in high-risk men (men with familial PCA and African American men) as subsets of these men undergoing screening have been diagnosed with PCA at ages < 55 years.(3, 4) Genetic markers of early-onset PCA may add clinical information beyond predicting early-onset disease among high-risk men useful in refining screening decisions. We performed this study in an ethnically-diverse longitudinal cohort of high-risk men enrolled in a PCA early detection program with significant African American participation. We took a broad approach to evaluating the clinical role of early-onset genetic markers in order to formulate hypotheses regarding the potential role of genetic markers in clinical PCA risk assessment.
METHODS
Prostate Cancer Risk Assessment Program (PRAP) cohort
The Prostate Cancer Risk Assessment Program (PRAP) at Fox Chase Cancer Center (FCCC) was established in 1996 to provide screening and perform research for men at high risk for PCA.(4) Eligibility criteria and cancer detection rates have been described previously.(4) Briefly, eligibility for PRAP include any man between ages 35–69 years without a previous diagnosis of PCA with one first-degree relative with PCA, two second-degree relatives with PCA on the same side of the family, any African American man regardless of family history, or men with known mutations in BRCA1 or BRCA2. Men with BRCA mutations (n= 13) were excluded from the clinical analysis. Accrual to PRAP is ongoing and participants are followed longitudinally for PCA screening and early detection. The PRAP study is approved by the Institutional Review Board at FCCC and at all previous and currently active community hospital sites that enrolled participants to PRAP.
PCA screening in PRAP
PCA screening procedures, biopsy criteria, PCA incidence, and PCA features have been described previously.(4) Briefly, screening tests include the total prostate specific antigen (PSA), percent free PSA (fPSA), digital rectal examination (DRE) by a PRAP physician, and the PSA velocity (PSAv).
Criteria for Biopsy
Until November 2005, the criteria for prostate biopsy were (i) PSA > 4 ng/mL, (ii) PSA 2.0–4.0 ng/mL with fPSA less than 27%, (iii) any abnormality on DRE, or (iv) PSAv of 0.75 ng/mL/year. After November 2005, the criteria for biopsy were changed to (i) PSA > 2.0 ng/mL, (ii) PSA 1.5–2.0 ng/mL with fPSA ≤25%, (iii) any abnormality on DRE, or (iv) PSAv of 0.75 ng/mL/year to investigate the detection of PCA at lower PSA values.(3) All biopsies are transrectal ultrasound-guided 5-region patterned prostate biopsies.(20, 21) If all screening parameters are within normal limits based on these biopsy criteria, no biopsy is performed and the participant is recommended to return to clinic in one year for repeat screening. Men diagnosed with PCA within 6 months after their first visit to PRAP were excluded (n=42) in order to eliminate pre-existing PCA from this analysis.
Genotyping of six early-onset genetic polymorphisms
Six genetic markers reported to be associated with early-onset PCA were chosen for this study: rs6983561 (8q24),(7) rs4430796 (17q12),(8) rs266849 (19q13),(9) rs10993994 (10q11),(10) rs1799950 (17q21),(17) and rs2171492 (7q32).(18) Genotyping was performed on genomic DNA using a fluorogenic 5′ nuclease allelic discrimination assay (TaqMan® SNP Genotyping Assay, Applied Biosystems). Reactions were prepared using TaqMan Universal PCR Mastermix, No AmpErase UNG or TaqMan Genotyping MasterMix (Applied Biosystems) according to manufacturer’s instructions. Thermal cycling and analysis were performed using an ABI7900 Sequence Detection System (Applied Biosystems). Control DNA samples with known genotypes were included in each run. In addition, a no template (water) control was included to assess DNA contamination. SNP assignment was achieved automatically with the SDS software (Applied Biosystems) using a proprietary algorithm. In addition, genotypes were confirmed on a random selection of 2% of the samples by standard sequencing with 100% concordance.
Statistical Methods
Distribution of early-onset markers was summarized by self-reported race and compared using the chi-squared test. In addition, Hardy-Weinberg equilibrium was tested for each allele using the chi-squared test.(22)
Time to PCA Diagnosis
For inferences on the relationship between time to PCA and the markers, individual genotype-specific Kaplan-Meier curves were constructed. Each SNP was examined as three genotypes, and then collapsed into the heterozygous or homozygous genotypes. Time from enrollment to PCA was analyzed. Participants were censored at the last follow-up if they were not diagnosed with PCA at the time of the analysis. Cox proportional hazards regressions were used with each marker tested separately among African American and Caucasian men, and adjusted for age and PSA at baseline. Continuous variables were entered as linear terms in models. The risk genotypes were determined as those that created the most separation in the Kaplan-Meier curve, had larger hazard ratios, and greater level of statistical significance.
PSA Prediction for PCA by genotype
To evaluate the effect of early-onset marker genotype on longitudinal prediction for PCA by PSA at baseline, the interaction terms between PSA at baseline and risk genotype were entered into each Cox model. To account for multiple hypotheses testing, we set the false discovery rate to 5% using Benjamini and Hochberg’s method.(23) Any significant interactions were considered as evidence of differential prediction ability of PSA for PCA by genotype. The hazard ratio of one point increase on PSA by genotype was also presented to show the differential prediction ability by genotype. To display the relationship of PSA with time to diagnosis in the models, age and PSA were entered into the Cox model as restricted cubic splines. The 3-year probabilities of being diagnosed with PCA were estimated for each participant. The estimated probabilities were plotted against the baseline PSA. Due to multiple age values, restricted cubic splines were fit to all estimated probabilities to represent the trend of the PSA prediction in each subpopulation.
Predictive accuracy of genetic markers
Harrell’s C index was used to evaluate the accuracy of longitudinal prediction for PCA by early-onset markers and PSA. We created a training dataset and validation dataset by randomly assigning each observation to either dataset and used the program somerd in STATA to compare Harrell’s C from different models.(24) Considering our outcome was longitudinal prediction for PCA and traditional area under the ROC curve analyses do not take into account the varied follow-up time, we used the C index which is a member of the “Kendall family” of rank parameters and is used to estimate the concordance probability with censored data. Like AUC, a value of 0.5 implies complete discordance. Higher values suggest higher concordance between the data and the predicted values from the model. All analyses were performed using STATA 10.1.
RESULTS
Genotype Distribution
At the time of this analysis, 778 participants were accrued to PRAP. Genotype distributions in the overall cohort for all six early-onset genetic markers differed significantly by race (Supplementary Table 1). Genotype distribution differences were also evaluated comparing those included in this analysis (n=460) and those excluded (n=310) for African American and Caucasian PRAP participants (Supplementary Table 2). Only rs4430796 was found have a statistically significant difference in distribution among African American men between those included vs. excluded from the analysis (p=0.001). No differences in genotype frequency were observed for African American or Caucasian men for the other five early-onset markers by inclusion in the analysis..
Demographic and clinical features
The sample size for the clinical analysis of the six early-onset genetic markers included 460 out of the 778 participants accrued. Exclusions from the overall cohort of 778 were as follows: race other than African American or Caucasian (n=8), BRCA1 or BRCA2 positive (n=13), missing clinical data (n= 4), diagnosed with PCA within 6 months of first PRAP visit (n=42) and no follow-up (either lost to follow-up after first visit or first visit was within 12 months of this analysis and participant was not due for follow-up) (n=251). Men diagnosed with PCA within 6 months were removed to exclude pre-existing PCA. The demographics of this sample set of 460 participants included in the analysis by self-reported race are shown in Table 1. Approximately 56% of participants in the analysis were African American. The mean age at diagnosis for Caucasian participants was 57.5 years (range 46.3–67.4 years) and for African American participants was 57.5 years (range 40.8–73.7 years). The mean time to PCA diagnosis for African American men was 37.9 months (range 6.4–92.0 months) and for Caucasian men was 54.7 months (range 9.4–131.7 months).
Table 1.
Demographics and Prostate Cancer Characteristics of 460 PRAP Participants Included in the Clinical Analysis by Self-reported Race
| African American (n=257) | Caucasian (n=203) | |||||
|---|---|---|---|---|---|---|
|
| ||||||
| N | Mean | Range | N | Mean | Range | |
| Age at entry (years) | 257 | 50.9 | 35–69 | 203 | 50.2 | 35–69 |
| Duration of follow-up (months) | 257 | 51.6 | 0.6–152.0 | 203 | 58.4 | 0.3–163.4 |
| PSA at baseline (ng/mL) | 257 | 1.6 | 0.1–27.2 | 203 | 1.5 | 0.2–9.8 |
| Percent Free PSA at baseline+ | 59 | 19.0 | 7.8–39.4 | 42 | 17.7 | 4.6–30.0 |
| DRE at baseline [28 missing] | ||||||
| Normal/BPH | 235(96.3) | 182 (96.8) | ||||
| Abnormal | 9 (3.7) | 6 (3.2) | ||||
| Biopsy history (reported at baseline) | ||||||
| No Prior Biopsy/Unknown | 230 (89.5) | 188(92.6) | ||||
| Had Prior Negative Biopsy | 27(10.5) | 15 (7.4) | ||||
| Number of Biopsy Sessions while in PRAP | 257 | 0.46 | 0–6 | 203 | 0.51 | 0–7 |
| PCA diagnosis * | 25(9.7) | 24 (11.8) | ||||
| Age at diagnosis (years) | 25 | 57.5 | 40.8–73.7 | 24 | 57.5 | 46.3–67.4 |
| Mean time to PCA diagnosis (months) | 25 | 37.9 | 6.4–92.0 | 24 | 54.7 | 9.4–131.7 |
| Last PSA prior to PCA diagnosis (ng/mL) | 25 | 4.3 | 0.9–31.6 | 24 | 3.3 | 1.5–9.8 |
| Gleason Score | 25 | 6.2 | 5–8 | 24 | 6.2 | 6–7 |
Note: Percent free PSA is only performed for men with a PSA 2.0–4.0 ng/mL by the previous criteria or a PSA 1.5–2.0ng/mL by the current criteria in PRAP. Therefore, not all men have a percent free PSA performed at baseline.
Percent of the race group
Analysis of clinical role of genetic markers of early-onset PCA
Out of the 460 participants included in this analysis, genotypes were able to be determined by marker as follows: rs6983561 (n=441), rs4430796 (n=430), rs266849 (n=436), rs10993994 (n=447), rs1799950 (n=444), and rs2171492 (n=432).
Time to PCA diagnosis
Among African American participants, rs6983561 genotype significantly predicted earlier time to PCA diagnosis for men carrying the CC vs. AA/AC genotype (hazard ratio = 3.34, 95% CI 1.45–7.70, p=0.005) (Table 2). Rs6983561 genotype was not predictive of time to PCA diagnosis among Caucasian PRAP participants, although no CC genotype was observed among this group. No difference was observed in mean Gleason score by rs6983561 genotype among African American men. None of the other five early onset markers were predictive of time to PCA diagnosis in either race groups. Figure 1 displays the Kaplan-Meier curves for time to PCA diagnosis for rs6983561 by race.
Table 2.
Time to PCA Diagnosis by rs6983561 Genotype Among PRAP Participants with ≥ 1 Follow-up Visit
| n | Genotype | Hazard Ratio£ | p-value§ | 95% CI | |
|---|---|---|---|---|---|
| African American* | |||||
| 247 | AC/CC vs. AA | 1.00 | 1.00 | 0.42–2.36 | |
| 247 | CC vs. AC/AA | 3.34 | 0.005 | 1.45–7.70 | |
| Caucasian* | |||||
| 194 | AC vs. AA | 1.66 | 0.42 | 0.48–5.73 | |
| No CC observed | -- | -- | -- |
Analysis by race adjusted for age at entry and PSA at entry.
Hazard ratio after controlling for age and PSA at enrollment.
p-values bolded are after controlling for multiple comparisons which included two group comparisons with false discovery rate of 5%.
Figure 1.
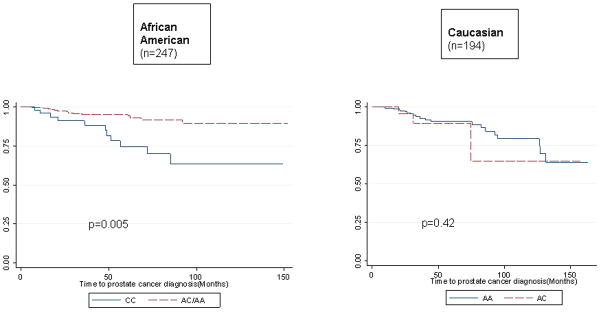
Time to PCA Diagnosis by rs6983561 Genotype
Prediction for PCA by PSA
Among African American participants, rs6983561 genotype influenced the 3-year prediction for PCA by the PSA (interaction p<0.001). The hazard ratio for PCA for African American participants carrying CC and AA/AC were 4.29 (95% CI 2.33–7.91) and 1.33 (95% CI 1.05–1.69), respectively. Among Caucasian participants, while there was no CC genotype observed, there was a trend toward AC genotype influencing PSA prediction for PCA (p=0.04) (Table 3). The predicted probability of developing PCA at three years by rs6983561 genotype is represented in Figure 2. None of the other five early-onset genetic markers influenced PSA prediction for PCA in this analysis.
Table 3.
Cox model results for PSA Prediction for Prostate Cancer by rs6983561 Genotype Among PRAP Participants with ≥ 1 Follow-Up Visit
| Genetic marker | n | Genotype | Hazard Ratio** | p-value | 95% CI |
|---|---|---|---|---|---|
| African American* | 168 | AC/CC | 1.42 | <.001 | 1.19–1.71 |
| 179 | AA | 1.32 | 0.13 | 0.92–1.90 | |
| 247+ | AC/CC vs. AA | 0.81 | |||
| 53 | CC | 4.29 | <.001 | 2.33–7.91 | |
| 194 | AC/AA | 1.33 | 0.018 | 1.05–1.69 | |
| 247+ | CC vs. AC/AA | <.001£ | |||
| Caucasian* | 20 | AC | 4.74 | 0.015 | 1.35–16.63 |
| 174 | AA | 1.23 | 0.033 | 1.02–1.49 | |
| 194+ | AC vs. AA | 0.04 |
Analysis by race adjusted for age at entry.
Hazard ratios are the genotype-specific hazard ratios for one point increase on PSA.
Two group comparison p-value is based on comparing the two genotype-specific hazard ratios.
Significant interaction between PSA and marker genotype is bold after controlling for FDR at 5%.
Figure 2. Race-Specific Three-Year Prediction for Prostate Cancer by PSA at Entry into PRAP by rs6983561 Genotype.
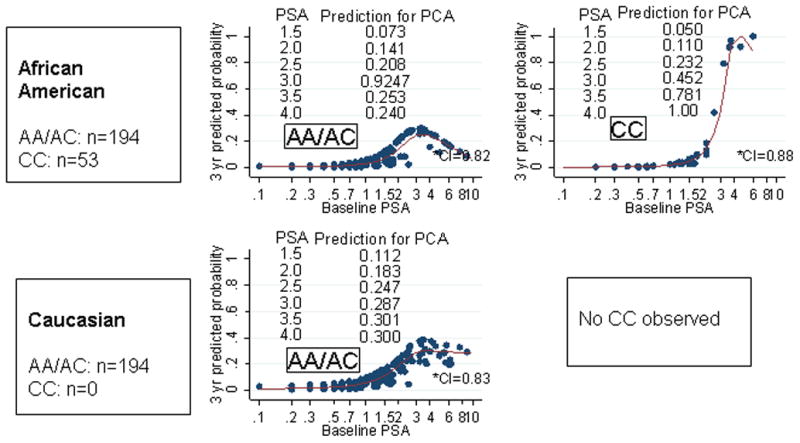
3-year prediction for prostate cancer was computed from Cox models where the covariates included the cubic spline of age and PSA at baseline. Probabilities were computed using STATA 10.0. Participant specific predictions were computed based on their individual characteristics (age and baseline PSA) and fitted model coefficient. Each participant that was used in the analysis received a unique prediction. Prostate cancer cases diagnosed within 6 months of enrollment into PRAP were excluded (n=42 cases excluded).
Assessing predictive accuracy for PCA
Analysis of the predictive accuracy for PCA was performed only for rs6983561 as this marker was associated with earlier time to PCA diagnosis and with higher prediction for PCA by the baseline PSA. Table 4 shows the Harrell’s C estimates as a measure of accuracy of prediction for PCA. Among African American men, there was improvement in predictive accuracy for PCA when rs6983561 genotype was added to PSA particularly when the interaction term was included (Harrell’s C: PSA alone= 0.57 vs. PSA+rs6983561 (CC) genotype + interaction term=0.75, p=0.03).
Table 4.
Accuracy of Longitudinal Prediction for PCA by PSA and rs6983561 Genotype among African American PRAP participants with ≥1 Follow-up Visit
| n | PSA | PSA + rs6983561 (CC) | PSA + rs6983561 (CC with interaction*) | |
|---|---|---|---|---|
| Harrell’s C | Harrell’s C (p-value**) | Harrell’s C (p-value**) | ||
| African American men | 247 | 0.5713 | 0.6679 (0.15) | 0.7540 (0.03) |
Interaction is the interaction term for rs6983561 (CC) genotype with PSA
p-values reflect pairwise comparisons of Harrell’s C in each column to the Harrell’s C for PSA.
Note: Caucasian participants not shown as no CC genotype for rs6983561 was observed.
DISCUSSION
Over the past 4 years, many genetic markers have been identified through genomewide association studies for association to PCA.(6) While many of these markers have a strong statistical association to PCA risk with p-values less than 10−7, the magnitude of risk for PCA is typically modest with odds ratios less than 2.0.(6) Therefore, the clinical role of these markers and assessing individual risk for PCA is unclear. A particular subgroup of men to benefit from efforts to understand individual risk and clinical role of genetic markers are men with familial PCA and African American men as these men are susceptible to early-onset disease which has greater potential impact for PCA-related morbidity and mortality.
Our study was performed to evaluate genetic markers associated with early-onset PCA and gain an understanding of the potential clinical information these markers may provide that can be useful to the individual patient for risk assessment. We assessed the clinical information gained from six genetic markers previously associated with early-onset PCA and evaluated these markers for their prediction of time to PCA diagnosis, influence on PSA prediction for PCA, and accuracy of these markers over the PSA in predicting PCA in an ethnically-diverse, longitudinal cohort of high-risk men enrolled in PCA early detection. Out of the six early-onset markers analyzed, our results show that rs6983561 is informative in predicting time to PCA diagnosis among African American men. In addition, this marker influences PSA prediction for PCA and adds predictive accuracy to PSA for longitudinal prediction for PCA. Our findings suggest that markers associated with early-onset PCA deserve further clinical study as they may add useful clinical information that can help guide clinicians and patients in making PCA screening recommendations in order to diagnose life-ending PCA at a curable point and decrease unnecessary biopsies in high-risk men.
Our study found that rs6983561 genotype has potential clinical use in PCA risk assessment. This polymorphism is located at chromosomal locus 8q24,(6) which is a gene-poor region. The actual function of rs6983561 is unknown; however the c-myc proto-oncogene is located downstream of this region. Studies to date have not found an effect on c-myc expression based on carrier status of markers at 8q24,(25) but research is ongoing to determine potential alternate mechanisms leading to PCA risk.(26)
There are some limitations to consider when interpreting the findings of this study. It is noted that there may be clinical utility from the other markers in this study which were not detected due to our sample size. Our results need further confirmation and validation in larger sample sets. Our study did not identify PCA of higher Gleason score. Further research will need to be performed to determine if identifying intermediate risk PCA will have clinical impact with reducing morbidity and mortality from PCA, particularly in younger men. The overall follow-up rate in the PRAP cohort is 66.8%, which may have limited the ability to detect the clinical impact of the other early-onset genetic markers. However, we did not find any difference in genotype distribution between those included vs. excluded from the analysis except for rs4430796 which was not associated with significant findings in the clinical analysis. Furthermore, our follow-up rate is close to follow-up rates of other screening cohorts.(3) In addition, prostate biopsies are not performed in all PRAP participants, only for those who meet biopsy criteria. Therefore, there may be undetected PCA among some participants which may have influenced our results. Confirmation of our findings is required in larger longitudinal screening studies. Finally, more genetic markers associated with early-onset PCA are being identified and will need study regarding their clinical role in PCA risk assessment.(27)
Overall, our study finds early-onset genetic marker rs6983561 to provide potentially important clinical information regarding time to PCA diagnosis and influences PSA prediction for PCA among African American men. Validation of these findings regarding rs6983561 is needed prior to drawing definitive conclusions regarding the incorporation of this marker in PCA risk assessment. In an era where genomic information is becoming rapidly available due to advances in technology, it is imperative to study the clinical information gained from genomic markers in order to develop optimal early detection approaches. Efforts to elucidate the clinical utility of genetic markers associated with early-onset PCA or clinically-meaningful PCA should inform future personalized risk assessment.
Supplementary Material
Acknowledgments
Funding: NIH 1R03CA150079-01 (V.N. Giri); Fox Chase Cancer Center Core Grant (P30 CA006927) from the National Cancer Institute.
We would like to thank the participants of the Prostate Cancer Risk Assessment Program at Fox Chase Cancer Center.
References
- 1.Cancer Facts and Figures. Atlanta: American Cancer Society; 2011. [accessed July 26 2011]. [Google Scholar]
- 2.Carter BS, Bova GS, Beaty TH, Steinberg GD, Childs B, Isaacs WB, et al. Hereditary prostate cancer: epidemiologic and clinical features. J Urol. 1993;150:797–802. doi: 10.1016/s0022-5347(17)35617-3. [DOI] [PubMed] [Google Scholar]
- 3.Catalona WJ, Antenor JA, Roehl KA, Moul JW. Screening for prostate cancer in high risk populations. J Urol. 2002;168:1980–1983. doi: 10.1016/S0022-5347(05)64276-0. discussion 3–4. [DOI] [PubMed] [Google Scholar]
- 4.Giri VN, Beebe-Dimmer J, Buyyounouski M, Konski A, Feigenberg SJ, Uzzo RG, et al. Prostate cancer risk assessment program: a 10-year update of cancer detection. J Urol. 2007;178:1920–1924. doi: 10.1016/j.juro.2007.07.010. discussion 4. [DOI] [PubMed] [Google Scholar]
- 5.Lin DW, Porter M, Montgomery B. Treatment and survival outcomes in young men diagnosed with prostate cancer: a Population-based Cohort Study. Cancer. 2009;115:2863–2871. doi: 10.1002/cncr.24324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kim ST, Cheng Y, Hsu FC, Jin T, Kader AK, Zheng SL, et al. Prostate cancer risk-associated variants reported from genome-wide association studies: meta-analysis and their contribution to genetic Variation. The Prostate. 2010;70:1729–1738. doi: 10.1002/pros.21208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Beebe-Dimmer JL, Levin AM, Ray AM, Zuhlke KA, Machiela MJ, Halstead-Nussloch BA, et al. Chromosome 8q24 markers: risk of early-onset and familial prostate cancer. Int J of Cancer. 2008;122:2876–2879. doi: 10.1002/ijc.23471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gudmundsson J, Sulem P, Manolescu A, Amundadottir LT, Gudbjartsson D, Helgason A, et al. Nat Genet. 2007;39:631–637. doi: 10.1038/ng1999. [DOI] [PubMed] [Google Scholar]
- 9.Eeles RA, Kote-Jarai Z, Giles GG, Olama AA, Guy M, Jugurnauth SK, et al. Multiple newly identified loci associated with prostate cancer susceptibility. Nat Genet. 2008;40:316–321. doi: 10.1038/ng.90. [DOI] [PubMed] [Google Scholar]
- 10.Camp NJ, Farnham JM, Wong J, Christensen GB, Thomas A, Cannon-Albright LA. Replication of the 10q11 and Xp11 prostate cancer risk variants: results from a Utah pedigree-based study. Cancer Epidemiol Biomarkers Prev. 2009;18:1290–1294. doi: 10.1158/1055-9965.EPI-08-0327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Amundadottir LT, Sulem P, Gudmundsson J, Helgason A, Baker A, Agnarsson BA, et al. A common variant associated with prostate cancer in European and African populations. Nat Genet. 2006;38:652–658. doi: 10.1038/ng1808. [DOI] [PubMed] [Google Scholar]
- 12.Yeager M, Chatterjee N, Ciampa J, Jacobs KB, Gonzalez-Bosquet J, Hayes RB, et al. Identification of a new prostate cancer susceptibility locus on chromosome 8q24. Nat Genet. 2009;41:1055–1057. doi: 10.1038/ng.444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang L, McDonnell SK, Slusser JP, Hebbring SJ, Cunningham JM, Jacobsen SJ, et al. Two common chromosome 8q24 variants are associated with increased risk for prostate cancer. Cancer Res. 2007;67:2944–2950. doi: 10.1158/0008-5472.CAN-06-3186. [DOI] [PubMed] [Google Scholar]
- 14.Savage SA, Greene MH. The evidence for prostate cancer risk loci at 8q24 grows stronger. J Natl Cancer Inst. 2007;99:1499–1501. doi: 10.1093/jnci/djm186. [DOI] [PubMed] [Google Scholar]
- 15.Gudmundsson J, Sulem P, Steinthorsdottir V, Bergthorsson JT, Thorleifsson G, Manolescu A, et al. Two variants on chromosome 17 confer prostate cancer risk, and the one in TCF2 protects against type 2 diabetes. Nat Genet. 2007;39:977–983. doi: 10.1038/ng2062. [DOI] [PubMed] [Google Scholar]
- 16.Levin AM, Machiela MJ, Zuhlke KA, Ray AM, Cooney KA, Douglas JA. Chromosome 17q12 variants contribute to risk of early-onset prostate cancer. Cancer Res. 2008;68:6492–6495. doi: 10.1158/0008-5472.CAN-08-0348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Douglas JA, Levin AM, Zuhlke KA, Ray AM, Johnson GR, Lange EM, et al. Common variation in the BRCA1 gene and prostate cancer risk. Cancer Epidemiol Biomarkers Prev. 2007;16:1510–1516. doi: 10.1158/1055-9965.EPI-07-0137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ross PL, Cheng I, Liu X, Cicek MS, Carroll PR, Casey G, et al. Carboxypeptidase 4 gene variants and early-onset intermediate-to-high risk prostate cancer. BMC Cancer. 2009;9:69. doi: 10.1186/1471-2407-9-69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Douglas JA, Zuhlke KA, Beebe-Dimmer J, Levin AM, Gruber SB, Wood DP, et al. Identifying susceptibility genes for prostate cancer--a family-based association study of polymorphisms in CYP17, CYP19, CYP11A1, and LH-beta. Cancer Epidemiol Biomarkers Prev. 2005;14:2035–2039. doi: 10.1158/1055-9965.EPI-05-0170. [DOI] [PubMed] [Google Scholar]
- 20.Eskew LA, Woodruff RD, Bare RL, McCullough DL. Prostate cancer diagnosed by the 5 region biopsy method is significant disease. J Urol. 1998;160:794–796. doi: 10.1016/S0022-5347(01)62789-7. [DOI] [PubMed] [Google Scholar]
- 21.Rosser CJ, Broberg J, Case D, Eskew LA, McCullough D. Detection of high-grade prostatic intraepithelial neoplasia with the five-region biopsy technique. Urology. 1999;54:853–856. doi: 10.1016/s0090-4295(99)00236-8. [DOI] [PubMed] [Google Scholar]
- 22.Granfeldt PBaJ. Statistics with applications in biology and geology. 1. Chapman and Hall/CRC; 2002. [Google Scholar]
- 23.Hochberg YBaY. Controlling the False Discovery Rate: A practical and powerful approach to multiple testing. Journal of the Royal Statistical Society Series B. 1995;57:289–300. [Google Scholar]
- 24.Newson R. Comparing the predictive power of survival models using Harrell’s c or Somers’ D. The Stata Journal. 2010;10:339–58. [Google Scholar]
- 25.Pomerantz MM, Beckwith CA, Regan MM, Wyman SK, Petrovics G, Chen Y, et al. Evaluation of the 8q24 prostate cancer risk locus and MYC expression. Cancer Res. 2009;69:5568–5574. doi: 10.1158/0008-5472.CAN-09-0387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wasserman NF, Aneas I, Nobrega MA. An 8q24 gene desert variant associated with prostate cancer risk confers differential in vivo activity to a MYC enhancer. Genome Res. 2010;20:1191–1197. doi: 10.1101/gr.105361.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lange EM, Salinas CA, Zuhlke KA, Ray AM, Wang Y, Lu Y, et al. Early onset prostate cancer has a significant genetic component. The Prostate. 2011 doi: 10.1002/pros.21414. (epub ahead of print) [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


