Abstract
Peroxisomes, single-membrane-bounded organelles with essentially oxidative metabolism, are key in plant responses to abiotic and biotic stresses. Recently, the presence of nitric oxide (NO) described in peroxisomes opened the possibility of new cellular functions, as NO regulates diverse biological processes by directly modifying proteins. However, this mechanism has not yet been analysed in peroxisomes. This study assessed the presence of S-nitrosylation in pea-leaf peroxisomes, purified S-nitrosylated peroxisome proteins by immunoprecipitation, and identified the purified proteins by two different mass-spectrometry techniques (matrix-assisted laser desorption/ionization tandem time-of-flight and two-dimensional nano-liquid chromatography coupled to ion-trap tandem mass spectrometry). Six peroxisomal proteins were identified as putative targets of S-nitrosylation involved in photorespiration, β-oxidation, and reactive oxygen species detoxification. The activity of three of these proteins (catalase, glycolate oxidase, and malate dehydrogenase) is inhibited by NO donors. NO metabolism/S-nitrosylation and peroxisomes were analysed under two different types of abiotic stress, i.e. cadmium and 2,4-dichlorophenoxy acetic acid (2,4-D). Both types of stress reduced NO production in pea plants, and an increase in S-nitrosylation was observed in pea extracts under 2,4-D treatment while no total changes were observed in peroxisomes. However, the S-nitrosylation levels of catalase and glycolate oxidase changed under cadmium and 2,4-D treatments, suggesting that this post-translational modification could be involved in the regulation of H2O2 level under abiotic stress.
Keywords: Abiotic stress, cadmium, 2, 4-dichlorophenoxy acetic acid, nitric oxide, S-nitrosylation, peroxisomes
Introduction
Peroxisomes are organelles that have an essentially oxidative type of metabolism (Huang et al., 1983; Fahimi and Sies, 1987; del Río et al., 2006). The important role of plant peroxisomes in a variety of metabolic reactions, including photorespiration, fatty acid β-oxidation, the glyoxylate cycle, and in the biosynthesis of plant hormones such as auxin and jasmonic acid are well known (del Río et al., 2009). Recent proteomic studies have identified new proteins that appear to have novel metabolic and regulatory functions of peroxisomes, such as the production of compatible osmosolute Gly betaine and the degradation of branched amino acids (Reumann et al., 2004). Furthermore, novel regulatory proteins in peroxisomes, such as heat-shock proteins, kinases, and phosphatases have been reported (Hayashi and Nishimura, 2006; Reumann et al., 2007).
Peroxisomes are also essential in plant responses to abiotic and biotic stress (Romero-Puertas et al., 1999; Koh et al., 2005; Rodríguez-Serrano et al., 2009), increasing even during different stress conditions such as xenobiotics, ozone, cadmium, H2O2, and light (del Río et al., 2009). In the last two decades, plant peroxisomes have been shown to produce reactive oxygen species (ROS) such as hydrogen peroxide (H2O2) and the superoxide radical (O2.−) as a consequence of their normal metabolism (reviewed by del Río, 2011). In addition, a complex battery of antioxidative systems had been described, including catalase (CAT), superoxide dismutase (Sandalio and del Río, 1987; del Río et al., 2002), and the ascorbate–glutathione cycle (Jimenez et al., 1997; Mittova et al., 2004; Kuzniak and Sklodowska, 2005; Romero-Puertas et al., 2006). Recently, the presence of an L-arginine-dependent nitric oxide synthase as well as nitric oxide (NO) in plant peroxisomes have also been shown (Barroso et al., 1999; Corpas et al., 2004). In plants, NO is a key signalling molecule involved in several physiological processes from the development to the defence response to both biotic and abiotic stress (Delledonne et al., 2004; Neill et al., 2008). The occurrence of NO in peroxisomes adds new cellular functions to these organelles related to oxygen and nitrogen reactive species (ROS/RNS). No function of NO in peroxisomes, however, has been discovered so far.
The way by which NO undertakes a plethora of physiological functions is still largely unknown. Some time ago in animal tissues, and very recently in plant tissues, it was shown that NO regulates diverse biological processes by directly modifying proteins. In fact, NO and RNS can oxidize, nitrate, or nitrosylate proteins. S-Nitrosylation refers to the binding of a NO group to a cysteine residue and it plays a significant role in NO-mediated signalling (Stamler et al., 2001). Recent evidence suggests that many proteins are S-nitrosylated in plants under physiological or stress conditions, leading to the first insights into S-nitrosylation-dependent regulation of protein function (Lindermayr and Durner, 2009). Although the mechanism remains unknown, several peroxisomal proteins have been shown to be regulated by NO. Thus, the activity and protein level of glutathione peroxidase is down-regulated by NO, whereas the activity of the peroxisomal H2O2-producing β-oxidation is stimulated by NO (Dobashi et al., 1997). Furthermore, in a reversible way, NO inhibits the activities of tobacco CAT and ascorbate peroxidase, whereas both are irreversibly inhibited by peroxynitrite (Clark et al., 2000).
In the present work, the occurrence of S-nitrosylation in plant peroxisomes is assessed. Six peroxisomal proteins are identified as a target for S-nitrosylation, using proteomic techniques (involving a specific biotin-switch method for detecting and purifying S-nitrosylated proteins) together with two other different mass spectrometry techniques: matrix-assisted laser desorption/ionization tandem time-of-flight (MALDI TOF/TOF) and two-dimensional nano-liquid chromatography coupled to two-dimensional ion-trap tandem mass spectrometry (2D-nLC-MS/MS). Additionally, the impact of S-nitrosylation on the functionality of three key peroxisomal proteins, CAT, glycolate oxidase (GOX), and malate dehydrogenase (MDH) has been analysed. Finally, the pattern of CAT and GOX S-nitrosylation under two different types of abiotic stress, metal toxicity by cadmium and treatment with the herbicide 2,4-dichlorophenoxy acetic acid (2,4-D) has also been studied.
Materials and methods
Plant material and growth conditions
Pea (Pisum sativum L. cv. Lincoln) plants were obtained from Royal Sluis (Enkhuizen, Holland). Plants were grown in greenhouse in aerated full-nutrient medium under optimum conditions for 14 d as indicated elsewhere (Sandalio et al., 2001). Then, the medium either remained unsupplemented (control plants) or was supplemented with 50 μM CdCl2 (cadmium-treated plants), and the plants were grown for 14 d. For 2,4-D treatment, plants were grown for 25 d and then herbicide was applied by spraying the leaves with 22.6 mM 2,4-D. The leaves were collected after 3 d of treatment. The concentrations of cadmium and 2,4-D were optimized in previous work to show that both of the selected concentrations induced an oxidative stress response in pea leaves (Sandalio et al., 2001; Romero-Puertas et al., 2004).
Purification of peroxisomes
Peroxisomes were purified from pea leaves by differential and sucrose density-gradient centrifugation (35–60%; w/w) and were recovered from gradients by puncture with a syringe as described by López-Huertas et al. (1995). Peroxisomes were broken by osmotic shock and sucrose was diluted (around 100 times) in 25 mM HEPES buffer (containing 1 mM EDTA and 0.1 mM neocuproine; pH 7.7). Finally, peroxisomes were concentrated by centrifugation (Amicon Ultra, Millipore).
Detection of S-nitrosylated proteins
This study used the biotin-switch method, which converts S-nitrosylated groups into biotinylated groups (Jaffrey et al., 2001) to detect S-nitrosylated proteins, and Western blotting in leaves and peroxisomes with slight modifications. Pea leaves were homogenized in MAE buffer (25 mM HEPES, 1 mM EDTA, 0.1 mM neocuproine, 0.2% Triton X-100; pH 7.7) containing complete protease inhibitor cocktail (Sigma, St. Louis, MO, USA). The extract was centrifuged at 4 °C for 30 min, and the protein concentration in the supernatant was measured by the Bradford assay (Bio-Rad, Hercules, CA, USA). When necessary, homogenates or peroxisomal proteins were pre-treated with the NO donors S-nitroso-N-acetylpenicillamine (SNAP, 1 mM, Calbiochem) and S-nitrosoglutathione (GSNO, 1 mM, Calbiochem) and the glutathionylating agent glutathione disulphide (GSSG, 1 mM, Sigma) in the dark at room temperature for 30 min with regular vortexing. Treatment with the reducing agent tris(2-carboxyethyl)phosphine (TCEP, 10 mM, Sigma) was carried out for 1 h under the same conditions. Reagents were removed by precipitation twice with two volumes of cold acetone and proteins were assayed by the biotin-switch method. Extracts were incubated with 20 mM methylmethanethiosulphonate and 2.5% sodium dodecyl sulphate (SDS) at 50 °C for 30 min with frequent vortexing to block free Cys. Methylmethanethiosulphonate was removed by protein precipitation with two volumes of cold acetone, proteins were resuspended in 0.1 ml RB buffer (25 mM HEPES, 1 mM EDTA, and 1% SDS, pH 7.7) per 1 mg protein. After the addition of 1 mM HPDP-biotin (Pierce, Rockford, IL, USA) and 1 mM ascorbic acid, the mixture was incubated for 1 h at room temperature in the dark with intermittent vortexing. S-Nitrosylated proteins were detected with an anti-biotin antibody (1:10,000, Sigma).
Purification of peroxisome S-nitrosylated proteins
Peroxisome S-nitrosylated proteins were purified by immunoprecipitation with anti-biotin antibody. Samples were prepared with a slight modification of the biotin-switch method (Jaffrey et al., 2001). In brief, 5 mg peroxisomes were incubated with 1 mM GSNO in the dark for 30 min with repeated vortexing. Samples treated with 20 mM dithiothreitol (DTT) were also kept for 1 h under the same conditions as a negative control. Peroxisomal proteins were recovered by precipitating with eight volumes of acetone for 20 min at –20 °C to remove excess GSNO/DTT and were assayed with the biotin-switch method as described before. Biotinylated proteins were purified by anti-biotin-Immobilized Protein A (IPA) as described elsewhere (Perazzolli et al., 2004). Briefly, biotinylated proteins were purified by immunoprecipitation overnight at 4 °C with 15 μl IPA (Pierce)/mg of protein, preincubated with 2 μl of anti-biotin antibody (Sigma). Beads were washed three times with phosphate-buffered saline. Bound proteins were eluted with 10 mM DTT in SDS-PAGE solubilization buffer, separated using 12% polyacrylimide gel, and visualized by Brilliant Blue-G Colloidal Stain (Sigma). Protein bands were analysed by MALDI TOF/TOF. Alternatively, purified proteins were analysed directly by 2D-nLC-MS/MS. When indicated, immunopurified proteins were transferred to a polyvinylidene fluoride (PVDF) membrane (Millipore, Bedford, MA, USA) to detect CAT and GOX proteins with their specific antibodies.
In-gel protein digestion and sample preparation
Bands of interest from Coomassie-stained gels were excised manually, deposited in 96-well plates, and processed automatically in a Proteineer DP (Bruker Daltonics, Bremen, Germany). The digestion protocol was based on Shevchenko et al. (1996) with minor amendments: gel plugs were washed first with 50 mM ammonium bicarbonate and second with acetonitrile (ACN) prior to reduction with 10 mM DTT in 25 mM ammonium bicarbonate solution; and alkylation was carried out with 55 mM iodoacetamide in 50 mM ammonium bicarbonate solution. Gel pieces were then rinsed first with 50 mM ammonium bicarbonate and second with ACN, and were dried under a stream of nitrogen. Modified porcine trypsin (sequencing grade, Promega, Madison WI, USA), at a final concentration of 16 ng/μl in 25% ACN/50 mM ammonium bicarbonate solution, was added and the digestion took place at 37 °C for 6 h. The reaction was stopped by adding 0.5% trifluoroacetic acid (TFA) for peptide extraction. The eluted peptides were dried by speed-vacuum centrifugation and were resuspended in 4 μl of MALDI solution (70% ACN/0.1% TFA aqueous solution). A 0.8 μl aliquot of each peptide mixture was deposited onto a 386-well OptiTOF plate (Applied Biosystems, Framingham, MA, USA) and allowed to dry at room temperature. A 0.8 μl aliquot of matrix solution (3 mg/ml α-cyano-4-hydroxycinnamic acid in MALDI solution) was deposited onto the dried digest and allowed to dry at room temperature.
MALDI peptide mass fingerprinting and MS/MS analysis
For MALDI TOF/TOF analysis, samples were automatically acquired in an ABI 4800 MALDI TOF/TOF mass spectrometer (Applied Biosystems) in positive ion reflector mode (the ion acceleration voltage was 25 kV to MS acquisition and 1 kV to MS/MS) and obtained spectra were stored into ABI 4000 Series Explorer Spot Set Manager. Peptide mass fingerprinting (PMF) and MS/MS fragment ion spectra were smoothed and corrected to zero baseline using routines embedded in ABI 4000 Series Explorer software v3.6. Each PMF spectrum was internally calibrated with mass signals of trypsin autolysis ions to reach a typical mass measurement accuracy of <25 ppm. Known trypsin and keratin mass signals, as well as potential sodium and potassium adducts (+21 Da and +39 Da) were removed from the peak list.
Two-dimensional nano-liquid chromatography and ion-trap tandem mass spectrometry
Alternatively, for 2D-nLC-MS/MS analysis the tryptic peptide mixtures were directly injected onto a strong cationic exchange micro-precolumn (500 mm ID615 mm BioX-SCX, LC Packings, Amsterdam, The Netherlands) with a flow rate of 30 ml/min as a first-dimension separation. Peptides were eluted from the column as fractions by injecting three salt steps of increasing concentration of ammonium acetate (10, 100, and 2000 mM). Each three fractions together with non-retained fraction was on line injected onto a C-18 reversed-phase micro-column (300 mm ID65 mm PepMap, LC Packings) to remove salts, and the peptides were analysed in a continuous ACN gradient consisting of 0–50% B in 45 min and 50–90% B in 1 min (B = 95% ACN, 0.5% acetic acid in water) on a C-18 reversed-phase self-packing nanocolumn (100 mm ID615 cm Discovery BIO Wide pore, Supelco, Bellefonte, PA, USA). A flow rate of about 300 nl/min was used to elute peptides from reversed-phase nanocolumn to a PicoTip emitter nano-spray needle (New Objective, Woburn, MA, USA) for real-time ionization and peptide fragmentation on an Esquire HCT ion-trap mass spectrometer (Bruker Daltonics). Every 1 s, the instrument cycled through the acquisition of a full-scan mass spectrum and one MS/MS spectrum. A 4 Da window (precursor m/z 62), an MS/MS fragmentation amplitude of 0.80 V, and a dynamic exclusion time of 0.30 min were used for peptide fragmentation. 2D-nLC-MS/MS was automatically performed on an advanced micro-column switching device (Switchos) coupled to an auto sampler (Famos) and a nano-gradient generator (Ultimate nano-HPLC, all from LC Packings). The software Hystar 2.3 was used to control whole analytical process.
Database analysis
To submit the combined PMF and MS/MS data to MASCOT version 2.2.04 software (Matrix Science, London, UK), GPS Explorer version 4.9 was used to search in a non-redundant NCBI protein database (NCBInr 20090406, http://blast.ncbi.nlm.nih.gov/Blast.cgi; 8,198,267 sequences, 2,824,199,726 residues). The following search parameters were used: enzyme, trypsin; allowed missed cleavages, 1; carbamidomethyl cysteine as fixed modification by the treatment with iodoacetamide; variable modifications, oxidation of methionine; mass tolerance for precursors was set to ± 50 ppm and for MS/MS fragment ions to ± 0.3 Da. The confidence interval for protein identification was set to ≥95% (P < 0.05) and only peptides with an individual ion score above the identity threshold were considered correctly identified.
Western blot analysis
Protein samples were separated on 12% polyacrylimide and transferred onto PVDF membranes (Sambrook and Russell, 2001). The membrane was stained with Ponceau red to check equivalency of protein loading. For detection of CAT and GOX, antibodies were produced against specific CAT and GOX peptides heterologously expressed in Escherichia coli (Sigma-Genosys) at 1:2000 dilution. Anti-biotin antibody (Sigma) was used at 1:10,000 dilution. Probing and detection of immunocomplexes were performed as described for the SuperSignal West Pico detection system (Pierce).
NO detection by confocal laser scanning fluorescence microscopy
For NO detection, pea-leaf segments (10 mm2) were incubated for 1 h at 25 °C in the dark with 10 μM 4,5-diaminoflorescein diacetate (DAF-2DA, Calbiochem; excitation 495 and emission 515) in 10 mM TRIS-HCl (pH 7.4), as described before (Rodríguez-Serrano et al., 2006).
Electron microscopy and immunolabelling
The samples were processed basically as described elsewhere (Sandalio et al., 1997). Specific polyclonal antibodies for GSNO and glutathione (GSH) were obtained from Calbiochem and Millipore Corporation, respectively. Immunolabelling was carried out by using goat anti-rabbit IgG conjugated to 15 nm gold particles (Sandalio et al., 1997). To avoid cross reactions, primary antibodies were used at minimal concentration for immunodetection and the pre-immune serum was used as a reaction control.
Enzyme assays
S-nitrosoglutathione reductase (GSNOR) activity was assayed in solution according to Sakamoto et al., (2002). Commercial purified proteins (CAT, EC 1.11.1.6; GOX, EC 1.1.3.1; and MDH, EC 1.1.1.37; Sigma) were incubated with two different NO donors, GSNO and diethylenetriamine (DETA) NONOate, at concentrations 0–1000 μM in the dark for 20 min. When indicated, after 1 mM GSNO and DETA NONOate treatment, proteins were incubated with DTT (10 mM) in the dark for 40 min. Pea extracts were incubated with 1 mM GSNO or SNAP for 45 min and activities were measured. GOX activity was assayed spectrophotometrically according to Kerr and Groves (1975), CAT activity was determined as described before (Aebi, 1984), and MDH activity was analysed as previously reported (Walk and Hock, 1977).
Results
NO production and GSNOR activity under 2,4-D treatment
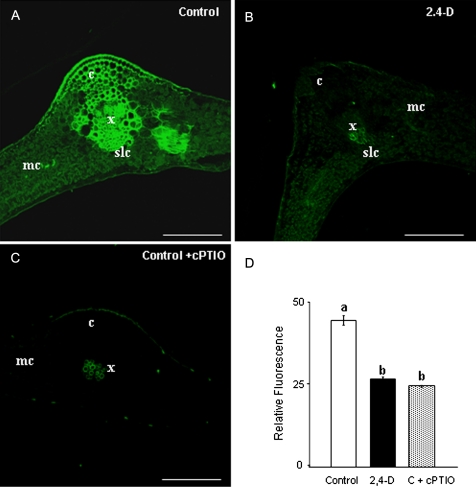
To check possible changes in NO metabolism which could affect S-nitrosylation pattern in pea leaf during abiotic stress, NO production was first studied under 2,4-D treatment (22.6 mM). The herbicide reduced NO accumulation in pea leaves as indicated by DAF-2DA fluorescence (Fig. 1A,B,D). In control plants, the main NO production was located in vascular tissue from the midrib (Fig. 1A) and was eliminated by 2-(4-carboxyphenyl)-4,4,5,5-tetramethylimidazoline-l-oxyl-3-oxide (Fig. 1C). Also, a reduction in NO production was observed previously under cadmium treatment (Barroso et al., 2006; Rodríguez-Serrano et al., 2009). NO can react with GSH, giving rise to GSNO, which is considered to be a NO reservoir (Liu et al., 2001). GSNOR metabolizes GSNO, an enzyme that regulates internal levels of nitrosothiols. Therefore, this study analysed the effect of 2,4-D on this activity and found that GSNOR was induced by 2,4-D treatment (Fig. 2). GSNOR activity was, however, reduced under cadmium treatment, as shown before (Barroso et al., 2006).
Fig. 1.
Nitric oxide (NO) production in pea leaves by confocal laser scanning fluorescence microscopy, after treatment with 22.6 mM 2,4-dichlorophenoxy acetic acid (2,4-D). (A–C) Maximum projections of several optical sections collected by confocal microscopy showing fluorescence due to 4,5-diaminoflorescein diacetate (excitation at 495 nm, emission at 520 nm) in leaf cross-sections of (A) control and (B) 2,4-D-treated plants; (C) negative control, leaves from control plants incubated with 300 μM carboxy 2-phenyl-4,4,5,5-tetramethylimidazoline-1-oxyl 3-oxide (cPTIO), which acts as NO scavenger. (D) Histogram showing relative fluorescence intensities corresponding to NO quantified in arbitrary units using LCS Lite software from Leica. Different letters indicate significant difference (P < 0.01) according to Student’s t-test. These results are representative of at least three independent experiments. c, collenchyma; mc, mesophyll cells; scl, sclerenchyma; x, xylem vessels. Bars, 250 μm.
Fig. 2.
Effect of 2,4-dichlorophenoxy acetic acid (2,4-D) on the activity of S-nitrosoglutathione reductase (GSNOR) in pea leaves. Each column represents the mean ± SE of three independent experiments. *, Differences were significant at P < 0.01 according to Student’s t-test.
S-nitrosylated proteins in pea-leaf extracts under abiotic stress
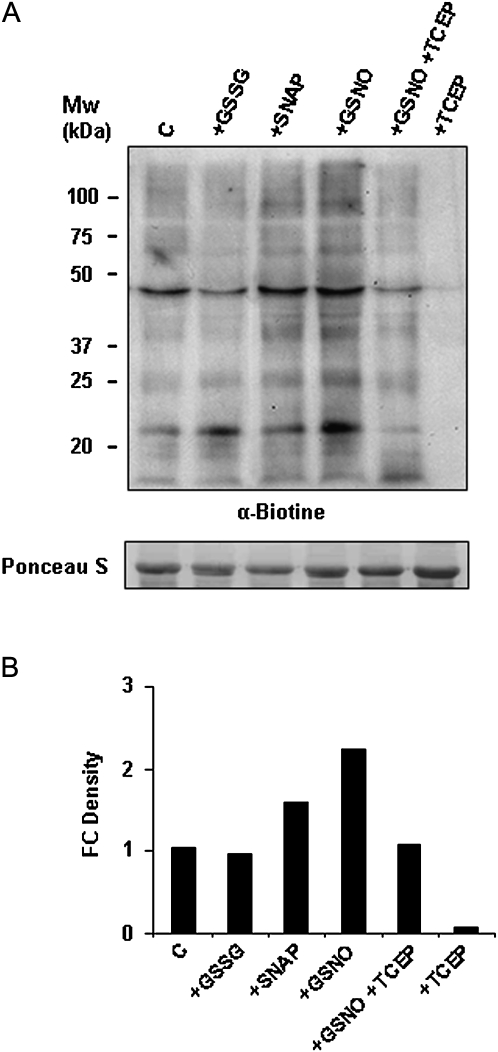
The biotin-switch assay (Jaffrey et al., 2001) that converts S-nitrosylated cysteine into biotinylated cysteine was first evaluated in pea-leaf extracts, for which no previous description is available. Pea-leaf extracts were incubated with the NO donors SNAP or GSNO to induce S-nitrosylation, with GSSG as the glutathionylating agent, or with the reducing agent TCEP as a negative control, subjected to the biotin-switch method, separated by SDS-PAGE, and immunoblotted into a PVDF membrane. Some endogenous S-nitrosylated proteins can be detected in control (non-treated) extracts. Meanwhile, the signals corresponding to S-nitrosylated proteins increased in both extracts treated with a NO donor, SNAP or GSNO, although the signal was higher with the trans-nitrosylating agent GSNO. A similar signal was detected in non-treated extracts, in proteins pre-incubated with glutathionylating agent GSSG, showing that the biotin-switch method does not detect glutathionylation (Fig. 3). Incubation with TCEP, which reduces all cysteine, eliminated the signal, demonstrating the specificity of the method. Based on these results, this method was considered suitable for studying S-nitrosylation in the pea and for analysing the S-nitrosylation pattern during abiotic stress. S-Nitrosylation in pea leaves was studied under cadmium (50 μM) and 2,4-D (22.6 mM) treatments. Both treatments altered NO metabolism in pea plants, although no data on S-nitrosylation has been previously provided. No differences in S-nitrosylation were found in pea-leaf extracts from cadmium-treated plants with respect to control (Fig. 4A), whereas an increase in this post-translational modification was found in plants after 2,4-D treatment (Fig. 4B).
Fig. 3.
Detection of S-nitrosylated proteins in pea-leaf (Pisum sativum L.) extracts. (A) Protein extracts (150 μg) from pea leaves were not treated (control, C) or treated with 1 mM glutathione disulphide (GSSG), 1 mM S-nitroso-N-acetylpenicillamine (SNAP), 1 mM S-nitrosoglutathione (GSNO), or 10 mM tris(2-carboxyethyl)phosphine (TCEP) previously treated or not with GSNO and were subjected to the biotin-switch assay, separated by SDS-PAGE, and immunoblotted with an anti-biotin antibody. Protein loading was verified by Ponceau staining. (B) Histogram showing relative quantification of the Western blot showed in (A). The intensity of bands was quantified using Quantity One Software (version 4.6.2). Band intensity was expressed as fold-change (FC) density. Each band density was first normalized by dividing it by the density of the Ponceau band in the same lane. Then, the relative increase or decrease in density was calculated by dividing the normalized band density of the problem sample by the normalized band density of the control sample. The results are representative of three different Western blots assayed.
Fig. 4.
Detection of S-nitrosylated proteins in pea-leaf (Pisum sativum L.) extracts under abiotic stress. Protein extracts (150 μg) from pea leaves were not treated (control, C) or treated with 50 μM cadmium or 22.6 mM 2,4-dichlorophenoxy acetic acid (2,4-D) and were subjected to the biotin-switch assay, separated by SDS-PAGE, and immunoblotted with an anti-biotin antibody. Protein loading was verified by Ponceau staining. B, Histogram showing relative quantification of Western blot showed in (A). The intensity of bands was quantified as described in the legend for Fig. 3. The results are representative of four different Western blots assayed.
Detection and identification of S-nitrosylated proteins in pea-leaf peroxisomes
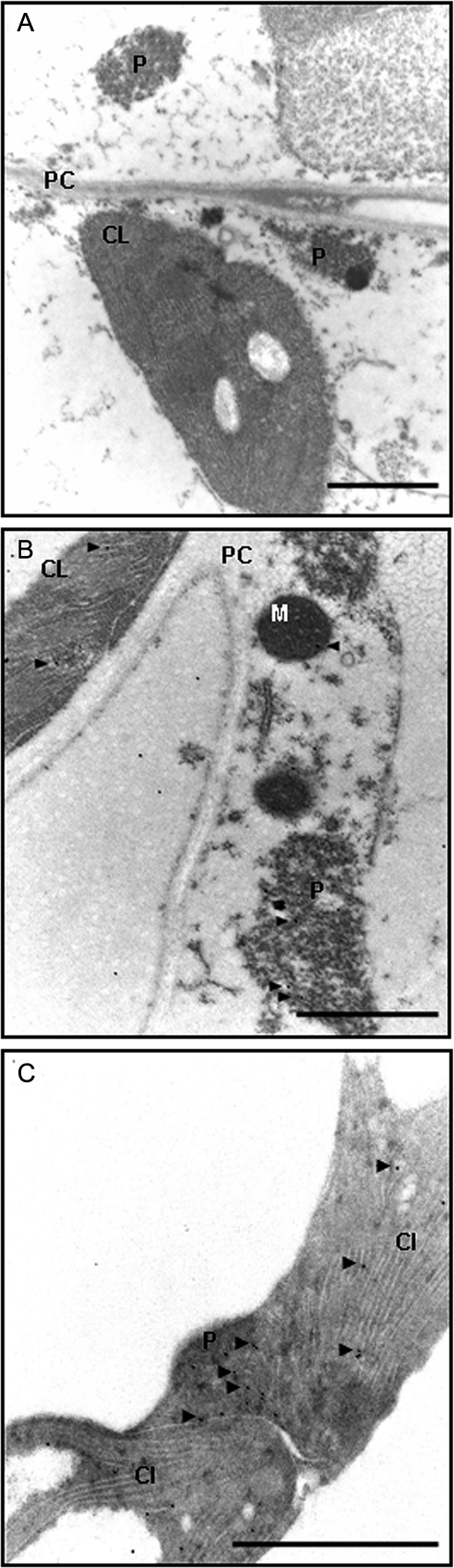
NO has been shown to be produced in pea peroxisomes and these organelles have been involved in stress/signalling related to oxygen and nitrogen species in response to abiotic stress (del Río, 2011), although no information about S-nitrosylation is available to date. First, by immunodetection, electron microscopy was used to check the presence of GSH in peroxisomes that, together with NO, could give rise to GSNO, a well-known S-nitrosylating agent. GSH has been previously described in peroxisomes by biochemical techniques (Jimenez et al., 1997) and by electron microscopy (Zechmann et al., 2008). In addition to GSH, the current study detected GSNO in peroxisomes under physiological conditions (Fig. 5B,C). The presence of GSNO suggests that S-nitrosylation could occur in peroxisomes. Also, GSNO labelling was observed to be located in the chloroplast (Fig. 5), but also located mainly in the collenchyma cell wall and xylem (data not shown).
Fig. 5.
Subcellular localization of S-nitrosoglutathione (GSNO) and glutathione (GSH) in pea leaves. (A) Pre-immune serum control. (B) Pea-leaf cells with anti-GSNO 1:250. (C) Pea-leaf cells with anti-GSH 1:250. Immunogold labelling of GSH and GSNO are indicated by arrowheads. CL, chloroplast; M, mitochondrion; P, peroxisome; PC, cell wall. Bars, 1 μm.
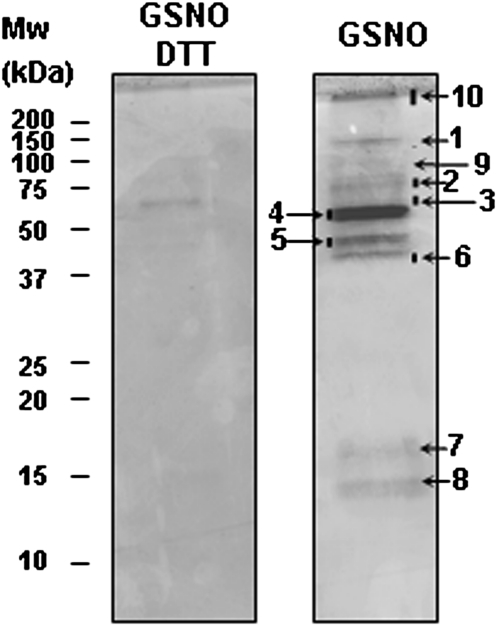
To study the pattern of S-nitrosylated proteins in peroxisomes, purified peroxisomes were treated with GSNO (1 mM) and S-nitrosylation was further evaluated by the biotin-switch assay. The analysis by immunoblots using anti-biotin antibodies showed a band of about 60 kDa in the absence of biotin, which probably corresponds to an endogenous biotinylated protein. Incubation with GSNO consistently intensified and increased the number of bands observed related to control peroxisomes, although some endogenous S-nitrosylated proteins were present under physiological conditions (Fig. 6). As the S-nitrosylation signal under physiological conditions was weak, the biotin-switch method after treating peroxisomal proteins with GSNO was used to identify S-nitrosylated proteins in pea-leaf peroxisomes. Additionally, peroxisomes were treated with DTT as a negative control. To identify S-nitrosylated proteins after the biotin-switch method, the biotinylated proteins were purified by immunoprecipitation with anti-biotin antibody-IPA while two other methods were used to identify the proteins. First, in the proteomic approach, 2D gels were made but there were difficulties in purifying adequate quantities of S-nitrosylated protein for identification by mass spectrometry. Next, samples were loaded on a 1D SDS-PAGE gel, which showed 10 protein bands stained with Coomassie blue, whereas just one band was slightly visible in the GSNO plus DTT-derived eluate (Fig. 7). Thus, with Coomassie staining, this study found bands >100 kDa (band 1), a band of about 75 kDa (band 2), another of 50–75 kDa (band 4), and two bands 37–50 kDa (bands 5 and 6). These presumably correspond to bands observed at these molecular weights by Western blotting (Fig. 6). In the Coomassie gel, the bands corresponding to 25–37 kDa observed in the Western blot were not detected, probably due to the low quantity of these proteins.
Fig. 6.
Detection of S-nitrosylated proteins in pea-leaf (Pisum sativum L.) peroxisomes. Peroxisomal proteins (250 μg) from pea plants were not treated (control, C), treated with 50 μM cadmium or 22.6 mM 2,4-dichlorophenoxy acetic acid (2,4-D), or incubated with 1 mM S-nitrosoglutathione (C+GSNO) or with 20 mM dithiothreitol previously incubated with GSNO (C+GSNO+DTT) and were subjected to the biotin-switch assay, separated by SDS-PAGE, and immunoblotted with an anti-biotin antibody. Protein loading was verified by Ponceau staining of the membrane. The experiment was repeated three times with similar results.
Fig. 7.
S-Nitrosylated proteins of peroxisomes from pea leaves (Pisum sativum L.). Peroxisomal proteins (5 mg) were treated with 1 mM S-nitrosoglutathione (GSNO) and then with 20 mM dithiothreitol (DTT) or not and were subjected to the biotin-switch assay. Biotinylated proteins were purified by immunoprecipitation with an anti-biotin antibody. Eluates were separated by SDS-PAGE and stained by Coomassie blue. The protein bands were identified by matrix-assisted laser desorption/ionization tandem time-of-flight (Table 1).
The Coomassie bands were digested with trypsin and the resulting peptides were subjected to MALDI TOF/TOF analysis (Table 1). To identify low-abundance peroxisomal targets of S-nitrosylation, GSNO-treated proteins were labelled with biotin, immunopurified as described above, and the eluates were subjected to 2D-nLC-MS/MS analysis. In this way, an additional peroxisomal protein could be identified as candidate for S-nitrosylation (Table 1). In total, this study identified 13 proteins having a significant score (score > 50). Two proteins, corresponding to bands 2 and 9, were albumin and immunoglobulin, respectively. Neither protein was a peroxisomal protein but both were involved in the process of obtaining peroxisomes and immunopurified S-nitrosylated proteins. Another protein was a chloroplast one, i.e. Rubisco, which has been shown to be inhibited by S-nitrosylation (Abat et al., 2008), and four proteins were mitochondrial protein contaminations. The mitochondrial proteins found as a target for S-nitrosylation were subunits of the glycine decarboxylase complex, important for the photorespiratory C2 cycle in plants, which was previously found to be a putative target of S-nitrosylation (Palmieri et al., 2010), a finding that strengthens the reliability of the current results. Finally, six peroxisomal proteins were identified as putative targets of S-nitrosylation (Table 1). Four of these, hydroxypyruvate reductase, glycolate oxidase, serine-glyoxylate aminotransferase, and aminotransferase 1, are enzymes that participate in photorespiration, the key physiological process involving peroxisomes. Another enzyme is CAT, one of the main enzymes involved in ROS detoxification. Finally, MDH has also been identified as one of the putative S-nitrosylated peroxisomal proteins. This enzyme, involved in fatty acid β-oxidation in germinating seeds (Pracharoenwattana et al., 2007), is also involved in supplying NADH to hydroxypyruvate reductase in the photorespiration pathway (Reumann and Weber, 2006).
Table 1.
Identified S-nitrosylated proteins from pea-leaf peroxisomes
| Protein namea | Accession number | Mol mass (Mr)/Cal pI | Identified peptides |
Cited as S-nitrosylated | |
| Sequence and scoreb | Sequence coverage (%) | ||||
| Catalase (CAT1) (3, 4, 10) | gi|115705 | 57594 / 6.72 | K.GFFEVTHDISHLTCADFLR.A (126) + 3 | >42 | Foster and Stamler (2004);eRhee et al. (2004);ePalmieri et al. (2010) |
| Aminotransferase 1 (5) | gi|18032028 | 44275 / 8.13 | K.LGSGVAAASAYLQNNIPLIPSR.I (144) | 16 | |
| Serine-glyoxylate aminotransferase (5) | gi|115477148 | 44466 / 8.19 | R.NHLFVPGPVNIPDQVIR.A (116) | 10 | |
| Glycolate oxidase (GOX1) (6) | gi|167961875 | 40825 / 9.16 | R.IAVQSGAAGIIVSNHGAR.Q (136) + 1 | 42 | Abat et al. (2008) |
| Hydroxypyruvate reductase (6) | gi|223528624 | 42192 / 6.97 | K.MNLIYFDLYQATR.L (63) + 1 | 23 | |
| Malate dehydrogenase, glyoxysomal precursor (•) | gi|3183079 | 37362 / 6.9 | R.LLGVTTLDVVR.A (41) | 25 | |
| Glycine dehydrogenase (1, 10)c | gi|121083 | 115411 / 7.17 | R.ESPYLTHPIFNTYQTEHELLR.Y (140) + 4 | 38 | Palmieri et al. (2010) |
| Chain A, dihydrolipoamide dehydrogenase (of Gly decarboxylase) (3)c | gi|9955321 | 49998 / 6.06 | K.LTVEPSAGGEQTIIEADVVLVSAGR.T (103) | 34 | Foster and Stamler (2004);eRhee et al. (2004)e |
| Aminomethyltransferase (5)c | gi|3915699 | 44656 / 8.79 | R.VLDINGSQCFLTR.T (95) + 2 | 45 | Palmieri et al. (2010) |
| H protein (7)c | gi|737595 | 13979 / 4.57 | K.IKPTSPDELESLLGAK.E (59) + 1 | 19 | Palmieri et al. (2010) |
| RuBisCO large subunit (3, 4, 10)d | gi|113374116 | 49312 / 6.46 | K.TFQGPPHGIQVER.D (79) + 1 | 35 | Lindermayr et al. (2005); Abat et al. (2008); Abat and Deswal (2009) |
| RuBisCO small subunit (8)d | gi|132097 | 20402 / 9.24 | K.ELDEVVAAYPQAFVR.I (125) + 5 | 69 | Abat et al. (2008) |
Peroxisomes treated with S-nitrosoglutathione were subjected to the biotin-switch method and analysed by matrix-assisted laser desorption/ionization tandem time-of-flight and two-dimensional nano-liquid chromatography coupled to ion-trap tandem mass spectrometry (filled circle).
Comassie bands are indicated in brackets.
The best-matching peptide identifying the protein is given, with the score in parentheses. If there were further peptides found, the number of the peptides is given as an additional number.
Protein localized in mitochondria
Protein localized in chloroplasts
In animal tissue or microbes.
Regulation of selected proteins activity by NO
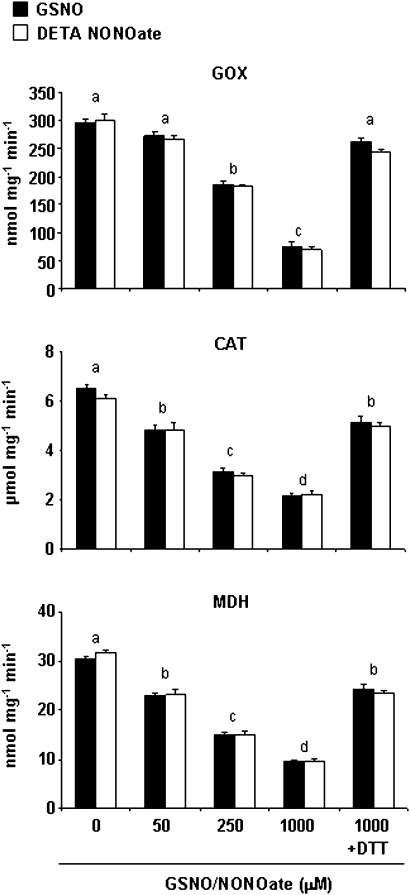
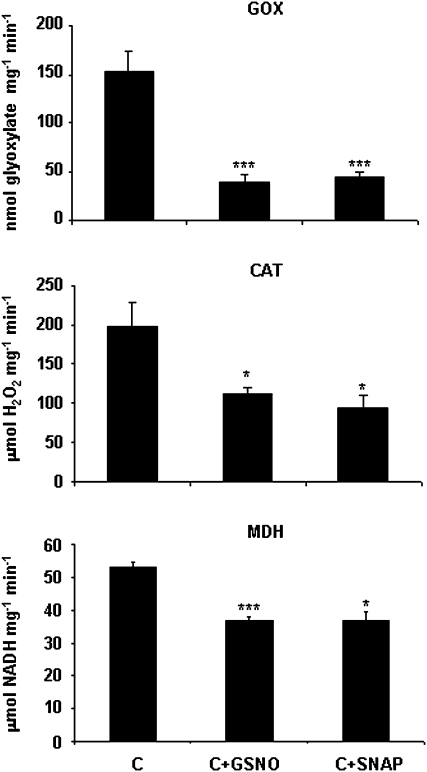
Approximately 90% of NO-Cys sites have both positively and negatively charged residues within 8 Å (Marino and Gladyshev, 2010) and therefore this study checked this feature for CAT and GOX. Although cytoplasmic MDH (Cys 154) lacks the acid–base motif, it seems that a His at 7.9 Å from the sulphur group of Cys is sufficient for direct nitrosylation of this Cys (Marino and Gladyshev, 2010). The structural model for AtGOX1 (http://swissmodel.expasy.org) gave insight into the spatial disposition of Cys residue in this enzyme and an acid–base motif was also found within 8 Å from the sulphur group of the Cys (Supplementary Fig. S1, available at JXB online). This reinforces the idea that this protein is a promising target for S-nitrosylation. All six cysteine residues that are present in CAT1 are free amino acids and therefore all are possible targets for NO. Using the computational prediction of the protein S-nitrosylation program (Xue et al., 2010), it was found that Cys-230 has a high probability of being the Cys that is potentially S-nitrosylated. The structural model for CAT1 from Arabidopsis thaliana (http://swissmodel.expasy.org) enabled the finding an acid–base motif within 8 Å from the sulphur group of this Cys (Supplementary Fig. S2), supporting the data gathered by this program. To verify the effects of S-nitrosylation on CAT, GOX, and MDH activities, their function was monitored after treatment with two different NO donors, GSNO and DETA NONOate (Fig. 8). Significant inhibition of these enzyme activities was found after incubation with both NO donors in a concentration-dependent manner, up to 25–30% of the total activity when treated with 1 mM GSNO or DETA NONOate, compared with untreated proteins. This effect could be counteracted by the thiol-specific reductant DTT (Fig. 8). To certify the effect of S-nitrosylation on pea CAT and GOX activities, pea extracts were incubated with the NO donors GSNO and SNAP, showing a similar reduction (Fig. 9). As MDH presents more than one isoform located in different compartments, pea-leaf peroxisomes were incubated with the NO donors, achieving a reduction in MDH activity as well (Fig. 9).
Fig. 8.
Effect of NO on glycolate oxidase (GOX), catalase (CAT), and malate dehydrogenase (MDH) activities. Commercial proteins were preincubated with different concentrations (0–1000 μM) of GSNO (black bars) or diethylenetriamine (DETA) NONOate (white bars) for 30 min at room temperature and then enzyme activities were determined (Materials and methods). Incubation with 10 mM dithiothreitol (DTT) after 1000 μM GSNO or DETA NONOate restored enzyme activities. For each concentration, measurements were made in triplicate. Different letters indicate significant differences (P < 0.001 for GOX and MDH and P < 0.01 for CAT), as determined by Student’s t-test.
Fig. 9.
Effect of NO on glycolate oxidase (GOX), catalase (CAT), and malate dehydrogenase (MDH) activities in pea leaves (Pisum sativum L.). Pea extracts for measuring CAT and GOX activity and pea peroxisomes for MDH activity were preincubated with 1 mM S-nitrosoglutathione (GSNO) and 1 mM S-nitroso-N-acetylpenicillamine (SNAP) for 45 min at room temperature and then enzyme activities were determined (Materials and methods). Three different extracts were used for each measurement. Asterisk indicates significant differences (*, P < 0.05; ***, P < 0.001) according to Student’s t-test.
S-nitrosylated proteins in pea-leaf peroxisomes under abiotic stress
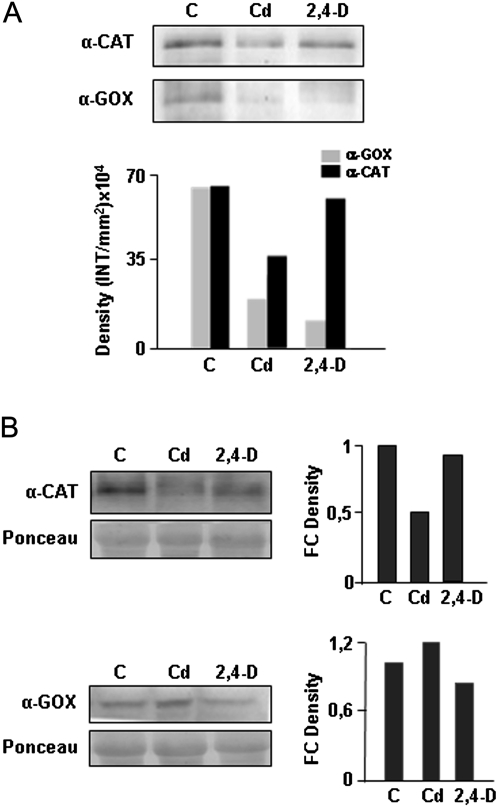
The peroxisomal targets for S-nitrosylation are key enzymes involved in important metabolic processes, so this study investigated their putative post-translational alteration in response to abiotic stress. Western blot analysis using an anti-biotin antibody showed no differences in the global pattern of S-nitrosylated proteins on peroxisomes from pea plants treated with cadmium or 2,4-D assayed with the biotin-switch method (Fig. 6). To gain further insight into the proteins identified previously, pea plants were treated with cadmium or 2,4-D, S-nitrosylated peroxisomal proteins were purified and blotted and specific antibodies used against CAT and GOX were used (Fig. 10). Both proteins were found to be S-nitrosylated under physiological conditions. A reduction of S-nitrosylated CAT under cadmium treatment was observed, while no differences were found under the 2,4-D treatment with respect to non-treated plants (Fig. 10A). Similar changes occurred in the total amount of this protein under both types of stress (Fig. 10B). Although an increase in total GOX protein occurred under cadmium treatment, S-nitrosylated GOX nearly disappeared under metal stress (Fig. 10). Under 2,4-D treatment, a slight reduction in total GOX was noted (Fig. 10B) and S-nitrosylation level of the protein practically vanished (Fig. 10A).
Fig. 10.
S-Nitrosylation level of catalase (CAT) and glycolate oxidase (GOX) during abiotic stress. (A) Peroxisomal proteins from pea plants were not treated (control, C) or treated with cadmium and 2,4-dichlorophenoxy acetic acid (2,4-D) and were subjected to the biotin-switch assay. S-Nitrosylated proteins were immunopurified with anti-biotin antibody and subjected to Western blot analysis with anti-CAT and anti-GOX antibodies. (B) Variation in CAT and GOX protein accumulation during abiotic stress was monitored by Western blot analysis of peroxisomal proteins, as described above and not subjected to the biotin-switch method. The intensity of bands was quantified using Quantity One version 4.6.2 software. Band intensity was expressed as density in (A) and as fold-change (FC) density in (B) as described in the legend for Fig. 3. The figure is representative of two independent experiments.
Discussion
In recent years, few proteomic studies have been made to identify targets for protein S-nitrosylation in plants. In A. thaliana cell-culture extracts treated with GSNO and in NO-treated plants, more than 100 proteins have been identified (Lindermayr et al., 2005) and 16 proteins have been found to be differentially S-nitrosylated in A. thaliana plants under a hypersensitive response (Romero-Puertas et al., 2008). Additionally, 19 and 20 proteins were identified in Kalanchoe pinnata and Brassica juncea, respectively, some being modulated by low temperature in Brassica (Abat et al., 2008; Abat and Deswal, 2009). The identification of the candidates was the starting point for the functional and biochemical characterization of S-nitrosylation in plants. Thus, different physiological processes, such as ethylene biosynthesis, defence response against both abiotic and biotic stress, antioxidant defence, proteolitic activity, DNA binding, photosynthesis, and the mitochondrial photorespiratory pathway has been shown to be regulated by S-nitrosylation of key enzymes involved in these processes (Perazzolli et al., 2004; Lindermayr et al., 2006; Belenghi et al., 2007; Romero-Puertas et al., 2007; Serpa et al., 2007; Abat et al., 2008; Tada et al., 2008; Wang et al., 2008; Palmieri et al., 2010). However, it should be borne in mind that S-nitrosylation leads to changes not only in protein activity but also in protein–protein interactions and/or subcellular localization (Benhar and Stamler, 2005), and therefore spatiotemporal distribution of S-nitrosylated proteins are key components for S-nitrosylation significance. This finding prompted this study to monitor the identification of the S-nitrosylated proteome of peroxisomes and its changes under abiotic stress.
Here, this study shows the presence of GSNO in peroxisomes by immunoelectron microscopy, suggesting that S-nitrosylation could occur in these organelles. An S-nitrosylation-dependent signal was also observed in pea peroxisomes by the biotin-switch method, although this signal was weak probably due to lability of this modification and the long process required to obtain the organelles. Peroxisomal proteins are, however, active and able to be S-nitrosylated after GSNO treatment despite the osmotic shock and dilution of peroxisomes. To identify the S-nitrosoproteome of peroxisomes, they were treated with GSNO, an effective trans-nitrosylating agent that increases S-nitrosylated proteins in the peroxisome. The biotin-switch method (Jaffrey et al., 2001) was used, coupled with affinity purification of biotinylated proteins that have previously been used in different plant species (Lindermayr et al., 2005; Abat et al., 2008; Romero-Puertas et al., 2008) and even in organelles such as the mitochondria (Foster and Stamler, 2004; Palmieri et al., 2010). The analysis of Coomassie bands found by MALDI TOF/TOF mass spectrometry and the direct analysis of the eluates by 2D-nLC-MS/MS resulted in the identification of six proteins belonging to peroxisomal metabolism related to ROS detoxification, β-oxidation, and photorespiration.
One of the main metabolic processes responsible for generation of H2O2 in peroxisomes is β-oxidation of fatty acids, exclusive of peroxisomes in plants and with several signalling molecules derived from it. In fact, jasmonic acid and its derivatives, salicylic acid and indolacetic acid, are generated from β-oxidation in peroxisomes (Nyathi and Baker, 2006). Additionally, β-oxidation of fatty acid converts the seed-lipid reserve into sugars to be used for germination and plant growth (Huang et al., 1983; Baker and Graham, 2002). The current analysis revealed that NAD-dependent MDH, the glyoxysomal precursor enzyme (peroxisomal MDH) that serves to reoxidize NADH produced from fatty acid β-oxidation (Pracharoenwattana et al., 2007), appears as a target of S-nitrosylation in pea-leaf peroxisomes. The mitochondrial MDH isoform has also been considered a candidate for S-nitrosylation in rat-liver mitochondrial extracts exposed to GSNO (Foster and Stamler, 2004) and in Arabidopsis leaves undergoing a hypersensitive response (Romero-Puertas et al., 2008). The effect of this modification on the activity of the protein, however, has not been studied up to now, although it is well known that the oxidation of -SH groups inactivates the enzyme (Varrone et al., 1970). The current study shows that different NO donors severely inhibit MDH activity in a concentration-dependent manner, and further studies are necessary to determine whether this post-translational modification could affect the NAD/NADH ratio in peroxisome and sugar bioavailability in the cell where MDH is involved. A key antioxidant enzyme such as CAT has been found to be one of the targets of S-nitrosylation in peroxisomes, which has been reported to undergo S-nitrosylation in the analysis of mitochondrial S-nitrosoproteomes in both animal and plant tissue. These mitochondrial fractions, however, seemed to be contaminated with peroxisomes (Foster and Stamler, 2004; Palmieri et al., 2010). Interestengly, CAT has not been described as a target for S-nitrosylation in proteomic studies from plant tissues, indicating that an analysis of isolated organelles can contribute to finding new targets that could go unnoticed in total extracts. It has been shown that S-nitrosylation inhibits the activity of a member of the peroxiredoxin family, peroxiredoxin II E (Romero-Puertas et al., 2007), which is located in chloroplast and reduces H2O2 and thereby protects cells from oxidative damage (Dietz, 2003). To test the functional effect of S-nitrosylation on CAT activity, this study incubated commercial enzyme and pea extracts with different NO donors and found an inhibition in activity. The results suggest that NO could regulate the antioxidant defence system and H2O2 levels in peroxisomes through S-nitrosylation. This effect is of particular interest because both molecules, NO and H2O2, have emerged as key players in plant-cell signalling, particularly under various types of stress. Starting from the same quantity of proteins, this study observed that under cadmium stress both the protein content and the S-nitrosylation level of CAT decreased with respect to non-treated plants. Apparently, the reduction observed in CAT activity in a previous work could be due to the reduction in protein content (Sandalio et al., 2001; Romero-Puertas et al., 2007) and not to changes in post-translational modifications or oxidation (Romero-Puertas et al., 2002). No changes were found in the content of CAT and S-nitrosylated CAT under 2,4-D treatment with respect to the control plants. In fact, CAT was not affected by the herbicide 2,4-D, either in its activity or its transcript levels (Pazmiño et al., 2011). It appears that ascorbate peroxidase could be the main enzyme involved in detoxifying H2O2 during the treatment with the herbicide (Pazmiño et al., 2011).
It bears mentioning that among the six peroxisomal proteins that were identified as targets of S-nitrosylation, four of them (five if MDH is included), were proteins of the photorespiration cycle, which is the main physiological process of peroxisomes. This metabolic pathway involves chloroplast, mitochondria, and peroxisomes, with more than 16 enzymes and at least six membrane channels distributed in the three organelles (Douce and Heldt, 2000; Reumann and Weber, 2006). It seems that photorespiration is a wasteful process: in this pathway, O2, ATP, and reducing equivalents are used for CO2, NH3, and H2O2 production (Buchanan et al., 2000). Although the functions of photorespiration are not clear, it is well accepted that this pathway affects a wide range of processes, such as carbon metabolism, nitrogen assimilation, bioenergetics, photosystem-II function, and respiration, and it is a critical source of H2O2 in photosynthetic cells (Foyer and Noctor, 2009). It seems that NO is able to modulate photorespiratory metabolism as the two enzymes involved in the mitochondrion step, glycine decarboxylase and serine hydroxymethyltransferase, which are responsible for the conversion of Gly to Ser, are targets of S-nitrosylation (Palmieri et al., 2010). Also, the current results show that all of the enzymes involved in the peroxisomal photorespiration steps are targets of S-nitrosylation as well. The key enzyme of the photorespiratory cycle in peroxisomes is GOX, which oxidizes glycolate, transfers the electrons to O2, and generates the product glyoxylate and H2O2 (Reumann, 2002). The current study has shown that GOX is inhibited by NO donors and this protein has been found to be S-nitrosylated under physiological conditions. These results suggest that NO could regulate H2O2 levels under physiological conditions, through S-nitrosylation by not only regulating the antioxidant defence system but also the H2O2-producing enzymes. Furthermore, this regulation could change, depending on the need to control the H2O2 level in the plants during stress conditions. In previous studies, an increase in H2O2 production in pea plants treated with cadmium has been shown (Romero-Puertas et al., 2004) probably due to the greater GOX activity under the metal stress (Romero-Puertas et al., 1999). Thus, the current study has shown that GOX is S-nitrosylated under physiological conditions while the S-nitrosylated protein almost disappears under cadmium stress, although an increase in GOX protein content is observed. This result agrees with the intensification of GOX activity found previously (Romero-Puertas et al., 1999), which was probably due to not only a slight increase in protein content but also a post-translational regulation through the reduction of S-nitrosylation of the enzyme. On the other hand, the reduction on S-nitrosylation of GOX in pea plants treated with 2,4-D (this work) is not correlated with the reduction of GOX activity observed after herbicide treatment (Pazmiño et al., 2011). This could be due to the reduction in the protein content observed with 2,4-D treatment. It has been shown that there is a reduction in the phosphorylation of GOX during 2,4-D treatment, which could be also responsible for the observed decrease in GOX activity (Pazmiño, 2009).
Although both abiotic stresses, cadmium (Barroso et al. 2006; Rodríguez-Serrano et al. 2009) and 2,4-D, reduced NO in pea plants, this study found no differences in the S-nitrosylation of proteins under cadmium treatment but an increase in the signal in the presence of the herbicide. This divergence is probably due to differences in NO metabolism under both types of stress. More intense GSNOR activity was found during 2,4-D treatment, while a reduction in this activity has previously been reported with the metal (Barroso et al., 2006). While the current results appear to contradict the findings reported in Arabidopsis mutants over-expressing AtGSNOR (obtained by expression of transgenes or T-DNA insertion mutants), which showed a lowering of total S-nitrosylation levels, no data on NO production, which could influence S-nitrosylation levels as well, are reported in these plants (Feechan et al., 2005; Rusterucci et al., 2007). Additionally, the methods used in these studies to analyse total SNOs in the plants are not the same as those used in the present work. In addition, this study examined the response of pea plants under specific stress conditions, while over-expression of a gene such as GSNOR could give rise to some pleiotropic effect involving hormone or nitrate impairment (Feechan et al., 2005; Lee et al., 2008) and cannot be compared with the response to a specific stress. In fact, it has been shown that GSNOR activity is necessary for the acclimation of plants to high temperatures and for normal development and fertility under optimal growth conditions (Lee et al., 2008). GSNOR controls both NO availability and intracellular S-nitrosylation levels, affecting protein S-nitrosylation and important metabolic pathways such as disease resistance (Liu et al., 2001; Feechan et al., 2005; Rusterucci et al., 2007). The current results suggest that this activity could be involved in cadmium and 2,4-D response as well.
In conclusion, six peroxisomal proteins have been identified as the target of S-nitrosylation and the activities of three proteins have been shown to be affected by this modification. The S-nitrosylation levels of CAT and GOX, the main H2O2-removing and -producing enzymes in peroxisomes, have been analysed in peroxisomes from pea leaves treated with two abiotic stresses. It was shown that S-nitrosylation could regulate the peroxisomal level of key signalling molecules such as H2O2, as explained in the model presented in Fig. 11. Future studies on the identified proteins will help in understanding the significance of S-nitrosylation in plant peroxisomes and the peroxisomal contribution throughout plant development and during plant responses to different stress conditions. Special interest on the photorespiration pathway is needed because it appears to be regulated by S-nitrosylation. Post-translational modifications fine tune the photorespiratory enzymes, which could be a fast adaptive mechanism to prevent oxidative damage by H2O2 accumulation, but also could regulate the flux of the metabolites shared by different pathways in peroxisomes. In addition to S-nitrosylation, other post-translational modifications such as phosphorylation or nitration could help to regulate peroxisomal metabolism. Preliminary results according to proteomic approaches have demonstrated that CAT, GOX, and MDH are also phosphorylated (Pazmiño, 2009), pointing to the relevance of post-translational regulation of photorespiration and paving the way for future studies on the relevance of post-translational modification cross-talk.
Fig. 11.
Model of the effect of cadmium and 2,4-dichlorophenoxy acetic acid (2,4-D) treatments on NO metabolism. Both types of abiotic stress reduced NO production, although a differential effect was observed in GSNOR activity. An increase in S-nitrosylated proteins under 2,4-D treatment was detected while no differences were found under cadmium treatment. However, a reduction in GOX S-nitrosylation was found in peroxisomes under both treatments while CAT S-nitrosylation was reduced only under cadmium treatment. The increase of GOX protein and the reduction of the S-nitrosylation pattern of this protein, which induced GOX activity under cadmium stress, with the reduction of CAT activity could in part be responsible for the increase in H2O2 observed under this stress. Under 2,4-D stress, however, the reduction of GOX protein led to a reduction in its activity, although GOX S-nitrosylation practically disappeared. It seems that the increase in H2O2 found with the herbicide treatment was not due to GOX activity but to ACX and XOD (Pazmiño et al., 2011). ACX, acyl CoA oxidase; CAT, catalase; GOX, glycolate oxidase; GSH, glutathione; GSNO, S-nitrosoglutathione; GSNOR, S-nitrosoglutathione reductase; NO, nitric oxide; SNO, S-nitrosylated; SNOS, S-nitrosylated proteins; XOD, xanthine oxidase.
Supplementary material
Supplementary data are available at JXB online.
Supplementary Fig. S1. Structure of Arabidopsis GOX1.
Supplementary Fig. S2. Structure of Arabidopsis CAT1.
Acknowledgments
This study was supported by Marie Curie Actions-European Re-integration Grants (FP7-PEOPLE-2007-2-2-ERG; project no. 208146), by the Proyecto Intramural Especial from Consejo Superior de Investigaciones Científicas (CSIC, project 200740I017) and by a European Regional Development Fund-cofinanced grant (BIO2008-04067) from the Ministerio de Ciencia e Innovación, Spain. Identification of proteins was carried out at Proteomic Services of Centro Nacional de Biotecnología (CSIC, Madrid), about which the authors appreciate the valuable advice given. Electron and confocal microscopy analysis were carried out at Technical Services from the Universities of Granada and Jaén, respectively.
References
- Abat JK, Deswal R. Differential modulation of S-nitrosoproteome of Brassica juncea by low temperature: change in S-nitrosylation of Rubisco is responsible for the inactivation of its carboxylase activity. Proteomics. 2009;9:4368–4380. doi: 10.1002/pmic.200800985. [DOI] [PubMed] [Google Scholar]
- Abat JK, Mattoo AK, Deswal R. S-Nitrosylated proteins of a medicinal CAM plant Kalanchoe pinnata – ribulose-1,5-bisphosphate carboxylase/oxygenase activity targeted for inhibition. FEBS Journal. 2008;275:2862–2872. doi: 10.1111/j.1742-4658.2008.06425.x. [DOI] [PubMed] [Google Scholar]
- Aebi H. Catalase in vitro. Methods in Enzymology. 1984;105:121–126. doi: 10.1016/s0076-6879(84)05016-3. [DOI] [PubMed] [Google Scholar]
- Baker A, Graham I. Plant Peroxisomes. Biochemistry, Cell Biology and Biotechnological Applications. Dordrecht, The Netherlands: Kluwer Academic Publishers; 2002. [Google Scholar]
- Barroso C, Romero LC, Cejudo FJ, Vega JM, Gotor C. Salt-specific regulation of the cytosolic O-acetylserine(thiol) lyase gene from Arabidopsis thaliana is dependent on abscisic acid. Plant Molecular Biology. 1999;40:729–736. doi: 10.1023/a:1006285016296. [DOI] [PubMed] [Google Scholar]
- Barroso JB, Corpas FJ, Carreras A, et al. Localization of S-nitrosoglutathione and expression of S-nitrosoglutathione reductase in pea plants under cadmium stress. Journal of Experimental Botany. 2006;57:1785–1793. doi: 10.1093/jxb/erj175. [DOI] [PubMed] [Google Scholar]
- Belenghi B, Romero-Puertas MC, Vercammen D, Brackenier A, Inze D, Delledonne M, Van Breusegem F. Metacaspase activity of Arabidopsis thaliana is regulated by S-nitrosylation of a critical cysteine residue. Journal of Biological Chemistry. 2007;282:1352–1358. doi: 10.1074/jbc.M608931200. [DOI] [PubMed] [Google Scholar]
- Benhar M, Stamler JS. A central role for S-nitrosylation in apoptosis. Nature Cell Biology. 2005;7:645–646. doi: 10.1038/ncb0705-645. [DOI] [PubMed] [Google Scholar]
- Buchanan BB, Gruissem W, Russell RL. Biochemistry and Molecular Biology of Plants. Rockville, MD: American Society of Plant Biologists; 2000. [Google Scholar]
- Clark D, Durner J, Navarre DA, Klessig DF. Nitric oxide inhibition of tobacco catalase and ascorbate peroxidase. Molecular Plant–Microbe Interactions. 2000;13:1380–1384. doi: 10.1094/MPMI.2000.13.12.1380. [DOI] [PubMed] [Google Scholar]
- Corpas FJ, Barroso JB, Carreras A, et al. Cellular and subcellular localization of endogenous nitric oxide in young and senescent pea plants. Plant Physiology. 2004;136:2722–2733. doi: 10.1104/pp.104.042812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- del Río LA. Peroxisomes as a cellular source of reactive nitrogen species signal molecules. Archives of Biochemistry and Biophysics. 2011;506:1–11. doi: 10.1016/j.abb.2010.10.022. [DOI] [PubMed] [Google Scholar]
- del Río LA, Corpas FJ, Sandalio LM, Plasama JM Gomez, M, Barroso JB. Reactive oxygen species, antioxidant systems and nitric oxide in peroxisomes. Journal of Experimental Botany. 2002;53:1255–1272. [PubMed] [Google Scholar]
- del Río LA, Sandalio LM, Corpas FJ, Palma JM, Barroso JB. Reactive oxygen species and reactive nitrogen species in peroxisomes. Production, scavenging, and role in cell signaling. Plant Physiology. 2006;141:330–335. doi: 10.1104/pp.106.078204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- del Río LA, Sandalio LM, Corpas FJ, Palma JM. Peroxisomes as a cellular source of reactive oxygen species signal molecules. In: del Río LA, Puppo A, editors. Reactive Oxygen Species in Plant Signaling. Dordrecht, Heidelberg, London, New York: Springer; 2009. pp. 95–111. [Google Scholar]
- Delledonne M, Perazzolli M, Dominici R, Romero-Puertas MC, Zago E. Non-symbiotic plant hemoglobin modulates nitric oxide bioactivity. Free Radical Biology and Medicine. 2004;36:S41–S42. doi: 10.1105/tpc.104.025379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dietz KJ. Plant peroxiredoxins. Annual Review of Plant Biology. 2003;54:93–107. doi: 10.1146/annurev.arplant.54.031902.134934. [DOI] [PubMed] [Google Scholar]
- Dobashi K, Chahal A, Singh I. Modulation of endogenous antioxidant enzymes by nitric oxide in rat C6 glial cells. Journal of Neurochemistry. 1997;68:1896–1903. doi: 10.1046/j.1471-4159.1997.68051896.x. [DOI] [PubMed] [Google Scholar]
- Douce R, Heldt HW. Photorespiration. In: Leegood RC, Sharkey TD, von Cammerer S, editors. Phothosynthesis: physiology and metabolism. Dordrecht: Kluwer Academic Publishers; 2000. pp. 115–136. [Google Scholar]
- Fahimi HD, Sies H. Peroxisomes in biology and medicine. 1987 Fahimi HD and Sies H, eds. Berlin, Springer-Verlag. [Google Scholar]
- Feechan A, Kwon E, Yun BW, Wang Y, Pallas JA, Loake GJ. A central role for S-nitrosothiols in plant disease resistance. Proceedings of the National Academy of Sciences USA. 2005;102:8054–8059. doi: 10.1073/pnas.0501456102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster MW, Stamler JS. New insights into protein S-nitrosylation. Mitochondria as a model system. Journal of Biological Chemistry. 2004;279:25891–25897. doi: 10.1074/jbc.M313853200. [DOI] [PubMed] [Google Scholar]
- Foyer CH, Noctor G. Redox regulation in photosynthetic organisms: signaling, acclimation, and practical implications. Antioxidants and Redox Signalling. 2009;11:861–905. doi: 10.1089/ars.2008.2177. [DOI] [PubMed] [Google Scholar]
- Hayashi M, Nishimura M. Arabidopsis thaliana: a model organism to study plant peroxisomes. Biochimica et Biophysica Acta. 2006;1763:1382–1391. doi: 10.1016/j.bbamcr.2006.08.014. [DOI] [PubMed] [Google Scholar]
- Huang AHC, Trelease RN, Moore TS. Plant Peroxisomes. New York: Academic Press; 1983. [Google Scholar]
- Jaffrey SR, Erdjument-Bromage H, Ferris CD, Tempst P, Snyder SH. Protein S-nitrosylation: a physiological signal for neuronal nitric oxide. Nature Cell Biology. 2001;3:193–197. doi: 10.1038/35055104. [DOI] [PubMed] [Google Scholar]
- Jimenez A, Hernandez JA, del Río LA, Sevilla F. Evidence for the presence of the ascorbate-glutathione cycle in mitochondria and peroxisomes of pea leaves. Plant Physiology. 1997;114:275–284. doi: 10.1104/pp.114.1.275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerr MV, Groves D. Purification and properties of glycolate oxidase from Pisum sativum leaves. Phytochemistry. 1975;14:359–362. [Google Scholar]
- Koh S, Andre A, Edwards H, Ehrhardt D, Somerville S. Arabidopsis thaliana subcellular responses to compatible Erysiphe cichoracearum infections. The Plant Journal. 2005;44:516–529. doi: 10.1111/j.1365-313X.2005.02545.x. [DOI] [PubMed] [Google Scholar]
- Kuzniak E, Sklodowska M. Compartment-specific role of the ascorbate-glutathione cycle in the response of tomato leaf cells to Botrytis cinerea infection. Journal of Experimental Botany. 2005;56:921–933. doi: 10.1093/jxb/eri086. [DOI] [PubMed] [Google Scholar]
- Lee U, Wie C, Fernandez BO, Feelisch M, Vierling E. Modulation of nitrosative stress by S-nitrosoglutathione reductase is critical for thermotolerance and plant growth in Arabidopsis. The Plant Cell. 2008;20:786–802. doi: 10.1105/tpc.107.052647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindermayr C, Durner J. S-Nitrosylation in plants: pattern and function. Journal of Proteomics. 2009;73:1–9. doi: 10.1016/j.jprot.2009.07.002. [DOI] [PubMed] [Google Scholar]
- Lindermayr C, Saalbach G, Bahnweg G, Durner J. Differential inhibition of Arabidopsis methionine adenosyltransferases by protein S-nitrosylation. Journal of Biological Chemistry. 2006;281:4285–4291. doi: 10.1074/jbc.M511635200. [DOI] [PubMed] [Google Scholar]
- Lindermayr C, Saalbach G, Durner J. Proteomic identification of S-nitrosylated proteins in Arabidopsis. Plant Physiology. 2005;137:901–930. doi: 10.1104/pp.104.058719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L, Hausladen A, Zeng M, Que L, Heitman J, Stamler JS, Steverding D. Nitrosative stress: protection by glutathione-dependent formaldehyde dehydrogenase. Redox Report. 2001;6:209–210. doi: 10.1179/135100001101536337. [DOI] [PubMed] [Google Scholar]
- López-Huertas E, Sandalio LM, del Río LA. Integral membrane polypeptides of pea leaf peroxisomes: characterization and response to plant stress. Plant Physiology and Biochemistry. 1995;33:295–302. [Google Scholar]
- Marino SM, Gladyshev VM. Structural analysis of cysteine S-nitrosylation: a modified acid-based motif and the emerging role of trans-nitrosylation. Journal Molecular Biology. 2010;395:844–859. doi: 10.1016/j.jmb.2009.10.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mittova V, Guy M, Tal M, Volokita M. Salinity up-regulates the antioxidative system in root mitochondria and peroxisomes of the wild salt-tolerant tomato species Lycopersicon pennellii. Journal of Experimental Botany. 2004;55:1105–1113. doi: 10.1093/jxb/erh113. [DOI] [PubMed] [Google Scholar]
- Neill S, Bright J, Desikan R, Hancock J, Harrison J, Wilson I. Nitric oxide evolution and perception. Journal of Experimental Botany. 2008;59:25–35. doi: 10.1093/jxb/erm218. [DOI] [PubMed] [Google Scholar]
- Nyathi Y, Baker A. Plant peroxisomes as a source of signalling molecules. Biochimica et Biophysica Acta. 2006;1763:1478–1495. doi: 10.1016/j.bbamcr.2006.08.031. [DOI] [PubMed] [Google Scholar]
- Palmieri MC, Lindermayr C, Bauwe H, Steinhauser C, Durner J. Regulation of plant glycine decarboxylase by S-nitrosylation and glutathionylation. Plant Physiology. 2010;152:1514–1528. doi: 10.1104/pp.109.152579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pazmiño DM. Ph.D thesis. Spain: University of Granada; 2009. Contribución de las especies de oxígeno y nitrógeno reactivo, y de los peroxisomas a la toxicidad del 2,4-D en plantas. [Google Scholar]
- Pazmiño DM, Rodríguez-Serrano M, Romero-Puertas MC, Archilla-Ruiz A, del Río LA, Sandalio LM. Differential response of young and adult leaves to the herbicide 2,4-dichlorophenoxyacetic acid in pea plants: role of reactive oxygen and nitrogen species. Plant, Cell and Environment. 2011;34:1874–1889. doi: 10.1111/j.1365-3040.2011.02383.x. [DOI] [PubMed] [Google Scholar]
- Perazzolli M, Dominici P, Romero-Puertas MC, Zago E, Zeier A, Sonoda M, Lamb C, Delledonne M. Arabidopsis nonsymbiotic hemoglobin AHb1 modulates nitric oxide bioactivity. The Plant Cell. 2004;16:2785–2794. doi: 10.1105/tpc.104.025379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pracharoenwattana I, Cornah JE, Smith SM. Arabidopsis peroxisomal malate dehydrogenase functions in beta-oxidation but not in the glyoxylate cycle. The Plant Journal. 2007;50:381–390. doi: 10.1111/j.1365-313X.2007.03055.x. [DOI] [PubMed] [Google Scholar]
- Reumann S. 2002 The photorespiratory pathway of leaf peroxisomes. In: Baker A, Graham I, eds, Plant peroxisomes: biochemistry, cell biology, and biotechnological applications. Dordrecht: Kluwer Academic Publishers, pp 141–189. [Google Scholar]
- Reumann S, Babujee L, Ma Ch, Wienkoop S, Siemsen T, Antonicelli GE, Rasche N, Luder F, Weckwerth W, Jahnd O. Proteome analysis of Arabidopsis leaf peroxisomes reveals novel targeting peptides, metabolic pathways, and defense mechanisms. The Plant Cell. 2007;19:3170–3193. doi: 10.1105/tpc.107.050989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reumann S, Ma C, Lemke S, Babujee L. AraPerox. A database of putative Arabidopsis proteins from plant peroxisomes. Plant Physiology. 2004;136:2587–2608. doi: 10.1104/pp.104.043695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reumann S, Weber AP. Plant peroxisomes respire in the light: some gaps of the photorespiratory C2 cycle have become filled others remain. Biochimica et Biophysica Acta. 2006;1763:1496–1510. doi: 10.1016/j.bbamcr.2006.09.008. [DOI] [PubMed] [Google Scholar]
- Rhee KY, Erdjument-Bromage H, Tempst P, Nathan CF. S-Nitrosoproteome of Mycobacterium tuberculosis: enzymes of intermediary metabolism and antioxidant defense. PNAS. 2004;102:467–472. doi: 10.1073/pnas.0406133102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodríguez-Serrano M, Romero-Puertas MC, Pazmiño DM, Testillano PS, Risueno MC, del Río LA, Sandalio LM. Cellular response of pea plants to cadmium toxicity: cross talk between reactive oxygen species, nitric oxide, and calcium. Plant Physiology. 2009;150:229–243. doi: 10.1104/pp.108.131524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodríguez-Serrano M, Romero-Puertas MC, Zabalza A, Corpas FJ, Gomez M, del Río LA, Sandalio LM. Cadmium effect on oxidative metabolism of pea (Pisum sativum) roots. Imaging of reactive oxygen species and nitric oxide accumulation in vivo. Plant, Cell and Environment. 2006;29:1532–1544. doi: 10.1111/j.1365-3040.2006.01531.x. [DOI] [PubMed] [Google Scholar]
- Romero-Puertas MC, Campostrini N, Matte A, Righetti PG, Perazzolli M, Zolla L, Roepstorff P, Delledonne M. Proteomic analysis of S-nitrosylated proteins in Arabidopsis thaliana undergoing hypersensitive response. Proteomics. 2008;8:1459–1469. doi: 10.1002/pmic.200700536. [DOI] [PubMed] [Google Scholar]
- Romero-Puertas MC, Corpas FJ, Sandalio LM, Leterrier M, Rodríguez-Serrano M, del Río L, Palma JM. Glutathione reductase from pea leaves: response to abiotic stress and characterization of the peroxisomal isozyme. New Phytology. 2006;170:43–52. doi: 10.1111/j.1469-8137.2006.01643.x. [DOI] [PubMed] [Google Scholar]
- Romero-Puertas MC, Laxa M, Matte A, Zaninotto F, Finkemeier I, Jones AM, Perazzolli M, Vandelle E, Dietz KJ, Delledonne M. S-Nitrosylation of peroxiredoxin II E promotes peroxynitrite-mediated tyrosine nitration. The Plant Cell. 2007;19:4120–4130. doi: 10.1105/tpc.107.055061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romero-Puertas MC, McCarthy I, Sandalio LM, Palma JM, Corpas FJ, Gomez M, del Río LA. Cadmium toxicity and oxidative metabolism of pea leaf peroxisomes. Free Radical Research. 1999;31:25–31. doi: 10.1080/10715769900301281. [DOI] [PubMed] [Google Scholar]
- Romero-Puertas MC, Palma JM, Gomez M, Del Río LA, Sandalio LM. Cadmium causes the oxidative modification of proteins in pea plants. Plant, Cell and Environment. 2002;25:677–686. [Google Scholar]
- Romero-Puertas MC, Rodríguez-Serrano M, Corpas F J, Gomez M, del Río L, Sandalio LM. Cadmium-induced subcellular accumulation of O2.− and H2O2 in pea leaves. Plant, Cell and Environment. 2004;27:1122–1134. [Google Scholar]
- Rusterucci C, Espunya MC, Diaz M, Chabannes M, Martinez MC. S-Nitrosoglutathione reductase affords protection against pathogens in Arabidopsis, both locally and systemically. Plant Physiology. 2007;143:1282–1292. doi: 10.1104/pp.106.091686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakamoto A, Ueda M, Morikawa H. Arabidopsis glutathione-dependent formaldehyde dehydrogenase is an S-nitrosoglutathione reductase. FEBS Letters. 2002;515:20–24. doi: 10.1016/s0014-5793(02)02414-6. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Russell D. Molecular Cloning: a laboratory manual. 3rd ed. New Yrok: Cold Spring Harbor Laboratory Press; 2001. [Google Scholar]
- Sandalio LM, Dalurzo HC, Gomez M, Romero-Puertas MC, del Río LA. Cadmium-induced changes in the growth and oxidative metabolism of pea plants. Journal of Experimental Botany. 2001;52:2115–2126. doi: 10.1093/jexbot/52.364.2115. [DOI] [PubMed] [Google Scholar]
- Sandalio LM, del Río LA. Localization of superoxide dismutase in glyoxysomes from Citrullus vulgaris. Functional implications in cellular metabolism. Journal of Plant Physiology. 1987;127:395–409. [Google Scholar]
- Sandalio LM, López-Huertas E, Bueno P, del Río LA. Immunocytochemical localization of copper, zinc superoxide dismutase in peroxisomes from watermelon. Citrullus vulgaris cotyledons. Free Radical Research. 1997;26:187–194. doi: 10.3109/10715769709097798. [DOI] [PubMed] [Google Scholar]
- Serpa V, Vernal J, Lamattina L, Grotewold E, Cassia R, Terenzi H. Inhibition of AtMYB2 DNA-binding by nitric oxide involves cysteine S-nitrosylation. Biochemical and Biophysical Research Communications. 2007;361:1048–1053. doi: 10.1016/j.bbrc.2007.07.133. [DOI] [PubMed] [Google Scholar]
- Shevchenko A, Wilm M, Vorm O, Mann M. Mass spectrometric sequencing of proteins silver-stained polyacrylamide gels. Analytical Chemistry. 1996;68:850–858. doi: 10.1021/ac950914h. [DOI] [PubMed] [Google Scholar]
- Stamler JS, Lamas S, Fang FC. Nitrosylation. the prototypic redox-based signaling mechanism. Cell. 2001;106:675–683. doi: 10.1016/s0092-8674(01)00495-0. [DOI] [PubMed] [Google Scholar]
- Tada Y, Spoel SH, Pajerowska-Mukhtar K, Mou Z, Song J, Wang C, Zuo J, Dong X. Plant immunity requires conformational changes of NPR1 via S-nitrosylation and thioredoxins. Science. 2008;321:952–956. doi: 10.1126/science.1156970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varrone S, Consiglio E, Covelli I. The nature of inhibition of mitochondrial malate dehydrogenase by thyroxine, iodine cyanide and molecular iodine. European Journal of Biochemistry. 1970;13:305–312. doi: 10.1111/j.1432-1033.1970.tb00932.x. [DOI] [PubMed] [Google Scholar]
- Walk S, Hock B. Glyoxysomal and mitochondrial malate dehydrogenase of watermelon (Citrullus vulgaris) cotyledons. Kinetic properties of the purified isoenzymes. Planta. 1977;136:221–228. doi: 10.1007/BF00385988. [DOI] [PubMed] [Google Scholar]
- Wang Y, Liu T, Wu C, Li H. A strategy for direct identification of protein S-nitrosylation sites by quadrupole time-of-flight mass spectrometry. Journal of The American Society for Mass Spectrometry. 2008;19:1353–60. doi: 10.1016/j.jasms.2008.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue Y, Liu Z, Gao X, Jin C, Wen L, Yao X, Ren J. GPS-SNO: computational prediction of protein S-nitrosylation sites with a modified GPS algorithm. PLoS One. 2010;5:e11290. doi: 10.1371/journal.pone.0011290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zechmann B, Mauch F, Sticher L, Müller M. Subcellular immunocytochemical analysis detects the highest concentrations of glutathione in motochondria and not in plastids. Journal of Experimental Botany. 2008;59:4017–4027. doi: 10.1093/jxb/ern243. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.