Abstract
During synaptogenesis, macromolecular protein complexes assemble at the pre- and postsynaptic membrane. Extensive literature identifies numerous transmembrane molecules sufficient to induce synapse formation and several intracellular scaffolding molecules responsible for assembling active zones and recruiting synaptic vesicles. However, little is known about the molecular mechanisms coupling membrane receptors to active zone molecules during development. Using C.elegans, we identify an F-actin network present at nascent presynaptic terminals required for presynaptic assembly. We unravel a sequence of events where specificity-determining adhesion molecules define the location of developing synapses and locally assemble F-actin. Next, an adaptor protein NAB-1/Neurabin binds to F-actin and recruits active zone proteins, SYD-1 and SYD-2/Liprin-α by forming a tripartite complex. NAB-1 localizes transiently to synapses during development and is required for presynaptic assembly. Together, we identify a role for the actin cytoskeleton during presynaptic development and characterize a molecular pathway where NAB-1 links synaptic partner recognition to active zone assembly.
Synapse formation is often initiated by membrane contacts between appropriate synaptic partner cells, which leads to intracellular assembly of the active zone and recruitment of synaptic vesicles underneath the presynaptic membrane. Numerous transmembrane molecules have been implicated in specifying synaptic connections1. Many pairs of these homotypic or heterotypic synaptic adhesion molecules including cadherins, EphrinB/EphB, Neurexin/Nlgn, synCAMs, Netrin-G/NGL2, LAR/NGL3 can induce synapse formation through trans-synaptic interactions2,3. While their synapse-inducing activities are robust in vitro, it is difficult to demonstrate their roles in synaptogenesis in vivo, possibly due to functional redundancy. For example, binding between neurexin and neuroligin triggers synapse assembly on both pre- and postsynaptic cells in dissociated cultured neurons, respectively4,5. However, the importance of this interaction for synapse formation in vivo appears to be restricted to certain systems. In Drosophila, mutations in the neurexin homolog, dnrx, cause reduced synapse number and defective active zone formation6. In the mammalian cerebellum, neurexin interacts trans-synaptically with GluRdelta2 through Cbln1 to mediate synapse formation7. Among the four neuroligins, neuroligin 2 found at inhibitory synapses is important for postsynaptic development8. It is still unclear whether these trans-synaptic interactions are important for synaptic target selection. At least in one case, heterologous binding between two Immunoglobulin superfamily proteins, SYG-1/NEPH1 and SYG-2/Nephrin is critical for selective synapse formation in Caenorhabditis elegans HSN (hermaphrodite specific neuron) neurons9. SYG-1 localizes to presynaptic sites prior to synapse formation and is necessary and sufficient for presynaptic development in vivo10.
While diverse membrane receptors induce synapse formation, it is thought that a common presynaptic assembly program constructs active zones and clusters synaptic vesicles. In mammals, many scaffolding and cytomatrix proteins are found at presynaptic terminals. For instance, one MAGUK family member CASK localizes to active zones and binds to neurexin and calcium channels11. In vertebrates, Piccolo and Bassoon are large multi-domain presynaptic cytomatrix proteins with long stretches of coiled-coil domains12. While both serve as excellent active zone markers, recent genetic analysis suggests that they are probably not essential for synaptic transmission, but might function redundantly in maintaining synaptic vesicles13.
Forward genetic approaches in worms and flies have identified three molecules as core active zone assembly genes, SYD-2/Liprin-α, SYD-1/dSYD-1 and ELKS-1/Bruchpilot (Brp). SYD-2/Liprin-α mutants showed complete loss of synaptic vesicles and active zone proteins in HSN synapses, and active zones were abnormal in size and shape in inhibitory synapses of worms and Drosophila neuromuscular junctions14-16. In flies and worms, syd-1/dsyd-1 mutants also display profound presynaptic assembly defects14,17. Brp was first shown to be required for active zone formation and calcium channel localization in Drosophila neuromuscular junctions18,19. The C. elegans homolog of Brp, ELKS-1 is also involved in presynaptic development, however, its role is only revealed in sensitized backgrounds20,21.
Of the three molecules, genetic epistasis analyses suggest that SYD-2/Liprin-α is the most important scaffold molecule while SYD-1 and ELKS-1 promote the activity of SYD-220,21. Numerous biochemical interactions between SYD-2 and other presynaptic proteins including UNC-10/RIM, GIT and ELSK-1/Brp support the notion that SYD-2 serves as the “hub” for active zone assembly17,22.
Actin networks decorate presynaptic terminals by forming a “ring-like” structure surrounding synaptic vesicles and active zones23. While filamentous-actin (F-actin) is not required for synaptic transmission, actin dynamics has been shown to participate in regulatory mechanisms modulating synapse efficacy24 and F-actin appears to be critical for synapse development25. Latrunculin A treatment of young synapses in hippocampal cultures leads to dramatic reduction of synapse numbers, conversely, mature synapses are resistant to actin depolymerization. Furthermore, regulators of actin dynamics such as the Rac GEF, Trio, have been shown to be critical for growth of presynaptic terminals26.
The molecular mechanisms linking the presynaptic actin network and the presynaptic assembly program remain unknown. NAB-1/Neurabin is an actin binding protein that localizes to both pre- and postsynaptic specializations27-29. In dendritic filopodia, neurabin regulates filopodia motility through its actin binding activity30-32. In C. elegans, nab-1 mutants exhibit reduced synapse density due to presynaptic defects33. In inhibitory neurons, NAB-1 appears to be involved in polarized trafficking of presynaptic components into axons through its interaction with SAD-1, an active zone serine/threonine kinase34.
Despite the wealth of knowledge we have on synapse-inducing membrane receptors and active zone assembly molecules, little is known how these two processes are coupled during development. Here we show that NAB-1 is required early during synapse formation to link the presynaptic actin network to active zone assembly proteins through its interaction with actin and to SYD-1 and SYD-2/Liprin-α proteins. NAB-1 functions downstream of specificity-determining transmembrane molecule, SYG-1, and upstream of active zone assembly genes. Hence, our data suggest that NAB-1 serves as an adaptor protein that links synaptogenic signals from transmembrane adhesion molecules to intracellular recruitment of active zones to specific subcellular domains.
Results
Assembly of nascent presynaptic sites requires F-actin
To understand the processes that underlie synapse formation in vivo, we studied the synapses formed by two bilaterally symmetrical motorneurons, HSN in C. elegans, that are involved in egg-laying behavior. HSN cell bodies are situated posterior to the vulva and each extends a single neuronal process anteriorly into the head. As the axon passes the vulva organ, HSN forms a cluster of 12-20 en passant synapses along a short stretch of the axon onto the vulva muscles and VC neurons (Fig. 1a). We visualize these presynaptic specializations in HSN by expressing fluorescently-tagged proteins using cell type-specific promoter, unc-8610. Due to the high density of HSN synapses, it is difficult to assign synaptic vesicle fluorescence signals to individual active zones. However, their highly stereotyped subcellular localization enabled us to unambiguously identify these synapses in wildtype and mutant animals.
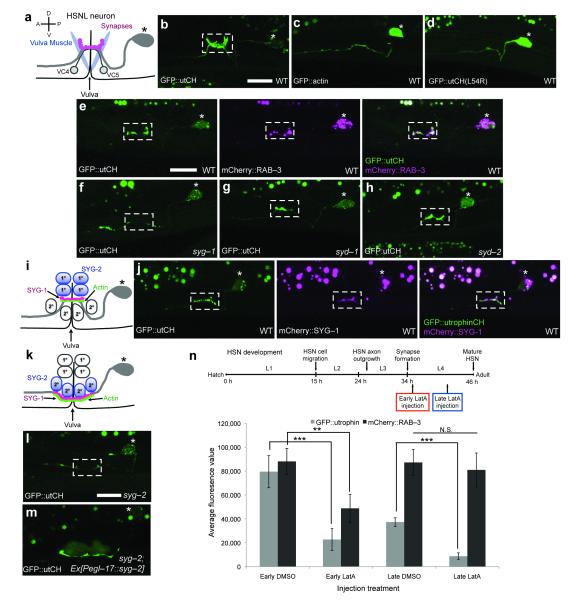
Figure 1. F-actin localizes to HSN synapses and is required for presynapse assembly.
(a) HSN neuron schematic. * Denotes the cell body and presynapses form onto the vulva muscles across the vulva slit. (b) GFP::utrophinCH labels stable F-actin that localizes to presynaptic specializations (white box) even though (c) GFP::actin labels the entire HSN neuron. (d) L54R mutation in utCH disrupts actin binding and abolishes its synaptic localization. (e) GFP::utCH labels F-actin in the region with mCherry::RAB-3 labeled synapses. Scale bars represent 10um. (f) utCH labeled F-actin is lost from synapses in syg-1 mutants, (g, h) but is unaffected in syd-1 or syd-2/liprin-α mutants. (i) Schematic of primary vulva epithelial cells expressing SYG-2 (blue) that recruits trans-synaptic binding partner SYG-1 (pink) and F-actin (green) along the axon. (j) SYG-1 and F-actin are enriched at synapses. (k) Diagram showing ectopic SYG-2 expression in secondary vulva cells in syg-2 mutants. (l) F-actin fails to localize to synapses in syg-2 mutants, (m) but form ectopically in regions contacting SYG-2 expressing secondary vulva cells (n) Timeline of HSN development relative to the worm’s larval stages (L1-L4) at 20°C. Graph quantifies the average fluorescence intensity for GFP::utCH and mCherry::RAB-3. Early LatA treatment reduced utCH and RAB-3 fluorescence by 71% and 45% respectively as compared to DMSO controls. Late LatA reduced utCH fluorescence by 76% but did not affect RAB-3 fluorescence. Bars represent average fluorescence value ±S.E.M. (**p<0.01, ***p<0.001 and N.S. p>0.05, Two-tailed Student’s t-test). For each treatment, utrophin and RAB-3 fluorescence were quantified from the same 15 animals.
F-actin can be found on both sides of the synaptic cleft30,35. At presynaptic terminals, F-actin surrounds the synaptic bouton and has been shown to be important for formation and modulation of synapses in vitro36-38. To understand the role of F-actin during synapse formation in vivo, we examined the distribution of stable F-actin using the utrophin calponin homology domain (utCH) fused to GFP. This F-actin probe has been shown to specifically bind subsets of stable F-actin39. Interestingly, stable F-actin labeled by GFP::utCH localizes to the synaptic region of HSN, while GFP::actin labeled the entire HSN neuron, consistent with the notion that the utCH binds to certain fractions of F-actin39 (Fig. 1b, c). The synaptic localization of the utCH probe was abolished when we introduced a mutation, L54R, which is postulated to destabilize the actin-binding domain based on structure prediction studies, indicating that synaptic enrichment of the utCH probe is due to actin binding40,41 (Fig. 1d). To verify the synaptic localization, we co-expressed GFP::utCH and mCherry::RAB-3, a synaptic vesicle associated protein, to visualize synapses in HSN and indeed, both proteins targeted to the same region (Fig. 1e). Thus, a network of stable F-actin is present at HSN synapses in vivo.
In HSN, initiation of synaptogenesis and synaptic specificity is determined by the presence of transmembrane immunoglobulin-superfamily protein, SYG-1/NEPH110. In the absence of SYG-1, synapses fail to form in the synaptic region at the vulva and instead form ectopic synapses anterior to the vulva. In syg-1 mutants, the F-actin network is lost from the synaptic region (Fig. 1f). Furthermore, localization of F-actin is unaffected by loss of SYD-1 or SYD-2 (Fig. 1g, h), two key active zone scaffolding proteins required for recruiting most other presynaptic proteins to assemble HSN synapses14, suggesting that the presynaptic F-actin network is most likely independent of the active zone structure.
Together with previous work, the data above are consistent with a hypothesis for HSN synaptogenesis where spatial placement of nascent synapses is determined by recruitment of SYG-1 along regions of the HSN axon where it contacts guidepost primary epithelial cells that express SYG-2/Nephrin, the trans-binding partner of SYG-1 (Fig. 1i)9. SYG-1 then recruits and assembles a stable F-actin network in the defined synaptic region. Consistent with this hypothesis, we observed that SYG-1 localizes together with utCH-labeled F-actin (Fig. 1j). To determine if SYG-1 is sufficient to assemble the F-actin network, we ectopically expressed SYG-2 in more ventrally and laterally located secondary vulva epithelial cells using the egl-17 promoter in syg-2 mutants and asked if aberrantly recruited SYG-1 would assemble an ectopic F-actin network (Fig. 1k)9. Like syg-1 mutants, F-actin localization is lost from HSN synapses of syg-2 mutants (Fig. 1l). In syg-2 mutants carrying the transgene, Ex[Pegl-17::syg-2], F-actin accumulates in regions of the HSN axon contacting SYG-2 expressing secondary epithelial cells (Fig. 1m). These results demonstrate that local axonal SYG-1 is necessary and sufficient to assemble a stable F-actin network along the HSN axon.
Finally, to determine if the F-actin network is required during presynaptic assembly, we disrupted F-actin by injecting latrunculin A into the vulva region of worms and assessed if perturbing F-actin would result in presynaptic assembly defects by visualizing mCherry::RAB-3 labeled synapses . Synapse formation begins as the growing HSN axon crosses the vulva during the late L3/early L4 stage and presynaptic material continues to accumulate throughout the L4 stage (Fig. 1n). These synapses become functional after the L4 molt. We performed latrunculin A injections at two stages: early L4 stage, during the onset of synapse formation and mid-late L4 stage, later in development. Latrunculin A treatment at both stages resulted in a decrease in fluorescence intensity of utCH labeled F-actin as compared to DMSO treated animals, suggesting that latrunculin A disrupted the F-actin network in vivo (Fig. 1n and Supplementary Fig. 1a-d). However, RAB-3 fluorescence intensity was significantly decreased only by early perturbation of F-actin with no significant effects on synapses when latrunculin A was injected late. This observation implies that the local F-actin network is required for presynaptic assembly early at the onset of HSN synapse formation. Similar effects of latrunculin have been reported on in vitro cultured hippocampal synapses suggestive of F-actin’s unique requirement during early stages of presynaptic development36. SYG-1 localization was unaffected by latrunculin A treatment thus SYG-1 functions upstream and independent of the F-actin network (Supplementary Fig. 1f).
NAB-1/Neurabin is required for HSN presynaptic assembly
Since the F-actin network is important for presynaptic assembly, there might be presynaptic actin-binding molecules involved in this process. We performed a candidate screen to identify presynaptic proteins containing actin-binding domains and showed defects in presynaptic assembly in HSN. We identified NAB-1/Neurabin, a neural tissue-specific actin binding protein, as a good candidate28. Firstly, NAB-1 localizes to synapses in HSN together with RAB-3 and F-actin (Fig. 2a, b). Secondly, two alleles of nab-1 mutants exhibited a dramatic loss of synaptic vesicles from synapses, labeled by synaptobrevin::YFP (Fig. 2c-e and i). This deficiency in synaptic vesicle recruitment is reflected behaviorally by defects in egg-laying behavior as nab-1 mutants significantly lay more late-staged eggs and less early-stage eggs as compared to wildtype animals (Fig. 2j). Both defects were enhanced by simultaneous loss of ELKS-1, an active zone scaffolding protein whose loss alone results in no apparent phenotype (Fig. 2f, i and Supplementary Fig. 2a, b). Loss of synaptic vesicle recruitment in nab-1;elks-1 double mutants is as severe as syd-1 mutants, suggesting that ELKS-1 functions redundantly with NAB-1 for presynaptic development (Fig. 2g-i).
Figure 2. NAB-1 localizes to presynaptic sites and nab-1 mutants display presynaptic assembly defects.
(a) NAB-1 is an active zone protein that is enriched at synapses together with utCH-labeled F-actin and (b) RAB-3 labeled synaptic vesicles. (c) A wildtype HSN neuron with synapses labeled by SNB-1::YFP. (d, e) nab-1(ok943) and nab-1(wy688) mutants show partial loss of SNB-1 fluorescence. (f) This defect is not observed in elks-1(tm1233) mutants, (g, h) but nab-1(ok943);elks-1 double mutants show an enhancement with almost complete lost of SNB-1 staining like syd-1 mutants. Scale bars represent 10um. (i) Quantification of SNB-1 fluorescence intensity at synapses normalized to wildtype animal controls. nab-1(ok943) (hypomorph) and nab-1(wy688) (functional null) has a 45 ± 4% and 68 ± 3% reduction in SNB-1 fluorescence which is enhanced in nab-1(ok943);elks-1 mutants that show a 90 ± 2% loss of synaptic vesicles. This defect in nab-1(ok943);elks-1 is completely rescued by cell-specific expression of nab-1 in HSN. Each bar represents the average fluorescence from n>20 and error bars are ± S.E.M. (***p<0.001, Two-tailed Student’s t-test). (i) Graph showing proportion of eggs classified in three developmental stages scored double blind in the egg-laying behavior assay. Both nab-1 mutants were significantly more defective in egg-laying behavior compared to wildtype controls. (n>50 per group, *p<0.05, **p<0.01, and ***p<0.001, Fisher’s exact test)
In mammals, neurabin has been described to regulate dendritic spine dynamics30,31. In C. elegans, NAB-1 has been shown to localize presynaptically and function in regulating synapse density and neuronal polarity33,34. To determine where NAB-1 functions, we performed cell-autonomous rescue experiments by expressing NAB-1 in HSN alone which was sufficient to rescue both synaptic vesicle recruitment as well as egg-laying defects in nab-1;elks-1 double mutants (Fig. 2i, j). Thus, NAB-1 is a putative actin-binding protein required for synapse assembly and functions cell-autonomously in HSN.
NAB-1 localizes to HSN synapses through actin binding
Mammalian neurabin has a functionally established N-terminal actin-binding domain (ABD), followed by a structurally predicted PDZ domain, coiled-coil region and a sterile-α motif (SAM) domain (Fig. 3a)28,31. C. elegans NAB-1 has a well conserved PDZ and SAM domain when aligned with human neurabin. However, NAB-1’s ABD is poorly conserved at the level of amino acid sequence. Since there is great diversity in ABD structures, we tested if NAB-1 binds actin. We expressed CFP::NAB-1 in heterologous HEK293T and NIH3t3 cells and found that fluorescence was enriched at subcellular F-actin structures, including subcortical regions under the plasma membrane and stress fibers (Supplementary Fig. 3). Furthermore, NAB-1’s subcellular localization in these cells was largely abolished with latrunculin A treatment. So, even though there is low conservation of NAB-1’s ABD, it interacts with F-actin.
Figure 3. NAB-1 is an actin-binding protein that localizes to presynapses by binding to F-actin.
(a) Diagram of NAB-1 protein highlighting its protein domains including an N-terminal actin-binding domain (ABD), a PDZ domain, a coiled-coil region and a C-terminal SAM domain. C. elegans amino acid sequence similarity of each domain to human Neurabin is shown in percentage below. List of NAB-1 truncation transgenes made for structure-function studies with a summary of each constructs’ ability to localize to presynaptic sites in HSN. (b) Proportion of animals showing wildtype or severe defects in synaptic vesicle recruitment with the expression of each NAB-1 truncation transgene. (c-k) Images showing the localization patterns of each YFP-tagged NAB-1 transgene. All constructs lacking the ABD failed to localize to synapses. White arrows  point to axon regions that should normally be devoid of NAB-1 labeling. (l) GFP::moesinABD is a commonly used in vivo F-actin probe from Drosophila and it labels the entire HSN neuron. (m) Making a chimeric protein by swapping out NAB-1’s ABD with moeABD [moeABD-NAB-1(ΔABD)] was sufficient to localize the protein, (n, o) and rescue synaptic vesicle recruitment, marked by SNB-1::YFP, in nab-1(ok943);elks-1 mutants. Scale bars represent 10um.
point to axon regions that should normally be devoid of NAB-1 labeling. (l) GFP::moesinABD is a commonly used in vivo F-actin probe from Drosophila and it labels the entire HSN neuron. (m) Making a chimeric protein by swapping out NAB-1’s ABD with moeABD [moeABD-NAB-1(ΔABD)] was sufficient to localize the protein, (n, o) and rescue synaptic vesicle recruitment, marked by SNB-1::YFP, in nab-1(ok943);elks-1 mutants. Scale bars represent 10um.
To determine if NAB-1’s actin binding ability is related to its function in mediating synapse assembly, we performed structure-function studies by making NAB-1 truncation constructs that deleted either one or combinations of various functional domains (Fig. 3a). These transgenes were expressed in HSN to assess their ability to localize to synapses as well as capacity to rescue synapse assembly defects in nab-1;elks-1 mutants (Fig. 3a, b). NAB-1 constructs lacking either PDZ, coiled-coil or SAM domains localized to punctate structures along the synaptic region like the full-length protein, but NAB-1’s localization became diffused throughout the HSN axon upon loss of the ABD (Fig. 3c-g). Thus, the ABD is necessary for proper localization of NAB-1 although it is insufficient to confer synaptic localization (Fig. 3h). Nevertheless, the N-terminal region comprising the ABD and PDZ showed partial synaptic localization (Fig. 3i), and this localization is not conferred by the PDZ alone as it failed to localize as well (Fig. 3j). Furthermore, the C-terminal half including the coiled-coil and SAM domains failed to localize and was diffused along the HSN neuron (Fig. 3k). Together, the data suggested that the ABD is required but not sufficient for localizing NAB-1 to presynaptic specializations.
When we assessed the ability of these truncated NAB-1 transgenes to rescue synapse assembly, all truncated forms of NAB-1 that failed to localize to synapses showed poor ability to rescue synaptic vesicle recruitment defects in nab-1;elks-1 mutants, consistent with the notion that NAB-1 functions at presynaptic terminals (Fig. 3b). Similarly, NAB-1 missing its PDZ or SAM domain displayed poor ability to rescue. (Fig. 3b). As these constructs are able to localize, this suggests that both the PDZ and SAM domain are required functionally for synapse assembly. Most animals expressing NAB-1 lacking the coiled-coil region showed complete rescue implying that the coiled-coils are relatively dispensable for synapse formation (Fig. 3b).
To show that F-actin binding is required for synaptic localization of NAB-1, we made a chimeric NAB-1 protein by replacing its ABD with the ABD of moesin. Moesin links the cortical actin cytoskeleton to the membrane in filopodia and microvilli, its C-terminal ABD is commonly used as an in vivo F-actin probe42. Expressing GFP-tagged moesin ABD in HSN results in partial enrichment at synapses (Fig. 3l). The chimeric protein (moeABD-NAB-1ΔABD) exhibits a synaptic localization pattern that is indistinguishable from full-length NAB-1 (Fig. 3m). This is in contrast to NAB-1ΔABD that showed no synaptic enrichment (Fig. 3g). Furthermore, moeABD-NAB-1ΔABD rescued synaptic defects in nab-1;elks-1 mutants (Fig. 3n, o). Since there is no sequence homology between NAB-1 ABD and moesin ABD, this experiment strongly argued that NAB-1 localizes and functions at synapses through binding to F-actin.
Together, the data suggest that NAB-1 localizes to synapses through actin binding and requires a combination of domains to localize correctly. Furthermore, the PDZ, SAM and ABD are functionally required for synapse assembly.
NAB-1 functions early in development downstream of SYG-1
If NAB-1’s synaptic localization requires actin binding, we expect NAB-1 to be dependent on SYG-1 since SYG-1 is required to recruit a stable F-actin network at synapses. Previous work has shown that SYG-1 localizes to developing synapses during the L4 stage but becomes diffused along the axon in adults due to loss of SYG-2 expression in primary vulva epithelial cells9. We expressed NAB-1::YFP and mCherry::SYG-1 in HSN and observed that both proteins localized to synapses during the L4 stage and both became more diffusely localized in adult animals whilst synapses were still present (Fig. 4a–d), suggesting that NAB-1 requires SYG-1 for proper localization. This was verified when we observed a loss of NAB-1 recruitment to synapses in syg-1 mutants and upon disruption of F-actin by latrunculin (Fig. 4e, f and Supplementary Fig. 1g). Given that F-actin is necessary for NAB-1 localization, F-actin assembly should be independent of NAB-1. Consistently, synaptic localization of F-actin was unperturbed in nab-1(wy688) mutants (Fig. 4g, h). Thus, NAB-1 functions downstream of SYG-1 and the F-actin network.
Figure 4. NAB-1 requires SYG-1 for synaptic localization and functions downstream of F-actin.
(a) NAB-1::YFP and SYG-1::mCherry are targeted to synapses in the early L4 larval stage, (b) but becomes diffuse along the entire neuron in later adult stages. White arrows  point to diffused NAB-1 and SYG-1 staining in the anterior axon of adults. (c) This is clearly shown when we examine NAB-1::YFP together with mCherry::RAB-3 where NAB is recruited to developing synapses in the L4 stage, (d) but is diffused in adult stages when RAB-3 labeled synapses have matured. (e, f) NAB-1::YFP synaptic localization is lost in syg-1 mutants. (g, h) In nab-1mutants, F-actin localization is unaffected suggesting F-actin recruitment is independent of NAB-1. Scale bars represent 10um.
point to diffused NAB-1 and SYG-1 staining in the anterior axon of adults. (c) This is clearly shown when we examine NAB-1::YFP together with mCherry::RAB-3 where NAB is recruited to developing synapses in the L4 stage, (d) but is diffused in adult stages when RAB-3 labeled synapses have matured. (e, f) NAB-1::YFP synaptic localization is lost in syg-1 mutants. (g, h) In nab-1mutants, F-actin localization is unaffected suggesting F-actin recruitment is independent of NAB-1. Scale bars represent 10um.
Both localization and perturbation experiments suggest that SYG-1 and F-actin function during the initial phase of presynaptic formation. NAB-1’s transient presynaptic localization also hints that it functions during early stages. To test when NAB-1 is required during synapse development, we induced expression of NAB-1 driven by a heatshock promoter in nab-1;elks-1 animals. We heatshock either during the early L4 stage when synapses are developing or in the young adult stage when synapses are mature, and assayed for rescue of synaptic vesicle recruitment in HSN (Fig. 5). Animals carrying the transgene that receive heatshock treatment during the L4 stage showed a significant increase in SNB-1::YFP fluorescence at synapses compared to control nab-1;elks-1 animals without the transgene (Fig. 5). This rescue was not observed when adult animals were heatshock. Therefore, NAB-1 is required early during synaptogenesis, which is consistent with the temporal localization pattern of NAB-1.
Figure 5. NAB-1 functions in the L4 stage early during synaptogenesis.
Timeline shows the development of HSN at 12.5°C and the timepoint when heatshock treatment was given to induce expression of NAB-1 driven by the heatshock promoter in nab-1(ok943);elks-1 double mutants. The graph plots the average fluorescence intensity of SNB-1::YFP fluorescence. Heatshock during the L4 stage showed rescue of synaptic vesicle recruitment between animals with and without the transgene. No significant differences were observed with heatshock treatment in adults. Each bar represents the average fluorescence from 20 individuals ± S.E.M. (***p<0.001 and N.S. is p>0.05, Two-tailed Student’s t-test).
NAB-1 recruits active zone molecules to instruct assembly
Prior work has established a hierarchical order in which various active zone molecules are recruited and assembled during development of HSN synapses14,20. Two core active zone scaffold molecules SYD-1 and SYD-2/Liprin-α are required to recruit multiple presynaptic components, including ELKS-1, GIT, UNC-10/RIM, calcium channels and synaptic vesicle markers to synapses. When we asked if NAB-1 is affected by loss of SYD-2 and SYD-1, importantly, we found that NAB-1’s synaptic localization is unaffected in syd-1;syd-2 double mutants (Fig. 6a). This is striking in contrast with the severe loss of multiple active zone and synaptic vesicle proteins in syd-2 or syd-1 mutants14, which demonstrates that NAB-1’s recruitment to synapses is independent of SYD-1 and SYD-2 and it probably functions upstream of these scaffold proteins. In addition, nab-1 mutants display loss of active zone proteins such as ELKS-1, GIT and SAD-1 similar to syd-1 and syd-2 mutants (Supplementary Fig. 2c, d)14.
Figure 6. NAB-1 functions upstream and is required to recruit both active zone scaffolding molecules SYD-1 and SYD-2.
(a) NAB-1::YFP localizes to synapses in syd-1;syd-2 double mutants. (b) Quantification of SYD-2::GFP fluorescence normalized to wildtype. (c, d) Images of GFP::SYD-2 localization in HSN. Scaffolding molecule SYD-2 localizes to punctate structures along the synaptic region in wildtype and elks-1 mutants. (e) syd-1 mutants show a 62 ± 6% decrease in SYD-2 recruitment compared to wildtype animals. (f) Similarly nab-1 mutants show a partial loss of SYD-2 (43 ± 7% and 52 ± 6% decrease in ok943 and wy688 respectively) and (g) a 40 ± 7% reduction in nab-1;elks-1. (h) nab-1;syd-1 double mutants display almost complete lost of SYD-2 (87 ± 7% reduction in fluorescence). (i) Quantification of SYD-1::GFP fluorescence normalized to wildtype. syd-2, nab-1(ok943) and nab-1(ok943);elks-1 mutants all show a ~50% reduction in SYD-1 recruitment to synapses. This lost is enhanced in nab-1;syd-2 mutants with 77 ± 3% loss of SYD-1 at synapses. (j–o) Images of GFP::SYD-1 localization in HSN. For both quantification of SYD-1 and SYD-2, each bar represents the average fluorescence from 20 individuals ± S.E.M. (***p<0.001 and **p>0.01, Two-tailed Student’s t-test). All scale bars represent 10um.
To understand the mechanism by which NAB-1 interacts with SYD-1 or SYD-2 to execute the presynaptic assembly program, we examined if recruitment of SYD-1 and SYD-2 is affected in nab-1 mutants. We found that fluorescence intensity of GFP::SYD-2 at synapses in both nab-1 mutants was decreased, suggesting that SYD-2 recruitment is partially dependent on NAB-1 (Fig. 6b, e and h). This reduction in SYD-2 localization is similar to syd-1 mutants (Fig. 6d, h) and is not enhanced by loss of ELKS-1 in nab-1(ok943);elks-1 mutants (Fig. 6c, f and h). However, almost complete loss of GFP::SYD-2 fluorescence was observed in nab-1(ok943);syd-1 double mutants (Fig. 6g, h) suggesting a model where SYD-1 and NAB-1 both recruit SYD-2 in parallel pathways to HSN synapses.
Likewise, we observed similar data for SYD-1 recruitment when we quantified the fluorescence intensity of GFP::SYD-1 in the various mutants. syd-2, nab-1(ok943) and nab-1(ok943);elks-1 mutants showed partial reduction in SYD-1 recruitment to synapses (Fig. 6i-m and o). In nab-1(ok943);syd-2 double mutants, almost complete loss of SYD-1 protein was observed (Fig. 6n). Thus, both SYD-2 and NAB-1 are required to recruit SYD-1 to synapses in HSN.
To address how NAB-1 recruits SYD-1 and SYD-2 to synapses, we used an in vitro single cell protein-protein interaction assay to assess potential interactions 21. We co-expressed NAB-1::YFP fused to a membrane-targeting sequence together with a potential interacting protein tagged with CFP in HEK293T cells. If the protein binds to NAB-1, it should localize to the plasma membrane. An active zone serine-threonine kinase SAD-1 has previously been found to bind NAB-134 and exhibits synapse assembly defects in HSN (Supplementary Fig. 5). To validate the in vitro binding assay, we used SAD-1::CFP as a positive control and observed recruitment of SAD-1 to the membrane when cotransfected with membrane-targeted NAB-1::YFP (Fig. 7a, b and g). When we expressed SYD-1 together with membrane-tethered NAB-1, we observed dramatic recruitment of SYD-1 to the plasma membrane (Fig. 7c, d and g). Structure function analyses show that SYD-1 interacts with the N-terminal regions of NAB-1 (Supplementary Fig. 4a-g). SYD-2 alone remained in the cytoplasm of cells co-expressing NAB-1 (Fig. 7e-g). However, we observed significant recruitment of SYD-2 to membrane-tethered NAB-1 when SYD-1 was co-expressed (Fig. 7h-l). These results suggest that while SYD-2 and NAB-1 do not show strong interactions alone, they can form a complex in the presence of SYD-1 (Supplementary Fig. 4h-j). Taken together, our genetic experiments suggest that NAB-1 recruits both SYD-1 and SYD-2 to presynaptic sites. The interaction data identifies SYD-1 as a new binding partner for NAB-1 and together may form a complex to recruit SYD-2 to nascent presynaptic sites to initiate downstream presynaptic assembly.
Figure 7. NAB-1 recruits downstream active zone molecules by interacting with SYD-1 and SYD-2.
Potential NAB-1 protein interactions were tested in a heterologous system using HEK293T cells. (a–c) Cells expressing control YFP fused to a membrane-targeting sequence and CFP-tagged proteins remain diffused in the cytoplasm of the cell. (d) When YFP::NAB-1 fused to a membrane-targeting sequence was co-expressed together with CFP::SAD-1, SAD-1 was recruited to the membrane (1.65 ± 0.05 fold increase) and co-localizes with NAB-1. SAD-1 was used as a positive control as the protein binds to NAB-1. (e) Protein interaction was observed between CFP::SYD-1 and NAB-1 (1.93 ± 0.09 fold increase in fluorescence intensity at the membrane). (f) No significant change in binding was observed between SYD-2 and NAB-1 (0.98 ± 0.04 fold change). (g) Quantification of fold change in CFP fluorescence pixel intensity at the membrane compared to cytoplasm. Each bar represents the average ratio from 20 cells ± S.E.M. (***p<0.001 and N.S. is p>0.05, Two-tailed Student’s t-test). (h, i) No binding was observed between SYD-2 and control vector YFP, when co-transfected with untagged SYD-1, similar to SYD-2 and NAB-1 alone (1.00 ± 0.02 and 0.93 ± 0.02 fold change respectively). (j-l) However, in the presence of untagged SYD-1, CFP::SYD-2 is recruited to the membrane by YFP::NAB-1 with 1.17 ± 0.04 fold increase in fluorescence intensity at the membrane.
Discussion
Using C. elegans HSN synapses, we investigated the molecular mechanisms of presynaptic assembly. Our results are consistent with a model in which transmembrane adhesion molecule SYG-1/NEPH1 defines a region of the axon where nascent synapses are induced to form during development. SYG-1 initiates synaptogenesis by recruiting a local F-actin network. NAB-1/neurabin links SYG-1 to the presynaptic assembly program by binding to the local F-actin network and functions as an adaptor for active zone molecules such as SYD-1 and SYD-2/liprin-α (Supplementary Fig. 6). Recruitment of downstream active zone molecules by NAB-1 potentially functions through the novel interaction between NAB-1, SYD-1 and SYD-2. Our results highlight the importance of local cytoskeletal rearrangements that can be mediated by transmembrane receptors during initiation of synaptogenesis.
Membrane receptors organize synapses through local F-actin
Recent studies on synapse formation present the notion that many molecules involved in axon guidance also play important roles in synapse formation43. This is not surprising since postsynaptic development requires dendritic spine motility in many experimental systems, which depends on dynamic actin mediated motility similar to axonal grow cone turning. Therefore, it is conceivable that molecules that affect actin dynamics might underlie both axon growth cone guidance and dendritic filopodia motility. However, it is harder to reconcile how axon guidance molecules can pattern and form presynaptic specializations because recruitment of synaptic vesicles and active zone components are fundamentally distinct cellular processes compared with growth cone turning through selective stabilization of subcellular F-actin.
Interestingly, the requirement of F-actin during early presynaptic development provides a conceptual framework for how guidance molecules can pattern presynaptic terminals. Actin has been found at the presynaptic terminals surrounding synaptic boutons23. In dissociated neuronal cultures, latrunculin A treatment blocks synaptogenesis in young cultures, but has little effect after the initial phase of synapse formation, suggesting that there is an early phase of synaptogenesis that requires F-actin36. Indeed, heparan sulfate proteoglycans, including syndecan-2, adsorbed onto beads or expressed on the axon surface, can assemble synapses via a mechanism dependent on the dynamic reorganization of F-actin25. This notion is strengthened by a recent report that an actin modification pathway involving Rac GEF, Trio, is required presynaptically for growth of neuromuscular junctions 26.
In this study, we provide several lines of evidence that transmembrane adhesion receptor SYG-1, which is necessary and sufficient to trigger synapse formation in HSN, patterns an actin network at presynaptic terminals. First, an F-actin network labeled by Utrophin-CH domain showed striking enrichment with SYG-1 in HSN (Fig. 1j). Second, this presynaptic F-actin is lost in the syg-1 mutants (Fig. 1f). Third, artificial targeting of SYG-1 to specific axonal domains is sufficient to recruit F-actin to ectopic sites, suggesting that SYG-1 is necessary and sufficient to build an F-actin network near nascent presynaptic clusters (Fig. 1l, m). Fourth, in vivo latrunculin treatment causes failure of synaptic vesicle clustering (Fig. 1n). Together, these results argue that SYG-1 might organize presynaptic terminals in HSN through building a F-actin network. Studies on Drosophila muscle fusion have convincingly showed that the SYG-1 homologs, Duf and Rst, both induce F-actin formation near the fusion pore44. Hence, the ability of SYG-1 family of genes to organize local F-actin networks is conserved while downstream functions of these genes have diversified in different contexts of development.
Is this relationship between F-actin and presynaptic assembly a general phenomenon? Considering the fact that latrunculin A treatment results in synaptic assembly defects both in C. elegans HSN neurons and hippocampal neurons, it seems plausible that local F-actin is a critical component of presynaptic development in many neurons. Interestingly, another study showed that UNC-40/DCC receptors can also induce local F-actin formation through rac proteins at presynaptic specializations in C. elegans AIY interneurons (personal communication), further suggesting that multiple synapse organizing membrane receptors can induce F-actin assembly and might play important roles during presynapse formation.
NAB-1 links presynaptic F-actin to assembly proteins
If local F-actin assembly is a critical event for presynapse development, how does F-actin lead to the construction of active zones and accumulation of synaptic vesicles? It is conceivable that adaptor proteins can couple the actin network to active zone components and therefore anchor the active zone at nascent synapses. Such adaptor proteins should have the following properties. First, it should bind to both F-actin and active zone proteins. Second, it should localize to nascent presynaptic terminals. Third, because F-actin is only required early during presynaptic development, such adaptors might also be required only during the development of synaptic structure but not the maintenance of synapses. Fourth, such adaptors should function upstream of active zone proteins to recruit them to developing synaptic terminals.
Our data suggest that NAB-1/neurabin fits the bill to function as such an adaptor. First, NAB-1 binds to F-actin. When expressed in HEK293T cells or NIH3t3 fibroblasts, NAB-1 localizes to F-actin structures such as cortical actin and stress fibers in a latrunculin A dependent manner (Supplementary Fig. 1). Through structure function analysis, we find that similar to the mammalian homolog, the N-terminal domain of NAB-1 is responsible for actin binding. Interestingly, the “actin-binding domain” of NAB-1 is also required for its presynaptic localization. Furthermore, an unrelated protein’s actin-binding domain can substitute for the N-terminal portion of NAB-1 to localize NAB-1 to synapses (Fig. 3l, m), strongly suggesting that NAB-1 is recruited to presynaptic terminals by the local F-actin network. We also show that NAB-1 interacts with core active zone assembly molecules, SYD-1 and SYD-2, suggesting that NAB-1 can couple actin and active zone.
Second, NAB-1 localizes to developing presynaptic specializations. Interestingly, unlike other known active zone proteins, NAB-1’s synaptic localization in HSN is transient. HSN synapses form in the L4 stage and become functional in adults when egg-laying behavior begins. In the adult stage, NAB-1’s synaptic localization is lost while active zone proteins such as SYD-2, SYD-1, ELKS-1 and GIT-1 continue to localize to synapses (Fig. 4d). This transient synaptic localization resembles SYG-1 and together with the fact that NAB-1 localizes with SYG-1 in developing synapses, these data argue that NAB-1 is transiently recruited to developing synapses possibly through SYG-1 or SYG-1 assembled F-actin.
Third, consistent with its transient localization, NAB-1 is required during early stages of synapse development (Fig. 5). This is again in striking contrast to other structural components of the active zone that are required throughout. Therefore, NAB-1 plays a unique role during early stages of active zone formation.
Finally, NAB-1 appears to function upstream of core active zone proteins SYD-2 and SYD-1. While almost all synaptic vesicle and active zone markers disappear from HSN synapses in syd-2 mutants14, NAB-1’s localization is unaffected in syd-1;syd-2 double mutants. In syd-1;nab-1 double mutants, SYD-2 almost completely fails to localize. Similarly, SYD-1 is largely absent from developing synapses in syd-2;nab-1 double mutants. While all of our data agree with this model, NAB-1 might not be the only adaptor protein because loss of SYD-2 showed a quantitatively stronger phenotype compared with the nab-1 single mutant. In addition, nab-1 mutants showed qualitatively weaker phenotypes in other synapses in worms, indicating that there might be other redundant proteins (data not shown).
How does NAB-1 recruit these active zone proteins? Our in vitro data suggest that NAB-1 interacts with SYD-1. Although it showed no significant interaction with SYD-2 alone, in the presence of SYD-1, both SYD-1 and SYD-2 appears to bind to NAB-1. From previous data, we could not detect any interaction between SYD-1 and SYD-2. Therefore, NAB-1 may catalyze the recruitment of SYD-1 and SYD-2, two molecules essential for presynaptic assembly in HSN.
A recent study showed that NAB-1 and the presynaptic kinase SAD-1 functions together in regulating the polarized distribution of synaptic components in the D-type inhibitory motorneurons34. They observed dendritic mislocalization of synaptic vesicles in nab-1 and sad-1 single mutants that was not enhanced in nab-1;sad-1 double mutants, hinting that they function in the same genetic pathway. In HSN, both nab-1 and sad-1 showed reduced levels of synaptic vesicles. However, the double mutants strongly enhanced each other and resulted in almost complete lost of synaptic vesilcles (Supplementary Fig. 5a-e), suggesting that NAB-1’s assembly function is distinct from its SAD-1-related function. Previous work showed that SAD-1 functions downstream of SYD-1 and SYD-214, consistent with our model that NAB-1 is required to recruit SYD-1 and SYD-2, we found that SAD-1’s localization is also dependent on NAB-1 and NAB-1’s localization is unaffected in sad-1 mutants (Supplementary. Fig 2d and 5f-h). This data suggest that SAD-1 functions downstream of NAB-1 in our model.
Collectively, these results present strong evidence that NAB-1 serves as an adaptor protein that localizes to the nascent presynaptic terminals to link the local F-actin network to developing active zones.
Methods
Strains
All worms strains were maintained at 20°C on OP50 E. coli seeded nematode growth medium plates. N2 Bristol stain worms were used as the wildtype reference and the following mutants were used: nab-1(ok943)I, nab-1(wy688::unc-119+)I (targeted deletion, method described below), syd-1(ju82)II, elks-1(tm1233)IV, syd-2(ju37)X, syg-1(ky652)X, syg-2(ky673)X, sad-1(ky330)X and unc-119(ed3)III. nab-1(ok943) mutation is a 1032 basepair deletion that removes exon 7 to part of exon 8 of nab-1’s genomic sequence resulting in a premature stop after the deleted region. It results in a truncated nab-1 sequence that still contains the putative actin binding domain and PDZ. This allele is a hypomorph as overexpression of the truncated protein can partially rescue synapse formation defects.
Mos transposon mediated targeted deletion of NAB-1
The targeted knockout nab-1(wy688::unc-119+) was made using the MosDEL method described by Frøkjær-Jensen et. al.45 with the Mos insertion line ttTi6300 requested from L. Ségalat46. Fifty ttTi6300;unc-119(ed3) animals were co-injected with the following plasmids: pJL43.1 (Pglh-2::MOStransposase) at 50ng/ul, pPH32 (targeting plasmid with positive selection marker, cb-unc-119(+), made with pRS413, see cloning for methods) at 50ng/ul, pCFJ90 (Pmyo-2::mCherry) at 2.5ng/ul and Punc-122::rfp at 40ng/ul. Plates with F1s carrying both co-injected fluorescent markers and rescued for unc-119 movement defects were identified and kept at 25°C till the plate is starved out. We identified animals with the deletion by screening plates for worms that have lost both co-injected fluorescent markers with no movement defects (due to unc-119 rescue). The deletion was verified by PCR genotyping with the primers (forward primer: ttcactttcaggtcttctcggcgt, reverse primer reading from inserted cb-unc-119(+): gcgccctaactttgagccaattca and reverse primer reading from deleted region of nab-1: tctgcaccaacacccatacctgaa) as well as sequencing. This deletion removes the entire nab-1 gene except the first 246 amino acids and is a functional null.
Transgenic lines
Utrophin label lines - wyEx3992 [Punc-86::gfp::utrophinCH; Podr-1::gfp], wyEx4095 [Punc-86::gfp::utrophinCH; Punc-86::mCherry::rab-3 ; Podr-1::gfp], wyEx4099 [Punc-86::gfp::utrophinCH; Punc-86::syg-1::mCherry ; Podr-1::gfp], wyEx4445 [Punc-86::gfp::utrophinCH; Punc-86::mCherry::nab-1 ; Podr-1::dsred], wyEx3840 [Punc-86::gfp::utrophinCH; Punc-86::mCherry::rab-3 ; Podr-1::gfp].
Other markers - kyIs235 [Punc-86::snb-1::yfp; Punc-4::lin-10::dsred; Podr-1::dsred], wyIs12 [Punc-86::gfp::syd-2; Podr-1::gfp], wyEx118 [Punc-86::gfp::syd-1; Podr-1::dsred], wyEx314 [Punc-86::syd-1; Podr-1::dsred], kyEx673 [Pegl-17::syg-2; Podr-1::gfp], wyEx196 [Punc-86::elks-1::yfp; Podr-1::dsred], wyEx163 [Punc-86::sad-1::yfp; Podr-1::dsred], wyEx146 [Punc-86::git-1::yfp; Podr-1::dsred].
Fluorescent-tagged NAB-1 localization lines - wyEx245 [Punc-86::nab-1::YFP; Podr-1::dsred], wyEx3353 [Punc-86::nab-1(1-623aa)::YFP; Podr-1::gfp], wyEx3733 [Punc-86::nab-1(1-197,370-722aa)::YFP; Podr-1::gfp], wyEx1492 [Punc-86::NAB-1(1-371,559-722aa)::YFP; Podr-1::dsred], wyEx3550 [Punc-86::nab-1(1-371aa)::YFP; Podr-1::gfp], wyEx1363 [Punc-86::nab-1(372-722aa)::YFP; Podr-1::dsred], wyEx3364 [Punc-86::nab-1(195-371aa)::YFP; Podr-1::gfp], wyEx3845 [Punc-86::nab-1(1-266aa)::YFP; Podr-1::gfp], wyEx4602 [Punc-86::nab-1(213-722aa)::YFP; Podr-1::dsred], wyEx3905 [Punc-86::nab-1::YFP; Punc-86::syg-1::mCherry; Podr-1::gfp], NAB-1 expression lines - wyEx3164 [Punc-86::nab-1; Podr-1::gfp], wyEx3329 [Punc-86::nab-1(1-623aa); Podr-1::gfp], wyEx3929 [Punc-86::nab-1(1-197,370-722aa)::YFP; Podr-1::gfp], wyEx3444 [Punc-86::nab-1(1-371,559-722aa); Podr-1::gfp], wyEx3333 [Punc-86::nab-1(1-371aa); Podr-1::gfp], wyEx3336 [Punc-86::nab-1(372-722aa); Podr-1::gfp], wyEx3907 [Punc-86::nab-1(1-266aa); Podr-1::gfp], wyEx4750 [Punc-86::nab-1(213-722aa); Podr-1::gfp], wyEx3297 [Pheatshock:nab-1; Podr-1::gfp], NAB-1 chimeric protein lines – wyEx4639 [Punc-86::moesinABD::nab-1(213-722aa)::YFP; Podr-1::dsred], wyEx4641 [Punc-86::moesinABD::nab-1(213-722aa); Podr-1::gfp],
Cloning of constructs
Expression plasmids for transgenic worm lines were made using the pSM vector, a derivative of pPD49.26 (A. Fire). The unc-86 promoter was cloned between SphI and XmaI and genes of interests were cloned between NheI and KpnI. Plasmids were injected into animals at 1 ng/μl together with co-injection markers Podr-1::gpf or Podr-1::dsred at 20ng/ul as previously described 47. MosDEL targeting construct, pPH32, was made using yeast homologous recombination methods using an EcoRI linearized pRS413 vector (ATCC) 48. The targeting construct includes a 2kb homologous sequence that flanks the genomic region left of the Mos insertion site in ttTi6300, followed by the cb-unc-119(+) positive selection marker replicated by PCR from pCFJ151 and a 2kb homologous region right of the nab-1 gene. Expression plasmids for HEK293T cells were made using CMV promoter mCerulean-C1 and eYFP-C1 vectors from Clontech. cDNAs of genes were cloned between HindIII and KpnI. KRAS membrane targeting sequence was cloned in between KpnI and XbaI sites. FLAG sequence was cloned between NheI and HindIII.
Fluorescence quantification and confocal imaging
All fluorescence images of HSNL synapses in L4 or young adults were taken with a 63X objective on a Zeiss Axioplan 2 Imaging System or a Plan-Apochromat 63X/1.4 objective on a Zeiss LSM710 confocal microscope using similar imaging parameters for the same marker across different genotypes. Total fluorescence intensity of across the synaptic region was determined using Image J software (NIH) by summing pixel intensity and the average fluorescence intensity was calculated for each group (n>20). Two-tailed Student’s t-test was used to assess statistical significance. Live cell images for in vitro assays were obtained using a 20X objective on a Zeiss Axioplan 2 Inverted Fluorescence Imaging System.
Latrunculin A injections
Late L3, early L4 or mid L4 animals were immobilized using 1mM levamisole (Sigma) and the developmental stage as well as expression levels of markers was verified under a Zeiss Axioplan 2 upright microscope before being isolated. Selected animals received microinjections of either 1mM or 400uM latrunculin A (Sigma) in 25% DMSO (Sigma) or 25% DMSO alone, into the pseudocoelom of the worm at a site slightly posterior of the developing vulva and then kept at room temperature to recover. Three hours post-injection, animals were imaged and the fluorescence intensity of markers was quantified. 15 animals were quantified for each treatment group and statistical significance was determined using two-tailed Student’s t-test.
Egg-laying assay
15 L4 stage animals were isolated and allowed to develop 28h at 20°C into gravid adults. The animals were transferred onto a fresh plate and the stage of each egg laid was scored double blind after an hour at 20°C. The eggs were classified into one of three developmental stages: 1-8 cell, 8-cell to comma, or post-comma. 2 independent assays were performed with at least 50 eggs quantified in total for each genotype. Fisher’s exact test was performed to compare statistical significance between 2 groups. Day to day variation was observed but each experimental round was performed together with wild type and mutant controls for comparison.
Heatshock rescue experiments
Animals were maintained at 12.5°C for multiple generations before being shifted to a different temperature. Experimental animals in the early L4 larval stage or young adults were shifted to 33°C for two hours 49 followed by a recovery period at 20°C for 6 hours prior to imaging. At least 20 animals were quantified for each heatshock treatment timepoint.
In vitro protein-protein interaction assay
As previously described 50, Hek293T cells were co-transfected with plasmid constructs driven by the CMV promoter with TransIT-LT1 reagent (Mirus) according to the manufacturer’s recommendations and cells were incubated at 37°C for 24h prior to imaging. To quantify the recruitment of protein to the cell membrane, we used ImageJ to perform line scans across the cell membrane and into cytoplasm. The fluorescence pixel intensity at the membrane and pixel intensity in the cytoplasm were used to obtain a ratio comparing the fold difference in intensities of the two compartments. (n>20 cells).
Immunocytochemistry
Transfected Hek293T and NIH3t3 cells were fixed with 4% paraformaldehyde and permeabilized with either 0.2% Triton X-100 (Sigma) or 0.05% saponin (Sigma). Samples were blocked with 10% BSA prior to antibody staining. Antibodies used included: rhodamine phalliodin (Invitrogen) diluted to 5U/ml, mouse monoclonal anti-FLAG M2 (Sigma) diluted 1:1000 and goat anti-mouse Alexa 568 diluted 1:1000.
Statistical analysis
Data are expressed as average mean ± S.E.M. All in vitro experiments were performed in duplicates. Statistical significance was assessed by two-tailed Student’s t-test when comparing between two groups and the Fisher’s exact test for cross-categorized frequency data.
Supplementary Material
Acknowledgements
This work was funded by the Howard Hughes Medical Institute. P. H. Chia is supported by the Agency for Science, Technology, and Research (Singapore). We thank the Caenorhabditis Genetic Center and Japanese National Bioresource Project for strains. We also thank C. Gao and Y. Fu for technical assistance and T. Clandinin, K. Zito, K. Mizumoto and C. Y. Ou and Shen Lab members for manuscript comments.
Footnotes
Author Contributions P.H.C. and K.S. designed experiments and wrote the paper. M.R.P. made initial observations and performed initial experiments. P.H.C performed experiments and analyzed the data.
References
- 1.Williams ME, de Wit J, Ghosh A. Molecular mechanisms of synaptic specificity in developing neural circuits. Neuron. 2010;68:9–18. doi: 10.1016/j.neuron.2010.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tallafuss A, Constable JR, Washbourne P. Organization of central synapses by adhesion molecules. The European journal of neuroscience. 2010;32:198–206. doi: 10.1111/j.1460-9568.2010.07340.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yamagata M, Sanes J, Weiner JA. Synaptic adhesion molecules. Current Opinion in Cell Biology. 2003;15:621–632. doi: 10.1016/s0955-0674(03)00107-8. [DOI] [PubMed] [Google Scholar]
- 4.Scheiffele P, Fan J, Choih J, Fetter R, Serafini T. Neuroligin expressed in nonneuronal cells triggers presynaptic development in contacting axons. Cell. 2000;101:657–669. doi: 10.1016/s0092-8674(00)80877-6. [DOI] [PubMed] [Google Scholar]
- 5.Dean C, et al. Neurexin mediates the assembly of presynaptic terminals. Nature Neuroscience. 2003;6:708–716. doi: 10.1038/nn1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Li J, Ashley J, Budnik V, Bhat MA. Crucial role of Drosophila neurexin in proper active zone apposition to postsynaptic densities, synaptic growth, and synaptic transmission. Neuron. 2007;55:741–755. doi: 10.1016/j.neuron.2007.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Uemura T, et al. Trans-synaptic interaction of GluRdelta2 and Neurexin through Cbln1 mediates synapse formation in the cerebellum. Cell. 2010;141:1068–1079. doi: 10.1016/j.cell.2010.04.035. [DOI] [PubMed] [Google Scholar]
- 8.Poulopoulos A, et al. Neuroligin 2 drives postsynaptic assembly at perisomatic inhibitory synapses through gephyrin and collybistin. Neuron. 2009;63:628–642. doi: 10.1016/j.neuron.2009.08.023. [DOI] [PubMed] [Google Scholar]
- 9.Shen K, Fetter RD, Bargmann CI. Synaptic specificity is generated by the synaptic guidepost protein SYG-2 and its receptor, SYG-1. Cell. 2004;116:869–881. doi: 10.1016/s0092-8674(04)00251-x. [DOI] [PubMed] [Google Scholar]
- 10.Shen K, Bargmann CI. The immunoglobulin superfamily protein SYG-1 determines the location of specific synapses in C. elegans. Cell. 2003;112:619–630. doi: 10.1016/s0092-8674(03)00113-2. [DOI] [PubMed] [Google Scholar]
- 11.Butz S, Okamoto M, S√°dhof TC. A tripartite protein complex with the potential to couple synaptic vesicle exocytosis to cell adhesion in brain. Cell. 1998;94:773–782. doi: 10.1016/s0092-8674(00)81736-5. [DOI] [PubMed] [Google Scholar]
- 12.tom Dieck S, et al. Bassoon, a novel zinc-finger CAG/glutamine-repeat protein selectively localized at the active zone of presynaptic nerve terminals. The Journal of Cell Biology. 1998;142:499–509. doi: 10.1083/jcb.142.2.499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mukherjee K, et al. Piccolo and bassoon maintain synaptic vesicle clustering without directly participating in vesicle exocytosis. Proceedings of the National Academy of Sciences. 2010;107:6504. doi: 10.1073/pnas.1002307107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Patel MR, et al. Hierarchical assembly of presynaptic components in defined C. elegans synapses. Nature Neuroscience. 2006;9:1488–1498. doi: 10.1038/nn1806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhen M, Jin Y. The liprin protein SYD-2 regulates the differentiation of presynaptic termini in C. elegans. Nature. 1999;401:371–375. doi: 10.1038/43886. [DOI] [PubMed] [Google Scholar]
- 16.Kaufmann N, DeProto J, Ranjan R, Wan H, Van Vactor D. Drosophila liprin-alpha and the receptor phosphatase Dlar control synapse morphogenesis. Neuron. 2002;34:27–38. doi: 10.1016/s0896-6273(02)00643-8. [DOI] [PubMed] [Google Scholar]
- 17.Owald D, et al. A Syd-1 homologue regulates pre- and postsynaptic maturation in Drosophila. The Journal of Cell Biology. 2010;188:565–579. doi: 10.1083/jcb.200908055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wagh DA, et al. Bruchpilot, a protein with homology to ELKS/CAST, is required for structural integrity and function of synaptic active zones in Drosophila. Neuron. 2006;49:833–844. doi: 10.1016/j.neuron.2006.02.008. [DOI] [PubMed] [Google Scholar]
- 19.Kittel RJ, et al. Bruchpilot promotes active zone assembly, Ca2+ channel clustering, and vesicle release. Science (New York, NY) 2006;312:1051–1054. doi: 10.1126/science.1126308. [DOI] [PubMed] [Google Scholar]
- 20.Dai Y, et al. SYD-2 Liprin-alpha organizes presynaptic active zone formation through ELKS. Nature Neuroscience. 2006;9:1479–1487. doi: 10.1038/nn1808. [DOI] [PubMed] [Google Scholar]
- 21.Patel MR, Shen K. RSY-1 is a local inhibitor of presynaptic assembly in C. elegans. Science (New York, NY) 2009;323:1500–1503. doi: 10.1126/science.1169025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Spangler SA, Hoogenraad CC. Liprin-alpha proteins: scaffold molecules for synapse maturation. Biochemical Society transactions. 2007;35:1278–1282. doi: 10.1042/BST0351278. [DOI] [PubMed] [Google Scholar]
- 23.Sankaranarayanan S, Atluri PP, Ryan TA. Actin has a molecular scaffolding, not propulsive, role in presynaptic function. Nature Neuroscience. 2003;6:127–135. doi: 10.1038/nn1002. [DOI] [PubMed] [Google Scholar]
- 24.Morales M, Colicos MA, Goda Y. Actin-dependent regulation of neurotransmitter release at central synapses. Neuron. 2000;27:539–550. doi: 10.1016/s0896-6273(00)00064-7. [DOI] [PubMed] [Google Scholar]
- 25.Lucido AL, et al. Rapid assembly of functional presynaptic boutons triggered by adhesive contacts. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2009;29:12449–12466. doi: 10.1523/JNEUROSCI.1381-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ball RW, et al. Retrograde BMP signaling controls synaptic growth at the NMJ by regulating trio expression in motor neurons. Neuron. 2010;66:536–549. doi: 10.1016/j.neuron.2010.04.011. [DOI] [PubMed] [Google Scholar]
- 27.Allen PB, Ouimet CC, Greengard P. Spinophilin, a novel protein phosphatase 1 binding protein localized to dendritic spines. Proceedings of the National Academy of Sciences of the United States of America. 1997;94:9956–9961. doi: 10.1073/pnas.94.18.9956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nakanishi H, et al. Neurabin: a novel neural tissue-specific actin filament-binding protein involved in neurite formation. The Journal of Cell Biology. 1997;139:951–961. doi: 10.1083/jcb.139.4.951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Satoh A, et al. Neurabin-II/spinophilin. An actin filament-binding protein with one pdz domain localized at cadherin-based cell-cell adhesion sites. The Journal of biological chemistry. 1998;273:3470–3475. doi: 10.1074/jbc.273.6.3470. [DOI] [PubMed] [Google Scholar]
- 30.Zito K, Knott G, Shepherd GM, Shenolikar S, Svoboda K. Induction of spine growth and synapse formation by regulation of the spine actin cytoskeleton. Neuron. 2004;44:321–334. doi: 10.1016/j.neuron.2004.09.022. [DOI] [PubMed] [Google Scholar]
- 31.Terry-Lorenzo RT, et al. Neurabin/protein phosphatase-1 complex regulates dendritic spine morphogenesis and maturation. Molecular biology of the cell. 2005;16:2349–2362. doi: 10.1091/mbc.E04-12-1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ryan XP, et al. The Rho-specific GEF Lfc interacts with neurabin and spinophilin to regulate dendritic spine morphology. Neuron. 2005;47:85–100. doi: 10.1016/j.neuron.2005.05.013. [DOI] [PubMed] [Google Scholar]
- 33.Sieburth D, et al. Systematic analysis of genes required for synapse structure and function. Nature. 2005;436:510–517. doi: 10.1038/nature03809. [DOI] [PubMed] [Google Scholar]
- 34.Hung W, Hwang C, Po MD, Zhen M. Neuronal polarity is regulated by a direct interaction between a scaffolding protein, Neurabin, and a presynaptic SAD-1 kinase in Caenorhabditis elegans. Development (Cambridge, England) 2007;134:237–249. doi: 10.1242/dev.02725. [DOI] [PubMed] [Google Scholar]
- 35.Dai Z, Peng HB. Dynamics of synaptic vesicles in cultured spinal cord neurons in relationship to synaptogenesis. Molecular and cellular neurosciences. 1996;7:443–452. doi: 10.1006/mcne.1996.0032. [DOI] [PubMed] [Google Scholar]
- 36.Zhang W, Benson DL. Stages of synapse development defined by dependence on F-actin. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2001;21:5169–5181. doi: 10.1523/JNEUROSCI.21-14-05169.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang W, Benson DL. Developmentally regulated changes in cellular compartmentation and synaptic distribution of actin in hippocampal neurons. Journal of neuroscience research. 2002;69:427–436. doi: 10.1002/jnr.10313. [DOI] [PubMed] [Google Scholar]
- 38.Cingolani LA, Goda Y. Actin in action: the interplay between the actin cytoskeleton and synaptic efficacy. Nature Reviews Neuroscience. 2008;9:344–356. doi: 10.1038/nrn2373. [DOI] [PubMed] [Google Scholar]
- 39.Burkel B, Von Dassow G, Bement W. Versatile fluorescent probes for actin filaments based on the actin-binding domain of utrophin. Cell Motility and the Cytoskeleton. 2007;64:822–832. doi: 10.1002/cm.20226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Keep NH. Structural comparison of actin binding in utrophin and dystrophin. Neurological sciences : official journal of the Italian Neurological Society and of the Italian Society of Clinical Neurophysiology. 2000;21:S929–937. doi: 10.1007/s100720070006. [DOI] [PubMed] [Google Scholar]
- 41.Norwood FL, Sutherland-Smith AJ, Keep NH, Kendrick-Jones J. The structure of the N-terminal actin-binding domain of human dystrophin and how mutations in this domain may cause Duchenne or Becker muscular dystrophy. Structure (London, England : 1993) 2000;8:481–491. doi: 10.1016/s0969-2126(00)00132-5. [DOI] [PubMed] [Google Scholar]
- 42.Edwards K, Demsky M, Montague R, Weymouth N, Kiehart D. GFP-Moesin Illuminates Actin Cytoskeleton Dynamics in Living Tissue and Demonstrates Cell Shape Changes during Morphogenesis inDrosophila* 1. Developmental biology. 1997;191:103–117. doi: 10.1006/dbio.1997.8707. [DOI] [PubMed] [Google Scholar]
- 43.Shen K, Cowan C. Guidance Molecules in Synapse Formation and Plasticity. Cold Spring Harbor Perspectives in Biology. 2010;2 doi: 10.1101/cshperspect.a001842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chen EH, Olson EN. Towards a molecular pathway for myoblast fusion in Drosophila. Trends in cell biology. 2004;14:452–460. doi: 10.1016/j.tcb.2004.07.008. [DOI] [PubMed] [Google Scholar]
- 45.Fr√Πkjaer-Jensen C, et al. Targeted gene deletions in C. elegans using transposon excision. Nature Methods. 7:451–453. doi: 10.1038/nmeth.1454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Granger L, Martin E, Ségalat L. Mos as a tool for genome-wide insertional mutagenesis in Caenorhabditis elegans: results of a pilot study. Nucleic acids research. 2004;32:e117. doi: 10.1093/nar/gnh111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mello C, Fire A. DNA transformation. Methods in cell biology. 1995;48:451–482. [PubMed] [Google Scholar]
- 48.Oldenburg KR, Vo KT, Michaelis S, Paddon C. Recombination-mediated PCR-directed plasmid construction in vivo in yeast. Nucleic acids research. 1997;25:451–452. doi: 10.1093/nar/25.2.451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Stringham E, Dixon D, Jones D, Candido E. Temporal and spatial expression patterns of the small heat shock (hsp16) genes in transgenic Caenorhabditis elegans. Molecular Biology of the Cell. 1992;3:221. doi: 10.1091/mbc.3.2.221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Patel MR, Shen K. RSY-1 is a local inhibitor of presynaptic assembly in C. elegans. Science. 2009;323:1500–1503. doi: 10.1126/science.1169025. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.