Abstract
As imaging, computing, and data storage technologies improve, there is an increasing opportunity for multiscale analysis of three-dimensional datasets (3-D). Such analysis enables, for example, microscale elements of multiple macroscale specimens to be compared throughout the entire macroscale specimen. Spatial comparisons require bringing datasets into co-alignment. One approach for co-alignment involves elastic deformations of data in addition to rigid alignments. The elastic deformations distort space, and if not accounted for, can distort the information at the microscale. The algorithms developed in this work address this issue by allowing multiple data points to be encoded into a single image pixel, appropriately tracking each data point to ensure lossless data mapping during elastic spatial deformation. This approach was developed and implemented for both 2-D and 3-D registration of images. Lossless reconstruction and registration was applied to semi-quantitative cellular gene expression data in the mouse brain, enabling comparison of multiple spatially registered 3-D datasets without any augmentation of the cellular data. Standard reconstruction and registration without the lossless approach resulted in errors in cellular quantities of ~ 8%.
I. Introduction
Dye-based in situ hybridization (ISH) is an experimental method that enables semi-quantitative detection of gene expression in every individual cell across an entire thin slice of tissue [1]. When combined with rapid tissue sectioning, robotic hybridization equipment, and automated microscale robotic imaging, the process becomes high-throughput (HT) ISH [2]. HT-ISH allows microscale components to be characterized for every cell throughout an entire organ or organism. In the past, comparing HT-ISH datasets required focusing on and delineating spatial areas of interest to compare between the different datasets [3]. The research presented here is motivated by the desire to accurately and automatically perform such localized comparisons across the entire HT-ISH datasets without human interaction. Critical to this goal is the ability to achieve spatial co-registration of the datasets to be compared. The data can differ to some degree in shape due to being acquired from different organisms. In addition, the sectioning process deforms that data. Standard approaches for 2-D and 3-D deformable registration move pixels or voxels to different locations in space, sometimes with multiple pixels or voxels attempting to re-locate to the same spot, and sometimes with certain destination points not being mapped to any particular pixel or voxel. These situations are normally handled by interpolation, which is generally fine for intensity-based data [4]. However, HT-ISH gene expression data has been quantified to identify what level of gene transcripts are expressed in each cell (strong, medium, weak, or not detected) using Celldetekt [5]. Comparing the relative number of strongly expressing cells between two datasets is highly informative, and this comparison would be significantly damaged if some cells duplicate or disappear due to deformation. In order to perform accurate comparisons, we developed an approach for lossless 2-D and 3-D registration of quantified cellular image data.
II. METHODS
A. Image Data
For developing and demonstrating lossless reconstruction and registration, we used two HT-ISH datasets representing Corticotropin releasing hormone (Crh) gene expression in both the wild-type and MECP2 mouse brains. MECP2 is a mouse model for Rett Syndrome, a genetic neurological disorder [6, 7]. Identifying local differences in Crh gene expression between normal and MECP2 brains can provide information about the molecular mechanisms underlying Rett Syndrome [3]. The preparation of the image data is described in McGill et al [3]. Briefly, each image dataset consisted of 350 images, with each of these representing a 25 μm thick tissue slice of serial sectioned adult mouse brain. Tissue slices were digitized at 1.6 μm per pixel, providing sufficient resolution to see cell bodies (~μm micron diameter) and to semi-quantitatively characterize levels of Crh gene expression within each cell. Using Celldetekt [5], each detected cell is assigned as having Strong (red), Moderate (blue), Weak (yellow), or Not Detected (grey) levels of gene expression. One type of output produced by Celldetekt is a downsampled representation of the tissue to the scale where each pixel represents one cell (~12 μm per pixel). Thus, counting pixels of different colors enables comparative analysis of cell populations.
B. Lossless encoding
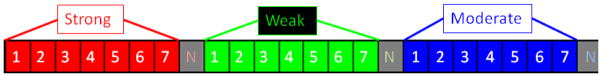
During the tissue sectioning process, distortions take place. In order to undo these distortions and reconstruct an accurate 3D volume of the mouse brain from 2D tissue slices, rigid and non-linear transformations are necessary. These transformations can force multiple pixels into the same pixel, or spread apart other pixels. If applied to quantitative data, this would potentially distort quantitative cell counts. We registered two Celldetekt images to determine the amount of disruption to cell counts. Then, we developed an approach that allows transformations to the Celldetekt images without loss of the quantitative cell counts. To accomplish this, we first substituted Green for Yellow to represent weak expression. Then we used the bitwise encoding depicted in Figure 1 to allow multiple cells to be represented in the same pixel.
Fig 1.
Encoding gene expression. Information for multiple cells can be stored within a single pixel or voxel.
For the new lossless encoding, each pixel in an RGB image is represented using 24-bits. Seven bits are allocated for strong expression, seven bits for weak expression, seven bits for moderate expression, and three bits for not detected expression. The encoding in this fashion allows visual interpretation of gene expression in its native form, while maintaining quantitative data without loss despite multiple cells occupying the same pixel or voxel. During 2-D and 3-D image transformations, cells are reassigned locations. Such transformations by nature will sometimes map two pixels to the same pixel space, or fail to map other pixels to the new space. In this lossless implementation of image transformations, if two pixels are mapped to the same space, they are encoded into that spot using the lossless encoding method. After initial transformation, pixels left behind and not mapped were detected. Then a breadth-first search was applied to find a nearby pixel that was mapped over to the new space, and the left behind pixel is also mapped to the new location of the nearby pixel using lossless encoding.
C. Image Processing Steps
To test lossless encoding in maintaining quantitative information during the volumetric reconstruction and volume-to-volume registration process, we proceeded through the steps depicted in Algorithms 1 and 2 with our two image datasets. New code for applying the steps to the lossless-data was written in the Python programming language (www.python.org) and utilized the Python Imaging Library (www.pythonware.com). The basic approach was to compute necessary image spatial deformations on rescaled raw images of the HT-ISH tissue sections, and then apply the spatial changes not only to these images, but also to the encoded Celldetekt quantified gene expression image data as well as to a set of artifact masks. First, Celldetekt and Deftekt [8] images were generated from the serial image data. Deftekt produced binary masks indicating where artifacts, such as dust particles, were in the image [8]. Deftekt also produced a set of deformations that will repair tears introduced to the tissue sections from the data preparation process. After applying the fixes to the tears, image-to-image rigid registrations (translations, rotations) and elastic registrations were computed and warps applied. 2-D elastic registrations were calculated using bUnwarpJ [9, 10] as described in Kindle et al [8]. Each image in the now aligned image series was downsampled 50% to achieve isotropic resolution at 25 μm, thus generating isotropic volumes of cellular gene expression for the mouse brain. This downsampling utilized the lossless encoding method for compressing the data. 3-D rigid registration minimized the sum of the squared distance for eight manually identified landmarks in the mouse brain. Elastic registration then utilized drop-3d software [11] to compute warps. 3-D transformations were also applied using the lossless encoding method.
Algorithm 1.
Lossless 2-D to 3-D Reconstruction
| L1: Start with set of serial 2-D images |
| L2: Quantify cellular gene expression in L1 images using Celldetekt |
| L3: Generate artifact mask images from L1 images using Deftekt |
| L4: Fix tears in L1 images using Deftekt |
| L5: Compute 2-D image-to-image rigid registration for L4 images |
| L6: Compute 2-D image-to-image elastic warps for L5 images using bUnwarpJ |
| L7: Compress L6 volume to isotropic resolution |
| L8: Use lossless encoding algorithms to apply transformations computed in steps L4 – L7 to L2 images. |
| L9: Use lossless encoding algorithms to apply transformations computed in steps L4 – L7 to L3 images. |
III. RESULTS
Cell counts for strong, medium, weak, and no gene expression were compared before and after image deformation without using the lossless approach (Table 1). Using nearest-neighbor assignment, 2.9% of pixels were vacant in warped result, and 2.4% were duplicated. In some images, these numbers were 8% and one pixel was duplicated 5 times. The lossless approach, on the other hand, when applied successfully maintained the exact same cell counts for 2-D and 3-D non-linear registration.
TABLE I.
Warping Quantitative Data without Lossless Approach
| Pixel Type | Before Warp | After Warp | # Gained (+) or Lost (−) |
|---|---|---|---|
| Strong Expression | 796 | 782 | −14 |
| Moderate Expression | 1020 | 1004 | −16 |
| Weak Expression | 7272 | 7117 | −155 |
| No Expression | 149338 | 147564 | −1774 |
| No Cell | 746790 | 748749 | +1959 |
Statistics for one example image on effects of warping on quantitative data without lossless approach.
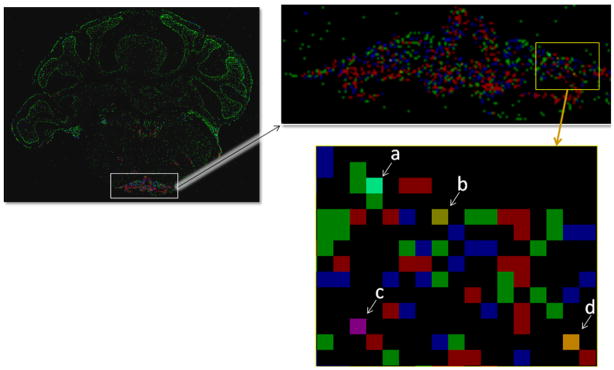
The lossless approach is designed to allow visualization of quantified cellular gene expression in addition to maintaining accurate cell counts after deformations. The visualization of a 2-D image after registration is shown in Figure 2. The different pixels clearly indicate gene expression in each cell and which pixels contain multiple cells. With a trained eye, the gene expression levels of the pixels containing multiple cells can be decoded in many common instances.
Fig 2.
Visualizing the data using lossless encoding of gene expression. Red is strong, blue is moderate, and green is weak cellular gene expression. Indicated pixels contain more than one cell: (a) 3 weak and 1 moderate; (b) 1 strong and 1 weak; (c) 1 strong and 1 moderate; and (d) 2 strong and 1 weak.
IV. DISCUSSION
The presented approach successfully reconstructed quantitative Celldetekt data from serial sections represented by 2-D images into a 3-D dataset using elastic registration, and performed 3-D registration upon the dataset to bring it into spatial co-alignment with another 3-D dataset. It performed these reconstructions using a lossless registration approach, resulting in conservation of cell counts in the spatial modified data. One of the limits of the approach is that only 7 cells of each gene expression strength could fit into each voxel (just 3 for Not Detected expression), with a possible total of 24 cells in a voxel. Switching to a binary encoding would allow for (127 + 127 + 127 + 7) = 388 cells to be in a single voxel using the same number of bits currently used for each cell type. The encoding may be able to be further optimized to allow for better viewing of non-expressing cells without relying on post-processing images.
The 3-D warp matrix computed as part of this work did not achieve sufficient alignment in the areas of anatomical interest. To be useful in this specific application, subregional features on the order of 500 μm in diameter (~5% total data dimension) need to align precisely in 3-D. The registration approach, while globally improving spatial alignment significantly, did not properly align these subregional features. Nevertheless, if a better warp matrix calculation is found, the proposed lossless encoding approach would be applicable to that method. One promising method that would bypass direct computation of a warp matrix is the use of deformable space, such as 3-D subdivision atlases [12]. This technology is in active development for multiregional atlases such as would be appropriate for the mouse brain. A potential future application of this work will be in comparing the accuracy of the 3-D non-linear registration with an atlas-based approach.
Algorithm 2.
Volumetric lossless registration
| V1: Compute 3-D translations between L7 volumes to minimize distance between landmarks |
| V2: Compute 3-D elastic warps using drop-3d between V1 volumes |
| V3: Using lossless encoding algorithms, apply transformations computed in steps V1 and V2 to L8 volumes to generate aligned artifact mask volumes |
| V4: Using lossless encoding algorithms, apply transformations computed in steps V1 and V2 to L9 volumes to output registered cellular gene expression data |
Acknowledgments
This work was supported in part by grants from the National Science Foundation (BDI 0743691), the National Institute of Neurological Disorders and Stroke (1R21NS058553-01), LDRD DE-AC05-76RL01830, and the Department of Energy Science Undergraduate Laboratory Internship Program.
Image data was graciously provided by Huda Zoghbi and Christina Thaller at Baylor College of Medicine. We thank Howard Durrant for his assistance.
Contributor Information
Matthew A. Enlow, Pacific Northwest National Laboratory, Richland, WA 99352 USA
Tao Ju, Email: taoju@wustl.edu, Department of Computer Science and Engineering, Washington University in St. Louis, St. Louis, MO USA.
Ioannis A. Kakadiaris, Email: ikakadia@uh.edu, Computational Biomedicine Laboratory, Department of Computer Science, University of Houston, Houston, TX USA
James P. Carson, Email: james.carson@pnl.gov, Biological Monitoring and Modeling Group, Pacific Northwest National Laboratory, Richland, WA 99352 USA. (phone: 509-371-6894; fax: 509-371-6978)
References
- 1.Carson J, Ju T, Bello M, Thaller C, Warren J, Kakadiaris IA, Chiu W, Eichele G. Automated pipeline for atlas-based annotation of gene expression patterns: application to postnatal day 7 mouse brain. Methods. 2010 Feb;50(2):85–95. doi: 10.1016/j.ymeth.2009.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Visel A, Thaller C, Eichele G. Genepaint.org: An Atlas of Gene Expression Patterns in the Mouse Embryo. Nucleic Acids Res. 2004 Jan 1;32(Database issue):D552–6. doi: 10.1093/nar/gkh029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.McGill BE, Bundle SF, Yaylaoglu MB, Carson JP, Thaller C, Zoghbi HY. Enhanced anxiety and stress-induced corticosterone release are associated with increased Crh expression in a mouse model of Rett syndrome. Proceedings of the National Academy of Sciences of the United States of America. 2006 Nov 28;103(48):18267–72. doi: 10.1073/pnas.0608702103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ju T, Warren J, Carson J, Bello M, Kakadiaris I, Chiu W, Thaller C, Eichele G. 3D volume reconstruction of a mouse brain from histological sections using warp filtering. J Neurosci Methods. 2006 Sep 30;156(1–2):84–100. doi: 10.1016/j.jneumeth.2006.02.020. [DOI] [PubMed] [Google Scholar]
- 5.Carson JP, Eichele G, Chiu W. A method for automated detection of gene expression required for the establishment of a digital transcriptome-wide gene expression atlas. J Microsc. 2005 Mar;217(Pt 3):275–81. doi: 10.1111/j.1365-2818.2005.01450.x. [DOI] [PubMed] [Google Scholar]
- 6.Ben-Shachar S, Chahrour M, Thaller C, Shaw CA, Zoghbi HY. Mouse models of MeCP2 disorders share gene expression changes in the cerebellum and hypothalamus. Hum Mol Genet. 2009 Jul 1;18(13):2431–42. doi: 10.1093/hmg/ddp181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fyffe SL, Neul JL, Samaco RC, Chao HT, Ben-Shachar S, Moretti P, McGill BE, Goulding EH, Sullivan E, Tecott LH, Zoghbi HY. Deletion of Mecp2 in Sim1-expressing neurons reveals a critical role for MeCP2 in feeding behavior, aggression, and the response to stress. Neuron. 2008 Sep 25;59(6):947–58. doi: 10.1016/j.neuron.2008.07.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kindle LM, Kakadiaris IA, Ju T, Carson JP. A semiautomated approach for artefact removal in serial tissue cryosections. Journal of Microscopy. 2010 Feb;241(2):200–6. doi: 10.1111/j.1365-2818.2010.03424.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Arganda-Carreras I, Sanchez CO, Marabini R, Carazo JM, Ortizde Solorzano C, Kybic J. Consistent and Elastic Registration of Histological Sections Using Vector-Spline Regularization. Lecture Notes in Computer Science - Computer Vision Approaches to Medical Image Analysis. 2006;4241:85–95. [Google Scholar]
- 10.Rasband WS. ImageJ. U. S. National Institutes of Health; 1997–2010. [Google Scholar]
- 11.Glocker B, Komodakis N, Tziritas G, Navab N, Paragios N. Dense image registration through MRFs and efficient linear programming. Med Image Anal. 2008 Dec;12(6):731–41. doi: 10.1016/j.media.2008.03.006. [DOI] [PubMed] [Google Scholar]
- 12.Ju T, Carson J, Liu L, Warren J, Bello M, Kakadiaris I. Subdivision meshes for organizing spatial biomedical data. Methods. 2010 Feb;50(2):70–6. doi: 10.1016/j.ymeth.2009.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]