Abstract
The discovery of new compounds for the pharmacological manipulation of protein function often embraces the screening of compound collections, and it is widely recognized that natural products offer beneficial characteristics as protein ligands. Much effort has therefore been focused on “natural product-like” libraries, yet the synthesis and screening of such libraries is often limited by one or more of the following: modest library sizes and structural diversity, conformational heterogeneity, and the costs associated with the substantial infrastructure of modern high-throughput screening centers. Here, we describe the design and execution of an approach to this broad problem by merging principles associated biologically-inspired oligomerization and the structure of polyketide-derived natural products. A novel class of chiral and conformationally-constrained oligomers is described (termed “chiral oligomers of pentenoic amides” – COPAs) that offers compatibility with split-and-pool methods and can be screened en masse in a batch mode. We demonstrate that a COPA library containing 160,000 compounds is a useful source of novel protein ligands by identifying a non-covalent synthetic ligand to the DNA-binding domain of the p53 transcription factor.
Oligomerization is the central synthetic strategy by which nature derives molecules with function. With only a small collection of monomeric units, and bond-forming processes compatible with the cellular environment, sequential union or modular assembly (oligomerization) results in products possessing great molecular and functional diversity. Examples include complex biological polymers like proteins, nucleic acids, and carbohydrates, as well as small molecule natural products (i.e. fatty acids, polyketides and terpenes). In contrast to Nature’s modular approach to molecular diversification, the impressive and elegant state-of-the-art laboratory solutions typically embrace strategic and divergent reactivity of complex organic intermediates (here, unique reactivity of library members is used to alter the structure of subpopulations within a library).(1–4) Attempts to emulate Nature’s modular process for establishing molecular diversity have delivered interesting compound classes (i.e. β-peptides, peptoids, and peptide nucleic acids; Figure 1a), whose bio-inspired oligomeric framework is compatible with split-and-pool solid phase synthesis (5) — a powerful technology for the creation of large and diverse chemical libraries.(6) However, a common limitation associated with such synthetic oligomers is that they uniformly lack the conformational constraints typical of small molecule natural products — a property that likely limits their affinity for protein targets due to entropic penalties incurred on assuming a particular bound conformation. Here, we describe a new class of natural product-inspired synthetic oligomers that overcome such limitations. In combination with solid-phase split-and-pool chemistry, and on-bead screening technology, the science described defines a powerful new approach to the discovery of small molecule protein-binding ligands.
Figure 1. Natural and synthetic oligomers, polyketide-derived natural products, and a polyketide-inspired class of chiral and conformationally rigid synthetic oligomer.
a, A selection of biological and biopolymer mimetics. b, A selection of polyketide-derived natural products (regions highlighted in red are substituted alkenes that impart conformational rigidification as a result of A1,3 strain). c, General structure of COPAs – chiral oligomers of N-substituted 5-amino-2,4-dialkyl-3-pentenoic amides. d, Structural features that lead to the rigidification of COPA oligomers.
Results
Design of a new class of chiral and conformationally rigid synthetic oligomer
Inspired by the power of modularity in synthetic design and the mathematical merits of oligomerization as a central theme for molecular diversification (7, 8), we hypothesized that a new class of chiral and conformationally constrained synthetic oligomers, where chirality could be readily addressed in concert with local conformational preferences, would define a substantial advance in discovery-oriented science. Being unbound by the desire to mimic three-dimensional motifs seen in biological macromolecules (9–20), we turned to the structure of polyketide-derived small molecule natural products for inspiration (Figure 1b).(21) Members of this class often contain relatively simple stereochemically-defined structural motifs that, in sum, dictate their global conformational preferences. With this molecular characteristic as a guiding principle, we have designed and executed a two-step iterative synthetic pathway to polyketide-inspired oligomers termed COPAs (chiral oligomers of pentenoic amides)(Figure 1c), where a central N-substituted 5-amino-2,4-dialkyl-3-pentenoic amide provides a rigid and chiral environment about each monomeric unit. The control of conformation resulting from this motif is substantial, and is based on the minimization of non-bonded steric interactions about the α-branched trisubstituted alkene and α-branched tertiary amide.(22) As illustrated in Figure 1d, each of these structural motifs imparts substantial rigidification, as the C2 proton is constrained to being in-plane with the C4-alkyl group (i.e. Me), and R2-amide substituent – defining a rigid chiral environment at each monomer, where the amide and alkyl substitution emerging from this core are precisely oriented in three-dimensional space.
Impact of stereochemistry on the structure of COPAs
While the local effect of stereochemistry and substitution within each COPA core unit is clear (Figure 1d), the combined influence of distinct chiral subunits on the gross conformational preferences of a COPA oligomer is more difficult to conceptualize/predict. For an initial assessment of the potential unique properties of chiral COPA oligomers, we turned to molecular mechanics calculations offered within Spartan-08 (Conformer Distribution, MMFF model/Monte-Carlo algorithm). At the outset, we embraced the notion that these calculations would only provide a uniform lens through which to observe differences associated with model oligomers, and would likely not depict relevant solution phase behavior. That said, analysis of homogeneously substituted oligomer backbones (COPA vs. peptoid) with this computational method leads to the identification of some striking differences. First, for the COPA oligomer, the three-dimensional structures of the lowest energy conformers found within each isomeric series (i.e. four random examples are depicted – RRRR, RRSS, RSRS, and RSSR) are unique with respect to one another, despite their identical polymethylated substitution pattern. Second, the analysis of each stereoisomer resulted in the identification of ≤ two conformers within 1.1 kcal/mol of the low energy conformer found. Using the identical molecular mechanics calculation for analysis of a polymethylated peptoid tetramer (MW = 371.4) and the larger peptoid octamer of related molecular weight to the COPA tetramers analyzed (MW = 655.7), seven and thirteen conformers were identified within 1.1 kcal/mol of the low energy conformation found for each (this analysis includes the mirror images of the conformations depicted). While we are mindful of not over-interpreting these results, the analysis described is consistent with the hypothesis that COPAs are more conformationally restricted than peptoids, and stereoisomeric COPA oligomers have disparate conformational preferences.
Design and execution of solution- and solid-phase organic chemistry for COPA library synthesis
Synthetically, COPAs have been designed to be accessible using a “sub-monomer” route akin to that employed in peptoid synthesis (Figure 3a; 2→3).(23–25) This allows simple primary amines, of which hundreds are commercially available, to be employed as one of the diversity elements in split-and-pool library synthesis – a critically important characteristic to enable the synthesis of large compound collections necessary to increase the likelihood of finding high affinity hits in unbiased screens.(26) Furthermore, this design was optimistically thought to be compatible with MS-MS analysis, where compound from a single bead would be sufficient to decode the precise structure of oligomer present. As such, one could avoid employing an encoding strategy to elucidate hit-structure.(27)
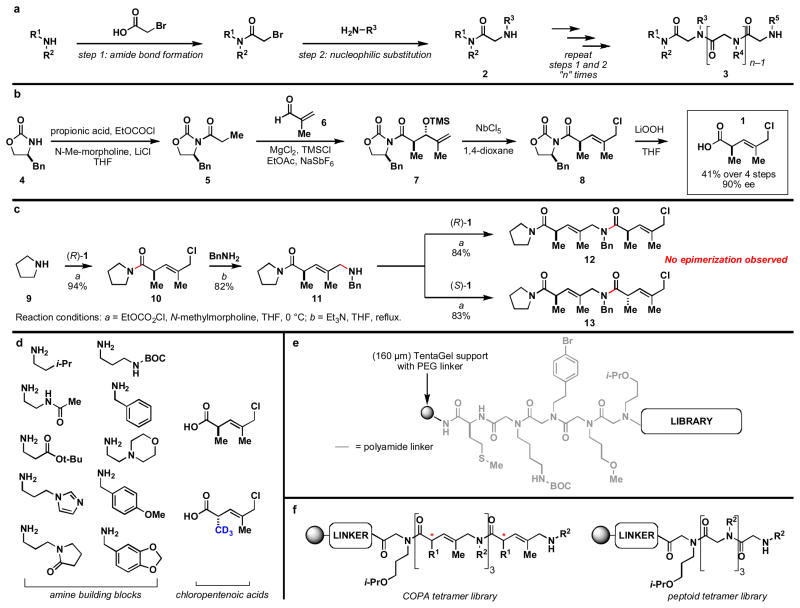
Figure 3. Chemical development of COPA oligomers: From general oligomerization strategy, asymmetric synthesis and library construction.
a , The “sub-monomer” style synthesis of peptoids. b, Asymmetric synthesis of 5-chloro-2,4-dimethyl-3-pentenoic acid 1. c, Use of 1 in solution phase oligomerization. d, Panel of monomers used in library synthesis. e, General information regarding resin and linker employed in solid-phase library synthesis. f, General structure of libraries prepared from building blocks depicted in Figure 3d – COPA and peptoid tetramers.
To affect this strategy, we required a practical and scalable synthesis of both antipodes of chloropentenoic acids like 1 (Figure 3b). Ideally, this synthesis would also be capable of delivering future analogs of 1 with diverse C2 and C4 substitution. Therefore, a convergent synthetic pathway was targeted to facilitate future analog generation that could avoid chromatographic purification at any step, proceed from readily available starting materials, limit the use of air and moisture sensitive reagents, and deliver the chiral monomers with high levels of stereochemical fidelity. Our solution to this problem is depicted in Figure 3b. Synthesis of the propionyl oxazolidinone 4 was accomplished without the requirement of a highly reactive base (28) and subsequent stereoselective aldol reaction with methacrolein (6) was achieved under reaction conditions that do not require pre-generation of a metal enolate.(29) Isolated by simple filtration through silica, the TMS-ether 7 was converted to the stereodefined allylic chloride 8 by Nb-mediated stereoselective halogenation (E:Z ≥ 20:1).(30) While seemingly difficult to accomplish in a highly selective fashion, hydrolysis of the imide proceeded uneventfully (without significant hydrolysis of the allylic chloride) and delivered 1 in 90% ee and 41% overall yield. Notably, this synthesis procedure delivers optically active 1 in acceptable yield and purity through a simple four-step sequence that does not require a single chromatographic operation.(31) As an indication of the robust nature of this sequence, routine syntheses of 1 (4g) were performed in just a few days from 10g of 4.
As depicted in Figure 3c, solution phase amide bond formation with a simple secondary amine (via the mixed anhydride) proceeded effectively, in this case delivering the chloroamide 10 in 94% yield. Unlike related coupling reactions for the synthesis of peptides, no evidence was found for epimerization of the potentially labile α-stereocenter of 10. Subsequent coupling with benzylamine proceeded in a similarly straightforward manner, delivering aminoamide 11 in 82% yield. This simple two-step sequence validated the central steps of the proposed oligomerization of 1 and confirmed that chiral chloroacids of this and related structure can be functionalized in a similar manner to α-bromoacetic acid in peptoid synthesis.
Subsequent homologation of 11 with either enantiomer of 1 leads to the production of the corresponding dimers 12 and 13 with similarly high levels of efficiency. This success indicates that double asymmetric relationships between amine 11 and acid 1 have little impact on chemical efficiency for this bond construction — a critically important virtue of this chemistry, as split-and-pool techniques will aim to prepare all combinations of stereochemistry along the growing COPA backbone. With regard to this later consideration associated with the projected application of this chemistry in split-and-pool format, we recognize that oligomerization of chiral monomers of 90% ee will result in the production of minor diastereomers on each bead. The combinatorial nature of this process however will ensure that the population contains beads that present these minor impurities as major constituents. In this way, a built in control mechanism exists to aid in the analysis of assay results as the minor component on any particular bead will be present as a major component on a different bead within the collection. That said, we anticipate that conditions can be found in future studies to crystallize intermediate 7 or 8 as a means to attain isomeric homogeneity.
Establishing the value of COPA libraries as a source of protein ligands
Moving forward to explore the utility of COPA oligomers as a potential source of protein ligands, a library of tetramers was prepared by split-and-pool methods. To be compatible with our on-bead screening platform (32), we selected 160 μm TentaGel beads that were functionalized with a tetrameric polyamide (Figure 3e), the structure of which was selected to optimize subsequent MS-based structure elucidation.(31) Targeting a library of 160,000 members, we employed ten primary amines and two pentenoic acids as depicted in Figure 3d. Since we planned to carry out structural elucidation by mass spectrometry, we employed a heavy atom label (CD3 at C2) to correlate differences in the mass of fragment ions with absolute stereochemistry of the chloropentenoic acid monomer. Alongside these efforts, a library of peptoid tetramers was prepared with the same amines used for the COPA library (Figure 3f) in an effort to establish a baseline for comparison between these two synthetic oligomer platforms. MALDI mass spectra revealed a single strong peak for the COPAs released from several individual beads chosen randomly from the library, indicating that each bead displays predominantly a single compound and that each synthetic step proceeded in high yield.
Having established that the library was of high quality, it was screened against the DNA-binding domain of p53, an important transcription factor that regulates a variety of genes involved in cell cycle control and apoptosis. More than half of human cancers express inactive p53 due to the presence of missense mutations in the DNA-binding domain (DBD) that destabilize the folding of the protein.(33) There is considerable interest in the identification of “chemical chaperones” whose binding to p53 might stabilize the wild-type, functional, folded conformation.(34) Since transcription factors are generally considered to be extremely challenging targets for small molecules (35), we considered p53 recognition a stringent test of the utility of this new class of compounds.
Purified, bacterially expressed, FLAG-tagged p53 DBD (10 μM) was incubated with the bead-displayed COPA library in the presence of high levels of competitor proteins to suppress non-specific binding events. The beads were then washed and treated with anti-FLAG antibody followed, after another washing step, by anti-IgG antibodies conjugated to red quantum dots. The beads were then examined under a low power fluorescent microscope. Several beads with a strong red halo surrounding them, indicating binding of the quantum dot via the p53-FLAG/anti-FLAG antibody/anti-IgG-QD sandwich complex, were observed (Figure 4a). These, as well as some beads with weaker staining, were picked using a micropipette. In all, 22 beads were collected. Six of these putative hits proved to be ligands for either anti-FLAG antibody or the secondary antibody-conjugated quantum dots. The same experiment was done with the peptoid library. In this case, no obvious “hits” with strong red halos were observed, but several more weakly fluorescent beads were picked. The beads were separated in wells of a microtiter plate and the oligomer was released from the resin via CNBr-mediated cleavage of a methionine residue in the linker.
Figure 4. COPA library synthesis, screening, structure elucidation and validation.
a , General scheme for on-bead screening of COPA library against DNA binding domain of p53 (p53-DBD, residues 94 to 312) expressed with an epitope tag FLAG. TentaGel beads bound to p53-DBD protein were visualized under fluorescent microscope by treating beads with anti-FLAG primary antibody and anti-IgG secondary antibody conjugated to Quantum dot emitting red fluorescent light at 655 nm. b, Sequence elucidation and identification of a COPA tetramer that binds to the p53-DBD. Sequence of the COPA tetramer was established by analysis of mass spectral data derived from ETD-based fragmentation. c, Fluorescence polarization assay for binding affinity of fluorescein conjugated COPA tetramer (14a) against p53-DBD, carbonic anhydrolase II (CAH II from bovine erythrocyte), platelet activating factor acetyl hydrolase (PAFAHIB3), and bromodomain containing 4 (BRD4) proteins. Error bars show the standard deviation (SD) of the data obtained from three independent experiments. The data analysis and curve fitting were performed with GraphPad Prism 5.0 (GraphPad Software, San Diego, CA) using a non-linear regression method (y=[x/(KD + x)](ymax − ymin) + ymin). A COPA tetramer with the same linker region and different side chains on the amide nitrogens was used as a control oligomer (co). The binding affinity of COPA tetramer to p53-DBD was determined as KD ~ 10μM. See Supporting Information for additional details.
While strong precursor ion signals were observed in the MALDI mass spectrum for the COPAs, sequence specific product ions were not produced via collision-induced dissociation. Therefore, we attempted to sequence these molecules via tandem ESI mass spectrometry with electron transfer dissociation (ETD).(36) This ETD approach proved reasonably successful and the sequences of 8 of the 16 COPA hits could be determined unequivocally (Figure S7). These eight molecules were re-synthesized with a fluorescein tag (Figure S8) and tested for binding to p53 by fluorescence anisotropy. Two of the eight COPA molecules showed clear binding to p53. The best of these, compound 14a shown in Figure 4c, bound to the p53 DBD with a KD of approximately 10 μM, but did not bind detectably (KD > 500 μM) to three control proteins. To the best of our knowledge, COPA 14a constitutes the first non-covalent (37) small molecule ligand for the wild-type (38) p53 DNA-binding domain. The addition of an oligonucleotide that binds p53 tightly did not disrupt the p53-COPA complex, indicating that the synthetic ligand does not recognize the DNA-binding surface of the p53 core domain (Supplementary Figure 13).
The same set of experiments was carried out for the peptoids collected as possible hits in the screening experiment. Not surprisingly, given the low intensity of QD fluorescence observed on the beads, none of the peptoids exhibited binding to the p53 DNA-binding domain (KDs > 500 μM). This is interesting in that the COPA and peptoid libraries contained exactly the same amine-derived side chains. Furthermore, controls with independently synthesized COPA diastereomers of 14a and an achiral peptoid bearing the identical amine side chains as 14a, all showed a substantial decrease of affinity for the p53 DBD (Figures S11 and S12). While more work will be required before definitive conclusions can be reached, the results of these experiments are consistent with the proposition that conformationally constrained COPAs may be superior to peptoids as a source of protein ligands.
Discussion
In summary, we have developed a new class of natural product-inspired oligomeric compounds that promises to be a valuable source of protein ligands. COPAs are unusual amongst synthetic small molecule oligomers in that their stereodefined structure is readily malleable and their backbone conformations are supported by concepts long utilized in organic synthesis for acyclic stereocontrol [such as minimization of A(1,3) interactions] — these features offer a unique and powerful means of addressing the disposition of all main chain substituents in three-dimensional space.(39) In essence, we describe a practical chemical solution to diversity-oriented library construction that couples building block diversity to substantial scaffold diversity. This is noteworthy, because the desirability of scaffold diversity in natural product-like libraries has been well documented (40), and current solutions to this problem require careful synthesis planning to accomplish strategic and divergent reactivity of complex organic intermediates in a library synthesis.(41–43) Further compounding the virtues of COPAs as a chemical foundation to discovery-oriented science, the synthetic strategy described here is completely compatible with split-and-pool solid phase synthesis, making very large libraries readily accessible. Moreover, the highly practical and scalable synthesis of either optically pure antipode of the chloropentenoic acid building blocks (i.e. 1), combined with the use of simple primary amines as the source of side chain diversity, allows the synthesis of potentially millions of compounds at a modest cost. Finally, COPA libraries synthesized on hydrophilic TentaGel beads can be employed in a variety of inexpensive, yet powerful, binding screens (32, 44–45), hence eliminating the requirement of substantial infrastructure to maintain diverse compound collections for use in traditional high-throughput screening. We look forward to future developments that explore the power of this chemistry in combination with available on-bead screening technologies as a platform for the discovery of bioactive synthetic molecules.
Supplementary Material
Figure 2. Stereochemistry of COPA backbone is anticipated to have a substantial impact on skeletal shape and the disposition of side chains in space.
Distribution of conformers found within 1.1 kcal/mol of the lowest energy conformation identified. These calculations were conducted with Spartan-08/MMFF model/Conformer Distribution option/Monte-Carlo algorithm. Colored spheres highlight the relative position of heteroatoms (green) and alkenes (blue). While these molecular mechanics calculations are not interpreted to predict the solution phase structure of these oligomers, the calculations provide a uniform lens through which to observe unique characteristics associated with this new class of synthetic oligomer.
Acknowledgments
G.C.M. acknowledges financial support from the Fidelity Biosciences Research Initiative, the Scripps Research Institute, Scripps Florida, and T.K.M. acknowledges support from 1 RO1 GM090294.
Footnotes
Author contributions
G.C.M. and T.K. conceived of and directed the project. C.A. and M.S. contributed equally to the execution of this work. C.A. prepared acid 1 and conducted all solution phase chemistry. M.S. and C.A. conducted all solid phase experiments, library synthesis, decoding experiments and biochemical evaluation of the hits. M.S. purified the p53-DBD, which was cloned and expressed by K.M., and conducted the biochemical characterization of the hit. M.C. developed the ETD-based method for compound decoding. G.C.M. and T.K. wrote the manuscript.
Competing Interests Statement
The authors declare no competing financial interests.
Supplementary information and chemical compound information accompany this paper at www.nature.com/nature. Reprints and permission information is available online at http://npg.nature.com/reprintsandpermissions/.
References
- 1.Schreiber SL. Target-oriented and diversity-oriented organic synthesis in drug discovery. Science. 2000;287:1964–1969. doi: 10.1126/science.287.5460.1964. [DOI] [PubMed] [Google Scholar]
- 2.Tan DS. Diversity-oriented synthesis: exploring the intersections between chemistry and biology. Nat Chem Biol. 2005;1:74–84. doi: 10.1038/nchembio0705-74. [DOI] [PubMed] [Google Scholar]
- 3.Spiegel DA, Schroeder FC, Duvall JR, Schreiber SL. An oligomer-based approach to skeletal diversity in small-molecule synthesis. J Am Chem Soc. 2006;128:14766–14767. doi: 10.1021/ja065724a. [DOI] [PubMed] [Google Scholar]
- 4.Nielsen TE, Schreiber SL. Towards the optimal screening collection: a synthesis strategy. Angew Chem Int Ed. 2008;47:48–56. doi: 10.1002/anie.200703073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Houghten RA. General method for the rapid solid-phase synthesis of large numbers of peptides: Specificity of antigen-antibody interaction at the level of individual amino acids. Proc Natl Acad Sci USA. 1985;82:5131–5135. doi: 10.1073/pnas.82.15.5131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Thompson LA, Ellman JA. Synthesis and applications of small molecule libraries. Chem Rev. 1996;96:555–600. doi: 10.1021/cr9402081. [DOI] [PubMed] [Google Scholar]
- 7.Khosla C, Kapur S, Cane DE. Revisiting the modularity of modular polyketide synthases. Curr Opinion Chem Biol. 2009;13:135–143. doi: 10.1016/j.cbpa.2008.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Walsh CT. The chemical versatility of natural-product assembly lines. Acc Chem Res. 2008;41:4–10. doi: 10.1021/ar7000414. [DOI] [PubMed] [Google Scholar]
- 9.Nielsen PE. Peptide nucleic acids (PNA) in chemical biology and drug discovery. Chem Biodivers. 2010;7:786–804. doi: 10.1002/cbdv.201000005. [DOI] [PubMed] [Google Scholar]
- 10.Seebach D, Gardiner J. β-Peptidic peptidomimetics. Acc Chem Res. 2008;41:1366–1375. doi: 10.1021/ar700263g. [DOI] [PubMed] [Google Scholar]
- 11.Gellman SH. Foldamers: a manifesto. Acc Chem Res. 1998;31:173–180. [Google Scholar]
- 12.Horne WS, Gellman SH. Foldamers with heterogeneous backbones. Acc Chem Res. 2008;41:1399–1408. doi: 10.1021/ar800009n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nowick JS. Exploring β-sheet structure and interactions with chemical model systems. Acc Chem Res. 2008;41:1319–1330. doi: 10.1021/ar800064f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Robinson JA. β-Hairpin peptidomimetics: design, structures and biological activities. Acc Chem Res. 2008;41:1278–1288. doi: 10.1021/ar700259k. [DOI] [PubMed] [Google Scholar]
- 15.Schafmeister CE, Brown ZZ, Gupta S. Shape-programmable macromolecules. Acc Chem Res. 2008;41:1387–1398. doi: 10.1021/ar700283y. [DOI] [PubMed] [Google Scholar]
- 16.Li X, Wu TD, Yang D. α-Aminoxy acids: new possibilities from foldamers to anion receptors and channels. Acc Chem Res. 2008;41:1428–1438. doi: 10.1021/ar8001393. [DOI] [PubMed] [Google Scholar]
- 17.Summerton J. Morpholino antisense oligomers: the case for an RNase H-independent structural type. Biochim Biophys Acta. 1999;1489:141–158. doi: 10.1016/s0167-4781(99)00150-5. [DOI] [PubMed] [Google Scholar]
- 18.Dervan PB. Molecular recognition of DNA by small molecules. Bioorg Med Chem. 2001;9:2215–2235. doi: 10.1016/s0968-0896(01)00262-0. [DOI] [PubMed] [Google Scholar]
- 19.Wuereb H, Maletic M, Gildersleeve J, Pelczer I, Kahne D. Design of an oligosaccharide scaffold that binds in the minor groove of DNA. J Am Chem Soc. 2000;122:1883–1890. [Google Scholar]
- 20.Davis JM, Tsou LK, Hamilton AD. Synthetic non-peptide mimetics of α-helices. Chem Soc Rev. 2007;36:326–334. doi: 10.1039/b608043j. [DOI] [PubMed] [Google Scholar]
- 21.Kumar K, Waldmann H. Synthesis of natural product inspired compound collections. Angew Chem Int Ed. 2009;48:3224–3242. doi: 10.1002/anie.200803437. [DOI] [PubMed] [Google Scholar]
- 22.Hoffmann RW. Flexible Molecules with Defined Shape—Conformational Design. Angew Chem Int Ed. 1992;31:1124–1134. [Google Scholar]
- 23.Simon RJ, et al. Peptoids: A modular approach to drug discovery. Proc Natl Acad Sci USA. 1992;89:9367–9371. doi: 10.1073/pnas.89.20.9367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zuckermann RN, Kerr JM, Kent SBH, Moos WH. Efficient method for the preparation of peptoids [oligo(N-substituted glycines)] by submonomer solid-phase synthesis. J Am Chem Soc. 1992;114:10646–10647. [Google Scholar]
- 25.Zuckermann RN, Kodadek T. Peptoids as potential therapeutics. Curr Opin Mol Ther. 2009;11:299–307. [PubMed] [Google Scholar]
- 26.Wilson DS, Keefe AD, Szostak JW. The use of mRNA display to select high-affinity protein-binding peptides. doi: 10.1073/pnas.061028198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Czarnik AW. Encoding strategies in combinatorial chemistry. Proc Natl Acad Sci USA. 1997;94:12738–12739. doi: 10.1073/pnas.94.24.12738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ho G-J, Mathre DJ. Lithium-initiated imide formation. A simple method for N-acylation of 2-oxazolidinones and bornane-2,10-sultam. J Org Chem. 1995;60:2271–2273. [Google Scholar]
- 29.Evans DA, Tedrow JS, Shaw JT, Downey CW. Diastereoselective magnesium halide-catalyzed anti-aldol reactions of chiral N-acyloxazolidinones. J Am Chem Soc. 2002;124:392–393. doi: 10.1021/ja0119548. [DOI] [PubMed] [Google Scholar]
- 30.Ravikumar PC, Yao L, Fleming FF. Allylic and allenic halide synthesis via NbCl5- and NbBr5-mediated alkoxide rearrangements. J Org Chem. 2009;74:7294–7299. doi: 10.1021/jo901287f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.See supporting information for details.
- 32.Xiao X, Yu P, Lim H-S, Sikder D, Kodadek T. Design and synthesis of a cell-permeable synthetic transcription factor mimic. J Comb Chem. 2007;9:592–600. doi: 10.1021/cc070023a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Levine AJ, Oren M. The first 30 years of p53: growing ever more complex. Nat Rev Cancer. 2009;9:749–758. doi: 10.1038/nrc2723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Brown CJ, Lain S, Verma CS, Fersht AR, Lane DP. Awakening guardian angels: drugging the p53 pathway. Nat Rev Cancer. 2009;9:862–873. doi: 10.1038/nrc2763. [DOI] [PubMed] [Google Scholar]
- 35.Cochran AG. Antagonists of protein-protein interactions. Chem & Biol. 2000;7:R85–R94. doi: 10.1016/s1074-5521(00)00106-x. [DOI] [PubMed] [Google Scholar]
- 36.Syka JEP, Coon JJ, Schroeder MJ, Shabanowitz J, Hunt DF. Peptide and protein sequence analysis by electron transfer dissociation mass spectrometry. Proc Natl Acad Sci USA. 2004;101:9528–9533. doi: 10.1073/pnas.0402700101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lambert JM, et al. PRIMA-1 reactivates mutant p53 by covalent binding to the core domain. Cancer Cell. 2009;15:376–388. doi: 10.1016/j.ccr.2009.03.003. [DOI] [PubMed] [Google Scholar]
- 38.Boekler FM, Joerger AC, Jaggi G, Rutherford TJ, Veprintsev DB, Fersht AR. Targeted rescue of a destabilized mutant of p53 by an in silico screened drug. Proc Natl Acad Sci USA. 2008;105:10360–10365. doi: 10.1073/pnas.0805326105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Coombs TC, Lushington GH, Douglas J, Aubé J. 1,3-Allylic strain as a strategic diversification element for constructing libraries of substituted 2-arylpiperidines. Angew Chem Int Ed. 2011;50:2734–2737. doi: 10.1002/anie.201007133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Clemons PA, et al. Small molecules of different origins have distinct distributions of structural complexity that correlate with protein-binding profiles. Proc Natl Acad Sci USA. 2010;107:18787–18792. doi: 10.1073/pnas.1012741107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Morton D, Leach S, Cordier C, Warriner S, Nelson A. Synthesis of natural-product-like molecules with over eighty distinct scaffolds. Angew Chem Int Ed. 2009;48:104–109. doi: 10.1002/anie.200804486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Luo T, Schreiber SL. Gold(I)-catalyzed coupling reactions for the synthesis of diverse small molecules using the build/couple/pair strategy. J Am Chem Soc. 2009;131:5667–5674. doi: 10.1021/ja900414s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Uchida T, Rodriguez M, Schreiber SL. Skeletally diverse small molecules using a build/couple/pair strategy. Org Lett. 2009;11:1559–1562. doi: 10.1021/ol900173t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Reddy MM, et al. Identification of candidate IgG biomarkers for Alzheimer’s Disease via combinatorial library screening. Cell. 2011;144:132–142. doi: 10.1016/j.cell.2010.11.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Udugamasooriya DG, Dineen SP, Brekken RA, Kodadek T. A peptoid “antibody surrogate” that antagonizes VEGF receptor 2 activity. J Am Chem Soc. 2008;130:5744–5752. doi: 10.1021/ja711193x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.