Abstract
Background and Aims
Studies of evolutionary diversification in the basal eudicot family Papaveraceae, such as the transition from actinomorphy to zygomorphy, are hampered by the lack of comparative functional studies. So far, gene silencing methods are only available in the actinomorphic species Eschscholzia californica and Papaver somniferum. This study addresses the amenability of Cysticapnos vesicaria, a derived fumitory with zygomorphic flowers, to virus-induced gene silencing (VIGS), and describes vegetative and reproductive traits in this species.
Methods
VIGS-mediated downregulation of the C. vesicaria PHYTOENE DESATURASE gene (CvPDS) and of the FLORICAULA gene CvFLO was carried out using Agrobacterium tumefaciens transfer of Tobacco rattle virus (TRV)-based vectors. Wild-type and vector-treated plants were characterized using reverse transcription–PCR (RT–PCR), in situ hybridization, and macroscopic and scanning electron microscopic imaging.
Key Results
Cysticapnos vesicaria germinates rapidly, can be grown at high density, has a short life cycle and is self-compatible. Inoculation of C. vesicaria with a CvPDS-VIGS vector resulted in strong photobleaching of green parts and reduction of endogenous CvPDS transcript levels. Gene silencing persisted during inflorescence development until fruit set. Inoculation of plants with CvFLO-VIGS affected floral phyllotaxis, symmetry and floral organ identities.
Conclusions
The high penetrance, severity and stability of pTRV-mediated silencing, including the induction of meristem-related phenotypes, make C. vesicaria a very promising new focus species for evolutionary–developmental (evo–devo) studies in the Papaveraceae. This now enables comparative studies of flower symmetry, inflorescence determinacy and other traits that diversified in the Papaveraceae.
Keywords: Agrobacterium tumefaciens; basal eudicots; Cysticapnos vesicaria; FLORICAULA; Papaveraceae; PHYTOENE DESATURASE; Ranunculales; Tobacco rattle virus; VIGS, zygomorphy
INTRODUCTION
Investigations in the Ranunculales (the earliest-branched eudicot order; APG III, 2009) are important for understanding core eudicot floral diversification, as early diverging eudicots precede the extensive canalization of floral bauplans observed in core eudicots (Soltis et al., 2002). The Ranunculales are also considered a ‘playground for floral diversification’, with, for example, more diverse perianth forms than in core eudicots (Litt and Kramer, 2010; Ronse de Craene, 2010). Within the order, Papaveraceae are especially notable for their range of flower and inflorescence morphologies (Hidalgo and Gleissberg, 2010, and references therein). The Papaveraceae (sensu lato) are a monophyletic lineage that consists of two subfamilies, Papaveroideae (the poppies) and Fumarioideae (the fumitories and bleeding hearts), together comprising about 770 species in 40 genera (Judd et al., 2007; APG III, 2009). The phylogenetic position and taxonomic status of a putative third subfamily, the monotypic Pteridophylloideae, are currently questioned (Wang et al., 2009). All poppies are united in having a dimerous perianth, which is actinomorphic (polysymmetric) in the subfamily Papaveroideae, and either disymmetric or zygomorphic (monosymmetric) in the subfamily Fumarioideae. Interestingly, the stepwise shifts in floral symmetry are paralleled by changes in inflorescence architecture. Actinomorphic or disymmetric flowers occur usually singly or clustered in determinate inflorescences, whereas zygomorphic flowers form in indeterminate inflorescences without a terminal flower. Further, unusual character combinations suggest incomplete coupling and/or reversals of the two traits (Hidalgo and Gleissberg, 2010). Other diversity patterns that qualify poppies for evolutionary–developmental (evo–devo) studies include a shift from oligandry to polyandry (Damerval and Nadot, 2007), diverse modes of leaf dissection (Gleissberg and Kadereit, 1999; Gleissberg, 2004) and alkaloid biosynthesis (Ziegler et al., 2006).
The California poppy, Eschscholzia californica, is an emerging model plant for basal eudicots (Carlson et al., 2006; Kramer, 2009; Zahn et al., 2010). Eschscholzia californica and Papaver somniferum are amenable to virus-induced gene silencing (VIGS) (Hileman et al., 2005; Wege et al., 2007), permitting the investigation of gene function in these species (Drea et al., 2007; Orashakova et al., 2009; Yellina et al., 2010; Bartholmes, 2011; Hands et al., 2011). VIGS is now a widely used experimental procedure that allows the transient interruption of gene function through a process similar to RNA interference (for a recent review, see Becker and Lange, 2010). The technique takes advantage of an inherent defence mechanism of plants against viruses (Baulcombe, 1999; Paroo et al., 2007). Engineered viruses carrying one or more target genes are introduced into the plant, and double-stranded RNA produced during virus replication triggers the degradation of any RNA with sequence similarity, including the endogenous transcripts of the target gene(s). The Tobacco rattle virus (TRV)-based binary vector system (Liu et al., 2002) is most commonly employed for VIGS in core eudicots, and it has also provided successful gene downregulation in some basal eudicots and monocots (Becker and Lange, 2010). This system is also the one used thus far in VIGS studies applied to the early-branched eudicot genera Aquilegia, Eschscholzia, Papaver and Thalictrum (Drea et al., 2007; Hileman et al., 2005; Gould and Kramer, 2007; Wege et al., 2007; Orashakova et al., 2009; Di Stilio et al., 2010; Yellina et al., 2010).
VIGS-based evo–devo studies in Papaveraceae would greatly benefit from a zygomorphic-flowered model species, to contrast with the actinomorphic-flowered E. californica and P. somniferum. Gene expression data provided evidence that CYCLOIDEA homologues may be implicated in the establishment of flower symmetry in Papaveraceae (Kölsch and Gleissberg, 2006; Damerval et al., 2007). However, these candidate genes still remain to be validated through functional studies. VIGS has been used to address the genetic control of floral zygomorphy in the core eudicot Pisum sativum (Wang et al., 2008), and the technique is currently being developed in other zygomorphic-flowered species, e.g. in the monocot lineages Orchidaceae (Lu et al., 2007) as well as in Zingiber officinale (Renner et al., 2009), and in Fedia cornucopiae, a species in the core eudicot order Dipsacales (Boyden et al., 2010). Here we report the application of VIGS in Cysticapnos vesicaria, a Papaveraceae with monosymmetric flowers. Cysticapnos belongs to a clade of derived fumarioid poppies that are characterized by monosymmetric flowers and open inflorescences (Hidalgo and Gleissberg, 2010, and references therein). Within this clade, molecular phylogenetic analysis placed Cysticapnos as sister to Discocapnos and Trigonocarpos, in the sub-tribe Discocapninae (Manning et al., 2009). Cysticapnos vesicaria is part of a small genus of three species endemic to South Africa (revised by Manning et al., 2009) of semi-succulent climbing plants with compound, apically tendrillate leaves. Stems end with terminal racemes of monosymmeric flowers presenting a dorsal nectary pouch. Cysticapnos vesicaria is polyploid, like most derived fumarioid poppies (Lidén, 1986); however, the species can be distinguished from its congeneric relatives by a chromosome number of 2n = 4x = 28 (2n = 4x = 32 for C. cracca and C. pruinosa) based on x = 7, whereas almost all Fumarioideae have x = 8 (Lidén, 1986).
Cysticapnos vesicaria was selected because it is easy to grow in a laboratory setting from readily germinating seeds produced in abundance from self-fertile plants. Our assessment of this potential fumarioid poppy model species includes: (1) demonstrating the feasibility of cultivating Cysticapnos under laboratory conditions. Some aspects of vegetative and reproductive morphology are characterized that can be used as a reference for future evo–devo studies of the species. (2) We show the applicability of standard molecular techniques such as reverse transcription–PCR (RT–PCR) and in situ hybridization to Cysticapnos. (3) Finally, we evaluate the amenability of Cysticapnos to functional studies through TRV-based VIGS, using the marker gene PHYTOENE DESATURASE (CvPDS). Further, we use the FLORICAULA/LEAFY gene CvFLO to test whether silencing of a floral regulator that is known to be expressed in meristematic tissues results in morphological defects.
MATERIALS AND METHODS
Plant cultivation
Cysticapnos vesicaria seeds were provided by the Botanical Garden of Göttingen, Germany (index seminum 2007-981) and were grown for several generations to generate a seed pool. Seeds were sown in trays of 48 pots (4 × 6 × 5·5 cm L × W × H) covered with a transparent lid and cold stratified at 4 °C. After 4 d, trays were transferred to a growth chamber at 22 °C in permanent light at 60–100 µmol of light m2 s−1. The first signs of germination were visible 6 d later. When the cotyledons were fully expanded or the first leaf had formed, seedlings were transplanted so as to have only one plant per pot, and the lid was removed. Two weeks after inoculating the plants with Agrobacterium tumefaciens, tap water was supplemented with 0·025 % liquid grow 7-9-5 (Dyna-Gro Co., San Pablo, CA, USA) as fertilizer.
Scanning electron microscopy
Flowers and inflorescences at different developmental stages were dissected under a stereomicroscope and fixed in 70 % ethanol. After dehydration through an ethanol series, samples were critical-point dried with CO2 using a Balzers (CPD030), mounted on aluminium stubs on carbon conductive adhesive tabs and gold-coated with a Balzers sputter coater (SCD050). Micrographs were taken with a Cambridge Instruments Stereoscan 240 scanning electron microscope (SEM) equipped with digital image capture and the Orion32 6·53 software.
Expression pattern of a Histone H4 gene by in situ hybridization
A 215 bp fragment of a Histone H4 gene, CvH4, was amplified with the primers H4F035 and H4R259 (Groot et al. 2005; GenBank accession no. JQ239046), and served as template to synthesize a DIG-11-UTP- (Roche, Indianapolis, IN, USA) labelled antisense RNA probe using T3 polymerase. In situ hybridization was carried out on seedling shoots, following the protocol described in Zachgo (2002). Development of signals with BCIP (5-bromo-4-chloro-3-indolyl phosphate) in 10 % polyvinyl alcohol was stopped after 2 d.
Isolation and sequencing of CvPDS and CvFLO
Total RNA from a tissue blend of C. vesicaria was isolated using the TRI Reagent (Sigma Aldrich, St Louis, MO, USA) according to the manufacturer's instructions. RNA was subsequently reverse transcribed to cDNA using the standard oligo(dT) primer AB05-R (5'-GACTCGAGTCGACATCTG(T)18-3') and the enzyme M-MLV (Promega, Madison, WI, USA). CvPDS (GenBank accession no. JQ239047) was amplified by semi-nested PCR with the forward primers PDS-4F (5'-GATGGAGATTGGTATGAGACTGG-3') and PDS-5F (5'-CGAGTAACTGATGAGGTGTTTATTGC-3') and the reverse primer PDS-7R (5'-AAGAGGGGACTTCTGCTGA-3'). These primers were designed for use throughout Papaveraceae and possibly other Ranunculales. CvFLO (GenBank accession no. JQ239044) was isolated using the forward primers FLO-41F (5'-GCTGAGTTAGGGTTTACTGTKAGCAC-3') and FLO-42F (5'-ACCCTATTGACGCMCTCTCTC-3') and the reverse primers FLO-43R (5'-GCCCWACCAAGGTGACRAAYC-3') and FLO-44R (5'-C GCATTTTCGGCTTGTTTATGTAACTAGC-3'). Sequencing was done in the Ohio University Genomics Facility. The CvFLO sequence was aligned with other angiosperm FLORICAULA/LEAFY homologues, and a Neighbor–Joining tree was constructed using SplitsTree (Huson and Bryant, 2006) based on a 267 bp data set (Supplementary Data Fig. S1).
Vector construction, preparation of the inoculation medium and infection
A 485 bp fragment of CvPDS was amplified using PDS-8F (5'-AATCTAGACGAGTAACTGATGAGGTGTTTATTGC-3') and PDS-9R (5'-CCCGGGAAGAGGGGACTTCTGCTGA-3') and cloned into the XbaI and SmaI sites of pTRV2 (Liu et al., 2002), resulting in the plasmid pTRV2-CvPDS. To test silencing of a meristematic gene, a 462 bp fragment of CvFLO was amplified with the primers FLO-42F and FLO-43R containing XbaI and XhoI restriction enzyme sites, respectively, and ligated at the corresponding sites of pTRV2, resulting in the plasmid pTRV2-CvFLO. The plasmids were sequenced to verify correct insertion of the fragment, and transformed into A. tumefaciens GV3101. Agrobacterium tumefaciens containing the appropriate plasmid (pTRV1, pTRV2-CvPDS, pTRV2-CvFLO, pTRV2-empty) were grown in standard LB medium for 24 h at 28 °C before harvesting them in a ratio of 1:1, pTRV1:pTRV2. Cells were resuspended in 1 mL of 5 % sucrose solution (i.e. half the volume of harvested cells), and the resulting inoculation media were left at room temperature for 30 min before infection.
Inoculation was carried out 25 d after sowing on seedlings at different developmental stages, with foliage leaf number between one and five. Leaves were considered when they reached an approximate petiole to blade length ratio of 1:1 and/or when the lamina was expanded flat. A 2 µL droplet of inoculation medium was applied onto needle scratches on the hypocotyl and leaf petiole base, allowing the Agrobacterium to enter the plant. Plants were left on the lab bench overnight and then returned to the growth chamber.
Expression analysis of CvPDS by RT–PCR
RNA of various tissues (vegetative shoot tips, whole inflorescences, floral buds and fruits) of PDS-VIGS and pTRV2-empty plants was extracted using an RNeasy Plant Mini Kit (Qiagen, Hilden, Germany). cDNA synthesis was conducted as described above with 250 ng of total RNA. In the subsequent PCRs, 5 µL of the 1:50 diluted cDNA was used in a 25 µL reaction. CvPDS expression profiling was carried out with PDS4F and PDS7R primers, as they target a portion of the gene not included in the VIGS construct and permit avoidance of the amplification of virus-derived sequences. Furthermore, those primers were chosen because they span introns and could be used to discriminate endogenous CvPDS transcripts from any genomic DNA amplification. CvPDS was amplified for 40 cycles of 30 s at 94 °C, 30 s at 55 °C and 60 s at 72 °C. The housekeeping gene GLYCERALDEHYDE 3-PHOSPHATE DEHYDROGENASE (GAPDH) was used as reference control and for calculating relative expression intensities (Czechowski et al., 2005). The amplification of a 377 bp region near the 3' end of CvGAPDH (GenBank accession no. JQ239045), carried out with the primers GAPDH-1F (5'-AAGGACTGGAGAGGTGG-3') and GAPDH-2R (5'-CCCCATTCGTTGTCGTACCA-3'), was done as described for CvPDS. Determination of pre-saturation cycles was assessed prior to quantification for the individual tissues and genes. A minimum of three technical replicates were run. ImageJ (Rasband 1997–2011) was used to convert gel images into relative intensities.
RESULTS
Cultivation of Cysticapnos vesicaria plants
Following a brief cold stratification, seeds germinated rather synchronously after 6 d, and could be readily transplanted to individual pots for further cultivation. Plants could be grown at high density in Conviron E8 growth chambers, and started to flower in permanent light conditions only a few weeks after germination. Plants started to die upon completion of fruit set around 3 months after germination. The dead capsules remain closed under our conditions, allowing easy and safe retrieval of seeds. A single plant produces several capsules, and each capsule contained 7–54 seeds (mean 24, n = 28). Seeds retained germination capability after storage at 4 °C for several months.
Morphological characterization of Cysticapnos vesicaria
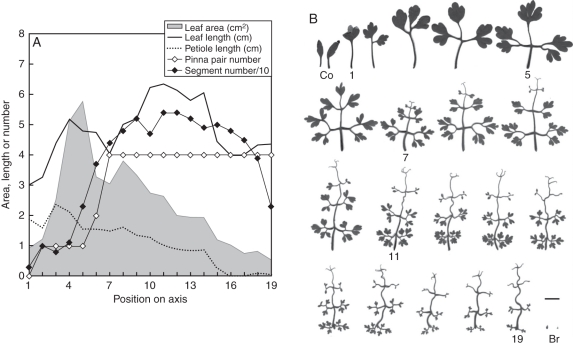
The morphology of wild C. vesicaria (Harvey and Sonder, 1894; Manning et al., 2009) is similar to that of plants cultivated in permanent light in growth chambers. Cysticapnos vesicaria is a small annual herb that uses tendriliform terminal leaflets, rachis and leaflet stalks for climbing (Fig. 1). Heteroblastic leaf series showed a characteristic pattern of increasing complexity also seen in other species (e.g. DeMason and Villani, 2001; Becker et al., 2005). First-order leaflet pair numbers reached a stable maximum at node 7, while total segment number, including second-order leaflets, serrations and tendrils, peaked together with leaf length around node 11, and declined afterwards. Interestingly, the maximum leaf area was already reached within the six early-formed tendril-less leaves which had larger leaflet blades. Petiole length declines steadily along the shoot, and the leaves preceding the first inflorescence are sessile. A sharp transition divides the last foliage leaf and the minute inflorescence bracts.
Fig. 1.
Heteroblastic leaf series of wild-type Cysticapnos vesicaria. (A) Some characteristics of leaves along a primary axis, corresponding to the series of leaf silhouettes illustrated in (B). Units for lengths are in cm and for area in cm2. (B) Some leaves are labelled: Co, cotyledons; 1, primary leaf; 5, leaf 5 with maximum area; 7, first leaf with tendrils; 11, leaf with maximum length and dissection; 19, leaf preceding terminal inflorescence; Br, inflorescence bracts subtending flowers. Scale bar = 1 cm.
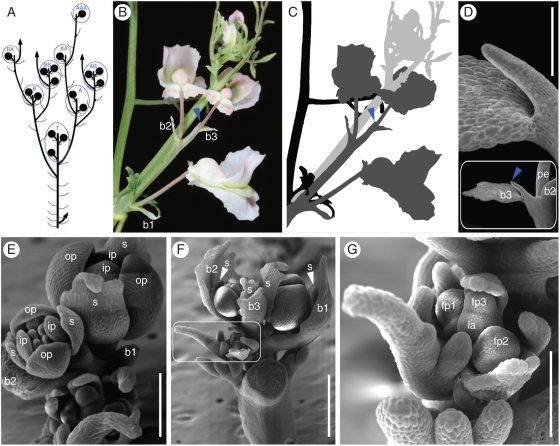
After formation of the primary terminal inflorescence (Fig. 2A), sympodial branching from the axils of the preceding one or two foliage leaves gives rise to higher order inflorescences (Fig. 2A). The inflorescence is sessile, with a short internode below the first bract and flower (Fig. 2B,C). Inflorescence bracts are small, scarious and lanceolate. Flowers cluster in a raceme of 1–3 light-pink, zygomorphic flowers. The small number of flowers identifies our accession as Cysticapnos vesicaria subspecies vesicaria (Manning et al., 2009). A pin-like structure forms the end of the inflorescence axis, and is sometimes preceded by an empty bract (Fig. 2D). Flowers initiate in a rapid sequence (Fig. 2G), and the sequence of maturation (Fig. 2E, F) and effloration (Fig. 2B) is acropetal. The calyx consists of two scale-like sepals that resemble bracts and are, like bracts, persistent (Fig. 3A). The corolla is composed of four petals in two whorls. Sepals are initiated in a medial position, in one plane with the subtending bract (Fig. 2E, F). Upon anthesis, pedicel torsion reorients the flower so that sepals assume a lateral position and the spurred outer petal is positioned upward (Figs 2B and 3A). Outer petals are identical at earlier developmental stages while in the lateral position (Fig. 2E), and epidermal cell shape remains similar even after zygomorphy establishment (data not shown). Inner petals are free at the base and partly connate at their tips, and have a translucent abaxial ridge (Fig. 3A). The androecium consists of six stamens in two bundles, and the bicarpellate gynoecium consists of a many-ovuled ovary, a linear style and a capitate stigma (Fig. 3A). Self-pollination occurs very effectively under our growth conditions, and plants produce many fruits and seeds. The fruit is an ovoid, membranous, vesicular and thin-walled capsule, that appears balloon-like inflated and grows up to about 30 mm in length. The numerous seeds are small lenticular, with a black shiny testa.
Fig. 2.
Morphology and ontogeny of reproductive structures in Cysticapnos vesicaria. (A) Branching architecture and naming system for inflorescence position. Inflorescences are circled in blue. (B, C) Details of the inflorescence. (C) Black, last foliage leaf preceding the terminal inflorescence; dark grey, terminal inflorescence, corresponding to ‘T’ in (A). Three flowers are subtended by bracts b1–b3. The pin-like inflorescence apex is marked with a blue arrowhead; light grey, sympodial shoot and inflorescence arising from the foliage leaf axil, corresponding to ‘A’ in (A). (D–G) SEM micrographs of inflorescence ontogeny. (D) Pin-like ending of the mature inflorescence axis that may be preceded by an empty bract. (E) Two-flowered inflorescence; subtending bracts b1 and b2 removed. Perianth organs are labelled. (F) Terminal inflorescence with bracts b1 and b2 subtending flowers. Bract three is empty; inflorescence apex not visible. Sympodial inflorescence in the axil of the last foliage leaf is framed and shown enlarged in (G). (G) Early-stage inflorescence with three flower primordia. The inflorescence apex is still meristematic. Abbreviations: b1–b3, consecutive bracts; fp1–fp3, consecutive flower primordia; ia, inflorescence apex; ip, inner petal; op, outer petal; pe, pedicel; s, sepal. Scale bars: (D, F) = 500 µm, (E) = 250 µm, (G) = 200 µm.
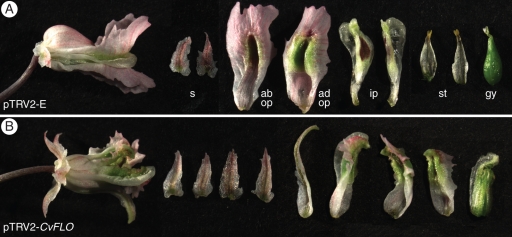
Fig. 3.
CvFLO-VIGS phenotypes (A) Control plants inoculated with pTRV2-E showed, from left to right, wild-type flowers that were zygomorphic and had two sepals, an abaxial and a spurred adaxial outer petal, two inner petals, two stamen bundles (adaxial faces shown) and a gynoecium. (B) A pTRV2-CvFLO flower with altered floral phyllotaxis and symmetry. Dissection of this flower revealed four sepals, one sepaloid organ, three organs with mixed outer and inner petal identities and a composite organ with carpel, stamen and inner petal characteristics. Abbreviations: ad op, adaxial outer petal; ab op, abaxial outer petal; gy, gynoecium; ip, inner petals; s, sepals; st, stamen bundles.
In situ hybridization of a Histone H4 gene
We tested whether Cysticapnos is amenable to gene expression characterization via in situ hybridization, an important component of evo–devo studies, including Ranunculales (e.g. Di Stilio et al., 2005, 2009; Shan et al., 2006; Liu et al., 2010; Ballerini and Kramer, 2011) and Papaveraceae (e.g. Busch and Gleissberg, 2003; Groot et al., 2005; Carlson et al., 2006; Damerval et al., 2007; Drea et al., 2007; Orashakova et al., 2009; Yellina et al., 2010; Hands et al., 2011). Vegetative shoot apices hybridized with an antisense CvH4 RNA probe strongly labelled CvH4 transcripts in individual cells in the shoot apical meristem and developing leaves (Supplementary Data Fig. S2), as expected (Groot et al., 2005). Neighbouring cells remained unstained, demonstrating a good signal-to-background ratio.
Fast, strong and long-lasting CvPDS silencing phenotype in infected Cysticapnos vesicaria plants
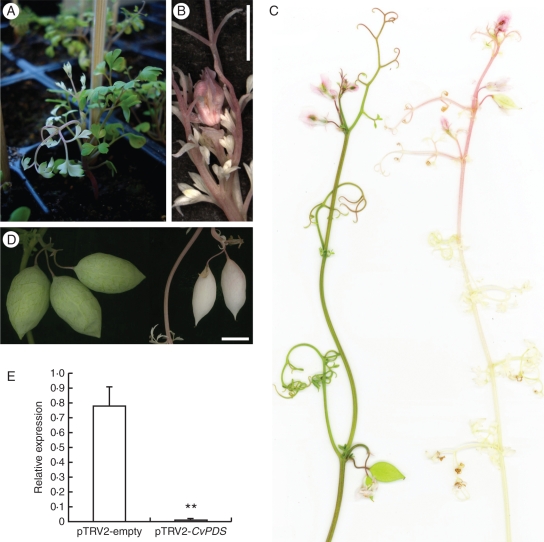
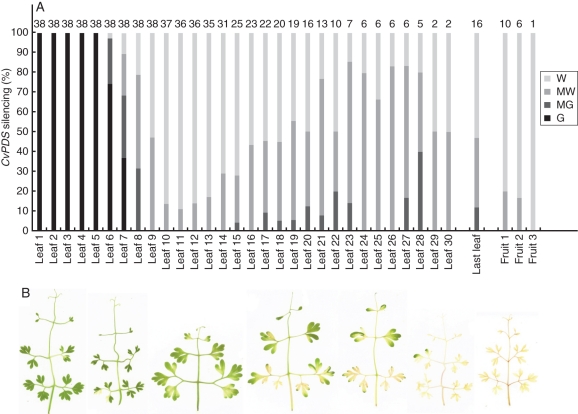
PDS-VIGS-mediated photobleaching was detected in the young plants 9 d after infection in the second or third leaf formed after inoculation (87 % of 68 plants in three experiments), more rarely in the first or fourth (Figs 4A and 5A). In one experiment in which detailed phenotypic scoring was undertaken, the proportion of individuals showing bleached parts was 100 % (n = 38; Fig. 5A), demonstrating a maximal penetrance capacity of the VIGS system in C. vesicaria. The transition from completely green to completely white leaves generally involved two intermediary leaves that were mostly green and mostly white, respectively (Fig. 5B). Not all individuals showed complete photobleaching, with some green leaflet and petiole parts occasionally formed. The PDS-VIGS phenotype remained remarkably strong and stable throughout further shoot development and included the stem, leaves and inflorescences (Figs 4B, C and 5A). Photobleached stem parts often appeared pinkish, probably due to exposed anthocyanin. Developing capsules were also photobleached, suggesting that silencing persisted throughout flower development (Fig. 4D). The photobleaching phenotype corresponded to effective downregulation of CvPDS transcripts, as shown by RT–PCR profiling (Fig. 4E). No difference between wild-type and negative control plants (pTRV2-empty) was noticeable for vegetative or reproductive parts, suggesting that TRV itself has minimal or no effects on development.
Fig. 4.
CvPDS silencing symptoms in vegetative and reproductive parts of Cysticapnos vesicaria. (A) Young plant showing beginning of photobleaching. (B) Young inflorescence of a CvPDS-VIGS plant with complete photobleaching. (C) Mature reproductive shoots of a control (pTRV2-empty; left) and a CvPDS-VIGS plant (right). (D) Capsules of a control (left) and a CvPDS-VIGS plant (right). (E) RT–PCR showing the strong reduction of CvPDS transcripts in C. vesicaria plants infected with pTRV2-CvPDS (n = 6), compared with a pTRV2-empty control (n = 2). Expression levels are shown relative to GAPDH. Asterisks indicate a significant difference compared with the control group at 99 % confidence intervals after ANOVA. Scale bars = 1 cm.
Fig. 5.
Degree of CvPDS silencing. (A) Intensity of leaf and fruit bleaching symptoms in the primary axis. ‘Last leaf’ = last leaf before inflorescence. Abbreviations: W, white; MW, mostly white; MG, mostly green; G, green. Sample sizes are indicated above each column. Note that CvPDS silencing has a negative effect on plant robustness and survival, leading to an over-representation of partially silenced plants. (B) Different intensities of CvPDS silencing are reflected in a gradient from almost completely green (left) to completely white (right).
Phenotypic effects of CvFLO silencing
To test whether VIGS of a developmental gene results in morphological alterations, we conducted a separate experiment in which we inoculated seedlings with a pTRV2-CvFLO construct aimed to silence the CvFLO gene. Of the plants inoculated with pTRV2-CvFLO (n = 48) 12 abnormal flowers were analysed that suggested successful silencing during meristematic stages of flower development (Fig. 3). No abnormal flowers were found in control plants inoculated with pTRV2-E (n = 40) grown side by side with pTRV2-CvFLO plants. The morphological defects indicated that CvFLO function was affected during initiation and differentiation of all floral organs, consistent with the strong silencing effects seen in pTRV2-CvPDS inflorescences (Fig. 3B). pTRV2-CvFLO flowers exhibited perturbed floral phyllotaxis and symmetry, altered organ numbers and mosaic organ identities, reflecting partial defects in flower meristem identity also known from FLORICAULA/LEAFY mutants in rosid core eudicots. Mutants in Arabidopsis and Lotus develop mosaic flower/shoots with sepals and carpels (Schultz and Haughn, 1991; Huala and Sussex, 1992; Dong et al., 2005), while equivalent mutations in the asterid core eudicots Antirrhinum and Solanum show a full conversions of flowers into shoots (Coen et al., 1990; Allen and Sussex, 1996). A more detailed characterization of the CvFLO-VIGS phenotype is currently underway that will allow a better understanding of the role of this developmental regulator in this zygomorphic-flowered basal eudicot.
DISCUSSION
Research in basal eudicots is crucial to help decipher major evolutionary transitions and associated histories of developmental genes between distant angiosperm lineages, such as basal angiosperms, monocots and core eudicots (Kramer, 2009; Zahn et al., 2010). In this sense the Papaveraceae are of special interest as this family occupies a basal position within Ranunculales (the sister clade to all other eudicots) together with the earliest-diverging monogeneric Eupteleaceae (Worberg, 2007; Wang et al., 2009). The Ranunculaceae, on the other hand, are considered a derived core Ranunculales family (Wang et al., 2009). Furthermore, Papaveraceae flowers exhibit a separate morphological canalization within Ranunculales, with whorled flowers, a clear differentiation of calyx and corolla, and well-defined expression domains of floral organ identity genes, potentially providing a model for core eudicot flower evolution (Chanderbali et al., 2009; Voelckel et al., 2010; Zahn et al., 2010). Finally, unique diversification patterns regarding floral symmetry and inflorescence (see Introduction) well qualify Papaveraceae for the study of correlated traits in a defined phylogenetic framework.
We have identified C. vesicaria as a new model system in the zygomorphic-flowered subfamily Fumarioideae that will enable comparative evolutionary developmental studies of reproductive traits in the poppy family to be conducted. The combination of relative phylogenetic proximity and contrasting morphology makes C. vesicaria and E. californica a very promising pair of species for comparative evo–devo studies. Recent VIGS-based studies in papaveroid poppies (Orashakova et al., 2009; Yellina et al., 2010; Bartholmes, 2011; Hands et al., 2011; S. Wreath et al., Ohio University, unpubl. res.) provide interesting starting points for comparative studies using C. vesicaria.
Extending developmental genetic studies from isolated model systems to morphologically divergent relatives minimizes phylogenetic noise in comparative studies (Baum et al., 2002; Kramer, 2009). For example, the Arabidopsis thaliana and Cardamine hirsuta models have been useful in the study of leaf dissection (Hay and Tsiantis, 2006; Canales et al., 2010). Within Ranunculales, VIGS technology has been recently developed for related species of the genera Aquilegia (Gould and Kramer, 2007) and Thalictum (Di Stilio, 2010). Our study provides proof of the value of a VIGS-based approach to comparative functional studies in basal eudicots.
The application of VIGS in Cysticapnos requires no vacuum infiltration, and results in maximal post-treatment survival and maximal phenotypic penetrance. In these respects, VIGS in Cysticapnos is more effective than in other Ranunculales systems such as P. somniferum (Hileman et al., 2005) or Aquilegia vulgaris (Gould and Kramer, 2007). The ability of TRV to trigger silencing in whole shoots and inflorescences, and the persistence of silencing until the fruit set will allow the study of a broad range of reproductive and vegetative developmental traits, with the only exclusion being pre-treatment stages of germination and early seedling establishment. In addition, the apparent absence of seed dormancy, easy cultivation under laboratory conditions at high density, a short life cycle and self-fertilization facilitate experimentation with Cysticapnos. The fact that Cysticapnos is tetraploid could potentially make knock-down studies more difficult in the presence of paralogues. In this study, we have identified only one copy of PDS and FLO, and silencing constructs based on these showed strong phenotypic consequences. This could be due to the absence of duplicated gene copies, or to co-silencing of paralogues. VIGS, in comparison with mutant-based studies, can be particularly suitable for studying polyploids (Scofield and Nelson, 2009) as it permits the simultaneous silencing of a set of paralogues, either directly when they share high sequence similarity, or by using a specific construct design that incorporates multiple genes. The copy number of target genes can be better assessed when genomic information become available for this and other fumarioid poppies.
Conclusions
We have demonstrated that Cysticapnos is amenable to laboratory culture and poses no challenge for gene isolation and expression studies. The emerging availability of transcriptomics data for Eschscholzia as well as other Papaveraceae and Ranunculales species (Chanderbali et al., 2009; Voelckel et al., 2010; Zahn et al., 2010) will greatly facilitate the identification of genes and their orthology in Cysticapnos. More importantly, Cysticapnos is the first species with zygomorphic flower symmetry in the Papaveraceae and in basal eudicots for which gene function studies are now available. We developed this system primarily to enable comparative functional studies of inflorescence determinacy and flower symmetry in basal eudicots. However, Cysticapnos is also a promising system with which to investigate other traits, including its climbing habit and compound, tendril-bearing leaves. Tendrilled compound leaves have thus far only been studied in P. sativum (garden pea) and to some extent in Lathyrus odoratus (sweet pea) (Gould et al., 1994; Hofer et al., 2009), and it is unknown whether any underlying genetic networks are shared between these rosid core eudicots and Cysticapnos. The addition of a first fumarioid poppy to the VIGS toolbox in poppies makes it likely that other Fumarioideae are amenable to this TRV-based silencing method as well. As comparative insights into the role of genetic networks in morphological evolution of poppies become available, future studies may extend to more basal fumitory species with disymmetric flowers.
SUPPLEMENTARY DATA
ACKNOWLEDGEMENTS
The authors thank members of the Gleissberg lab (Ohio University) for technical help: Anandi Bhattacharya, Thomas Ingram, Eden Kinkaid, Alastair Plant, Celeste Taylor and Mariah Thrush, and especially Avery Tucker for his work on Fig. 1. The Botanical Garden of Göttingen is greatly acknowledged for providing the seeds of Cysticapnos vesicaria. This work was funded by Ohio University. O.H. was financially supported by MICINN and Juan de la Cierva postdoctoral contracts from the Ministerio de Ciencia e Innovación, Spain, and C.B. by a Donald Clippinger Graduate Fellowship.
LITERATURE CITED
- Allen KD, Sussex IM. Falsiflora and anantha control early stages of floral meristem development in tomato (Lycopersicon esculentum Mill.) Planta. 1996;200:254–264. [Google Scholar]
- APG III. An update of the Angiosperms Phylogeny Group classification for the orders and families of flowering plants: APG III. Botanical Journal of the Linnean Society. 2009;161:105–121. [Google Scholar]
- Ballerini ES, Kramer EM. Environmental and molecular analysis of the floral transition in the lower eudicot Aquilegia formosa. EvoDevo. 2011;2(4) doi: 10.1186/2041-9139-2-4. http://dx.doi.org/10.1186/2041-9139-2-4 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartholmes C. Regulation of morphogenesis of lateral organs in the basal eudicot. 2011 Eschscholzia californica. PhD dissertation, Ohio University, USA. [Google Scholar]
- Baulcombe DC. Fast forward genetics based on virus-induced gene silencing. Current Opinion in Plant Biology. 1999;2:109–113. doi: 10.1016/S1369-5266(99)80022-3. [DOI] [PubMed] [Google Scholar]
- Baum DA, Doebley J, Irish VF, Kramer EM. Response: Missing links: the genetic architecture of flower and floral diversification. Trends in Plant Science. 2002;7:31–34. doi: 10.1016/s1360-1385(01)02098-2. [DOI] [PubMed] [Google Scholar]
- Becker A, Lange M. VIGS – genomics goes functional. Trends in Plant Science. 2010;15:1–4. doi: 10.1016/j.tplants.2009.09.002. [DOI] [PubMed] [Google Scholar]
- Becker A, Gleissberg S, Smyth DR. Floral and vegetative morphogenesis in California poppy (Eschscholzia californica Cham.) International Journal of Plant Sciences. 2005;166:537–555. [Google Scholar]
- Boyden GS, Hardej D, Howarth DG. Virus-induced gene silencing of CYCLOIDEA-like genes in Fedia cornucopiae. 2010 Botany 2010 meeting abstract. http://2010.botanyconference.org/engine/search/index.php?func=detail&aid=397 . [Google Scholar]
- Busch A, Gleissberg S. EcFLO, a FLORICAULA-like gene from Eschscholzia californica is expressed during organogenesis at the vegetative shoot apex. Planta. 2003;217:841–848. doi: 10.1007/s00425-003-1046-z. [DOI] [PubMed] [Google Scholar]
- Canales C, Barkoulas M, Galinha C, Tsiantis M. Weeds of changes: Cardamine hirsuta as a new model for studying dissected leaf development. Journal of Plant Research. 2010;123:24–33. doi: 10.1007/s10265-009-0263-3. [DOI] [PubMed] [Google Scholar]
- Carlson JE, Leeens-Mack JH, Wall PK, et al. EST database for early flower development in Calfornia poppy (Eschscholzia californica Cham., Papaveraceae) tags over 6,000 genes from a basal eudicot. Plant Molecular Biology. 2006;62:351–369. doi: 10.1007/s11103-006-9025-y. [DOI] [PubMed] [Google Scholar]
- Chanderbali AS, Albert VA, Leebens-Mack J, Altman NS, Soltis DE, Soltis PS. Transcriptional signatures of ancient floral developmental genetics in avocado (Persea americana; Lauraceae) Proceedings of the National Academy of Sciences, USA. 2009;106:8929–8934. doi: 10.1073/pnas.0811476106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coen ES, Romero JM, Doyle S, Elliott R, Murphy G, Carpenter R. floricaula: a homeotic gene required for flower developmment in Antirrhinum majus. Cell. 1990;63:1311–1322. doi: 10.1016/0092-8674(90)90426-f. [DOI] [PubMed] [Google Scholar]
- Czechowski T, Stitt M, Altmann T, Udvardi MK, Scheible WR. Genome-wide identification and testing of superior references genes for transcript normalization in Arabidopsis. Plant Physiology. 2005;139:5–17. doi: 10.1104/pp.105.063743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damerval C, Nadot S. Evolution of perianth and stamen characteristics with respect to floral symmetry in Ranunculales. Annals of Botany. 2007;100:631–640. doi: 10.1093/aob/mcm041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damerval C, Le Guilloux M, Jager M, Charon C. Diversity and evolution of CYCLOIDEA-like TCP genes in relation to flower development in Papaveraceae. Plant Physiology. 2007;143:759–772. doi: 10.1104/pp.106.090324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeMason DA, Villani PJ. Genetic control of leaf development in pea (Pisum sativum) International Journal of Plant Sciences. 2001;162:493–511. [Google Scholar]
- Di Stilio VS, Kramer EM, Baum DA. Floral MADS box genes and homeotic gender dimorphism in Thalictrum dioicum (Ranunculales) – a new model for the study of dioecy. The Plant Journal. 2005;41:755–766. doi: 10.1111/j.1365-313X.2005.02336.x. [DOI] [PubMed] [Google Scholar]
- Di Stilio VS, Martin C, Schulfer AF, Connelly CF. An ortholog of MIXTA-like2 controls epidermal cell shape in flowers of Thalictrum. New Phytologist. 2009;183:718–728. doi: 10.1111/j.1469-8137.2009.02945.x. [DOI] [PubMed] [Google Scholar]
- Di Stilio VS, Kumar RA, Oddone AM, Tolkin TR, Salles P, McCarty K. Virus-induced gene silencing as a tool for comparative studes in Thalictrum. PLoS ONE. 2010;5:e12064. doi: 10.1371/journal.pone.0012064. http://dx.doi.org/10.1371/journal.pone.0012064 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong Z-C, Zhao Z, Liu C-W, et al. Floral patterning in Lotus japonicus. Plant Physiology. 2005;137:1272–1282. doi: 10.1104/pp.104.054288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drea S, Hileman LC, de Martino G, Irish VF. Functional analyses of genetic pathways controlling petal specification in poppy. Development. 2007;134:4157–4166. doi: 10.1242/dev.013136. [DOI] [PubMed] [Google Scholar]
- Gleissberg S. Comparative analysis of leaf shape development in Eschscholzia californica and other Papaveraceae–Eschscholzioideae. American Journal of Botany. 2004;91:306–312. doi: 10.3732/ajb.91.3.306. [DOI] [PubMed] [Google Scholar]
- Gleissberg S, Kadereit JW. Evolution of leaf morphogenesis: evidence from developmental and phylogenetic data in Papaveraceae. International Journal of Plant Sciences. 1999;160:787–794. [Google Scholar]
- Gould B, Kramer EM. Virus-induced gene silencing as a tool for functional analyses in the emerging model plant Aquilegia (columbine, Ranunculaceae) Plant Methods. 2007;3(6) doi: 10.1186/1746-4811-3-6. http://dx.doi.org/doi:10.1186/1746-4811-3-6 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gould KS, Cutter EG, Young JPW. The determination of pea leaves, leaflets, and tendrils. American Journal of Botany. 1994;81:352–360. [Google Scholar]
- Groot EP, Sinha N, Gleissberg S. Expression patterns of STM-like KNOX and Histone H4 genes in shoot development of the dissected-leaved basal eudicot plants Chelidonium majus and Eschscholzia californica (Papaveraceae) Plant Molecular Biology. 2005;58:317–331. doi: 10.1007/s11103-005-4548-1. [DOI] [PubMed] [Google Scholar]
- Hands P, Vosnakis N, Betts D, Irish VF, Drea S. Alternate transcripts of a floral developmental regulator have both distinct and redundant functions in opium poppy. Annals of Botany. 2011;107:1557–1566. doi: 10.1093/aob/mcr045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harvey WH, Sonder OW. Ranunculaceae to Connaraceae. Ashford, Kent, UK: L. Reeve & Co; 1894. Flora capensis: being a systematic description of the plants of the Cape Colony, Caffraria, & Port Natal. Vol. I. [Google Scholar]
- Hay A, Tsiantis M. The genetic basis for differences in leaf form between Arabidopsis thaliana and its wild relative Cardamine hirsuta. Nature Genetics. 2006;38:942–947. doi: 10.1038/ng1835. [DOI] [PubMed] [Google Scholar]
- Hidalgo O, Gleissberg S. Evolution of the reproductive architecture in the bleeding hearts and poppies (Papaveraceae s.l.) International Journal of Plant Developmental Biology. 2010;4(S1):76–85. [Google Scholar]
- Hileman DC, Drea S, De Martino G, Litt A, Irish VF. Virus-induced gene silencing is an effective tool for assaying gene function in the basal eudicot species Papaver somniferum (opium poppy) The Plant Journal. 2005;44:334–341. doi: 10.1111/j.1365-313X.2005.02520.x. [DOI] [PubMed] [Google Scholar]
- Hofer J, Turner L, Moreau C, et al. Tendril-less regulates tendril formation in pea leaves. The Plant Cell. 2009;21:420–428. doi: 10.1105/tpc.108.064071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huala E, Sussex M. LEAFY interacts with floral homeotic genes to regulate Arabidopsis floral development. The Plant Cell. 1992;4:910–913. doi: 10.1105/tpc.4.8.901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huson DH, Bryant D. Application of phylogenetic networks in evolutionary studies. Molecular Biology and Evolution. 2006;23:254–267. doi: 10.1093/molbev/msj030. [DOI] [PubMed] [Google Scholar]
- Judd WS, Campbell CS, Kellogg EA, Stevens PS. Plant systematics: a phylogenetic approach. 3rd edn. Sunderland, MA: Sinauer Associates; 2007. [Google Scholar]
- Kölsch A, Gleissberg S. Diversification of CYCLOIDEA-like TCP genes in the basal eudicot families Fumariaceae and Papaveraceae s.str. Plant Biology. 2006;8:680–687. doi: 10.1055/s-2006-924286. [DOI] [PubMed] [Google Scholar]
- Kramer EM. New model systems for the study of developmental evolution in plants. Current Topics in Developmental Biology. 2009;86:67–105. doi: 10.1016/S0070-2153(09)01004-7. [DOI] [PubMed] [Google Scholar]
- Lidén M. Synopsis of Fumarioideae (Papaveraceae) with a monograph of the tribe Fumarieae. Opera Botanica. 1986;88:1–133. [Google Scholar]
- Litt A, Kramer EM. The ABC model and the diversification of floral organ identity. Seminars in Cell Development and Biology. 2010;21:129–137. doi: 10.1016/j.semcdb.2009.11.019. [DOI] [PubMed] [Google Scholar]
- Liu C, Zhang J, Zhang N, et al. Interactions among proteins of floral MADS-box genes in basal-eudicots: implications for evolution of the regulatory network for flower development. Molecular Biology and Evolution. 2010;27:1598–1611. doi: 10.1093/molbev/msq044. [DOI] [PubMed] [Google Scholar]
- Liu Y, Schiff M, Dinesh-Kumar SP. Virus-induced gene silencing in tomato. The Plant Journal. 2002;31:777–786. doi: 10.1046/j.1365-313x.2002.01394.x. [DOI] [PubMed] [Google Scholar]
- Lu H-C, Chen H-H, Tsai W-C, et al. Strategies for functional validation of genes involved in reproductive stages of orchids. Plant Physiology. 2007;143:558–569. doi: 10.1104/pp.106.092742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manning JC, Goldblatt P, Forest F. A revision of Fumariaceae (Fumarioideae) in southern Africa, including naturalized taxa. Bothalia. 2009;39:47–65. [Google Scholar]
- Orashakova S, Lange M, Lange S, Wege S, Becker A. The CRABS CLAW ortholog from California poppy (Eschscholzia californica, Papaveraceae), EcCRC, is involved in floral meristem termination, gynoecium differentiation and ovule initiation. The Plant Journal. 2009;58:682–693. doi: 10.1111/j.1365-313X.2009.03807.x. [DOI] [PubMed] [Google Scholar]
- Paroo Z, Liu Q, Wang X. Biochemical mechanisms of the RNA-induced silencing complex. Cell Research. 2007;17:187–194. doi: 10.1038/sj.cr.7310148. [DOI] [PubMed] [Google Scholar]
- Rasband WS. ImageJ. Bethesda, Maryland, USA: US National Institutes of Health; 1997–2011 http://imagej.nih.gov/ij/ [Google Scholar]
- Renner T, Bragg J, Driscoll HE, Cho J, Jackson AO, Specht CD. Virus-induced gene silencing in the culinary ginger (Zingiber officinale): an effective mechanism for down-regulating gene expression in tropical monocots. Molecular Plant. 2009;2:1084–1094. doi: 10.1093/mp/ssp033. [DOI] [PubMed] [Google Scholar]
- Ronse de Craene LP. Floral diagrams. An aid to understand flower morphology and evolution. Cambridge: Cambridge University Press; 2010. [Google Scholar]
- Schultz EA, Haughn GW. LEAFY, a homeotic gene that regulates inflorescence development in Arabidopsis. The Plant Cell. 1991;3:771–781. doi: 10.1105/tpc.3.8.771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scofield SR, Nelson RS. Resources for virus-induced gene silencing in the grasses. Plant Physiology. 2009;149:152–157. doi: 10.1104/pp.108.128702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shan H, Su K, Lu W, Kong H, Chen Z, Meng Z. Conservation and divergence of candidate class B genes in Akebia trifoliata (Lardizabalaceae) Development Genes and Evolution. 2006;216:785–795. doi: 10.1007/s00427-006-0107-2. [DOI] [PubMed] [Google Scholar]
- Soltis DE, Soltis PS, Albert VA, et al. Floral Genome Project Research Group. Missing links: the genetic architecture of flower and floral diversification. Trends in Plant Science. 2002;7:22–31. doi: 10.1016/s1360-1385(01)02098-2. [DOI] [PubMed] [Google Scholar]
- Voelckel C, Borevitz JO, Kramer EM, Hodges SA. Within and between whorls: comparative transcriptional profiling of Aquilegia and Arabidopsis. PLoS One. 2010;5:e9735. doi: 10.1371/journal.pone.0009735. http://dx.doi.org/10.1371/journal.pone.0009735 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W, Lu A-M, Ren Y, Endress ME, Chen Z-D. Phylogeny and classification of Ranunculales: evidence from four molecular loci and morphological data. Perspectives in Plant Ecology, Evolution and Systematics. 2009;11:81–110. [Google Scholar]
- Wang Z, Luo Y, Li X, et al. Genetic control of floral zygomorphy in pea (Pisum sativum L.) Proceedings of the National Academy of Sciences, USA. 2008;105:10414–10419. doi: 10.1073/pnas.0803291105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wege S, Scholz A, Gleissberg S, Becker A. Highly efficient virus-induced gene silencing (VIGS) in California poppy (Eschscholzia californica Cham.): an evaluation of VIGS as a strategy to obtain functional data from non-model plants. Annals of Botany. 2007;100:641–649. doi: 10.1093/aob/mcm118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Worberg A, Quandt D, Barniske A-M, Löhne C, Hilu KW, Borsch T. Phylogeny of basal eudicots: insights from non-coding and rapidly evolving DNA. Organisms, Diversity and Evolution. 2007;7:55–77. [Google Scholar]
- Yellina AL, Orashakova S, Lange S, Erdmann R, Leebens-Mack J, Becker A. Floral homeotic C function genes repress specific B function genes in the carpel whorl of the basal eudicot California poppy (Eschscholzia californica) EvoDevo. 2010;1(13) doi: 10.1186/2041-9139-1-13. http://dx.doi.org/10.1186/2041-9139-1-13 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zachgo S. In situ hybridization. In: Gilmartin PM, Bowler C, editors. Molecular plant biology. Vol. 2. Oxford: Oxford University Press; 2002. pp. 41–63. [Google Scholar]
- Zahn ML, Ma X, Altman NS, et al. Comparative transcriptomics among floral organs of the basal eudicot Eschscholzia californica as reference for floral evolutionary developmental studies. Genome Biology. 2010;11 doi: 10.1186/gb-2010-11-10-r101. R101. http://dx.doi.org/10.1186/gb-2010-11-10-r101 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziegler J, Voigtlander S, Schmidt J, et al. Comparative transcript and alkaloid profiling in Papaver species identifies a short chain dehydrogenase/reductase involved in morphine biosynthesis. The Plant Journal. 2006;48:177–192. doi: 10.1111/j.1365-313X.2006.02860.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.