Abstract
Small heat shock proteins (sHSPs) play an important role in the cellular defense of prokaryotic and eukaryotic organisms against a variety of internal and external stressors. In this study, a cDNA clone encoding a member of the α-crystallin/sHSP family, termed AccHsp27.6, was isolated from Apis cerana cerana. The full-length cDNA is 1,014 bp in length and contains a 708-bp open reading frame encoding a protein of 236 amino acids with a calculated molecular weight of 27.6 kDa and an isoelectric point of 7.53. Seven putative heat shock elements and three NF-κB binding sites were present in the 5′-flanking region, suggesting a possible function in immunity. A semi-quantitative RT-PCR analysis indicated that AccHsp27.6 was expressed in all tested tissues and at different developmental stages. Furthermore, expression of the AccHsp27.6 transcript was induced by exposure to heat shock, H2O2, a number of different chemicals (including SO2, formaldehyde, alcohol, acetone, chloroform, and the pesticides phoxime and acetamiprid), and the microbes Staphylococcus aureus and Micrococcus luteus. In contrast, the mRNA expression could be repressed by CO2, the pesticides pyriproxyfen and cyhalothrin, and the microbes Bacillus subtilis and Pseudomonas aeruginosa. Notably, the recombinant AccHsp27.6 protein exhibited significant in vitro molecular chaperone activity and antimicrobial activity. Taken together, these results suggest that AccHsp27.6 might play an important role in the response to abiotic and biotic stresses and in immune reactions.
Electronic supplementary material
The online version of this article (doi:10.1007/s12192-012-0330-x) contains supplementary material, which is available to authorized users.
Keywords: Apis cerana cerana, Expression, Gene cloning, Immunological function, Small heat shock proteins
Introduction
Members of the heat shock protein (HSP) superfamily are ubiquitously present in most organisms. The HSPs play important roles as protein chaperones (Hendrick and Hartl 1993) and are also involved in a number of biological processes, including embryogenesis, diapause, and morphogenesis. Based on their molecular weights, HSPs have been classified into four categories, including HSP90, HSP70, HSP60, and a family of small HSPs (sHSPs) (Mizrahi et al. 2010). The sHSPs, also called small stress proteins, range in molecular weight from approximately 15 to 43 kDa. Compared to other types of HSPs, the sHSPs exhibit the greatest variation in sequence, size, and function (de Jong et al. 1998). However, there is a conserved domain of 80–100 residues, called the α-crystallin domain, that is bordered by variable amino- and carboxy-terminal extensions (Sun and MacRae 2005). In the higher-order structure, the α-crystallin domain forms a conserved β-sandwich composed of two antiparallel β-sheets (Pérez-Morales et al. 2009). This conserved structure facilitates the assembly of the sHSPs into oligomeric complexes of up to 800 kDa, which is crucial for their primary function as molecular chaperones that bind partially denatured proteins to prevent irreversible protein aggregation during stresses, such as extreme temperatures, oxidation, UV irradiation, heavy metals, and chemical intoxication (Reineke 2005). The chaperone function of the sHSPs is not only involved in stress conditions but also in normal development. Indeed, these proteins are also involved in a number of biological processes, including cell growth, differentiation, apoptosis (Arrigo 1998), membrane fluidity (Tsvetkova et al. 2002), diapause (Gkouvitsas et al. 2008), and lifespan (Morrow et al. 2004a).
There have been numerous studies concerning sHSPs in bacteria, algae, plants, amphibians, birds, and mammals. The four primary sHSPs of Drosophila melanogaster, Hsp22, Hsp23, Hsp26, and Hsp27, have been studied in detail. They share significant sequence similarity and are coordinately expressed following stress, but they have distinct developmental expression patterns and intracellular localizations (Michaud et al. 2002). Recently, a few members of the α-crystallin/sHSP family have been cloned from other insect species, including Plutella xylostella (Sonoda et al. 2006), Mamestra brassicae (Sonoda et al. 2007), Ceratitis capitata (Kokolakis et al. 2008), Sesamia nonagrioides (Gkouvitsas et al. 2008), Liriomyza sativa (Huang et al. 2009), and Macrocentrus cingulum (Xu et al. 2010).
The Chinese honeybee, Apis cerana cerana, is an important indigenous species that plays a critical role in agricultural economic development as the pollinator of flowering plants (Yang 2005). To date, no sHSP has been identified in the Chinese honeybee. In this study, a cDNA clone, AccHsp27.6, encoding a putative sHSP gene was isolated and characterized. Our results indicate that the expression of AccHsp27.6 responds to a number of chemical and biological inducers. Moreover, a recombinant AccHsp27.6 protein exhibited significant in vitro molecular chaperone activity and antimicrobial activity. We speculate that AccHsp27.6 might play an important role in regulating biotic and abiotic stress responses and immune reactions.
Experimental procedures
Animals and treatments
The worker bees of A. cerana cerana obtained from the Technology Park of Shandong Agricultural University were reared on an artificial diet at 34°C and 80% humidity. The entire bodies of second (L2), fourth (L4), fifth (L5), and sixth (L6) instar larvae with white (Pw), pink (Pp), and dark (Pd)-eyed pupae were obtained from the hive, and the adult workers (1 and 10 day after emergence) were collected at the entrance of the hive upon their return to the colony after foraging (Bitondi et al. 2006). The adult bees (12 day) were divided into groups (n = 40) and exposed to abiotic and biotic stresses. The abiotic stresses consisted of heat shock (37°C), UV-induced oxidative stress (30 mj/cm2), H2O2 treatment (30% H2O2 was delivered in 0.5 μL of distilled water to the thoracic notum of the worker bees at a final concentration of 2 mM), and chemical stress through treatment with four different pesticides (imidacloprid, cyhalothrin, pyriproxyfen, and phoxime; the pesticides were delivered in 0.5 μL of distilled water to the thoracic notum of the worker bees at final concentrations of 10, 12.5, 20, and 20 mg/L, respectively), CO2, SO2, formaldehyde, alcohol, acetone, and chloroform (a gas generator in the incubator was used to generate the CO2 and SO2, and the organic reagents were volatilized and added to the incubator containing the honeybees). The biotic stress was microbial infection. The 3-day-old larvae were maintained in a 24-well plate and inoculated with microbes (Staphylococcus aureus, Micrococcus luteus, Bacillus subtilis, and Pseudomonas aeruginosa). At 6 days after inoculation, the bees were flash-frozen in liquid nitrogen at the indicated time points and stored at −80°C.
RNA extraction, cDNA synthesis, and DNA isolation
Total RNA was extracted using Trizol reagent (Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s instructions. To remove genomic DNA, the RNA was treated with RNase-free DNase-I. Subsequently, the cDNA was synthesized using a reverse transcription system (TransGen Biotech, Beijing, China). The genomic DNA was isolated using an EasyPure Genomic DNA Extraction Kit in accordance with the manufacturer’s instructions (TransGen Biotech).
PCR primers and conditions
The primers used in this study are listed in Table 1. The amplification conditions are shown in Table 2.
Table 1.
The primers used in this study
| Abbreviation | Primer sequence (5′–3′) | Description |
|---|---|---|
| HP1 | TTCGGAAAGTGGGGAAACATTC | cDNA sequence primer of AccHsp27.6, forward |
| HP2 | AAGGGCAGTAAAGGTGTATAAGTCA | cDNA sequence primer of AccHsp27.6, reverse |
| 5P1 | CATAAACGCGGCAAAAGACATCA | 5′ RACE reverse primer of AccHsp27.6, outer |
| 5P2 | GGATCAGCATTTTGGCACAGC | 5′ RACE reverse primer of AccHsp27.6, inner |
| 3P1 | GATCAAATGAGAGAACAGTGAAGG | 3′ RACE forward primer of AccHsp27.6, outer |
| 3P2 | AAGGGCAGTAAAGGTGTATAAGTCA | 3′ RACE forward primer of AccHsp27.6, inner |
| AAP | GGCCACGCGTCGACTAGTAC(G)14 | Abridged Anchor Primer |
| AUAP | GGCCACGCGTCGACTAGTAC | Abridged universal amplification primer |
| B26 | GACTCTAGACGACATCGA(T)18 | 3′ RACE universal adaptor primer |
| B25 | GACTCTAGACGACATCGA | 3′ RACE universal primer |
| QP1 | TTCGGAAAGTGGGGAAACATTC | Full-length cDNA primer, forward |
| QP2 | GACATGCAATAAATATTGTATTAATTATTTC | Full-length cDNA primer, reverse |
| GP1 | TTCGGAAAGTGGGGAAACATTC | Genomic sequence primer of AccHsp27.6, forward |
| GP2 | AAGGGCAGTAAAGGTGTATAAGTCA | Genomic sequence primer of AccHsp27.6, reverse |
| PS1 | CAGCAAAATCAGCAATTAC | Inverse PCR forward primer, outer |
| PX1 | CGTCATAATCATCATCC | Inverse PCR reverse primer, outer |
| PS2 | CAACAACATCAGCGTTGT | Inverse PCR forward primer, inner |
| PX2 | CGATCTAGATTCACTTGG | Inverse PCR reverse primer, inner |
| QS1 | GCATTTGCATGTAAATAGATG | Promoter special primer, forward |
| QS2 | GTGGGGAAACATTCCTAAC | Promoter special primer, reverse |
| RP1 | GATGTTTTCCGACTGGTGG | RT-PCR primer of AccHsp27.6, forward |
| RP2 | GAACATGGTTGGGTATCCAG | RT-PCR primer of AccHsp27.6, reverse |
| β-actin-s | GTTTTCCCATCTATCGTCGG | Standard control primer, forward |
| β-actin-x | TTTTCTCCATATCATCCCAG | Standard control primer, reverse |
| EP1 | GGTACCTTCGGAAAGTGGGGAAACATTC | Protein expression primer, forward |
| EP2 | AAGCTTTGACTTATACACCTTTACTGCCCTT | Protein expression primer, reverse |
Table 2.
PCR amplification conditions in the study
| Primes pair | Amplification conditions |
|---|---|
| HP1/HP2 | 6 min at 94°C, 40 s at 94°C, 40 s at 52°C, 1 min at 72°C for 35 cycles, 10 min at 72°C |
| 5P1∕B26 | 6 min at 94°C, 40 s at 94°C, 40 s at 53°C, 30 s at 72°C for 28 cycles, 5 min at 72°C |
| 5P2∕B25 | 6 min at 94°C, 40 s at 94°C, 40 s at 58°C, 30 s at 72°C for 35 cycles, 5 min at 72°C |
| 3P1∕AAP | 6 min at 94°C, 40 s at 94°C, 40 s at 49°C, 40 s at 72°C for 28 cycles, 5 min at 72°C |
| 3P2∕AUAP | 6 min at 94°C, 40 s at 94°C, 40 s at 55°C, 40 s at 72°C for 35 cycles, 5 min at 72°C |
| QP1/QP2 | 6 min at 94°C, 40 s at 94°C, 40 s at 52°C, 1 min at 72°C for 35 cycles, 10 min at 72°C |
| GP1∕GP2 | 6 min at 94°C, 40 s at 94°C, 40 s at 52°C, 1.5 min at 72°C for 35 cycles, 10 min at 72°C |
| PS1∕PX1 | 6 min at 94°C, 50 s at 94°C, 50 s at 50°C, 2 min at 72°C for 30 cycles, 5 min at 72°C |
| PS2∕PX2 | 6 min at 94°C, 50 s at 94°C, 50 s at 50°C, 2 min at 72°C for 30 cycles, 5 min at 72°C |
| QS1/QS2 | 6 min at 94°C, 40 s at 94°C, 40 s at 52°C, 2 min at 72°C for 35 cycles, 10 min at 72°C |
| RP1∕RP2 | 6 min at 94°C, 40 s at 94°C, 40 s at 52°C, 40 s at 72°C for 28 cycles, 5 min at 72°C |
| β-actin-s, x | 6 min at 94°C, 30 s at 94°C, 30 s at 53°C, 30 s at 72°C for 28 cycles, 5 min at 72°C |
| EP1/EP2 | 6 min at 94°C, 40 s at 94°C, 40 s at 53°C, 1.5 min at 72°C for 28 cycles, 10 min at 72°C |
Isolation of the AccHsp27.6 gene
The internal conserved fragment was cloned by RT-PCR using the primers HP1 and HP2, which were designed based on the conserved DNA sequence among the homologous genes from Apis mellifera, Nasonia vitripennis, and Macrocentrus cingulum. Using the sequence of the internal fragment, two sets of primers, 5P2 and 5P1 and 3P2 and 3P1, were designed and synthesized for 5′ and 3′ RACE (rapid amplification of cDNA ends), respectively. Following RACE-PCR, the 5′- and 3′-end sequences and the internal fragment were assembled, and the full-length cDNA sequence was derived. Subsequently, the specific primers QP1 and QP2 were used to amplify the full-length cDNA sequence using end-to-end PCR. The genomic DNA was cloned using the primers GP1 and GP2 and genomic DNA as a template. All PCRs were performed as previously described (Wang et al. 2008).
Cloning of the 5′-flanking region of AccHsp27.6
The sequence of the 5′-flanking region was cloned using inverse PCR and the restriction endonuclease SspI. Genomic DNA digested with SspI at 37°C overnight and ligated using T4 DNA ligase (TaKaRa, Dalian, China) was used as the template. Based on the genomic sequence of AccHsp27.6, two pairs of primers (PS1 + PX1 and PS2 + PX2) were designed to obtain the 5′-flanking region, and two specific sequencing primers (QS1 and QS2) were designed to confirm the amplified product. The PCR conditions are shown in Table 2. The PCR products were cloned into the pMD18-T vector and sequenced. The TFBind software (http://tfbind.hgc.jp/) was used to search for transcription factor binding sites in the 5′-flanking region (Tsunoda and Takagi 1999).
Bioinformatic and phylogenetic analyses
The NCBI bioinformatics tools (available at http://blast.ncbi.nlm.nih.gov/Blast.cgi) were used to detect conserved domains in AccHsp27.6. In addition, the DNAman software was used for the prediction of secondary structure and to search for open reading frames (ORFs). An online protein 3D structure prediction tool (http://swissmodel.expasy.org/) was also used in the structural analysis. Multiple sequence alignments were performed using the Clustal X program with its default parameters (Kokolakis et al. 2008). Phylogenetic and molecular evolutionary analyses were performed using the neighbor-joining method with the Molecular Evolutionary Genetics Analysis (MEGA version 4.1) software.
Expression analysis using semi-quantitative RT-PCR
A pair of specific primers was designed to amplify a fragment of AccHsp27.6. The housekeeping gene β-actin (GenBank accession number: XM640276) was used to estimate equal amounts of RNA among the samples. RT-PCR of β-actin was performed using the primers RP1 and RP2 and the same conditions used to amplify the AccHsp27.6 fragment. The PCRs were repeated three times, and the electrophoresis results were normalized against the β-actin results using the Quantity-One™ image analysis software implemented in VersaDoc 4000 (Bio-Rad, Hercules, CA, USA).
Construction of expression plasmids, recombinant protein expression, and purification
To express the recombinant AccHsp27.6 protein in BL21, a pair of specific primers (EP1 and EP2) was designed to amplify the 708-bp fragment encoding the entire ORF. The ORF sequence was cloned into pEasy-T3 and digested with the restriction endonucleases Kpn I and Hind III. After extraction and purification using the Gel Extraction Kit (Solarbio, Beijing, China), the ORF sequence was ligated into the expression vector pET-30a (+), which was digested with the same restriction endonucleases. The expression plasmid pET-30a (+)-AccHsp27.6 was then transformed into the BL21 (DE3) Escherichia coli strain. After intermediate culture and isopropyl-β-d-thiogalactopyranoside (IPTG) induction at a final concentration of 0.6 mM, the bacterial cells were harvested by centrifugation, resuspended, sonicated, and centrifuged again. The final pellet was resuspended in PBS and subjected to 15% SDS-PAGE. The AccHsp27.6 in the medium was purified using the 6XHis-Tagged Protein Purification Kit (CW Biotechnology, Beijing, China) according to the manufacturer’s instructions, and the purified protein was examined by 15% SDS-PAGE. In addition, AccCPR26-6XHis fusion protein (A. cerana cerana cuticle protein R&R 26) was purified by the same method.
Molecular chaperone activity and antimicrobial activity of the recombinant AccHsp27.6 protein
The capacity of AccHsp27.6 to suppress the thermal aggregation of the MDH (mitochondrial malate dehydrogenase) from pig heart (EC 1.1.1.37; Amresco) was examined. Three samples (A, MDH, B, MDH + AccHsp27.6, and C, MDH + BSA) were incubated at 43°C. The BSA (bovine serum albumin) was used as control to eliminate the effect of nonspecific chaperone activity. Absorbance was monitored at 360 nm at regular intervals (10 min) for 1 h (Pérez-Morales et al. 2009).
Sterile paper disks were saturated with a solution of recombinant AccHsp27.6 protein and dried at 30°C. Separate tubes of LB broth were inoculated with overnight cultures of four bacterial strains, M. luteus, S. aureus, B. subtilis, and P. aeruginosa, to give an initial bacterial density of 106 cells/mL and overlaid onto agarose medium plates. The AccHsp27.6-saturated paper disks were placed on the plates and incubated at 37°C for 24 h.
Results
AccHsp27.6 cloning and sequence analysis
Based on the conserved DNA sequence of the sHSPs from A. mellifera, N. vitripennis, and M. cingulum, a fragment of approximately 785 bp was amplified using the primers HP1 and HP2 (Fig. 1). Specific primers were designed according to the obtained internal sequence and subsequently used for the amplification of 3′ (primers 3P1 and 3P2) and 5′-end (primers 5P1 and 5P2) cDNA, resulting in 244- and 161-bp fragments corresponding to the 3′- and 5′-end sequences, respectively. The full-length AccHsp27.6 cDNA sequence was deduced and amplified by RT-PCR using the full-length cDNA primers QP1 and QP2 and confirmed by sequencing. The full-length AccHsp27.6 cDNA (GenBank accession number GQ254650) was 1,014 bp, with a 76-bp 5′ untranslated region (UTR), a 223-bp 3′ UTR and a 708-bp ORF encoding a protein of 236 amino acids with a calculated molecular weight of 27.6 kDa and an isoelectric point of 7.53.
Fig. 1.
Nucleotide sequence of AccHsp27.6 and primers. The cDNA sequence is indicated on the top line, and the names of the primers and their corresponding orientations are shown on the second line
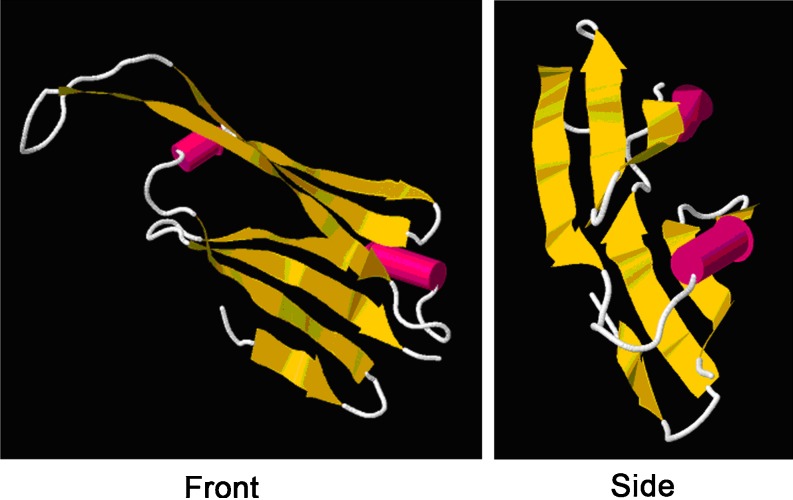
The NCBI Conserved Domain (CD) search revealed that the α-crystallin domain (ACD) of metazoan α-crystallin-type sHSPs was present in the deduced amino acid sequence of AccHsp27.6, suggesting that the AccHsp27.6 gene might belong to the sHSP family. The analysis of the secondary structure indicated that the helix content was 10.55%, which is a characteristic of the structure of sHSPs (Augusteyn 2004). β-sheets occurred frequently throughout the structure of the protein, which is consistent with previous reports that the secondary structure of the sHSPs is rich in β-sheets (de Jong et al. 1998). In addition, there were only two cysteines in the entire protein sequence, which is consistent with previous data showing that cysteine residues were rarer in the sequences of molecular chaperones than in other protein families (Fu et al. 2003). Moreover, the tertiary structure was also analyzed (Fig. 2). The human alphaB crystallin (PDB code is 2WJ7) was the best template for homology modeling of the deduced protein. This model consisted of a single polypeptide chain of approximately 94 amino acids from residue 67 to residue 157. In the 3D structure, the conserved 188-residue α-crystallin domain was enriched in β-strands that were organized into a β-sheet sandwich. These sheets are the basic structural unit of sHSPs (Sun and MacRae 2005) and help facilitate dimer formation and the subsequent oligomerization of sHSPs.
Fig. 2.
The tertiary structure of α-crystallin domain residues 67–157
The predicted amino acid sequence of AccHsp27.6 exhibited 43.88% and 44.07% similarity to the sHSP from M. cingulum (GenBank accession number EU624206) and Hsp21.7 from N. vitripennis (GenBank accession number XP001604512), respectively. The protein sequence comparison showed that AccHsp27.6 contains two prominent conserved regions (Fig. 3). In addition to the one including the α-crystallin domain, the region of the initial 27 amino acids in the amino-terminal section is of relatively high homology and this domain is highly hydrophobic and might be involved in modulating oligomerization, subunit dynamics, and substrate binding. sHSP oligomerization and chaperoning are influenced by the amino-terminus in several, but not all sHSPs (Sun and MacRae 2005). Usually, the amino-terminal extension is variable in length and sequence. However, according to Kokolakis et al. (2008), the 14-amino acid domain of Hsp27, at the very amino-terminal region of the three proteins, is very hydrophobic and shows 57% amino acid sequence identity among Drosophila, medfly, and Sarcophaga crassipalpis. And analogous hydrophobic domain is also conserved in Drosophila Hsp23 and Hsp26 but not in Drosophila Hsp22 (Southgate et al. 1983). The relatively higher homology of AccHsp27.6 in the amino-terminus suggests the conservation of sHSP in the structure and function.
Fig. 3.
Amino acid sequence comparison of sHSP homologs from Apis cerana cerana, Macrocentrus cingulum, and Nasonia vitripennis. The asterisks, double dots, and single dots denote fully, strongly, and weakly conserved residues, respectively. The conserved regions are shown in gray. The α-crystallin domain is underlined. The degree of identity relative to AccHsp27.6 is shown at the end of each sequence. The secondary structure assignment of the AccHsp27.6 is shown above the sequence
To investigate the regulation of AccHsp27.6 expression, we cloned its 5′-flanking region and obtained a 1,376-bp DNA fragment upstream of the translation start site using inverse PCR (GenBank accession number: JF317273). The TFBIND software was used to search the transcription factor binding sites. Seven heat shock elements (HSEs) and three NF-κB binding sites were detected. In addition, a TATA-BOX, which represents the putative core promoter element upstream of the transcription start site, was found (Fig. 4).
Fig. 4.
The nucleotide sequence and putative transcription factor binding sites of the 5′-flanking region of AccHsp27.6. The transcription and translation start sites are indicated with an arrowhead. The underlined sections indicate the putative NF-κB binding sites, and the boxes indicate the putative HSEs. The TATA-box is highlighted in light gray
Phylogenetic analysis
To study the evolutionary relationships among different sHSPs from various insects, a phylogenetic tree was constructed using the MEGA (version 4.1) software, and all of the amino acid sequences used were obtained from GenBank. As shown in Fig. 5, the sHSPs from insects of the same order were phylogenetically closer than homologous sHSPs from other insect orders. However, an orthologous cluster containing several sHSPs from different insect orders was found, suggesting that these sHSPs evolved before the divergence of the species (Kokolakis et al. 2008).
Fig. 5.
Phylogenetic analysis of sHSP amino acid sequences from different insect species (Diptera, Hymenoptera, and Lepidoptera). The full names of species and the accession numbers of the genes are designated by the following abbreviations: DmHsp26 (Drosophila melanogaster, NM079273), DmHsp27 (Drosophila melanogaster, NM079276), DmCG14207 (Drosophila melanogaster, NM134482), LhHsp20 (Liriomyza huidobrensis, DQ452370), LsHsp21.7 (Liriomyza sativae, DQ452372), McsHsp (Macrocentrus cingulum, EU624206), NvHsp21.7 (Nasonia vitripennis, XP001604512), NvHsp20.6 (Nasonia vitripennis, XM001607625), AccHsp27.6 (Apis cerana cerana, GQ254650), BmHsp20.1 (Bombyx mori, AB195971), BmHsp23.7 (Bombyx mori, AB195973), BmHsp20.4 (Bombyx mori, AF315318), BmHsp22.6 (Bombyx mori, FJ602788), BmHsp21.4 (Bombyx mori, AB195972), AgHsp20.9 (Anopheles gambiae, XM560153), TcHsp21.8 (Tribolium castaneum, XM968592), AmHsp25.6 (Apis mellifera, XM392405), LmHsp20.6 (Locusta miratoria, DQ355964)
Developmental and tissue-specific expression patterns of AccHSP27.6
To determine the developmental expression patterns of the AccHSP27.6 gene, semi-quantitative RT-PCR was employed to detect mRNA accumulations at different developmental stages and in various tissues. As shown in Fig. 6a, the expression level of AccHSP27.6 showed a gradual decrease from the second to the fifth instar larvae stages followed by a remarkable increase at the sixth stage. A similar expression pattern was observed at the pupal stage, with a rapid decline from the white to the pink-eyed pupae followed by a significant increase in the dark-eyed pupae. In newly emerged adults, the expression of AccHsp27.6 was low, and it peaked in 10-day-old adults. Analysis of the spatial expression profiles of AccHsp27.6 indicated that its transcripts were abundant in the thorax and abdomen and scarce in the head (Fig. 6b).
Fig. 6.
Expression patterns of AccHsp27.6 at different developmental stages and in various tissues. a Expression patterns of AccHsp27.6 at different developmental stages. Total RNA was isolated from larvae (2d second instars, 4d fourth instars, 5d fifth instars, 6d sixth instars), pupae (pw white-eyed pupae, pp pink-eyed pupae, pd dark-eyed pupae), and adults (a1 1 day post-emergence adults, a10 10 day post-emergence adults). The transcripts of AccHsp27.6 were detected throughout the entire development process. b Expression patterns of AccHsp27.6 in various tissues. Total RNA was extracted from the brain, thorax, and abdomen of the adult bees (12 day) at the indicated times, and semi-quantitative RT-PCR was used to examined the transcript level of AccHsp27.6 under different treatments. The β-actin gene was used as an internal control. The histograms indicate the relative expression levels. Values on the y-axis are the normalization units relative to β-actin levels. In addition, the values on the x-axis represent the lanes. The signal intensities (INT/mm2) of the bands were assigned using the Quantity-One™ image analysis software implemented in VersaDoc 4000 (Bio-Rad). Each value is given as the mean (SD) of three replicates
Expression profiles of AccHSP27.6 in response to abiotic stress
The sHSPs are products of heat shock and other stress responses. Previous studies have reported that sHSPs provide a protective function under conditions of high temperature, oxidation, UV irradiation, and chemical intoxication. (Reineke 2005). This idea prompts us to examine the responses of AccHSP27.6 to different environmental stressors. Twelve-day-old bees were subjected to heat shock, oxidative stress (UV and H2O2), and exposure to a number of chemicals (pesticides, CO2, SO2, formaldehyde, alcohol, acetone, and chloroform). As shown in Fig. 7, when treated with heat stress, AccHSP27.6 transcripts quickly accumulated to a maximum level at 2 h and then declined at 4 h. UV treatment induced only a mild increase in AccHSP27.6 expression at 4 h. Under H2O2 stress conditions, an increase in the accumulation of AccHSP27.6 transcripts was detected 1 h after treatment, and the transcript levels reached a maximum at 2 h. Interestingly, the AccHSP27.6 gene exhibited different responses to the application of various pesticides. Treatments with pyriproxyfen and cyhalothrin downregulated AccHSP27.6 expression, whereas the application of phoxime and acetamiprid greatly upregulated gene expression, with maximum expression levels at 3 and 1 h, respectively. The application of other chemicals, including SO2, formaldehyde, alcohol, acetone, and chloroform, induced the expression of AccHSP27.6 to various degrees. In contrast, the CO2 challenge suppressed its transcription to nearly zero.
Fig. 7.
Expression profiles of AccHsp27.6 under abiotic stress. Total RNA was extracted from adult bees (12 day) treated at the indicated times with heat (a), UV irradiation (b), H2O2 (c), and pesticides (d Pyriproxyfen, e Cyhalothrin, f Phoxime, g Acetamiprid) and hazardous chemicals (h). Semi-quantitative RT-PCR was used to examine the transcript levels of AccHsp27.6 under different treatments. FA formaldehyde, alc alcohol, acet acetone, chl chloroform. The β-actin gene was used as an internal control. The line CK indicates the control. The histograms show the relative expression levels. The values on the y-axis are the normalization units relative to β-actin levels. In addition, the values on the x-axis represent the lanes. The signal intensities (INT/mm2) of the bands were assigned using the Quantity-One™ image analysis software implemented in VersaDoc 4000 (Bio-Rad). Each value is given as the mean (SD) of three replicates
AccHSP27.6 mRNA accumulation under biotic stress
Bacterial diseases of honeybee larvae seriously affect the growth of the honeybee, the quality of the bee products, and the beekeeping industry. S. aureus, P. aeruginosa, and M. luteus are opportunistic pathogens that are widely distributed in the natural environment, and B. subtilis could stimulate the development of animal immune organs. To investigate whether Hsp27.6 is involved in the immune system, these four bacterial strains were injected into the body surface of 3-day-old larvae. Three days later, RT-PCR analysis revealed that AccHsp27.6 expression was induced by S. aureus and M. luteus and suppressed by B. subtilis and P. aeruginosa (Fig. 8). These results suggest an important role for AccHsp27.6 in honeybee microbe resistance.
Fig. 8.
Expression profiles of AccHsp27.6 in response to microbe infection. SA Staphylococcus aureus, ML Micrococcus luteus, BS Bacillus subtilis, PA Pseudomonas aeruginosa. The 3-day-old larvae fed in the 24-well plates were inoculated with microbes and maintained until they were 6 days old. Total RNA was extracted at the indicated times, and semi-quantitative RT-PCR was used to examine the transcript levels of AccHsp27.6. The β-actin gene was used as an internal control. The line CK indicates the control. The histograms show the relative expression levels. The values on the y-axis are the normalization units relative to β-actin levels. In addition, the values on the x-axis represent the lanes. The signal intensities (INT/mm2) of the bands were assigned using the Quantity-One™ image analysis software implemented in VersaDoc 4000 (Bio-Rad). Each value is given as the mean (SD) of three replicates
Expression and antimicrobial activity of a recombinant AccHsp27.6 protein
The AccHsp27.6 gene was expressed in BL21 E. coli strain as a 6XHis-tagged fusion protein. After induction with IPTG at 37°C, SDS-PAGE analysis detected a band corresponding to the HSP27.6-6XHis fusion protein (Fig. 9, lane 3). No protein induction was observed in the non-induced control (Fig. 9, lane 2). The data indicated that the AccHsp27.6 gene was expressed, and a purified Hsp27.6-6XHis fusion protein was obtained (Fig. 9, lane 1).
Fig. 9.
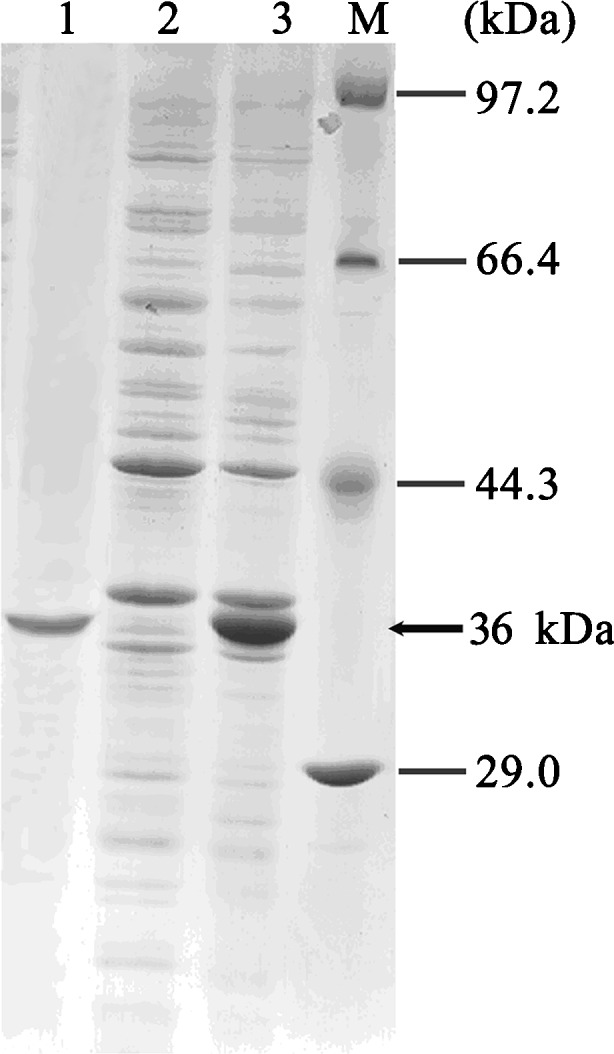
Expression in E. coli and purification of the AccHsp27.6 recombinant protein as analyzed by SDS-PAGE. Lane 1 purified AccHsp27.6 recombinant protein; lane 2 non-induced; lane 3 induced; lane M molecular mass standard
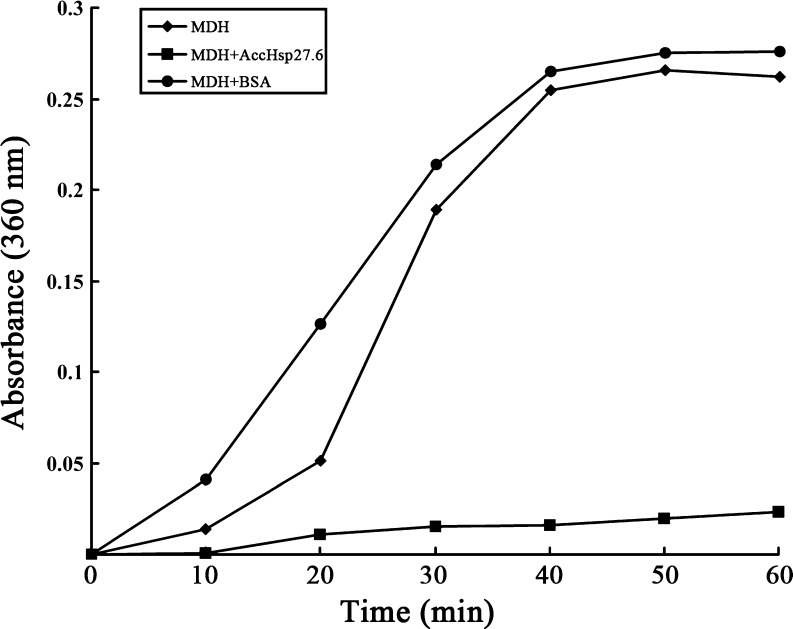
The molecular chaperone activity in vitro of the recombinant AccHsp27.6 was investigated by the malate dehydrogenase thermal aggregation assay. As shown in Fig. 10, MDH aggregated when incubation at 43°C, which was revealed by the significant increase of the absorbance at 360 nm (Pérez-Morales et al. 2009). In addition, MDH aggregated faster when mixed with BSA that has no chaperone activity. However, the addition of an equivalent of AccHsp27.6 to MDH almost totally suppressed MDH aggregation (Fig. 10). Therefore, the recombinant protein could have significant molecular chaperone activity in vitro.
Fig. 10.
Molecular chaperone activity of AccHsp27.6 shown by the malate dehydrogenase thermal aggregation assay. MDH was heated at 43°C (filled diamond), mixed with AccHsp27.6 in a molar ratio 1:1 (filled square), or mixed with BSA (1:1; filled circle). Absorbance was monitored at 360 nm at regular intervals (10 min) for 1 h
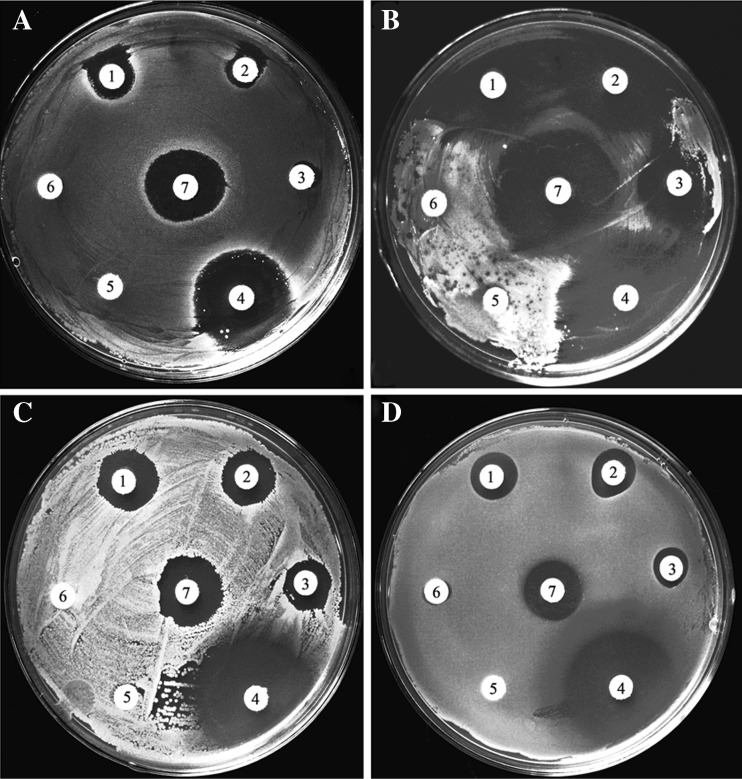
We investigated the antimicrobial activity of the bioactive recombinant AccHsp27.6 protein against several bacteria using the disk diffusion method. As shown in Fig. 11, the protein exhibited significant antimicrobial activity against S. aureus, M. luteus, B. subtilis, and P. aeruginosa. In addition, AccCPR26 purified by the same method with AccHsp27.6 was used as the control to exclude the effect of the 6XHIS tag and the toxic that accumulate during the purification steps. And as shown in Fig. 12 in enclosure, AccCPR26 had no clearly antimicrobial activity.
Fig. 11.
Antimicrobial activity of AccHsp27.6 shown by the disk diffusion method. aStaphylococcus aureus. bMicrococcus luteus. cBacillus subtilis. dPseudomonas aeruginosa. 1 twofold dilution of the AccHsp27.6 protein solution; 2 fivefold dilution of the AccHsp27.6 protein solution; 3 tenfold dilution of the AccHsp27.6 protein solution; 4 positive control (kanamycin, 250 mg/ml); 5 negative control (the total protein from BL21 (DE3) E. coli, 0.5 mg/ml); 6 PBS; 7 the original concentrated solution of purified AccHsp27.6 protein (0.5 mg/ml)
Discussion
Although the molecular and biochemical characterization of sHSP genes in insects has been extensively studied, most studies are performed in model insect species, such as D. melanogaster. However, the expression of sHSPs in the Chinese honeybee (A. cerana cerana) has not been previously reported. This is the first study to investigate the existence of sHSPs in the Chinese honeybee and their possible involvement in response to abiotic and biotic stresses. The results of experiments involving the cloning, characterization, and expression of a Chinese honeybee sHSP gene suggests the possibility of an sHSP-mediated resistance against adverse environments in the honeybee.
The HSEs are DNA sequences recognized by heat shock factors (HSFs), transcription factors that are responsible for the expression of HSPs following stress. HSEs are the most critical elements in HSP gene transcription. We detected seven putative HSEs in the 5′-flanking region of AccHsp27.6. Previous studies on the Drosophila Hsp22 promoter revealed three functional HSEs that are required for the proper expression of Hsp22 following stress (Morrow et al. 2004b). The HSE responsible for the heat-induction of the AccHsp27.6 gene has not yet been determined.
The sHSPs not only function as molecular chaperones to protect proteins from denaturation under high-temperature stress but also develop protective functions under other stress conditions, such as cold, drought, oxidation, hypertonic stress, heavy metals, and even high population densities (Li et al. 2009). In the present study, the expression of AccHsp27.6 in response to abiotic stress was analyzed by RT-PCR. The results showed that AccHsp27.6 could be markedly induced by heat shock and H2O2 treatments as well as the application of pesticides (phoxime and acetamiprid) and several other chemicals (including SO2, formaldehyde, alcohol, acetone, and chloroform). However, gene expression was suppressed by CO2 and the application of pyriproxyfen and cyhalothrin.
Interestingly, the expression of the AccHSP27.6 gene exhibited different responses to the application of various microbes. S. aureus and M. luteus upregulated AccHSP27.6 expression, whereas B. subtilis and P. aeruginosa downregulated the gene expression. These data suggest that AccHsp27.6 is related to the immune system.
Insects lack an acquired immune system, and their defense mechanisms mainly rely on innate immune responses. Extensive work over the past 10 years suggests that the HSP family might be potential activators of the innate immune system (Wu et al. 2011); however, information concerning immunity and the potential role of the sHSPs is relatively limited and has emerged only recently. An sHSP gene from shrimp, Hsp21, was downregulated during a white spot syndrome viral infection, which might be related to host cell apoptosis (Huang et al. 2008). The genome-wide analysis of host gene expression in silkworm cells infected with Bombyx mori nucleopolyhedrovirus revealed that the expression of Hsp20.1 was decreased to approximately 50% at 2 h post-infection, which suggested that silkworm Hsp20.1 is involved in anti-BmNPV immunity at the early phase of infection (Sagisaka et al. 2010). More recently, an sHSP gene from the bloody clam (Tegillarca granosa) was reported to be involved in the immune response against Vibrio parahaemolyticus and lipopolysaccharides (Bao et al. 2011). A microarray analysis of the gene expression profile in the midgut of silkworms infected with cytoplasmic polyhedrosis virus revealed that a number of HSPs (including sHsp23.7) were upregulated, suggesting that these HSPs might be involved in anti-BmCPV immunity (Wu et al. 2011). However, previous studies have focused primarily on expression analysis at the transcriptional level. In this study, several lines of evidence indicate that AccHsp27.6 might be involved in immune reactions. First, three nuclear factor-κB (NF-κB) binding sites were observed in the 5′-flanking region of AccHsp27.6. NF-κB is a ubiquitous transcription factor that plays critical roles in host defense and chronic inflammatory diseases by regulating the expression of multiple inflammatory and immune genes (Barnes 1997). Second, the transcription of AccHsp27.6 was greatly up- or downregulated upon microbial infection. Direct evidence came from the analysis of the recombinant AccHsp27.6 protein, which showed that the protein exhibited in vitro antimicrobial activity. Therefore, it is reasonable to speculate that AccHsp27.6 could be related to the immune response of the honeybee.
In this study, we only examined microbial infections in the larvae because bacterial diseases of the honeybee larvae seriously affect the growth of the honeybee, the quality of bee products and the beekeeping industry. Moreover, larvae may have the least developed immune systems, thus requiring HSP protection and eliciting a strong response to microbial challenge. Therefore, it would have been useful to examine the effects of a honey bee-specific microbe, such as Paenebacilis larvae, which causes American Foulbrood. Further investigation is of great importance to uncover a more comprehensive picture of the functional roles and mechanisms of AccHsp27.6 in the immune response.
Electronic supplementary material
The additional experiment of antimicrobial activity of AccHsp27.6. aStaphylococcus aureus; bMicrococcus luteus; cBacillus subtilis; dPseudomonas aeruginosa. K, kanamycin (250 mg/ml); A, ampicillin (250 mg/ml); 1, 2 purified AccHsp27.6 protein (0.5 mg/ml); 3 purified AccCPR26 protein (0.5 mg/ml) (JPEG 49 kb)
Acknowledgments
This work was financially supported through earmarked funds from China’s Agricultural Research System (no. CARS-45), special funds for Agro-scientific Research in the Public Interest (no. 200903006), and the Promotive Research Fund for Excellent Young and Middle-aged Scientists of Shandong Province (BS2011SW001).
Footnotes
Zhaohua Liu and Dongmei Xi contributed equally to this paper.
References
- Arrigo AP. Small stress proteins: chaperons that act as regulators of intracellular redox state and programmed cell death. Biol Chem. 1998;379:19–26. [PubMed] [Google Scholar]
- Augusteyn RC. α-Crystallin: a review of its structure and function. Clin Exp Optom. 2004;87:356–366. doi: 10.1111/j.1444-0938.2004.tb03095.x. [DOI] [PubMed] [Google Scholar]
- Bao Y, Wang Q, Liu H, Lin Z. A small HSP gene of bloody clam (Tegillarca granosa) involved in the immune response against Vibrio parahaemolyticus and lipopolysaccharide. Fish Shellfish Immunol. 2011;30:729–733. doi: 10.1016/j.fsi.2010.12.002. [DOI] [PubMed] [Google Scholar]
- Barnes PJ. Molecules in focus nuclear factor-κB. Int J Biochem Cell Biol. 1997;29:867–870. doi: 10.1016/S1357-2725(96)00159-8. [DOI] [PubMed] [Google Scholar]
- Bitondi MM, Nascimento AM, Cunha AD, Guidugli KR, Nunes FM, Simões ZL. Characterization and expression of the hex 110 gene encoding a glutamine-rich hexamerin in the honey bee, Apis mellifera. Arch Insect Biochem Physiol. 2006;63:57–72. doi: 10.1002/arch.20142. [DOI] [PubMed] [Google Scholar]
- Jong WW, Caspers GJ, Leunissen JA. Genealogy of the alpha-crystallin-small heat-shock protein superfamily. Int J Biol Macromol. 1998;22:151–162. doi: 10.1016/S0141-8130(98)00013-0. [DOI] [PubMed] [Google Scholar]
- Fu X, Li W, Mao Q, Chang Z. Disulfide bonds convert small heat shock protein Hsp16.3 from a chaperone to a non-chaperone: implications for the evolution of cysteine in molecular chaperones. Biochem Biophys Res Commun. 2003;308:627–635. doi: 10.1016/S0006-291X(03)01450-5. [DOI] [PubMed] [Google Scholar]
- Gkouvitsas T, Kontogiannatos D, Kourti A. Differential expression of two small Hsps during diapause in the corn stalk borer Sesamia nonagrioides (Lef.) J Insect Physiol. 2008;54:1503–1510. doi: 10.1016/j.jinsphys.2008.08.009. [DOI] [PubMed] [Google Scholar]
- Hendrick JP, Hartl FU. Molecular chaperone functions of heat-shock proteins. Annu Rev Biochem. 1993;62:49–384. doi: 10.1146/annurev.bi.62.070193.002025. [DOI] [PubMed] [Google Scholar]
- Huang P, Kang S, Chen W, Hsu TC, Lo CF, Liu K, Chen L. Identification of the small heat shock protein, HSP21, of shrimp Penaeus monodon and the gene expression of HSP21 is inactivated after white spot syndrome virus (WSSV) infection. Fish Shellfish Immunol. 2008;25:250–257. doi: 10.1016/j.fsi.2008.06.002. [DOI] [PubMed] [Google Scholar]
- Huang L, Wang C, Kang L. Cloning and expression of five heat shock protein genes in relation to cold hardening and development in the leafminer, Liriomyza sativa. J Insect Physiol. 2009;55:279–285. doi: 10.1016/j.jinsphys.2008.12.004. [DOI] [PubMed] [Google Scholar]
- Kokolakis G, Tatari M, Zacharopoulou A, Mintzas AC. The hsp27 gene of the Mediterranean fruit fly, Ceratitis capitata: structural characterization, regulation and developmental expression. Insect Mol Biol. 2008;17:699–710. doi: 10.1111/j.1365-2583.2008.00840.x. [DOI] [PubMed] [Google Scholar]
- Li Z, Li X, Yu Q, Xiang Z, Kishino H, Zhang Z. The small heat shock protein (sHSP) genes in the silkworm, Bombyx mori, and comparative analysis with other insect sHSP genes. BMC Evol Biol. 2009;9:215. doi: 10.1186/1471-2148-9-215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaud S, Morrow G, Marchand J, Tanguay RM. Drosophila small heat shock proteins: cell and organelle-specific chaperones? Prog Mol Subcell Biol. 2002;28:79–101. doi: 10.1007/978-3-642-56348-5_5. [DOI] [PubMed] [Google Scholar]
- Mizrahi T, Heller J, Goldenberg S, Arad Z. Heat shock proteins and resistance to desiccation in congeneric land snails. Cell Stress Chaperones. 2010;15:351–363. doi: 10.1007/s12192-009-0150-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrow G, Battistini S, Zhang P, Tanguay RM. Decreased lifespan in the absence of expression of the mitochondrial small heat shock protein Hsp22 in Drosophila. J Biol Chem. 2004;279:43382–43385. doi: 10.1074/jbc.C400357200. [DOI] [PubMed] [Google Scholar]
- Morrow G, Samson M, Michaud S, Tanguay RM. Overexpression of the small mitochondrial Hsp22 extends Drosophila life span and increases resistance to oxidative stress. FASEB J. 2004;18:598–599. doi: 10.1096/fj.03-0860fje. [DOI] [PubMed] [Google Scholar]
- Pérez-Morales D, Ostoa-Saloma P, Espinoza B. Trypanosoma cruzi SHSP16: characterization of an a-crystallin small heat shock protein. Exp Parasitol. 2009;123:182–189. doi: 10.1016/j.exppara.2009.06.019. [DOI] [PubMed] [Google Scholar]
- Reineke A. Identification and expression of a small heat shock protein in two lines of the endoparasitic wasp Venturia canescens. Comp Biochem Physiol A Mol Integr Physiol. 2005;141:60–69. doi: 10.1016/j.cbpb.2005.04.001. [DOI] [PubMed] [Google Scholar]
- Sagisaka A, Fujita K, Nakamura Y, Ishibashi J, Noda H, Imanishi S, Mita K, Yamakawa M, Tanaka H. Genome-wide analysis of host gene expression in the silkworm cells infected with Bombyx mori nucleopolyhedrovirus. Virus Res. 2010;147:166–175. doi: 10.1016/j.virusres.2009.10.015. [DOI] [PubMed] [Google Scholar]
- Sonoda S, Ashfaq M, Tsumuki H. Cloning and nucleotide sequencing of three heat shock protein genes (hsp90, hsc70, and hsp19.5) from the Diamondback moth, Plutella xylostella (L.) and their expression in relation to developmental stage and temperature. Arch Insect Biochem Physiol. 2006;62:80–90. doi: 10.1002/arch.20124. [DOI] [PubMed] [Google Scholar]
- Sonoda S, Ashfaq M, Tsumuki H. A comparison of heat shock protein genes from cultured cells of the Cabbage armyworm, Mamestra brassicae, in response to heavy metals. Arch Insect Biochem Physiol. 2007;65:210–222. doi: 10.1002/arch.20178. [DOI] [PubMed] [Google Scholar]
- Southgate R, Ayme A, Voellmy R (1983) Nucleotide sequence analysis of the Drosophila small heat shock gene cluster at locus 67B. Journal of Molecular Biology 165:35–57 [DOI] [PubMed]
- Sun Y, MacRae TH. Small heat shock proteins: molecular structure and chaperone function. Cell Mol Life Sci. 2005;62:2460–2476. doi: 10.1007/s00018-005-5190-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsunoda T, Takagi T. Estimating transcription factor bindability on DNA. Bioinformatics. 1999;15:622–630. doi: 10.1093/bioinformatics/15.7.622. [DOI] [PubMed] [Google Scholar]
- Tsvetkova NM, Horváth I, Török Z, Wolkers WF, Balogi Z, Shigapova N, Crowe LM, Tablin F, Vierling E, Crowe JH, Vigh L. Small heat-shock proteins regulate membrane lipid polymorphism. Proc Natl Acad Sci U S A. 2002;99:13504–13509. doi: 10.1073/pnas.192468399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Wang X, Liu C, Zhang J, Zhu C, Guo X. The NgAOX1a gene cloned from Nicotiana glutinosa is implicated in the response to abiotic and biotic stresses. Biosci Rep. 2008;28:259–266. doi: 10.1042/BSR20080025. [DOI] [PubMed] [Google Scholar]
- Wu P, Wang X, Qin G, Liu T, Jiang Y, Li M, Guo X. Microarray analysis of the gene expression profile in the midgut of silkworm infected with cytoplasmic polyhedrosis virus. Mol Biol Rep. 2011;38:333–341. doi: 10.1007/s11033-010-0112-4. [DOI] [PubMed] [Google Scholar]
- Xu P, Xiao J, Liu L, Li T, Huang D. Molecular cloning and characterization of four heat shock protein genes from Macrocentrus cingulum (Hymenoptera: Braconidae) Mol Biol Rep. 2010;37:2265–2272. doi: 10.1007/s11033-009-9715-z. [DOI] [PubMed] [Google Scholar]
- Yang G. Harm of introducing the western honeybee Apis mellifera L. to the Chinese honeybee Apis cerana F. and its ecological impact. Acta Entomol Sin. 2005;3:401–406. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The additional experiment of antimicrobial activity of AccHsp27.6. aStaphylococcus aureus; bMicrococcus luteus; cBacillus subtilis; dPseudomonas aeruginosa. K, kanamycin (250 mg/ml); A, ampicillin (250 mg/ml); 1, 2 purified AccHsp27.6 protein (0.5 mg/ml); 3 purified AccCPR26 protein (0.5 mg/ml) (JPEG 49 kb)