Abstract
Longitudinal image analysis has become increasingly important in clinical studies of normal aging and neurodegenerative disorders. Furthermore, there is a growing appreciation of the potential utility of longitudinally acquired structural images and reliable image processing to evaluate disease modifying therapies. Challenges have been related to the variability that is inherent in the available cross-sectional processing tools, to the introduction of bias in longitudinal processing and to potential over-regularization. In this paper we introduce a novel longitudinal image processing framework, based on unbiased, robust, within-subject template creation, for automatic surface reconstruction and segmentation of brain MRI of arbitrarily many time points. We demonstrate that it is essential to treat all input images exactly the same as removing only interpolation asymmetries is not sufficient to remove processing bias. We successfully reduce variability and avoid over-regularization by initializing the processing in each time point with common information from the subject template. The presented results show a significant increase in precision and discrimination power while preserving the ability to detect large anatomical deviations; as such they hold great potential in clinical applications, e.g. allowing for smaller sample sizes or shorter trials to establish disease specific biomarkers or to quantify drug effects.
Keywords: Unbiased longitudinal image processing, MRI biomarkers, Reliability and power, Within-subject template, FreeSurfer
Introduction
Progressive brain atrophy can be observed in a variety of neurodegenerative disorders. Several longitudinal studies have demonstrated a complex, regionally and temporally dynamic series of changes, that occur in normal aging and that are uniquely distinct in neurodegenerative disorders, such as Alzheimer's disease, Huntington's disease, and schizophrenia. The availability of large, high quality longitudinal datasets, has already begun to significantly expand our ability to evaluate selective, progressive anatomical changes. One of the major caveats in these studies is the use of tools that were originally designed for the analysis of data collected cross-sectionally. Inherent noise in cross-sectional methods, based on a single common template or atlas, often shadow individual differences and result in more heterogeneous measurements. However, by exploiting the knowledge that within-subject anatomical changes are usually significantly smaller than inter-individual morphological differences, it is possible to reduce within-subject noise without altering the between-subject variability. As such, the development of unbiased longitudinal analytical approaches are critical in fully elucidating phenotypic variability, and in the construction of imaging based biomarkers to quantify response in clinical trials and to evaluate disease modifying therapies. In particular, these tools can be expected to increase the sensitivity and reliability of the measurements sufficiently to require smaller sample sizes and fewer time points or shorter follow-up periods.
The novel longitudinal methodologies described in this paper are designed to overcome the most common limitations of contemporary longitudinal processing methods: the introduction of processing bias, over-regularization, and the limitation to process only two time points. In addition, building on FreeSurfer (Fischl, in press; Fischl et al., 2002), our methods are capable of producing a large variety of reliable imaging statistics, such as segmentations of subcortical structures, cortical parcellations, pialand white matter surfaces as well as cortical thickness and curvature estimates.
Bias
Longitudinal image processing aims at reducing within subject variability, by transferring information across time, e.g. enforcing temporal smoothness or informing the processing of later time points with results from earlier scans. These approaches, however, are susceptible to processing bias. It is well documented that especially interpolation asymmetries can influence downstream processing and subsequent analyses (Thompson and Holland, 2011; Yushkevich et al., 2010) and can result in severe underestimation of sample sizes due to overestimation of effect sizes. Interpolation asymmetries occur when, for example, resampling follow-up images to the baseline scan and thus smoothing only the follow-up images while keeping the baseline image untouched. As described in Reuter and Fischl (2011) and as demonstrated below, interpolation asymmetries are not the only source of bias. Consistently treating a single time point, usually baseline, differently from others, for instance, to construct an atlas registration or to transfer label maps for initialization purposes, can already be sufficient to introduce bias. Bias is a problem that often goes unnoticed, due to large measurement noise, imprecise methods, small sample sizes or insufficient testing. Not treating all time points the same can be problematic as the absence of bias cannot simply be proven by not finding it. Furthermore, the assumption that group effects are not (or only mildly) influenced by processing bias is usually incorrect. It is rather unlikely that bias affects all groups equally, considering that one group usually shows only little longitudinal change, while the other undergoes significant neurodegeneration. For these reasons, we carefully designed and implemented our longitudinal methods to treat all time points exactly the same. Another potential source of bias may be induced when constraining sequential results to be smooth. Temporal regularization can limit the power of an algorithm to detect large changes. We aim at avoiding this kind of over-regularization by initializing the processing in each time point with common information, but allowing the methods to evolve freely.
It should be noted, that different types of bias, not induced by the image analysis software but rather related to pre-processing or image acquisition steps, can already be present in the images, equally affecting both longitudinal and independent (cross-sectional) processing. Examples include the use of different scanner hardware, different scanner software versions, different calibration, acquisition parameters or protocols across time. These biases cannot easily be removed by downstream processing, although they can possibly be reduced. Other types of bias are related to intrinsic magnetic properties of the tissue (e.g. T1, T2*) across time (aging) or across groups (neurodegenerative disease) potentially introducing bias in measures of thickness or volume (Salat et al., 2009; Westlye et al., 2009). However, since age and disease level are usually very similar within-subject, the rate of change in a longitudinal study will be less affected than cross-sectional volume or thickness analysis.
Related work
In SIENA, Smith et al. (2001, 2002) introduced the idea of transforming two input images into a halfway space, to ensure both undergo the same resampling steps to avoid interpolation bias. However, traditionally, the baseline image is treated differently from the follow-up images. Often longitudinal processing is approached by employing higher order registration methods to compute and analyze the deformation field that aligns baseline to a follow-up scan, e.g. SPM2 uses high dimensional warps Ashburner et al. (2000)). These procedures are usually not inverse consistent and resample only the follow-up images. SPM, for example, has been employed in longitudinal studies of neurodegeneration in two time points (Chételat et al., 2005; Kipps et al., 2005) without specifically attempting to avoid asymmetry-related bias. The longitudinal segmentation algorithm CLASSIC (Xue et al., 2006) jointly segments a 4D image via longitudinal high-order warps to the baseline scan using an elastic warping algorithm. Also Avants et al. (2007) work in the baseline space as a reference frame. In that work, first a spatiotemporal parameterization of an individual's image time series is created via nonlinear registration (SyN). The underlying diffeomorphism is then resampled at the one year location and compared to baseline to quantify the annual atrophy. Qiu et al. (2006) present a method for longitudinal shape analysis of brain structures, quantifying deformations with respect to baseline and transporting the collected information from the subject baseline to a global template. Other authors focus on cortical measures. Han et al. (2006) describe a method to initialize follow-up surface reconstruction with surfaces constructed from the baseline scans. Li et al. (2010) register follow-up images to the baseline (rigidly and nonlinearly based on CLASSIC) and then keep the directions fixed across time along which they locally compute thickness in the cortex.
Over the last few years, several authors attempt to avoid processing bias. In 2009, initial software versions of our methods, relying on unbiased within-subject templates as described in this paper, were made publicly available (Reuter, 2009; Reuter et al., 2010a). Related efforts, however, aim primarily at removing only interpolation bias. Avants et al. (2010), for example, similarly utilize within-subject templates, while still treating the baseline image consistently differently from follow-up time points. Nakamura et al. (2011) avoid bias only in the registration procedure by combining forward and inverse linear registrations to construct symmetric pairwise maps. Also combining forward and backward transformations, Holland and Dale (2011) use a nonlinear pairwise registration and intensity normalization scheme to analyze the deformation in follow-up images by measuring volume changes of labels defined in baseline space.
Approach
In this work we present an automated longitudinal processing pipeline that is designed to enable a temporally unbiased evaluation of an arbitrary number of time points by treating all inputs the same. First an unbiased, within-subject template is generated by iteratively aligning all input images to a median image using a symmetric robust registration method (Reuter et al., 2010b). Because of the simultaneous co-registration of all time points, processing can be performed in a spatially normalized voxel space across time reducing variability of several procedures. Furthermore, the median image functions as a robust template approximating the subject's anatomy, averaged across time, and can be used as an estimate to initialize the subsequent segmentations.
Cortical and subcortical segmentation and parcellation procedures involve many complex nonlinear optimization problems, such as topology correction, nonlinear atlas registration, and nonlinear spherical surface registration. These nonlinear problems are typically solved with iterative methods. The final results can thus be sensitive to the selection of a particular starting point. However, by initializing the processing of a new data set in a longitudinal series with common information, the variations in the processing procedures can be efficiently reduced and the robustness and sensitivity of the overall longitudinal analysis significantly improved. Increased reliability often comes at the cost of over-regularization by enforcing temporal smoothness. Our methods do not add explicit constraints such as temporal smoothness or higher-order within-subject warps to transfer labels, nor do they incorporate the order of time points at all. Higher precision is achieved solely by common initialization while segmentation and surface reconstruction procedures are allowed to evolve freely. We demonstrate that the resulting measurements are significantly more reliable in both healthy controls (in test–retest, simulated noise and simulated atrophy) as well as in neurodegeneration studies. We show that the increased precision enables greater power to evaluate more subtle disease effects or to reduce sample sizes. This longitudinal processing stream is made available as part of FreeSurfer (Fischl, in press; Fischl et al., 2002; Reuter, 2009). The FreeSurfer software package is an open access resource that has gained popularity in evaluating cortical and subcortical measures.
Impact
An early version of the methods described in this paper has been successfully employed in a variety of studies analyzing progressive changes in Alzheimer's disease (Chiang et al., 2010, 2011; Desikan et al., 2010; Sabuncu et al., 2011), Huntington's disease (Rosas et al., 2011), memory training (Engvig et al., 2010) and for the validation of prospective motion correction (Tisdall et al., in press). The Alzheimer's Disease Neuroimaging Initiative (ADNI), for instance, makes available2 their raw image data and derived measures, processed with the initial version of our longitudinal method (FS 4.4). ADNI is one of the largest publicly available longitudinal image data sets, consisting of more than 3000 scans, released with the goal to determine in-vivo biomarkers for the early detection of AD. Although our initial processing methods that were used for the derived measures are less powerful than the newer version presented in this paper, the available results are still of great importance to researchers without the possibility to locally process the raw images, as well as to function as a benchmark for method development and comparison (Holland et al., 2011).
Currently, large datasets such as ADNI are under consideration for other neurological diseases. As such, the highly sensitive, reliable and fully automated unbiased longitudinal methods described in this paper have the potential to help us understand natural progression of regionally and spatially selective neurodegeneration as occurs in distinct neurological disorders. The resulting, subject specific, morphometric measurements yield biomarkers that potentially serve as surrogate endpoints in clinical trials, where the increase of statistical power is of most immediate importance.
Methods
Overview of longitudinal processing pipeline
The proposed processing of longitudinal data consists of the following three steps:
[CROSS]: First all time points of all subjects are processed independently. This is also called cross-sectional processing. Here a full image segmentation and surface reconstruction for each time point is constructed individually. Some of this information is needed later during the longitudinal processing and to construct the subject template in the next step.
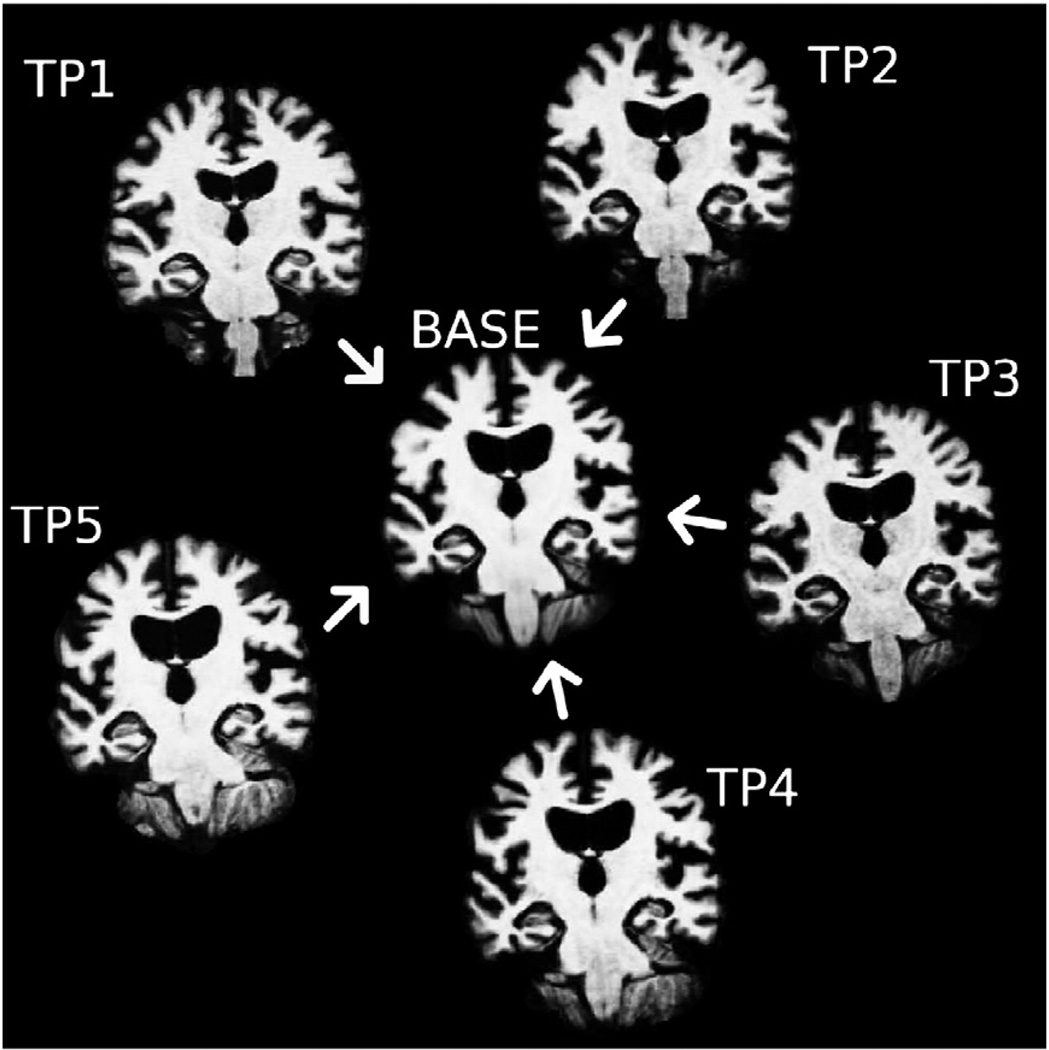
[BASE]: For each subject a template is created from all time points to estimate average subject anatomy. Often the within-subject template is also referred to as the subject ‘base’ (different from baseline scan!) or simply as the template. Here an unbiased median image is used as the template and a full segmentation and surface reconstruction is performed. We describe the creation of the subject template in the following sections.
[LONG]: Finally each time point is processed “longitudinally”, where information from the subject-template [BASE] and from the individual runs [CROSS] is used to initialize several of the algorithms. A [LONG] process usually takes about half the time to complete than a [CROSS] or [BASE] run.
The improved and more consistent results from the final set of [LONG] runs (step 3) provide the reliable input for post-processing or subsequent statistical analysis.
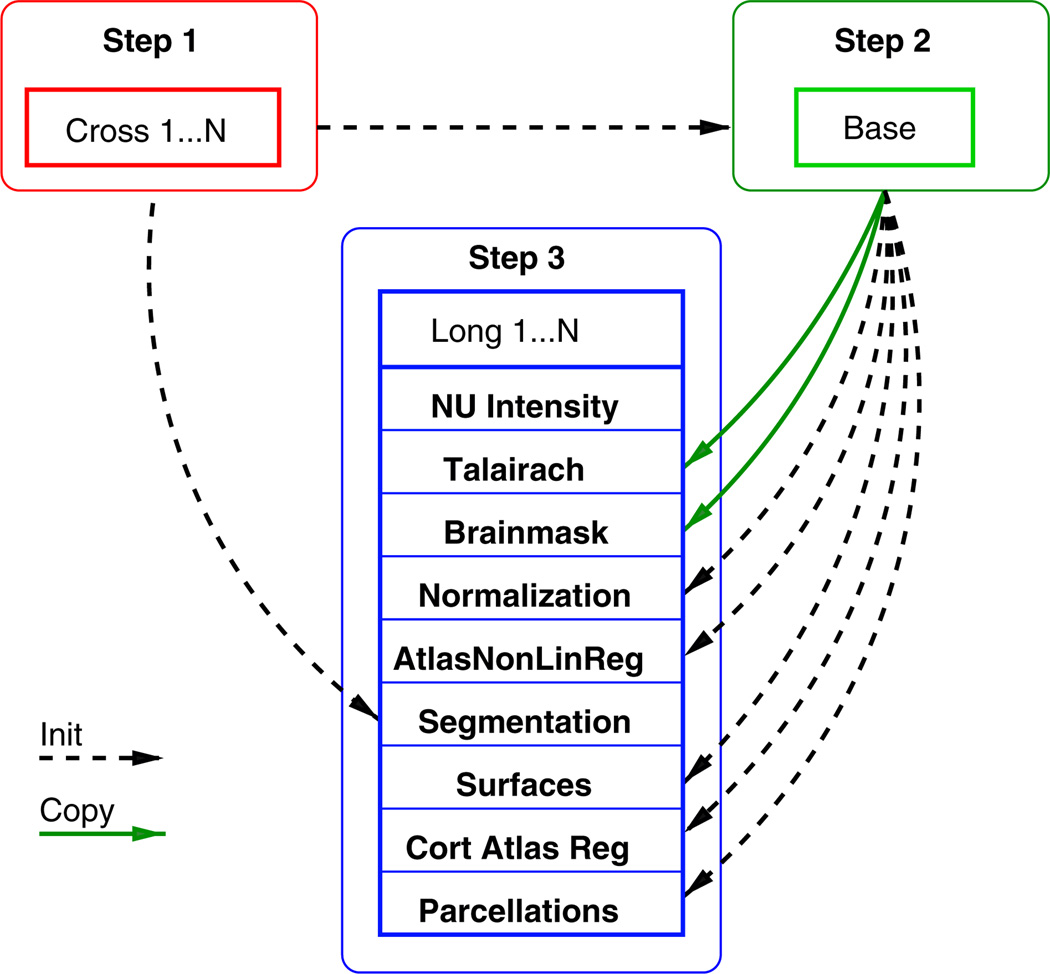
In step 3 above, the longitudinal processing of each time point is initialized with information from the subject template [BASE] and the [CROSS] results to reduce variability. However, depending on the flexibility of the individual algorithms, this general procedure may sacrifice accuracy and potentially underestimate changes of greater magnitude. Whenever information is transferred across time, e.g. to regularize or explicitly smooth results, methods can become biased towards underestimating change and accuracy may suffer particularly when measuring longitudinal change over long periods of time. While a conservative estimate of change is often preferable in a power analysis, than an overestimation, we focus on avoiding asymmetries and over-regularization to remain as accurate and unbiased as possible. The longitudinal processing step (see also Fig. 1) mainly consists of the following procedures (more details can be found online (Reuter, 2009)).
Fig. 1.
Simplified diagram of the three steps involved in longitudinal processing, showing information flow at a single longitudinal run. Dashed line: information is used for initialization. Solid line: information is copied.
Spatial normalization and NU intensity
All inputs are resampled to the unbiased template voxel space to further reduce variability (since FS 5.1). This can be achieved during the motion correction and conforming steps by composing the linear transforms and only resampling once to avoid additional resampling/smoothing. For this paper, we employ linear interpolation, but recently switched to cubic B-spline interpolation for future releases to reduce interpolation artifacts (Thevenaz et al., 2000). Then acquisition bias fields in [LONG] are independently corrected using a non-parametric non-uniform intensity normalization (N3) (Sled et al., 1998).
Talairach registration
The affine map from the robust template [BASE] to the Talairach space is fixed across time. It can be assumed that a single global affine transformation to the Talairach coordinate system is appropriate since the bulk of the anatomy within-subject is not changing. The advantages of this approach lie in the noise reduction obtained by avoiding the use of individual intensity volumes for each time point and in consistent intra-cranial volume estimation. Data is only copied from the subject template if fixing it across time is meaningful, for example the affine Talairach registration or the brain mask (see green solid arrows in Fig. 1).
Brainmask creation
The brain mask, including some cerebrospinal fluid (CSF), is kept constant for all time points (in [LONG] and in the subject template [BASE]) to reduce variability under the assumption that the location and size of the intracranial vault are not changing (although of course the contents may be). The brain mask is constructed as the union (logical OR) of the registered brain masks across time. In other words, a voxel is included in the brain mask if it is included in any of the time points, to ensure no brain is accidentally clipped. Although the brain mask is fixed across time by default, it can be adjusted in individual time points manually if necessary (e.g. by editing it or mapping it from the initial [CROSS] results).
Normalization and atlas registration
For the second intensity correction (pre-segmentation filter) (Dale et al., 1999), each [LONG] run is initialized with the common set of control points that were constructed in the [BASE], to encourage consistency across time. Similarly for the normalization to the probabilistic atlas (Fischl et al., 2002, 2004a), the segmentation labels of the [BASE] are passed as a common initialization in each time point (dashed arrows in Fig. 1). Also the nonlinear atlas registration is initialized with results from the [BASE]. However, these procedures are intrinsically the same as before and are still performed for each time point to allow for the necessary flexibility to detect larger departures from the average anatomy of the [BASE]. Starting these procedures without a common initialization would only increase variability as more time points may terminate in different local minima.
Subcortical segmentation
Specifically for the subcortical segmentation we allow even more flexibility. Instead of initializing it with the template segmentation, a fused segmentation is created for each time point by an intensity based probabilistic voting scheme. Similar to Sabuncu et al. (2010), where training label maps are fused for the segmentation, we also use a weighted voting scheme to construct a fused segmentation of each time point from the initial segmentations of all time points obtained via independent processing [CROSS] (using the standard atlas based registration procedure in FreeSurfer). Based on local intensity differences between the time points at each voxel we employ a kernel density estimation (Parzen window) using a Gaussian kernel on the registered and intensity normalized input images and initial labels. In other words, if the intensity at a given location is similar at another time point, the corresponding label is highly probable. This weighted majority voting (including votes from all time points) yields the labels to construct the fused segmentation. Since this procedure is driven by all time point's initial segmentations rather than the template's segmentation, it allows for more flexibility. For each time point, it represents the anatomy more accurately than the segmentation of the template. In order to correct for potential remaining noise, each fused label map is used to initialize a final run through the regular atlas based segmentation procedure. This fine-tuning step usually converges quickly.
Surfaces reconstruction
The regular cortical surface construction in FreeSurfer starts with the tessellation of the gray matter / white matter boundary, automated topology correction (Fischl et al., 2001; Ségonne et al., 2007), and surface deformation following intensity gradients to optimally place the gray/white and gray/CSF borders at the location where the greatest shift in intensity defines the transition to the other tissue class (Fischl and Dale, 2000). In the longitudinal stream, the white and pial surfaces in each time point are initialized with the surfaces from the template [BASE] and are allowed to deform freely. This has the positive effect that surfaces across time demonstrate an implicit vertex correspondence. Furthermore, manual editing to fix topology or surface placement is in many cases only necessary in the subject's template [BASE] instead of in each individual time point, reducing potential manual intervention (and increasing reliability).
Cortical atlas registration and parcellations
Once the cortical surface models are complete, a number of deformable surface procedures are usually performed for further data processing. The spherical registration (Fischl et al., 1999b) and cortical parcellation procedures (Desikan et al., 2006; Fischl et al., 2004b) establish a coordinate system for the cortex by warping individual surface models into register with a spherical atlas in a way that aligns the cortical folding patterns. In the longitudinal analysis we make the first-order assumptions that large-scale folding patterns are not substantially altered by disease, and thus assume the spherical transformation of the subject template to be a good initial approximation. In [LONG] the surface inflation with minimal metric distortion (Fischl et al., 1999a) is therefore copied from the [BASE] to all time points. The subsequent non-linear registration to the spherical atlas and also the automatic parcellation of the cerebral cortex in each time point are initialized with the results from the [BASE]. This reliably assigns a neuroanatomical label to each location on the surface by incorporating both geometric information derived from the cortical model (gyral and sulcal structure as well as curvature), and neuroanatomical convention.
Note that no temporal smoothing is employed, also the order of time points is not considered. The above steps are meaningful under the assumptions that head size is relatively constant across time which is reasonable for most neurodegenerative diseases but not, for example, for early childhood development. However, users can optionally keep individual brainmasks or introduce manual edits to accommodate for special situations, at the cost of decreased reliability.
Within-subject template
Atlas construction, usually creating a template of several subjects, has been an active field of research. For example, Joshi et al. (2004), Avants and Gee (2004) or Ashburner (2007) approach unbiased nonlinear atlas construction by iteratively warping several images to a mean. In order to create a robust within-subject template of several longitudinal scans, we need to make several design decisions. All time points need to be treated the same to avoid the possible introduction any asymmetries. Furthermore, we use only a rigid transformation model to remove pose (or optionally an affine transformation to additionally remove scanner calibration differences). Currently we avoid higher order warps to not introduce any temporal smoothing constraints or worse, incorrect correspondence that is relied upon by downstream processing (e.g. when transferring labels). We do not assume exact tissue correspondence in our model. Finally, instead of the commonly used intensity mean, a voxel-wise intensity median is employed to create crisper averages and remove outliers such as motion artifacts from the template.
Template estimation of N images Ii is usually stated as a minimization problem of the type:
| (1) |
where the template Î and the transformations φ̂i, that map each input image to the common space, need to be estimated. For robustness and other reasons as described below, we set the image dissimilarity metric E(I1, I2) = ∫Ω|I1(x) − I2(x)|dx where Ω denotes the coordinate space. Thus, for fixed transformations φi the minimizing Î is given by the voxel-wise median of all Ii. For a rigid transformation consisting of translation and rotation φ = (t⃗, r) ∈ℝ3 × SO(3), we choose the following squared distance with respect to identity , where we compute the Frobenius norm of the difference between the identity I and the 3 × 3 rotation matrix R representing the rotation r. Since the inputs Ii are rather similar, Î can be approximated by the following iterative algorithm:
Compute the median of the N input images Î.
Register and resample each input image to Î.
Continue with step 1 until the obtained transforms φi converge.
The registration step adjusts the location of the inputs closer to Î, so that the next average can be expected to improve. This iterative algorithm is performed on a Gaussian pyramid of the images, with differently many iterations on each resolution. The inexpensive low resolutions are iterated more often to quickly align all images approximately, while the more time consuming higher resolutions only need to refine the registration in a few steps.
Robustness
For the registration of two images at the core of the template estimation, we use a robust and inverse consistent registration method as described in Reuter et al. (2010b). Inverse consistency means that the registration φij of imagesIi to Ij is exactly the inverse of , which is usually not guaranteed. This property keeps individual registrations unbiased and is achieved by a symmetric resampling step in the algorithm to the halfway space between the two input images, as well as a symmetric model, to avoid estimation and averaging of both forward and backward transformations. This approach, incorporating the gradients of both inputs, additionally seems to be less prone to local optima. While pairwise symmetry is not strictly necessary to keep the template unbiased it avoids unnecessary iterative averaging in the common case of only two input images, where both can be resampled at the halfway space after a single registration step. Another advantage of the robust registration algorithm is its ability to reduce the influence of outlier regions resulting in highly accurate brain registrations, even in the presence of local differences such as lesions, longitudinal change, skull stripping artifacts, remaining dura, jaw/neck movement, different cropping planes or gradient nonlinearities. Reuter et al. (2010b) showthe superiority of this method over standard registration tools available in the FSL (Jenkinson et al., 2002; Smith et al., 2004) or SPM packages (Ashburner and Friston, 1999; Collignon et al., 1995) with respect to inverse consistency, noise, outlier data, test–retest analysis and motion correction.
In spite of the fact that in the above template estimation algorithm convergence is not guaranteed, the procedure works remarkably well even if significant longitudinal change is contained in the images (see Fig. 2), due to the robustness of the median image. In this context see also related work in Fletcher et al. (2009) who construct a different intrinsic median for atlas creation by choosing “metamorphosis” as the metric on the space of images that accounts for both geometric deformations as well as intensity changes. In a mean image outlier regions and longitudinal structural change will introduce blurring. Strong motion artifacts in specific time points may corrupt the whole image. The median, however, suppresses outlier artifacts, ghosts and blurry edges and seems to be a good choice as normality cannot be assumed due to longitudinal change, motion artifacts etc. Only for the special case of two time points, the median reduces to the mean and may contain two ghost images. Registration with this ill-defined average can be avoided by computing the mid-space directly from the registration of the two inputs. In general, the use of the median leads to crisper within-subject templates. It is therefore well suited for constructing initial estimates of location and size of anatomical structures averaged across time or for creating white matter and pial surfaces. As described above this information is used to inform the longitudinal processing of all individual time points.
Fig. 2.
Unbiased template estimation for a subject with neurodegenerative disease and significant atrophy: All time points are iteratively aligned to their median image with an inverse consistent robust registration method, resulting in a template image and simultaneously a co-registration of all time points.
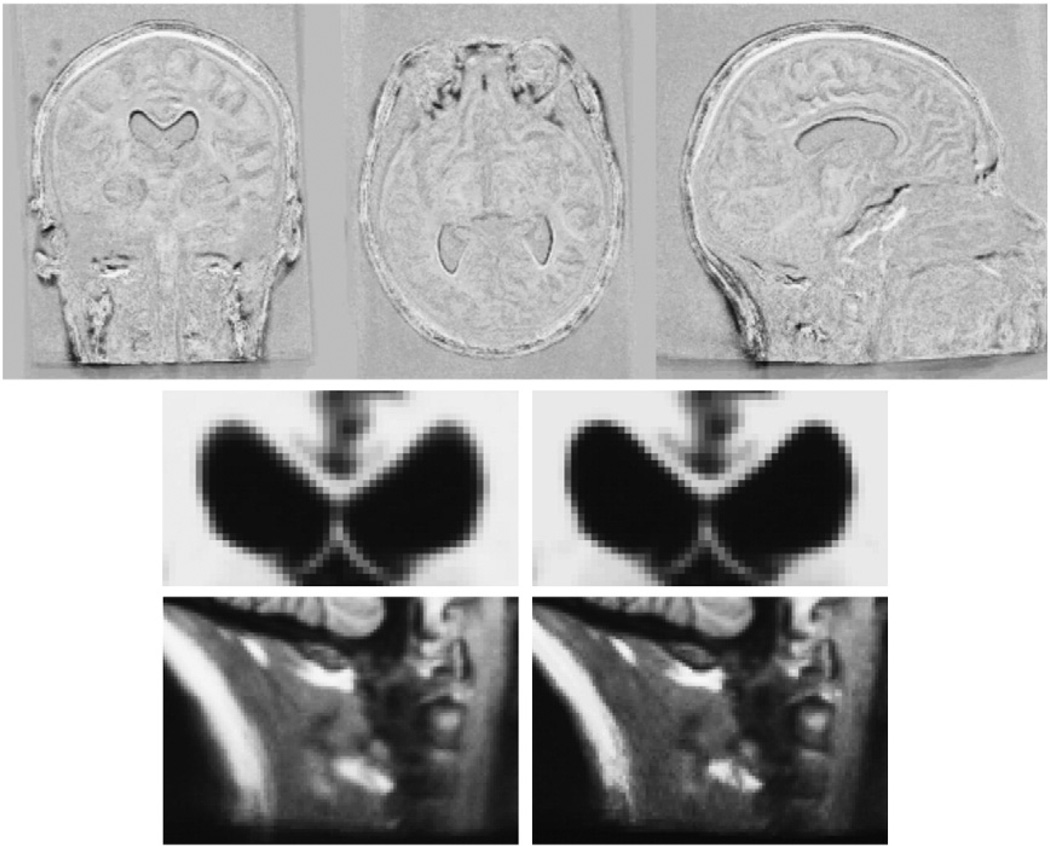
To demonstrate the effect of the median, Fig. 3 shows the difference between the mean and median template of a series of 18 time point images of a Huntington's disease subject, taken over a span of 7 years. Some of the time points contain strong motion artifacts. Additionally, this subject exhibits significant atrophy, i.e. approximately 8% volume loss in the caudate and significant thinning of the corpus callosum. Due to the robustness of the median the template image remains crisp with well defined anatomical boundaries, in spite of the longitudinal change, as opposed to the smoother mean image. In a two bin histogram of the gradient magnitude images, the bin with larger gradient magnitudes contains 4.4% of the voxels in the median and 3.7% in the mean image, indicating crisper edges in the median. The difference images in the top row of Fig. 3 localize the differences between the mean and median image mainly in regions with large longitudinal change such as the ventricles, corpus callosum, eyes, neck and scalp. Because of the crispness of the median image, co-registration of all 18 inputs needed only three global iterations, while it took five iterations to converge for the mean at a higher residual cost.
Fig. 3.
Comparison of mean and median template image for a series of 18 images (7 years) of a subject with neurodegenerative disorder (Huntington's disease). The difference image (top row) between median and mean reveals large differences in regions that change over time (e.g. ventricles, corpus callosum, eyes, neck, scalp). Below are close-ups of the mean image (left, softer edges) and the median image (right, crisper edges).
Improved template estimation
We found in our tests that the template estimation converges without pre-processing. However, it may need a large number of iterations to converge in specific cases and thus a considerable amount of processing time (> 1 h). If the early average images are very distorted the corresponding registrations will be inaccurate. Once the average becomes crisper the convergence is fast. The following procedure is designed to speed up computations, by initially mapping all images to a mid-space and starting the algorithm there:
First the registration of each image Ii to a randomly selected image Ĩ is computed, to get estimates of where the head/brain is located in each image with respect to the location in Ĩ. These registrations do not need to be highly accurate as they are only needed to find an approximation to the mid-space for the initialization. However, highly accurate registration in this step will reduce the number of iterations later.
Then the mid-space is computed from the set of transformation maps and all images are resampled at that location. See Appendix B for details on how the average space is constructed.
The iterative template estimation algorithm (Within-subject template section) is initialized with the images mapped to the mid-space and, in most cases, needs only two further iterations to converge: One to register all images to the average image and another one to check that no significant improvements are possible.
Since all images are remapped at the mid-space location, including image Ĩ, they all undergo a common resampling step removing any interpolation bias. The random asymmetry that may be introduced when selecting image Ĩ as the initial target is further reduced due to the fact that the registration method is inverse consistent, so the order of registration (image Ĩ to image Ii or vice versa) is irrelevant. Alternatively it is possible to construct all pairwise registrations and compute the average location considering all the information (e.g. by constructing average locations using each input as initial target independently and averaging the N results). This, however, significantly increases computational cost unless N is very small and seems unnecessarily complicated given that the above algorithm already removes resampling asymmetries and randomizes any potential remaining asymmetry possibly induced by choosing an initial target.
Global intensity scaling
If the input images show differences in global intensity scales, the template creation needs to adjust individual intensities so that all images have an equal weight in the final average. This can be done by computing a global intensity scale parameter in each individual registration. Once we know the intensity scale si of each image with respect to the target (initially image Ĩ but later average template), the individual intensities can be adjusted to their geometric mean
| (2) |
by scaling each image intensity values with . Note, in the longitudinal processing stream presented in this paper, intensity normalized skull stripped images (norm.mgz in FreeSurfer) are used as input to construct the co-registration, thus global intensity scaling during the registration step is not necessary, however, it can be important in other applications.
Affine voxel size correction
It may be desired to adjust for changes in scanner voxel sizes, possibly induced by the use of different scanners, drift in scanner hardware, or different scanner calibrations, which are frequently performed especially after software/hardware upgrades. Takao et al. (2011) find that even with scanners of the exact same model, inter-scanner variability affects longitudinal morphometric measures, and that scanner drift and inter-scanner variability can cancel out actual longitudinal brain volume change without correction of differences in imaging geometry. Clarkson et al. (2009) compare phantom based voxel scaling correction with correction using a 9 degrees of freedom (DOF) registration and show that registration is comparable to geometric phantom correction as well as unbiased with respect to disease status. To incorporate automatic voxel size correction, we design an optional affine [BASE] stream, where all time points get mapped to an average affine space by the following procedure:
First perform the rigid registration as described above on the skull stripped brain images to obtain an initial template image and average space.
Then use the rigid transforms to initialize an iterative affine registration employing the intensity normalized full head images.
Fine tune those affine registrations by using the affine maps to initialize a final set of registrations of the skull stripped brain images to the template, where only rigid parameters are allowed to change.
Since the registration of the brain-only images in the final step is rather quick (a few minutes), especially with such a high quality initialization, the fine tuning step comes at little additional effort and ensures accurate brain alignment. Note, that we also propose a full affine (12 DOF) registration as two non-uniform orthogonal scalings and a rotation/translation in the middle generally cannot be represented by 9 DOF.
Adding time points
Opposed to independent processing, longitudinal processing evaluates concurrently scans that have been collected at different time points in order to transfer information across time. As a result, it always implicitly requires a delay in processing until all of the time points are available to remain unbiased, independent of the longitudinal method used. While this would be standard in a clinical therapeutic study, it is less optimal in observational studies. However, due to the robust creation of the subject median template, the subsequent addition of time points, assuming that they are not collected significantly later in time, would not be expected to have a large influence on the analyses if sufficient temporal information is contained in the template already. The purpose of the subject template is mainly to remove interpolation bias and to initialize processing of the individual time points in an unbiased way. Similar to atlas creation, where a small number of subjects is usually sufficient for convergence, it can be expected that within subject template estimation converges even faster, due to the smaller variability. Nevertheless, in order to avoid being potentially biased with respect to a healthier/earlier state, it is recommended, if possible, to recreate the template from all time points and reprocess all data until further studies investigate this issue more thoroughly.
Results and discussion
Data
TT-115
Two different sets of test–retest data are analyzed below. The first set consists of 115 controls scanned twice within the same session and will be referred to as TT-115. Two full head scans (1 mm isotropic, T1-weighted multi-echo MPRAGE (van der Kouwe et al., 2008), Siemens TIM Trio 3T, TR 2530 ms, TI 1200 ms, multi echo with BW 650 Hz/px and TE=[1.64 ms, 3.5 ms, 5.36 ms, 7.22 ms], 2 × GRAPPA acceleration, total acq. time 5:54 min) were acquired using a 12 channel head coil and then gradient unwarped. The two scans were separated by a 60 direction 2 mm isotropic EPI based diffusion scan and accompanying prior gradient echo field map (2:08 min, 9:45 min), not used here. The two multi-echo MPRAGE images are employed to evaluate the reliability of the automatic segmentation and surface reconstruction methods. It can be assumed that biological variance and variance based on the acquisition is minimized, therefore, this data will be useful to reveal differences in the two processing streams (independent processing versus longitudinal processing).
TT-14
We also evaluate a second test–retest set consisting of 14 healthy subjects with two time points acquired 14 days apart (TT-14). The images are T1-weighted MPRAGE full head scans (dimensions 1 mm× 1 mm×1.33 mm, Siemens Sonata 1.5T, TR 2730 ms, TI 1000 ms, TE 3.31 ms). Each time point consists of two within session scans that were motion corrected and averaged to increase signal to noise ratio (SNR). This data set exhibits a lower SNR than the TT-115 above, since it was acquired using a volume coil and at a lower field strength of 1.5T. Because of this and the larger time difference it therefore better reflects the expected variability of a longitudinal study and will be used for power analyses below.
OA-136
To study group discrimination power in dementia in both processing streams we analyze a disease dataset: the Open Access Series of Imaging Studies (OASIS)3 longitudinal data. This set consists of a longitudinal collection of 150 subjects aged 60 to 96. Each subject was scanned on two to five visits, separated by approximately one year each for a total of 373 imaging sessions. For each subject and each visit, 3 or 4 individual T1-weighted MPRAGE scans (dimensions 1 mm× 1 mm× 1.25 mm, TR 9.7 ms, TI 20 ms, TE 4 ms) were acquired in single sessions on a Siemens Vision 1.5T. For each visit two of the scans were selected based on low noise level in the background (indicating high quality, i.e., no or little motion artifacts). These two scans were then registered and averaged to increase signal to noise ratio and used as input. The subjects are all right-handed and include both men and women. 72 of the subjects were characterized as non-demented for the duration of the study and 64 subjects as demented. Here we do not include the third group of 14 converters who were characterized as non-demented at the time of their initial visit and were subsequently characterized as demented at a later visit. The dataset therefore only includes 136 subjects and will be called OA-136.
HD-54
To highlight improvements in situations with less statistical power, a second and smaller disease data set is employed containing 10 healthy controls (C), 35 pre-symptomatic Huntington's disease subjects of which 19 are near (PHDnear) and 16 far (PHDfar) from estimated onset of symptoms, and 9 progressed symptomatic Huntington's patients (HD). The near and far groups where distinguished based on the estimated time to onset of symptoms using CAG repeat length and age (Langbehn et al., 2004) where far means expected onset in more than 11 years. Each subject has image data from three visits approximately half a year to a year apart (dimensions 1 mm× 1 mm× 1.33 mm, T1-weighted MPRAGE, Siemens Avanto 1.5T, TR 2730 ms, TI 1000ms, TE 3.31 ms, 12 channel head coil). Scanner software versions have changed for several subjects between the visits and in the HD group two subjects even have 1–2 of their scans on an older Siemens Sonata scanner, which adds potential sources of variability. This data will be referred to as HD-54.
Bias in longitudinal processing
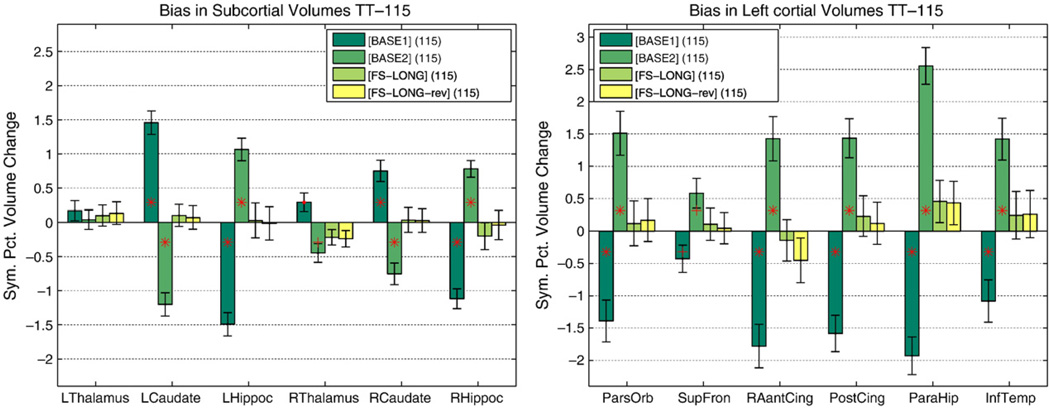
Yushkevich et al. (2010) demonstrated that interpolation asymmetries can bias longitudinal processing. As mentioned above, our method prevents interpolation and other asymmetry induced bias by treating all time points equivalently. In Reuter and Fischl (2011) we argue that avoiding only resampling bias may not be sufficient as different sources of bias exist, such as informing processes in follow-up images with information from baseline. To demonstrate the effect we introduce asymmetry by using information from one of the time points (segmentation and surfaces) to initialize processing of the other on the TT-115 dataset. To remove any potential change in the images, the order of time points is previously randomized for this test. Note, that none of the inputs are mapped or resampled to baseline or a common space but stay in their native spaces, only label maps and surfaces are transferred. As a dimensionless measure of change we compute the symmetrized percent change (SPC) of the volume of a structure with respect to the average volume,4 defined as:
| (3) |
where Vi is the volume at time point i. Fig. 4 shows the SPC for different structures when processing the test–retest data both forward (initializing time point 2 with results from time point one) [BASE1] and backward [BASE2]. One can expect average zero change in each structure as both images are from the same session, but instead the processing bias can clearly be seen, even for the cortical volumes where no interpolation is used at all when mapping the surfaces. It can be assumed that the bias is introduced by letting the results evolve further in the other time point. It should be noted that the bias affects different structures differently, and although it is strong, it cannot, for example, be detected in the left thalamus. Furthermore, it can be observed in Fig. 4 that the proposed processing stream, [FS-LONG] and [FS-LONG-rev] where time points are passed in reversed order when constructing the template, shows no bias. The remaining small differences can be accounted to subtle numerical instabilities during the template estimation.
Fig. 4.
Initializing time point 2 with results from time point 1 [BASE1] and vice-versa [BASE2] shows a bias in symmetrized percent change. Using our method [FS5.1] and passing time points in reverse order [FS5.1rev] does not show a processing bias. Significant differences from zero are denoted by a red plus: p<0.01 and red star: p<0.001 in Wilcoxon signed rank test. Error bars show a robust standard error where standard deviation is replaced by σ≈ 1.4826 median absolute deviation. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
Robustness, precision and accuracy
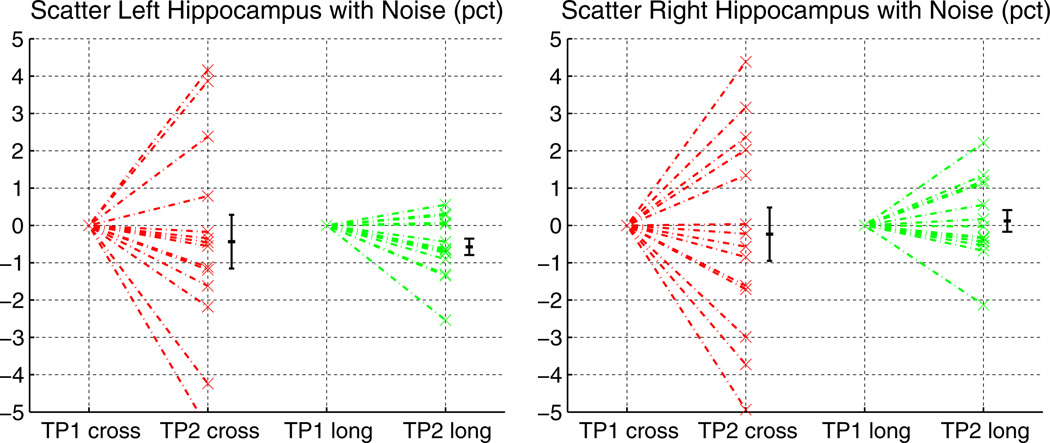
For the following synthetic tests only the first time point of data set TT-14 is taken as baseline. It is resliced to 1 mm isotropic voxels, intensity scaled between 0 and 255. The synthetic second time point is a copy of that image, but artificially modified to test robustness with respect to noise and measurement precision of the longitudinal stream. As Rician noise is nearly Gaussian at larger signal to noise ratios, robustness with respect to noise is tested by applying Gaussian noise (σ= 1) to the second time point. Fig. 5 shows the percent change with respect to the original in the hippocampal volume for both hemispheres for cross-sectional and longitudinal processing. The longitudinal stream is more robust and reduces the variability (increased precision).
Fig. 5.
Effect of simulated noise (σ= 1) on hippocampal volume measurements. The longitudinal processing is less affected.
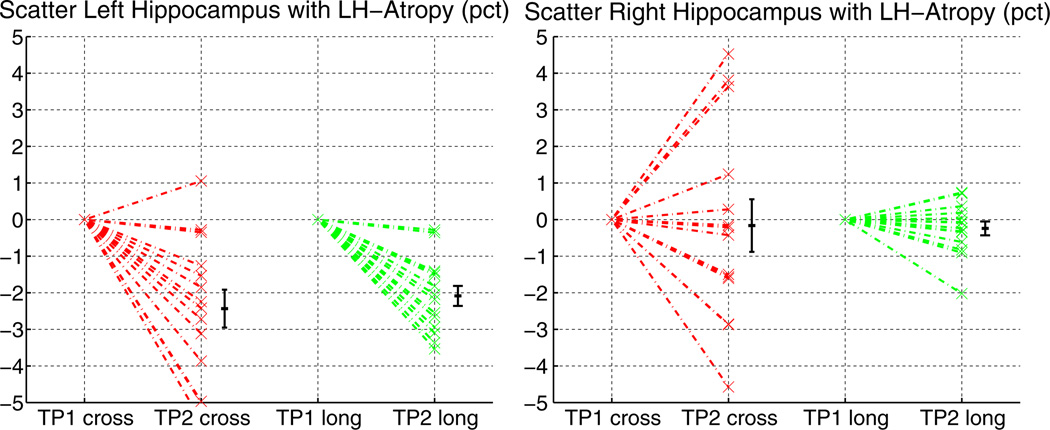
In order to assess precision and accuracy of the longitudinal analysis we applied approximately 2% simulated atrophy to the hippocampus in the left hemisphere and took this synthetic image as the second time point. The atrophy was automatically simulated by reducing intensity in boundary voxels of the hippocampus with partial ventricle volume (labels as reported by FreeSurfer). See Appendix C for details. Note, that real atrophy is more complex and variates among individuals and diseases. The well defined atrophy used here should be accurately detected by any automatic processing method. It can be seen in Fig. 6 that the longitudinal analysis detects the atrophy more precisely and also shows less variability around the zero mean in the right hemisphere. Based on these results neither method can be determined to be biased and both accurately find the ground truth, but longitudinal processing with higher precision.
Fig. 6.
Simulated 2% atrophy in the left hippocampus. The longitudinal processing manages to detect the change more precisely and at the same time reduces the variability in the right hemisphere.
Test–retest reliability
In order to evaluate the reliability of the longitudinal scheme we analyze the variability of the test–retest data sets (focusing on TT-115). Variability of measurements can have several sources. Real anatomical change can occur in controls, e.g. due to dehydration, but is unlikely in within-session scans. More likely, there will be variability due to acquisition procedures (motion artifacts, change of head position inside the scanner etc.). In TT-115, for example, head position has changed significantly as subjects sink into the pillow and relax their neck muscles (mean: 1.05 mm, p < 10−8) during the 12 min diffusion scan separating the test–retest. Acquisition variability is of course identical for both processing methods. Finally there is variability due to the image processing methods themselves, that we aim to reduce.
As a dimensionless measure of variability, similar to Eq. (3), we compute the absolute symmetrized percent change (ASPC) of the volume of a structure with respect to the average volume:
| (4) |
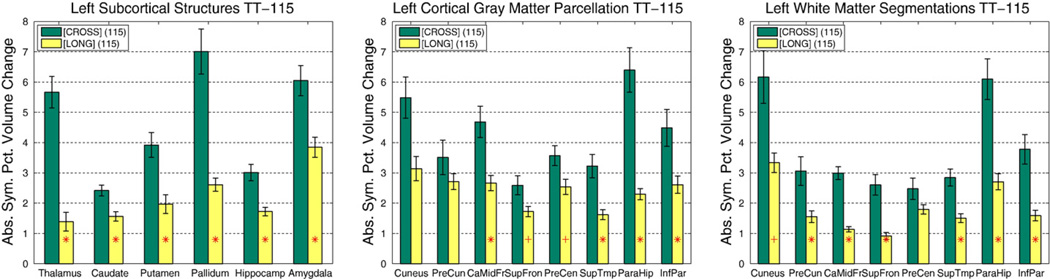
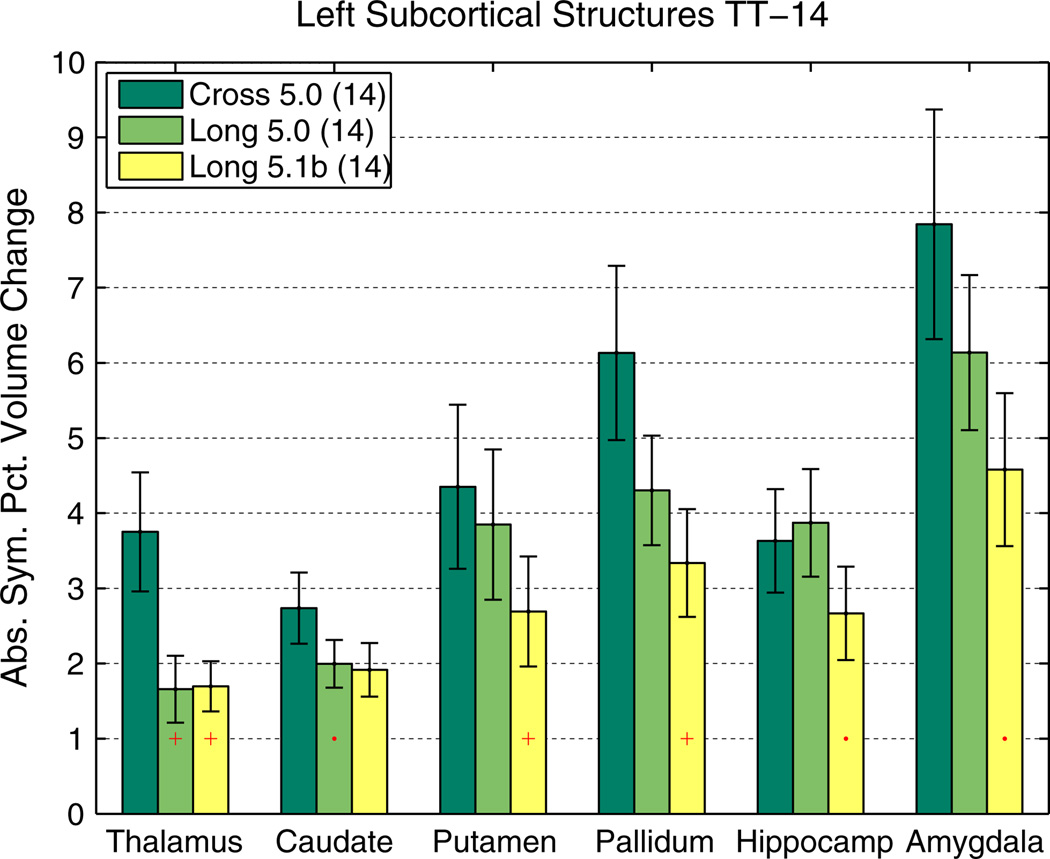
The reason is that estimating a mean (here of ASPC) is more robust than estimating the variance of differences or symmetrized percent change when not taking the absolute value. Fig. 7 shows the reliability of subcortical, cortical and white matter segmentation on the TT-115 data set, comparing the independently processed time points [CROSS] and the longitudinal scheme [LONG]. It can be seen that [LONG] reduces variability significantly in all regions. Morey et al. (2010) also report higher scan–rescan reliability of subcortical volume estimates in our method compared to independent processing in FreeSurfer and compared to FSL/FIRST (Patenaude et al., 2011), even before we switched the longitudinal processing to a common voxel space. Instead of processing time points in their native spaces (Long 5.0, and earlier versions), having all images co-registered to the common template space (Long 5.1b) significantly improves reliability in several structures (see Fig. 8 for a comparison using TT-14).
Fig. 7.
Test–retest comparison of independent [CROSS] versus longitudinal [LONG] processing on TT-115 data (left hemisphere).Subcortical (left), cortical gray matter (middle), and white matter segmentations (right). The mean absolute volume difference (as percent of the average volume) is shown with standard error. [LONG] significantly reduces variability. Red dot: p<0.05, red plus: p<0.01, red star: p<0.001. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
Fig. 8.
Test–retest comparison of [CROSS] versus [LONG] on TT-14 data (subcortical volumes, left hemisphere). See also Fig. 7 for description of symbols. Additionally here reliability improvements of using a common voxel space (Long 5.1b) over previous method (Long 5.0) can be seen.
Note, that often the intraclass correlation coefficient (ICC) (Shrout and Fleiss, 1979) is reported to assess reliability, which is nearly identical to Lin's concordance correlation coefficient (Lin, 1989; Nickerson, 1997), another common reliability measure. Table A.3 in the Appendix, reports ICC among other measures for subcortical volumes in TT-14. We decided to report ASPC here, as it has a more intuitive meaning.
A comparison of Dice coefficients to test for overlap of segmentation labels L1 and L2 at the two time points is reported in Table 1. Dice's coefficient:
| (5) |
measures the amount of overlap with respect to the average size (reaching 1 for perfect overlap). For the Dice computation the segmentation labels need to be aligned across time. In [LONG] the time points are automatically resampled to the subject template space during processing and labels can be directly compared. For the [CROSS] results the same transforms are used with nearest neighbor interpolation on the label map to also align both segmentations in the subject template space for the Dice computation. Therefore it is likely that resampling the label map has an additional detrimental effect on the [CROSS] results due to partial voluming effects. The longitudinal stream improves the Dice in all regions (see Table 1) and in each individual subject (not shown). The reported differences are all significant (p < 0.001 based on the Wilcoxon signed rank test).
Table 1.
Dice coefficient averaged across subjects for subcortical structures for both test–retest data sets (TT-14 and TT-115). Longitudinal processing (L-prefix) improves results significantly in all instances.
| Structure | C-14 | L-14 | C-115 | L-115 |
|---|---|---|---|---|
| L WM | 0.904 | 0.948 | 0.904 | 0.955 |
| R WM | 0.902 | 0.948 | 0.902 | 0.956 |
| L corticalGM | 0.888 | 0.944 | 0.909 | 0.962 |
| R corticalGM | 0.885 | 0.944 | 0.909 | 0.963 |
| Subcortical GM | 0.889 | 0.948 | 0.887 | 0.957 |
| L lat vent | 0.922 | 0.968 | 0.904 | 0.966 |
| R lat vent | 0.916 | 0.966 | 0.896 | 0.964 |
| L hippocampus | 0.872 | 0.933 | 0.875 | 0.948 |
| R hippocampus | 0.870 | 0.936 | 0.880 | 0.952 |
| L thalamus | 0.906 | 0.956 | 0.910 | 0.971 |
| R thalamus | 0.908 | 0.957 | 0.915 | 0.974 |
| L caudate | 0.849 | 0.928 | 0.861 | 0.943 |
| R caudate | 0.840 | 0.928 | 0.858 | 0.944 |
| L putamen | 0.864 | 0.929 | 0.874 | 0.943 |
| R putamen | 0.868 | 0.932 | 0.875 | 0.948 |
| L pallidum | 0.829 | 0.916 | 0.796 | 0.934 |
| R pallidum | 0.830 | 0.927 | 0.800 | 0.937 |
| L amygdala | 0.815 | 0.895 | 0.850 | 0.919 |
| R amygdala | 0.802 | 0.881 | 0.848 | 0.923 |
| 3rd ventricle | 0.860 | 0.949 | 0.868 | 0.957 |
| 4th ventricle | 0.860 | 0.929 | 0.847 | 0.941 |
| L inf lat vent | 0.700 | 0.843 | 0.705 | 0.860 |
| R Inf lat vent | 0.684 | 0.841 | 0.715 | 0.864 |
| Mean | 0.853 | 0.927 | 0.859 | 0.942 |
| StD | 0.062 | 0.034 | 0.058 | 0.030 |
Finally we analyze reliability of cortical thickness maps. In order to compare repeated thickness measures at each vertex, a pointwise correspondence needs to be constructed on each hemisphere within each subject and across time. Similar to Han et al. (2006) the surfaces in [CROSS] are rigidly aligned first, but here using the robust registration of the images as created for the [BASE] (described above). In [LONG], images and thus surfaces are in the same geometric space across time and don't need to be aligned. Then, to construct correspondence on the surfaces, the nearest neighbor for each vertex is located on the neighboring surface. This is done for both [CROSS] and [LONG] to treat them the same for a fair comparison, in spite of the fact that [LONG] implicitly creates a point-wise correspondence of surfaces across time. The nearest neighbor approach makes use of the fact that surfaces are very close, which can be assumed for the same subject across time in this test–retest study, and thus avoids the complex nonlinear spherical registration, that is commonly used when registering surfaces across subjects.
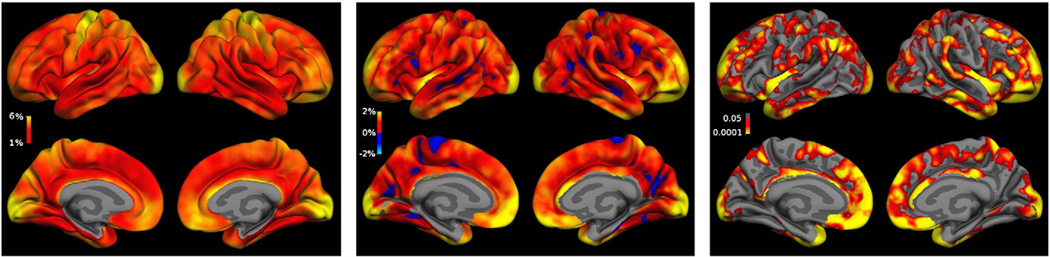
Fig. 9 (left) depicts the average vertex wise ASPC as a measure of variability in [CROSS] (in yellow regions at 6%). Still the thickness differences between the two time points is mainly around 0.1 mm and rarely above 0.2 mm (not shown) and therefore smaller than in Han et al. (2006), which can be expected as the processing methods have improved and the TT-115 data is higher quality (multi-echo MPRAGE 3T as compared to MPRAGE 1.5T, volume coil). Fig. 9 (middle) shows plots of the difference ([CROSS]-[LONG]) of absolute symmetrized percent change for each vertex averaged across all subjects (smoothing kernel with 15 mm full width at half maximum). The orange/yellow color indicates regions where the longitudinal stream improves reliability, most prominently in the frontal and lateral cortex (where improvements are more than 4%). Dark red and blue regions are small magnitudes and basically noise. They have not been clipped to present the full picture and are not significant. The false discovery rate corrected (at p = 0.05) significance maps in Fig. 9 (right) demonstrate that only the improvements are significant.
Fig. 9.
Left: Average absolute symmetrized percent thickness change at each vertex for TT-115 using [CROSS]. Some regions (yellow) show 6% ASPC and above. Middle: Comparison: ([CROSS]-[LONG]) of average absolute symmetrized percent thickness change at each vertex for TT-115. Blue: [LONG] has larger variability, red/yellow [CROSS] has larger variability. [LONG] improves reliability in most regions, especially in the frontal and lateral cortex (yellow: more than 2% reduction of variability, frontal and lateral even more than 4%). Blue and red regions are mainly noise. Right: corresponding significance map, false discovery rate corrected at 0.05. [LONG] improves reliability significantly in most regions.
Improvements in reliability are summarized in Table 2, showing the average of the absolute symmetrized percent change in each hemisphere across all subjects. LONG(cor) is based on the implicit correspondence across time constructed in the longitudinal steam, and LONG(reg) uses nearest neighbor registration. [LONG] improves reliability significantly. The difference of [LONG] (either version) and [CROSS] yields a p < 0.001 in the Wilcoxon signed rank test. Furthermore, nearest neighbor surface registration (reg) in [LONG] and the implicit correspondence constructed by the longitudinal stream (cor) yields almost the same results. The remaining difference is significant only for TT-115 (at p < 0.01 based on Wilcoxon signed rank test). Note, that the nearest neighbor registration (reg) is used here for a fair comparison of the test–retest results across methods ([CROSS] vs. [LONG]) and is not recommended in general, as it is not constrained along the surface. Surfaces may move due to atrophy, potentially causing the nearest neighbor approach to incorrectly pair vertices from different sides of a sulcus.
Table 2.
Average across subjects of the average vertex wise absolute symmetrized percent thickness change between time points 2 and 1 for the different methods in both data sets. Skipping the surface registration (reg) in [LONG] and using the implicit correspondence across time constructed by the longitudinal stream (cor) yields similar results.
| Hemi | CROSS(reg) | LONG(reg) | LONG(cor) |
|---|---|---|---|
| L TT-14 | 4.04 | 3.39 | 3.44 |
| R TT-14 | 4.60 | 3.76 | 3.80 |
| L TT-115 | 4.00 | 3.21 | 3.26 |
| R TT-115 | 4.07 | 3.29 | 3.33 |
Sample size reduction
The lower variability in the longitudinal processing is particularly important for detecting small effects, such as in drug trials, or for studies with a small number of subjects.
Instead of reporting an exemplary power analysis, we will more generally provide the fraction of subjects needed in the [LONG] method compared to [CROSS]. The reason is that such fractions will be valid independent of the specific underlying parameters, which can differ depending on the specific situation, e.g. number of time points, their variance, effect size, p-value, power. According to Diggle et al. (2002) longitudinal sample size calculations of a continuous response for detecting a difference in the rate of change across time in two groups (each containing m subjects) are usually of the form:
| (6) |
where σ2 denotes the assumed common variance measuring the unexplained variability in the response,
ρ the correlation of the repeated observations,
N the number of time points (assumed to contain no missing values and to be spaced the same for all subjects),
zp the pth quantile of a standard Gaussian distribution,
α the type I error rate (the probability to reject the null hypothesis when it is correct),
d = βB−βA smallest meaningful difference in the mean slope (rate of change) between group A and B to be detected (effect size),
P the power of the test (the probability to reject the null hypothesis when it is incorrect),
and sx = Σj (xj−x̄)2 /N the within-subject variance of the time points (more specifically of the explanatory variable, usually the duration between the first and the j-th visit, assumed to be the same for all subjects).
Eq. (6) shows that the sample size can be reduced by increasing the number of time points N, by increasing the correlation of repeated measures or by reducing the variability of the response. As the variability between subjects is usually fixed and cannot be influenced, one aims at decreasing within-subject variability by using a more reliable measurement instrument or method. To analyze the effect of switching from independent image processing [CROSS] to longitudinal processing [LONG], it becomes clear that all values except σ and ρ are fixed for the two different methods and the requisite number of subjects decreases with decreasing variance and increasing correlation. For a power analysis usually these values are estimated from earlier studies with similar samples. Here we can compute them based on the test–retest results TT-14, as 14 days between the scans sufficiently model the variability of follow-up scans on a different day (using the same scanner).
The fraction of necessary sample sizes when choosing [LONG] over [CROSS] is determined by the fraction of variances and correlation:
| (7) |
This ratio specifies what percent of subjects is needed when processing the data longitudinally as opposed to independent processing.
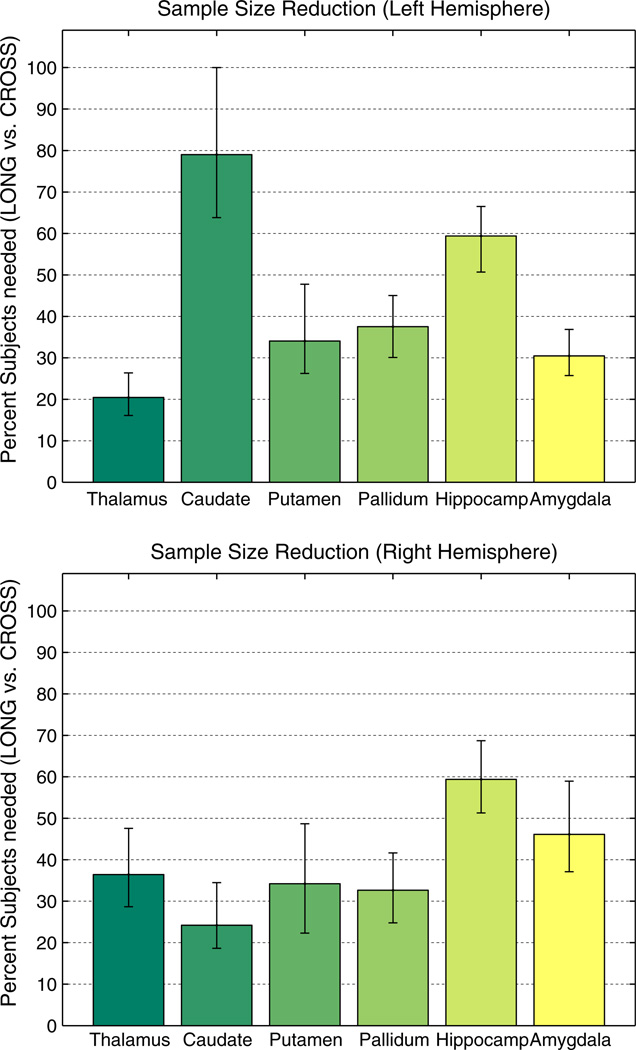
Based on variance and correlation results from TT-14, Fig. 10 shows the ratio for several subcortical regions. Given this single test population to compute variance and correlation, we estimate stability of these results via bootstrapping (1000 resamples). Fig. 10 therefore depicts the median, the error bars extend from the 1st to the 3rd quartile. The results indicate that sample size can usually be reduced in [LONG] to less than half the size assuming same power, p-value, effect size and number and variance of time points. The small reduction in the left caudate is due to the fact that in [CROSS] the correlation of the measures is very high and almost the same as in [LONG], which is not true for the other hemisphere and most of the other structures where the correlation in [CROSS] is usually much lower. Even larger improvements can be expected when switching to modern acquisition hardware and methods, for example as used in the TT-115 dataset (see improvements in Fig. 7). However, we cannot base this sample size estimation onTT-115 since within-session scans do not model the noise induced by removing and re-positioning a subject in the scanner, nor variability due to hydration levels, etc.
Fig. 10.
Percent of subjects needed in [LONG] with respect to [CROSS] to achieve same power at same p to detect same effect size. In most regions less than 40% of the subjects are needed. Equivalently this figure shows the reduction in necessary time points when keeping the number of subjects and within-subject variance of time points the same. Variance of measurements and correlation were estimated based on TT-14 using bootstrap with 1000 samples. Bars show median and error bars depict 1st and 3rd quartile.
Note that the number of subjects m and the number of time points n can be swapped in Eq. (6), thus Fig. 10 can also be understood as showing the reduction in the necessary number of time points in a longitudinal design when keeping the number of subjects (and variance of time points) constant. The reduced number of subjects or necessary time points in the longitudinal stream can constitute a significant reduction in costs for longitudinal studies such as drug trials. Several other relevant statistics on TT-14 are reported in Table A.3 in the Appendix for different structures to establish individual power analyses. Of course these results are specific to the acquisition in TT-14 and may not be transferable to other studies.
Sensitivity and specificity in neurodegeneration
Since no longitudinal data set with manual labels is freely available that could be taken as “ground truth”, we analyze a set of images of different disease groups and demonstrate that longitudinal processing improves discrimination among the groups. Here we are interested in detecting differences in the yearly volume percent change.
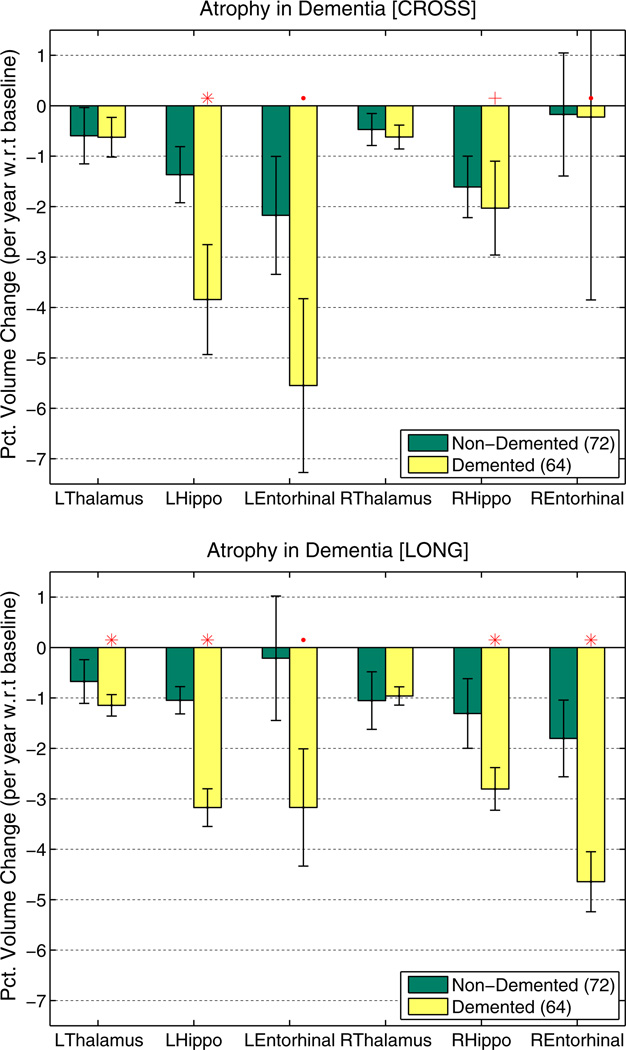
The longitudinal OASIS dataset OA-136 was selected to analyze behavior of the processing streams in a disease study where subjects have differently many visits (2–5). Fig. 11 highlights the improvements of longitudinal processing: more power due to higher precision to distinguish the demented from the non-demented group based on the percent volume change with respect to baseline volume mainly in the hippocampus and entorhinal cortex. Baseline volume was not taken directly from the results of the first time point, but instead we used the value of the linear fit within each subject at baseline to obtain more robust baseline volume estimates for the percent change computation (for both [CROSS] and [LONG]). Again the red ‘.’ denotes a p ≤ 0.05, the ‘+’: p ≤ 0.01 and the ‘*’: p ≤ 0.001 in the Mann–Whitney-U (also Wilcoxon rank-sum) test. Note that the Mann–Whitney-U test is closely related to the area under the Receiver Operator Characteristic (ROC) (Mason and Graham, 2002). For a binary classifier the ROC curve plots the sensitivity vs. the false positive rate (1-specificity). The area under the curve therefore measures the performance of the classifier. Thus the significant differences across the groups above imply both improved sensitivity and specificity to distinguish the different disease stages based on atrophy rate.
Fig. 11.
Percent volume change per year with respect to baseline of the OA-136 dataset (2 to 5 visits per subject) for both independent [CROSS] (top) and longitudinal [LONG] (bottom) processing. [LONG] shows greater power to distinguish the two groups and smaller error bars (higher precision).
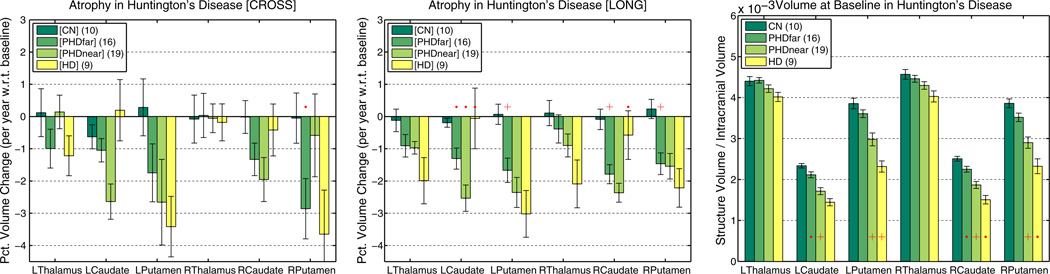
The other disease data set HD-54 was selected as it describes a small study with images from different scanner software versions, where statistical power is relatively low. Fig. 12 (left and middle) shows plots of percent change averages (and standard errors) for thalamus, caudate and putamen in both hemispheres. Percent change is computed with respect to the baseline volume here, where baseline volume is taken from the linear fit within each subjects as a more robust estimate. For the PHDfar we test difference from controls, for PHDnear difference from PHDfar and for the HD difference from PHDnear. Because of the large variability in the measurements, the cross-sectional stream cannot distinguish well between the groups. [LONG], however, is capable of differentiating PHDfar from controls based on atrophy rates in the caudate and putamen and PHDfar from PHDnear based on the left caudate. Caudate and putamen are structures that are affected very early (in PHDfar more than 11 years from expected onset of symptoms) while other structures such as the thalamus seem to be affected later in the disease and show a faster atrophy rate in the HD group. In HD the small atrophy rate in the caudate seem to indicate a floor effect (or difficulties with the automatic segmentation as most of the caudate is lost).
Fig. 12.
Symmetric percent volume change per year of several subcortical structures. Left: [CROSS] almost no significant differences due to high variability (small group sizes). Middle: [LONG] significant differences between pre-symptomatic (PHD far from onset) and controls and between PHD far and PHD near (left caudate). Right: Volume means (ICV normalized) at baseline (tp1). While baseline volume distinguishes groups in several cases, the significant difference between controls and PHDfar in atrophy rate in the putamen cannot be detected in the baseline volumes.
To visualize group volume differences Fig. 12 (right) depicts the mean volumes of thalamus, caudate and putamen at baseline (tp1) after intracranial volume (ICV) normalization. Even though here we analyze volume at a single time point, each structure's volume and ICV are taken from the results obtained via longitudinal processing and should therefore be more robust than independent processing. Due to large between-subject variability in anatomical structures, it is often not possible to distinguish groups simply based on structure size (even after head size correction). In longitudinal studies, however, the additional temporal information within each subject (atrophy rate) is computed with respect to average or baseline structure size (i.e. percent change) within each subject. This removes between subject variability and, at the same time, increases power to distinguish groups based on the anatomical change in addition to size. For example the atrophy rate in the putamen differs significantly between controls and pre-symptomatic subjects far from disease onset, while baseline putamen volume does not.
Conclusion
The robust subject template yields an initial unbiased estimate of the location of anatomical structures in a longitudinal scheme. We demonstrated that initializing processing of individual time points with common information from the subject template improves reliability significantly as compared to independent processing. Furthermore, our approach to treat all inputs the same removes asymmetry induced processing bias. This is important as the special treatment of a specific time point such as baseline, e.g. to inform follow-up processing, induces bias even in the absence of resampling asymmetries. Moreover we avoid imposing regularization or temporal smoothness constraints to keep the necessary flexibility for detecting large deviations. Therefore, in our framework, the order of time points is not considered at all and individual segmentation and deformation procedures are allowed to evolve freely. This reduces over-regularization and thus the risk of consistently underestimating change.
We have shown, that our method significantly improves precision of the automatically constructed segmentations with respect to volume and location, and of the cortical thickness measures. Thus, statistical power is increased, i.e. the necessary number of subjects or time points reduced (at same effect size and significance level). This may have a profound clinical impact particularly in drug trials where small effect sizes need to be detected or disease processes quantified early in the course, when therapeutic intervention is still possible. The presented methodology is capable of precisely and accurately detecting differences as demonstrated in simulated hippocampal atrophy and in evaluating complex, subtle changes that occur in neurodegenerative disorders.
A common challenge of longitudinal analyses is change in scanner hardware or software. Due to scanner drift and calibration, images cannot be assumed to be sized exactly the same. Any change in preprocessing can bias results and potentially void a study trying to establish absolute measures such as the rate of change in a specific disease. Group comparisons may still be possible, if both groups underwent the same processing changes, but even then it is likely that the processing change influences one group more than the other and that influences are regional. To account for calibration effects, we include optional affine template creation into our framework. However, potential image contrast changes cannot easily be removed retrospectively. This is of course true for both longitudinal and independent processing, where variability will be even higher. A consistent change in input images, independent of the source, is supposed to be detected by an accurate and precise analysis tool. Longitudinal methods may actually reveal these kinds of consistent acquisition differences, because they are more sensitive and need less subjects to detect them. It is therefore essential to control scanner hardware and software or to model upgrades as potential shifts when running a statistical analysis on the results.
Future work will include procedures to jointly estimate or optimize results in all time points simultaneously without necessarily relying on the subject template. In unbiased simultaneous processing memory usage is scaled at least linearly by the number of time points (Gaussian classifiers scale with the square if full covariance estimation is used), which implies that hardware requirements may not be met by standard desktop computers. However, this is a direction we intend to pursue. For example, a joint intensity normalization and bias correction can employ all the imaging data across time to estimate a high SNR image at each time point while retaining regions that display temporal change. It is also possible to generate an unbiased initial estimate of the average surface locations for both the gray/white and the pial surfaces by minimizing the distance to each of the individual cross-sectional surfaces directly. Furthermore, it is expected that variational approaches for the thickness computation in each time point will improve reliability compared to the current method, which estimates and averages the shortest distance from the gray to white matter surface and vice versa.
The presented longitudinal scheme is freely available in the software package FreeSurfer at www.freesurfer.net and has been successfully applied in our lab and by others in various studies of e.g. Huntington's, Alzheimer's disease and aging.
Acknowledgments
Support for this research was provided in part by the National Center for Research Resources (P41RR14075, and the NCRR BIRN Morphometric Project BIRN002, U24RR021382), the National Institute for Biomedical Imaging and Bioengineering (R01EB006758), the National Institute on Aging (R01AG022381, U01AG024904), the National Institute for Neurological Disorders and Stroke (R01NS052585, R01NS042861, P01NS058793, R21NS072652, R01NS070963). Additional support was provided by The Autism & Dyslexia Project funded by the Ellison Medical Foundation, the National Center for Alternative Medicine (RC1AT005728), and was made possible by the resources provided by Shared Instrumentation Grants (S10RR023401, S10RR019307, S10RR023043). For the OASIS dataset we acknowledge the following grants (P50AG05681, P01AG03991, R01AG021910, P50MH071616, U24RR021382, R01MH56584). The authors would also like to thank Mert Sabuncu and Gheorghe Doros for helpful discussions and advice.
Appendix A. Probabilistic fusion
For the following discussion we assume for each subject the N normalized and skull stripped images {Ii} to be registered and resampled to the common template space, together with their initial label maps {Li}, i = 1, …,N, where we use nearest neighbor interpolation. The goal is to construct a fused segmentation L̃i for each time point containing the label with the highest probability at each location based on all inputs {Ii} and initial segmentations {Li}. This procedure is designed for N ≥ 3 time points, for N = 2 it reduces to selecting the label from the initial label map of the specific time point. Here we just discuss one selected time point and call its image I without subscript, note that it is an element of the set {Ii} although we assume it was generated from all inputs later:
| (A.1) |
| (A.2) |
| (A.3) |
where Ω denotes the set of all voxels and assuming that the labels at each voxel are conditionally independent from each other. This allows us to work on each voxel separately. By assuming that the current image I is generated with equal probability from the {Ii} and dropping 1/N we obtain:
| (A.4) |
| (A.5) |
| (A.6) |
where we further assumed I and L to be conditionally independent (not meaning independent, see Sabuncu et al., 2010). We specify a simple voting model for the label prior:
| (A.7) |
While for the image likelihood we choose a normal distribution with stationary variance σ2:
| (A.8) |
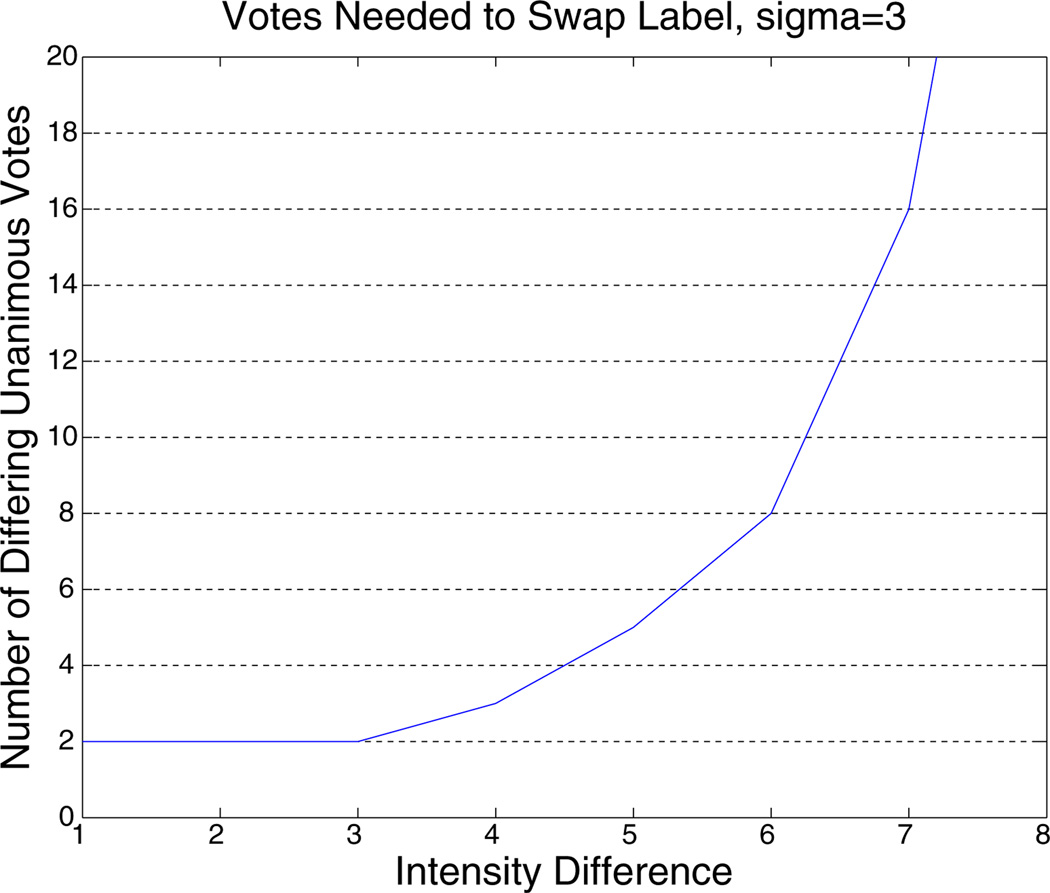
The above procedure basically describes a kernel density estimation with a Gauss kernel (Parzen window). Each initial label map votes on the label based on the intensity difference to the current image. With constant image likelihood (σ→∞) this reduces to majority voting. The image likelihood that we use above can be seen as temporal smoothing of the label maps. In the kernel density estimation the free smoothing parameter σ is called the bandwidth and has been selected to be σ= 3 here based on the possible intensity values and noise level. This value is relative conservative, for example it needs 5 time points that agree on a different label and have an intensity difference of at most 5 out of 0…255 to convince a label to switch over. For larger intensity differences this number rapidly increases, see Fig. A.13.
Fig. A.13.
Votes that need to agree on a different label to convince a time point to swap at σ= 3 for a given intensity difference.
Note that, as mentioned above, the fused segmentation L̃ needs to be constructed for each time point. It is not the final segmentation, but is used only to initialize the segmentation algorithm in the longitudinal processing. We presented the probabilistic approach above to highlight what design choices have been made. Possible modifications are:
to use a non rigid higher dimensional warp to align the images for this purpose.
to allow for different probabilities of the {Ii}, for example, to account for distances in time and/or neighborhood intensities.
to employ a more complex model for the label prior, e.g., based on neighboring voxels or the signed distance transform.
However, since the fused segmentation is only an initialization, the above approach is sufficient in that it allows flexibility of detecting large change across time, as opposed to using the segmentation of the template to initialize all time points.
Appendix B. Mean space
Here we discuss, how to compute the average location from a set of N-1 rigid transformations (step 2 in Improved template estimation section). For this a new coordinate system is defined with its origin at the center of the random target image Ĩ with axes aligned to the right, anterior, superior directions (RAS). The first step is to find the location and orientation (translation and rotation) of each of the other images in this space, so that the average location can be computed.
A rigid transformation consists of a rotation and translation and is usually written as φ (x⃗) ≔ Rx⃗ + t⃗, where R is a 3 × 3 rotation matrix. R and t⃗ are returned when registering image Ii to image Ĩ. Equivalently the order can be reversed, so that the translation will be executed before the rotation:
| (B.1) |
Note that the inverse of the rotation matrix is simply the transpose R−1 = RT. The rotation remains the same, while the translation becomes (registering image i to the first via transform φi). The − v⃗i directly give the translation offset of each image with respect to image Ĩ (located at the origin). Therefore the average:
| (B.2) |
marks the mean of all locations. For rotations different averages can be defined (Moakher, 2002; Sharf et al., 2010). Since rotational differences are rather small, it will be sufficient to compute the projected arithmetic mean. This is the usual arithmetic mean of 3 × 3 matrices, orthogonally projected back to SO (3), the space of rotation matrices, via a polar decomposition. To find the rotation from Ĩ to the average position, the inverse rotations obtained from the registration above are averaged:
| (B.3) |
Note that both sums run over all images, where the translation of Ĩ with respect to itself is of course zero and the rotation matrix is identity. Since the matrix mean Q is not a rotation matrix in general, its polar decomposition Q = US into an orthogonal rotation matrix U and a symmetric matrix S needs to be computed. S is always unique and given by . Because the head positions in the images are sufficiently close to each other,5 Q is invertible and then U is also unique. It can be computed through a singular value decomposition of Q = WDVT and is given6 by U = WVT.
Once the mean location p⃗ and orientation U are determined, we construct the transform φ̂ (x⃗) ≔ U x⃗ + p⃗ from image Ĩ to the average location. The other transforms of each image to the average location are then created by composition φ̂i ≔ φ̂∘φi. All images are averaged at that location and serve as high quality input to the intrinsic mean algorithm.
Appendix C. Simulated atrophy
In order to simulate atrophy in the hippocampus (see Robustness, precision and accuracy section) we reduce the intensity of boundary voxels adjacent to ventricle CSF. Let H denote the set of all hippocampus voxels and B the subset of boundary voxels containing partial ventricle, then we have Vhippo (B) = Σx∈B Vhippo (x) the sum of partial hippocampus volumes for all voxels in B, and total hippocampal volume Vhippo (H). For atrophy rate p (here p = 0.02) we compute the boundary scaling factor as:
| (C.1) |
necessary for adjusting partial volume of boundary voxels to achieve the desired volume reduction. For this we first estimate local mean hippocampus intensity IH (x) and ventricle intensity IV(x) in a 153 box centered at x. Then we compute the partial hippocampal volume at x
| (C.2) |
and update intensity according to
| (C.3) |
Table A.3
Statistics based on test–retest data (14 subjects, two time points). Columns: mean and standard deviation at both time points, correlation across time, intraclass correlation coefficient icc(2,1), standard deviation of the difference (tp2 − tp1),standard deviation of the symmetrized percent change (diff/avg). The icc and std of SPC are reported also for cross-sectional processing to show improvements of the longitudinal stream.
| Volume stats. | [LONG]: | [CROSS]: | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Structure | Mean 1 | std 1 | Mean 2 | std 2 | corr | icc | std (diff) | std (spc) | icc | std (spc) |
| L corticalGM | 192,866 | 20,512 | 190,918 | 20,157 | 0.987 | 0.983 | 3336.69 | 1.760 | 0.983 | 2.070 |
| R corticalGM | 194,939 | 21,068 | 192,368 | 21,966 | 0.978 | 0.972 | 4571.14 | 2.449 | 0.971 | 2.825 |
| L thalamus | 5680.7 | 616.4 | 5672.4 | 583.1 | 0.979 | 0.979 | 128.03 | 2.153 | 0.873 | 4.861 |
| R thalamus | 5733.6 | 544.5 | 5705.0 | 526.9 | 0.951 | 0.953 | 167.89 | 2.841 | 0.841 | 4.810 |
| L caudate | 3224.4 | 378.9 | 3197.7 | 393.1 | 0.982 | 0.980 | 74.55 | 2.208 | 0.967 | 2.558 |
| R caudate | 3333.0 | 440.2 | 3331.3 | 443.5 | 0.993 | 0.994 | 51.56 | 1.551 | 0.971 | 3.056 |
| L putamen | 4664.1 | 898.9 | 4694.7 | 865.4 | 0.985 | 0.985 | 155.35 | 3.808 | 0.951 | 6.048 |
| R putamen | 4534.5 | 701.4 | 4536.1 | 729.4 | 0.991 | 0.991 | 98.15 | 2.266 | 0.962 | 3.536 |
| L pallidum | 1632.2 | 219.4 | 1625.4 | 196.1 | 0.943 | 0.941 | 73.99 | 4.372 | 0.823 | 7.320 |
| R pallidum | 1442.9 | 205.8 | 1436.6 | 205.0 | 0.948 | 0.951 | 65.96 | 4.534 | 0.861 | 8.158 |
| L hippocampus | 3075.2 | 384.2 | 3089.6 | 390.2 | 0.958 | 0.960 | 112.07 | 3.583 | 0.922 | 4.537 |
| R hippocampus | 3173.1 | 421.8 | 3190.0 | 438.8 | 0.981 | 0.981 | 85.14 | 2.781 | 0.964 | 3.352 |
| L amygdala | 1142.8 | 150.0 | 1167.0 | 176.5 | 0.923 | 0.907 | 69.06 | 5.801 | 0.776 | 9.810 |
| R amygdala | 1168.9 | 185.7 | 1161.4 | 183.4 | 0.933 | 0.937 | 67.48 | 5.967 | 0.860 | 7.954 |
Footnotes
See Berry and Ayers (2006) for advantages (such as symmetry and increase of power) of using the average for normalization rather than the volume at baseline to compute the percent change.
i.e. sufficiently less than 180° away from each other to prevent the average from becoming ill conditioned or even singular.
using VTV = WTW = I
References
- Ashburner J. A fast diffeomorphic image registration algorithm URL. NeuroImage. 2007;38(1):95–113. doi: 10.1016/j.neuroimage.2007.07.007. http://www.sciencedirect.com/science/article/pii/S1053811907005848. [DOI] [PubMed] [Google Scholar]
- Ashburner J, Friston K. Nonlinear spatial normalization using basis functions. Hum. Brain Mapp. 1999;7(4):254–266. doi: 10.1002/(SICI)1097-0193(1999)7:4<254::AID-HBM4>3.0.CO;2-G. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashburner J, Andersson J, Friston K. Image registration using a symmetric prior–in three dimensions. Hum. Brain Mapp. 2000;9(4):212–225. doi: 10.1002/(SICI)1097-0193(200004)9:4<212::AID-HBM3>3.0.CO;2-#. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avants B, Gee JC. Geodesic estimation for large deformation anatomical shape averaging and interpolation. NeuroImage. 2004;23(1):139–150. doi: 10.1016/j.neuroimage.2004.07.010. [DOI] [PubMed] [Google Scholar]
- Avants B, Anderson C, Grossman M, Gee JC. Spatiotemporal normalization for longitudinal analysis of gray matter atrophy in frontotemporal dementia. Medical Image Computing and Computer-Assisted Intervention - MICCAI 2007: Vol. 4792 of Lecture Notes in Computer Science. 2007:303–310. doi: 10.1007/978-3-540-75759-7_37. [DOI] [PubMed] [Google Scholar]
- Avants B, Cook PA, McMillan C, Grossman M, Tustison NJ, Zheng Y, Gee JC. Sparse unbiased analysis of anatomical variance in longitudinal imaging. MICCAI 2010, Part I. Vol. 6361 of LNCS. Springer. 2010:324–331. doi: 10.1007/978-3-642-15705-9_40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berry DA, Ayers GD. Symmetrized percent change for treatment comparisons. Am. Stat. 2006;60:27–31. [Google Scholar]
- Chételat G, Landeau B, Eustache F, Mézenge F, Viader F, de la Sayette V, Desgranges B, Baron J-C. Using voxel-based morphometry to map the structural changes associated with rapid conversion in MCI: a longitudinal MRI study. NeuroImage. 2005;27(4):934–946. doi: 10.1016/j.neuroimage.2005.05.015. URL http://www.sciencedirect.com/science/article/pii/S1053811905003277. [DOI] [PubMed] [Google Scholar]
- Chiang GC, Insel PS, Tosun D, Schuff N, Truran-Sacrey D, Raptentsetsang S, Jack CR, Aisen P, Petersen RC, Weiner MW ADNI. Hippocampal atrophy rates and CSF biomarkers in elderly APOE2 normal subjects. Neurology. 2010;75(22):1976–1981. doi: 10.1212/WNL.0b013e3181ffe4d1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiang GC, Insel PS, Tosun D, Schuff N, Truran-Sacrey D, Raptentsetsang S, Jack CR, Weiner MW ADNI. Identifying cognitively healthy elderly individuals with subsequent memory decline by using automated MR temporoparietal volumes. Radiology. 2011;259(3):844–851. doi: 10.1148/radiol.11101637. URL http://dx.doi.org/10.1148/radiol.11101637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarkson MJ, Ourselin S, Nielsen C, Leung KK, Barnes J, Whitwell JL, Gunter JL, Hill DL, Weiner MW, Jack CR, Jr, Fox NC. Comparison of phantom and registration scaling corrections using the adni cohort. NeuroImage. 2009;47(4):1506–1513. doi: 10.1016/j.neuroimage.2009.05.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collignon A, Maes F, Delaere D, Vandermeulen D, Suetens P, Marchal G. Information Processing in Medical Imaging. Kluwer; 1995. Automated multi-modality image registration based on information theory; pp. 263–274. [Google Scholar]
- Dale A, Fischl B, Sereno MI. Cortical surface-based analysis: I. segmentation and surface reconstruction. NeuroImage. 1999;9(2):179–194. doi: 10.1006/nimg.1998.0395. [DOI] [PubMed] [Google Scholar]
- Desikan RS, Ségonne F, Fischl B, Quinn BT, Dickerson BC, Blacker D, Buckner RL, Dale AM, Maguire RP, Hyman BT, Albert MS, Killiany RJ. An automated labeling system for subdividing the human cerebral cortex on MRI scans into gyral based regions of interest. NeuroImage. 2006;31(3):968–980. doi: 10.1016/j.neuroimage.2006.01.021. [DOI] [PubMed] [Google Scholar]
- Desikan RS, Sabuncu MR, Schmansky NJ, Reuter M, Cabral HJ, Hess CP, Weiner MW, Biffi A, Anderson CD, Rosand J, Salat DH, Kemper TL, Dale AM, Sperling RA, Fischl B ADNI. Seletive disruption of the cerebral neocortex in Alzheimer's disease. PLoS One. 2010;5(9):e12853. doi: 10.1371/journal.pone.0012853. URL http://dx.doi.org/10.1371/journal.pone.0012853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diggle PJ, Heagerty PJ, Liang K-Y, Zeger SL. Analysis of Longitudinal Data. 2nd Edition. Oxford University Press; 2002. [Google Scholar]
- Engvig A, Fjell AM, Westlye LT, Moberget T, Sundseth Ø, Larsen VA, Walhovd KB. Effects of memory training on cortical thickness in the elderly. NeuroImage. 2010;52(4):1667–1676. doi: 10.1016/j.neuroimage.2010.05.041. [DOI] [PubMed] [Google Scholar]
- Fischl B. FreeSurfer. NeuroImage. doi: 10.1016/j.neuroimage.2012.01.021. in press. URL http://dx.doi.org/10.1016/j.neuroimage.2012.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischl B, Dale AM. Measuring the thickness of the human cerebral cortex from magnetic resonance images. Proc. Natl. Acad. Sci. U. S. A. 2000;97(20):11050–11055. doi: 10.1073/pnas.200033797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischl B, Sereno MI, Dale A. Cortical surface-based analysis: II. Inflation, flattening, and a surface-based coordinate system. NeuroImage. 1999a;9(2):195–207. doi: 10.1006/nimg.1998.0396. [DOI] [PubMed] [Google Scholar]
- Fischl B, Sereno MI, Tootell RB, Dale AM. High-resolution intersubject averaging and a coordinate system for the cortical surface. Hum. Brain Mapp. 1999b;8(4):272–284. doi: 10.1002/(SICI)1097-0193(1999)8:4<272::AID-HBM10>3.0.CO;2-4. URL http://dx.doi.org/10.1002/(SICI)1097-0193.1999.8.4<272::AID-HBM10>3.0.CO;2-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischl B, Liu A, Dale AM. Automated manifold surgery: constructing geometrically accurate and topologically correct models of the human cerebral cortex. IEEE Trans. Med. Imaging. 2001;20(1):70–80. doi: 10.1109/42.906426. [DOI] [PubMed] [Google Scholar]
- Fischl B, Salat DH, Busa E, Albert M, Dieterich M, Haselgrove C, van der Kouwe A, Killiany R, Kennedy D, Klaveness S, Montillo A, Makris N, Rosen B, Dale AM. Whole brain segmentation: automated labeling of neuroanatomical structures in the human brain. Neuron. 2002;33(3):341–355. doi: 10.1016/s0896-6273(02)00569-x. [DOI] [PubMed] [Google Scholar]
- Fischl B, van der Kouwe A, Destrieux C, Halgren E, Ségonne F, Salat DH, Busa E, Seidman LJ, Goldstein J, Kennedy D, Caviness V, Makris N, Rosen B, Dale AM. Automatically parcellating the human cerebral cortex. Cereb. Cortex. 2004a;14(1):11–22. doi: 10.1093/cercor/bhg087. [DOI] [PubMed] [Google Scholar]
- Fischl B, Salat DH, van der Kouwe A, Makris N, Ségonne F, Quinn BT, Dale AM. Sequence-independent segmentation of magnetic resonance images. NeuroImage. 2004b;23(Suppl. 1):69–84. doi: 10.1016/j.neuroimage.2004.07.016. [DOI] [PubMed] [Google Scholar]
- Fletcher PT, Venkatasubramanian S, Joshi S. The geometric median on Riemannian manifolds with application to robust atlas estimation. NeuroImage. 2009;45(Suppl. 1)(1):143–152. doi: 10.1016/j.neuroimage.2008.10.052. mathematics in Brain Imaging. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han X, Jovicich J, Salat D, van der Kouwe A, Quinn B, Czanner S, Busa E, Pacheco J, Albert M, Killiany R, Maguire P, Rosas D, Makris N, Dale A, Dickerson B, Fischl B. Reliability of MRI-derived measurements of human cerebral cortical thickness: the effects of field strength, scanner upgrade and manufacturer. NeuroImage. 2006;32(1):180–194. doi: 10.1016/j.neuroimage.2006.02.051. [DOI] [PubMed] [Google Scholar]
- Holland D, Dale AM. Nonlinear registration of longitudinal images and measurement of change in regions of interest. Med. Image Anal. 2011;15(4):489–497. doi: 10.1016/j.media.2011.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holland D, McEvoy LK, Dale AM ADNI. Unbiased comparison of sample size estimates from longitudinal structural measures in ADNI. Hum. Brain Mapp. 2011 doi: 10.1002/hbm.21386. online URL http://dx.doi.org/10.1002/hbm.21386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenkinson M, Bannister PR, Brady JM, Smith SM. Improved optimization for the robust and accurate linear registration and motion correction of brain images. NeuroImage. 2002;17:825–841. doi: 10.1016/s1053-8119(02)91132-8. [DOI] [PubMed] [Google Scholar]
- Joshi S, Davis B, Jomier BM, Gerig G. Unbiased diffeomorphic atlas construction for computational anatomy. NeuroImage. 2004;23:151–160. doi: 10.1016/j.neuroimage.2004.07.068. [DOI] [PubMed] [Google Scholar]
- Kipps CM, Duggins AJ, Mahant N, Gomes L, Ashburner J, McCusker EA. Progression of structural neuropathology in preclinical huntington's disease: a tensor based morphometry study. J. Neurol. Neurosurg. Psychiatry. 2005;76(5):650–655. doi: 10.1136/jnnp.2004.047993. URL http://jnnp.bmj.com/content/76/5/650.full.pdf. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langbehn D, Brinkman R, Falush D, Paulsen J, Hayden M. A new model for prediction of the age of onset and penetrance for Huntington's disease based on CAG length. Clin. Genet. 2004;65(4):267–277. doi: 10.1111/j.1399-0004.2004.00241.x. URL http://dx.doi.org/10.1111/j.1399-0004.2004.00241.x. [DOI] [PubMed] [Google Scholar]
- Li Y, Wang Y, Xue Z, Shi F, Lin W, Shen D. Consistent 4D cortical thickness measurement for longitudinal neuroimaging study. In: Jiang T, Navab J, Pluim J, Max V, editors. Medical Image Computing and Computer-Assisted Intervention - MICCAI 2010.: Vol. 6362 of Lecture Notes in Computer Science. Berlin/Heidelberg: Springer; pp. 133–142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin LIK. A concordance correlation coefficient to evaluate reproducibility. Biometrics. 1989;45(1):255–268. [PubMed] [Google Scholar]
- Mason S, Graham N. Areas beneath the relative operating characteristics (ROC) and relative operating levels (ROL) curves: statistical significance and interpretation. Q. J. R. Meteorol. Soc. 2002;128(584):2145–2166. [Google Scholar]
- Moakher M. Means and averaging in the group of rotations. SIAM J. Matrix Anal. Appl. 2002;24(1):1–16. URL http://dx.doi.org/10.1137/S0895479801383877. [Google Scholar]
- Morey RA, Selgrade ES, Wagner HR, Huettel SA, Wang L, McCarthy G. Scan-rescan reliability of subcortical brain volumes derived from automated segmentation. Hum. Brain Mapp. 2010;31(11):1751–1762. doi: 10.1002/hbm.20973. URL http://dx.doi.org/10.1002/hbm.20973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura K, Fox R, Fisher E. CLADA: cortical longitudinal atrophy detection algorithm. NeuroImage. 2011;54(1):278–289. doi: 10.1016/j.neuroimage.2010.07.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nickerson CAE. A note on “A concordance correlation coefficient to evaluate reproducibility”. Biometrics. 1997;53(4):1503–1507. [PubMed] [Google Scholar]
- Patenaude B, Smith SM, Kennedy DN, Jenkinson M. A Bayesian model of shape and appearance for subcortical brain segmentation. NeuroImage. 2011;56(3):907–922. doi: 10.1016/j.neuroimage.2011.02.046. URL http://dx.doi.org/10.1016/j.neuroimage.2011.02.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu A, Bitouk D, Miller MI. Smooth functional and structural maps on the neocortex via orthonormal bases of the Laplace–Beltrami operator. IEEE Trans. Med. Imaging. 2006;25(10):1296–1306. doi: 10.1109/tmi.2006.882143. [DOI] [PubMed] [Google Scholar]
- Reuter M. Longitudinal Processing in FreeSurfer. 2009 URL http://freesurfer.net/fswiki/LongitudinalProcessing. [Google Scholar]
- Reuter M, Fischl B. Avoiding asymmetry-induced bias in longitudinal image processing. NeuroImage. 2011;57(1):19–21. doi: 10.1016/j.neuroimage.2011.02.076. URL http://dx.doi.org/10.1016/j.neuroimage.2011.02.076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reuter M, Rosas HD, Fischl B. Unbiased robust template estimation for longitudinal analysis in FreeSurfer; Proceedings of the 16th Annual Meeting of the Organization for Human Brain Mapping; 2010a. [Google Scholar]
- Reuter M, Rosas HD, Fischl B. Highly accurate inverse consistent registration: a robust approach. NeuroImage. 2010b;53(4):1181–1196. doi: 10.1016/j.neuroimage.2010.07.020. URL http://dx.doi.org/10.1016/j.neuroimage.2010.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosas HD, Reuter M, Doros G, Lee SY, Triggs T, Malarick K, Fischl B, Salat DH, Hersch SM. A tale of two factors: what determines the rate of progression in Huntington's disease? A longitudinal MRI study. Mov. Disord. 2011;26(9):1691–1697. doi: 10.1002/mds.23762. URL http://dx.doi.org/10.1002/mds.23762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabuncu MR, Yeo BT, Van Leemput K, Fischl B, Golland P. A generative model for image segmentation based on label fusion. IEEE Trans. Med. Imaging. 2010;29(10):1714–1729. doi: 10.1109/TMI.2010.2050897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabuncu MR, Desikan RS, Sepulcre J, Yeo BT, Liu H, Schmansky NJ, Reuter M, Weiner MW, Buckner RL, Sperling RA, Fischl B ADNI. The dynamics of cortical and hippocampal atrophy in Alzheimer's disease. Arch. Neurol. 2011;68(8):1040–1048. doi: 10.1001/archneurol.2011.167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salat D, Lee S, van der Kouwe A, Greve DN, Fischl B, Rosas HD. Age-associated alterations in cortical gray and white matter signal intensity and gray to white matter contrast. NeuroImage. 2009;48(1):21–28. doi: 10.1016/j.neuroimage.2009.06.074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ségonne F, Pacheco J, Fischl B. Geometrically accurate topology-correction of cortical surfaces using nonseparating loops. IEEE Trans. Med. Imaging. 2007;26:518–529. doi: 10.1109/TMI.2006.887364. [DOI] [PubMed] [Google Scholar]
- Sharf I, Wolf A, Rubin M. Arithmetic and geometric solutions for average rigid-body rotation. Mech. Mach. Theory. 2010;45(9):1239–1251. [Google Scholar]
- Shrout PE, Fleiss JL. Intraclass correlations: uses in assessing rater reliability. Psychol. Bull. 1979;86(2):420–428. doi: 10.1037//0033-2909.86.2.420. [DOI] [PubMed] [Google Scholar]
- Sled J, Zijdenbos A, Evans A. A nonparametric method for automatic correction of intensity nonuniformity in MRI data. IEEE Trans. Med. Imaging. 1998;17(1):87–97. doi: 10.1109/42.668698. [DOI] [PubMed] [Google Scholar]
- Smith SM, De Stefano N, Jenkinson M, Matthews P. Normalized accurate measurement of longitudinal brain change. J. Comput. Assist. Tomogr. 2001;25(3):466–475. doi: 10.1097/00004728-200105000-00022. [DOI] [PubMed] [Google Scholar]
- Smith SM, Zhang Y, Jenkinson M, Chen J, Matthews P, Federico A, Stefano ND. Accurate, robust, and automated longitudinal and cross-sectional brain change analysis. NeuroImage. 2002;17(1):479–489. doi: 10.1006/nimg.2002.1040. [DOI] [PubMed] [Google Scholar]
- Smith SM, Jenkinson M, Woolrich MW, Beckmann CF, Behrens TE, Johansen-Berg H, Bannister PR, De Luca M, Drobnjak I, Flitney DE, Niazy RK, Saunders J, Vickers J, Zhang Y, De Stefano N, Brady JM, Matthews P. Advances in functional and structural MR image analysis and implementation as FSL. NeuroImage. 2004;23:208–219. doi: 10.1016/j.neuroimage.2004.07.051. URL http://dx.doi.org/10.1016/j.neuroimage.2004.07.051. [DOI] [PubMed] [Google Scholar]
- Takao H, Hayashi N, Ohtomo K. Effect of scanner in longitudinal studies of brain volume changes. J. Magn. Reson. Imaging. 2011;34(2):438–444. doi: 10.1002/jmri.22636. [DOI] [PubMed] [Google Scholar]
- Thevenaz P, Blu T, Unser M. Interpolation revisited. IEEE Trans. Med. Imaging. 2000;19(7):739–758. doi: 10.1109/42.875199. [DOI] [PubMed] [Google Scholar]
- Thompson WK, Holland D. Bias in tensor based morphometry Stat-ROI measures may result in unrealistic power estimates. NeuroImage. 2011;57(1):1–4. doi: 10.1016/j.neuroimage.2010.11.092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tisdall MD, Hess AT, Reuter M, Meintjes EM, Fischl B, van der Kouwe AJW. Volumetric navigators for prospective motion correction and selective reacquisition in neuroanatomical MRI. Magn. Reson. Med. doi: 10.1002/mrm.23228. in press. http://dx.doi.org/10.1002/mrm.23228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Kouwe A, Benner T, Salat DH, Fischl B. Brain morphometry with multiecho MPRAGE. NeuroImage. 2008;40(2):559–569. doi: 10.1016/j.neuroimage.2007.12.025. URL http://www.sciencedirect.com/science/article/pii/S1053811907011457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westlye LT, Walhovd KB, Dale AM, Espeseth T, Reinvang I, Raz N, Agartz I, Greve DN, Fischl B, Fjell AM. Increased sensitivity to effects of normal aging and Alzheimer's disease on cortical thickness by adjustment for local variability in gray/white contrast: a multi-sample MRI study. NeuroImage. 2009;47(4):1545–1557. doi: 10.1016/j.neuroimage.2009.05.084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue Z, Shen D, Davatzikos C. CLASSIC: consistent longitudinal alignment and segmentation for serial image computing. NeuroImage. 2006;30:388–399. doi: 10.1016/j.neuroimage.2005.09.054. [DOI] [PubMed] [Google Scholar]
- Yushkevich PA, Avants B, Das SR, Pluta J, Altinay M, Craige C ADNI. Bias in estimation of hippocampal atrophy using deformation-based morphometry arises from asymmetric global normalization: an illustration in ADNI 3 Tesla MRI data. NeuroImage. 2010;50(2):434–445. doi: 10.1016/j.neuroimage.2009.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]