Abstract
The coat protein of positive-stranded RNA viruses often contains a positively-charged tail that extends toward the center of the capsid and interacts with the viral genome. Electrostatic interaction between the tail and the RNA has been postulated as a major force in virus assembly and stabilization. The goal of this work is to examine the correlation between electrostatic interaction and the amount of RNA packaged in the tripartite Brome Mosaic Virus (BMV). Nanoindentation experiment using atomic force microscopy showed that the stiffness of BMV virions with different RNAs varied by a ten-fold higher range than would be predicted by electrostatics. BMV mutants with decreased positive charges encapsidated lower amounts of RNA while mutants with increased positive charges packaged additional RNAs up to ~900 nucleotides. However, the extra RNAs included truncated BMV RNAs, an additional copy of RNA4, potential cellular RNAs, or a combination of the three, indicating that change in the charge of the capsid could result in several different outcomes in RNA encapsidation. In addition, mutant with specific arginines changed to lysines in the capsid also exhibited defects in the specific encapsidation of one or more of the four BMV positive-strand RNAs. The experimental results indicate that electrostatics is a major component in RNA encapsidation, but was unable to account for all of the observed effects on RNA encapsidation. Thermodynamic modeling incorporating the electrostatics was able to predict the approximate length of the RNA to be encapsidated for the majority of mutant virions, but not for a mutant with extreme clustered positive charges. Cryo-electron microscopy of virions that encapsidated an additional copy of RNA4 revealed that, despite the increase in RNA encapsidated, the capsid structure was minimally changed. These results experimentally demonstrated the impact of electrostatics and additional restraints in the encapsidation of BMV RNAs, which could be applicable to other viruses.
Keywords: Brome Mosaic Virus, coat protein, capsid, RNA encapsidation, electrostatic interaction, thermodynamic modeling, virions, cryo-EM
Introduction
Viruses have evolved distinct mechanisms to ensure the proper encapsidation of their highly negatively-charged genomes. Among DNA viruses, SV40 forms minichromosomes using cellular histones while adenovirus complexes its DNA with an adenovirus-encoded protein that is basic in overall charge.1,2 Negative-stranded RNA viruses have nucleocapsid proteins bind their RNAs through a positively-charged cleft.3
In positive-stranded RNA viruses, a common motif in the viral coat proteins (CPs) is a relatively unstructured peptide tail rich in basic amino acid residues. These tails extend into the center of the virions and are largely unresolved in the crystallographic maps, implicating their interactions with the RNA genome.4–9 The nonspecific electrostatic interaction between these tails and RNA had been proposed to constrain the length of viral genome. For instance, Belyi and Muthukumar, and also Hu and Shklovskii reported a striking linear correlation between the total number of positive charges in the tails and the length of the encapsidated RNA.10,11 In studies of various viruses, partial deletions of the positively-charged residues have resulted in virions that packaged a reduced amount of viral RNAs.12–15 Here we use Brome Mosaic Virus (BMV) to systematically examine the influence of electrostatics on RNA packaging.
BMV is well suited for analysis of selective encapsidation of RNA.16 The BMV RNAs are composed of four RNAs, named RNA1 (3.2 kb), RNA2 (2.9 kb), RNA3 (2.1 kb), and RNA4 (0.9 kb). During infection, RNA1 and RNA2 encode, respectively, the 1a and 2a proteins that function in the replicase complex. The dicistronic RNA3 is used to translate movement protein for the systemic infection of BMV in plants. RNA4 is transcribed from a subgenomic promoter in minus-strand RNA3 and directs the translation of the CP.
Infectious BMV contains three separate particles.17 RNA3 is encapsidated at a 1:1 ratio with RNA4 while RNA1 and RNA2 are individually encapsulated into separate capsids. The three types of BMV particles are indistinguishable, with 180 copies of the CP arranged with a T=3 icosahedral symmetry. Furthermore, the division of the RNAs in the particles allows each to contain a total RNA length of between 2.9 and 3.2 kb (the sum of RNA3 and RNA4 is 3.0 kb). The ratios of the three particles, as identified by the relative abundances of the four BMV RNAs, are also kept fairly constant in a typical infection; in N. benthamiana plants, particles contain RNA1 and RNA2 are approximately equal in abundance while the particles containing RNA3/4 are in several fold excess relative to the first two particles.
The CP is 189 residues in length with 8 positively-charged residues among the first 26. The density of the entire C subunit in an asymmetric trimer was elucidated, but residues 1–40 and 1–25 were respectively missing for the A and B subunits, presumably unresolved due to their interaction with the RNA.18 Deletion of residues 2–25 of the BMV CP failed to yield viable virions in plants and a truncated CP without residues 1–35 only formed empty T=1 capsids in vitro.19,20 Other BMV mutants with selectively removed positively-charged residues in the N-terminal tail have reduced packaging of RNA4, or RNA1.21–23
In this study, we compared the stiffness of BMV virions that package different RNAs through atomic force microscopy nanoidentation. We also examined the effects of altered capsid charge on BMV RNA packaging. In addition, we compared the empirical results to the theoretical optimal RNA lengths for the mutant virions under thermodynamic equilibrium. Our results suggest that the number of positive charges is an important, but not the sole factor in controlling both the abundance and the species of the RNAs packaged.
Results
BMV exhibits heterogeneous stiffness among particles packaging different RNAs
The stiffness of virus, as measured by elastic constant, is the most important factor in evaluating the viral physical stability.24,25 Assuming the electrostatic interaction between the positively charged N-terminal tail of BMV CP and RNA is the fundamental force in virus assembly and stabilization, we expect a narrow distribution of the stiffness of BMV as the RNAs packaged in each particle is 3±0.2 kb. To compare the stiffness of the BMV particles that package different RNAs, we produced two forms of virus in the geranium tobacco plant (N. benthamiana) through agroinfiltration. The first used replication-competent BMV RNAs to yield the three subpopulations of particles that typified wild-type BMV (henceforth referred to as the three-particle virions). The second was made by expressing the 1a and 2a replication proteins with RNA3 to produce virions that package only RNA3 and RNA4 (henceforth referred to as the R3/4 virions).26
Atomic force microscopy imaging and nanoindentation were used, respectively, to measure the height over the surface support and the elastic constants of R3/4 particles and three-particle BMV. The 2D histogram of the virions allowed testing for possible correlations between height and stiffness (Fig. 1A). The heights of both sets of particles were between 25 nm to 29 nm, the expected size for BMV, and ruled out the possibility of us analyzing incomplete or broken particles. However, a graphic comparison of elastic constant distributions regardless the particle diameter clearly showed the broader distribution of three-particle BMV with respect to that of R3/4 BMV (Fig. 1B), shifting towards stiffer particles. Such a significant difference in the distribution breadth of ~0.12 N/m far exceeded the estimated instrumental broadening (0.04 N/m) (sFig. 1), and argued against the difference being solely due to capsid stabilization by electrostatic interaction, which should be less than 0.003 N/m for BMV particles packaging different RNAs, if one assumes a direct proportionality between the number of charges and the force constant. These results suggested that force(s) in addition to electrostatics would affect the encapsidation of BMV RNAs.
Fig. 1. BMV exhibits heterogeneous stiffness among particles packaging different RNAs.

A) Joined histograms of particle height and elastic constants measured from 108 three-particle virions and 80 R3/4 virions. The three-particle BMV contains a significant subpopulation of stiffer particles than R3/4 BMV and a few that are softer. B) Boxplot representation of elastic constants distribution of three-particle and R3/4 BMV. The whiskers were set at 5% and 95% of the total range.
Capsids with an additional 360 positive charges form virions of different densities
To better understand the role of the electrostatics in packaging of BMV RNAs, we increased the number of positive charges in the capsid and examined its effects on RNA packaging. One turn of an α-helix encompassing two positive-charged residues was inserted after residue 15 (Fig. 2A). Using this design, we should minimally perturb the N-terminal tail that may form an α-helix upon interaction of BMV RNA.27 Virions produced from this construct, named 1H, would increase the number of basic residues within the capsid by 360. For the 1H and the majority of the virus preparations, both the three-particle and the R3/4 particle were generated and characterized. However, the results for the R3/4 virions will be presented while the results for the three-particle virions are provided as supplemental information.
Fig. 2. Capsids with an additional 360 positive charges form two subpopulations of virions.

A) Sequence of the BMV capsid residues 1–26. All positively-charged residues are in bold and underlined. The triangle indicates the position of extra inserted residues in mutant 1H. B) The R3/4 1H virions separated into two bands in a cesium chloride gradient. The names of the two preparations, 1H-L and 1H-H, are shown to the right of a photograph of the density gradient illuminated with white light. C) Spectrophotometric analysis of the number of nucleotides per BMV capsid. The change in percentage was shown to allow normalization of the spectroscopic results. D) RNAs encapsidated in the 1H-L and 1H-H virions. The left panel shows an ethidium bromide stained glyoxal gel. The right panel shows the RNA from the same gel blotted by the riboprobe specific to BMV 3′UTR. The quantification at the bottom shows the ratio of RNA4 to RNA3 in each sample. R3 and R4 are short for RNA3 and RNA4. E) A demonstration that RNAs from the heated virions generate a noncovalent complex of RNA3 and RNA4. The upper panel shows a native TBE gel and lower panel a glyoxal gel. Both gels were stained with ethidium bromide. RNA from the WT three-particle virion serves as a molecular standard. The 3 kb RNA3-RNA4 complex is denoted by the asterisk. F) Analysis of the RNA complex formed by heating of 1H-L and 1H-H virions. The image shows the ethidium bromide stained native TBE gel.
Strikingly, the 1H R3/4 virions segregated into two bands of approximately equal abundance in a cesium chloride (CsCl) gradient (Fig. 2B), henceforth named 1H-L and 1H-H (for light and heavy, respectively). Northern blot analysis for BMV-specific RNAs among the total RNAs extracted from N. benthamiana confirmed that the insertion in 1H did not cause a defect in viral RNA replication (sFig. 2). In negative-stained electron micrographs, 1H-L and 1H-H virions were indistinguishable from the WT in their diameters and shapes (sFig. 3A). The CPs from both 1H-L and 1H-H exhibited electrophoretic mobilities in SDS-PAGE that were commensurate with the presence of the four-residue insertion (sFig. 3B). In a differential scanning fluorometry (DSF) assay that examined the denaturation profile of the capsids as a function of increasing temperature,28 both 1H-L and 1H-H had one dominant transition at 78°C, the Tmapp was comparable to the WT R3/4 virions (sFig. 4A, 4B). These results indicated that 1H-L and 1H-H shared the same capsid despite having distinct densities.
We postulate that the difference in density of the 1H-L and 1H-H particles is due to a change in the RNA packaged by the virions. The average amount of RNA in each virion population was examined by UV-spectrophotometry.29 The number of nucleotides (nt) present per virion was calculated from the corrected absorption of virions at wavelengths of 260 and 280 nm using the theoretical extinction coefficients of the CP and RNA. The 1H-L virions contained nearly identical amounts of nucleotides as the WT, while the 1H-H virions packaged 13% more RNA (Fig. 2C). We noted that the calculations based on the spectroscopic readings for the WT R3/4 virions were ~1000 nt higher than what would be expected for the sum of RNA3 and RNA4, despite adjustments for the buffer. This discrepancy is likely due to the theoretical extinction coefficients used. Nonetheless, the percentage changes unambiguously suggest that the 1H-H virions package more RNAs than a WT virion.30
The RNAs extracted from 1H-L and 1H-H virions were subjected to gel electrophoresis followed by Northern blot analysis. The 1H-L and 1H-H virions both packaged approximately equal amounts of RNA3 and RNA4 as the WT, but 1H-H also contained a larger amount of truncated RNAs (Fig. 2D). The truncated RNAs were detected in Northern blots probed with a BMV-specific riboprobe that is complementary to the nearly identical 3′UTR of all BMV RNAs, indicating that they were derived from BMV RNAs.
We sought to determine whether the truncated BMV RNAs are present in the same particles as RNA3 and RNA4 in the 1H-H virions. Krishna, et al. had shown that the co-packaged RNAs in Flock House Virus migrated as a complex in non-denaturing gels when the RNAs were extracted after a brief heating of the virions.31 We applied the same approach to the R3/4 WT BMV by heating it to 45, 55 or 65°C for 10 minutes, extracted the RNAs, and subjected them to electrophoresis. In a non-denaturing gel, BMV virions heated to 55 or 65°C unambiguously generated a ca. 3 kb band, the size expected for an RNA3-RNA4 complex (Fig. 2E, top). In a denaturing gel, the RNA complex was resolved to the individual bands (Fig. 2E, bottom). This result demonstrates that heating BMV virions can provide information about whether RNAs are located within a virion.
Heating the 1H-H virions to 65°C resulted in an RNA smear that migrated slightly slower than BMV RNA1 (3.2 kb), suggesting that the truncated RNAs were co-packaged in the 1H-H virions with BMV RNA3 and RNA4. In contrast, the 1H-L virions treated in the same manner resulted in a 3 kb RNA band (Fig. 2F). The results with the 1H virions suggest that 360 extra positive charges could lead to two equally probable outcomes of either packaging or not packaging additional truncated RNAs, a significant portion of which are recognized by BMV-specific probes.
Mutants with 720 additional positive charges packaged extra RNAs
The results with the 1H mutant prompted us to further examine the relationship between charges in the capsid and the amount and types of RNA encapsidated. Three mutants that increased the number of positive charges in the capsid by 720 were constructed, along with a control for each (Fig. 3A). Mutant 2H15 contains an eight residue insertion after residue 15 in the CP that could form two α-helical turns containing four positively-charged residues. The control, 2HA15, has an eight residues insertion, but with alanines replacing the arginines. To examine whether the location of the insertions affected RNA encapsidation, the insertions within 2H15 and 2HA15 were placed after residue 7 to generate 2H7 and 2HA7. In addition, to keep the length of the N-terminal tail unchanged from the WT, we made 4R that had four arginines substituted for the uncharged residues at positions 9, 12, 16, and 17. Mutant 4S that has the same four residues replaced with serines serves as a control. All six mutants produced in the R3/4 particle form did not exert major adverse effect in viral RNA accumulation in N. benthamiana.
Fig. 3. Analysis of mutant virions that had an additional 720 positive charges.

A) Schematic of the N-terminal sequences of the six mutants characterized. The basic residues are in bold and underlined. The triangles balanced on lines denote the locations of the insertions. Only the substituted residues were typed out in 4R and 4S. B) Spectroscopic analysis of the number of nucleotides per virion. C) RNAs from the six purified BMV mutants. For each set of mutants, the left image was ethidium bromide stained glyoxal gel and the right was Northern blot images that detect BMV RNAs. The ratios of the full-length RNA4 to RNA3 are shown at the bottom of the Northern blot image. The white and black asterisks denote an unknown cellular RNA in the 4R virion. D) RNA complex formed by heating the virions. The RNAs were separated on a TBE gel and stained by ethidium bromide.
All six virions purified as a single band in CsCl density gradients and produced particles that were indistinguishable from the WT in negative-stained electron microscopy images (sFig. 3A and data not shown). The mutant CPs also exhibited the expected electrophoretic mobilities in SDS-PAGE (sFig. 3B). In Northern blots examining RNA accumulation, all of the mutants accumulated the expected BMV RNAs by 3dpi, suggesting that they are not significantly defective in RNA synthesis (sFig. 2). In the DSF assay, all mutants exhibited predominant denaturation at 78°C, the same as the WT virions, and varying extents of a minor peak at 68°C (sFig. 4A, 4B). The presence of a minor peak with a lower-than-normal denaturing temperature is likely associated with aberrant packaging of RNAs in these mutant virions.
Spectroscopic analysis showed that 2H7, 2H15 and 4R had, respectively, an average of 19%, 21% and 9% higher amounts of nucleotides per capsid relative to the WT virions (Fig. 3B). In contrast, the RNAs within the three corresponding control mutants were within 3% of that of the WT virion. These results show that increasing positive charges on the capsid increased the amount of RNA encapsidated.
Gel electrophoresis and Northern blot analyses showed that 2H7 and 4R have a 1:1 ratio of RNA3 to RNA4 (Fig. 3C). However, these virions packaged a higher abundance of truncated RNAs that annealed to the BMV-specific probe when compared to the corresponding controls or WT BMV. Mutant 2H15 contained comparable amounts of the truncated RNAs as 2HA15, but had a close to 2:1 ratio of RNA4 to RNA3, suggesting that an extra RNA4 (0.9 kb) was packaged in the 2H15 virion. Ethidium bromide stained RNAs from the 4R virion also revealed a band slightly larger than RNA1, which is likely of cellular origin because it did not hybridize to the BMV-specific RNA probe (Fig. 3C, see asterisk). 4R also contained more truncated RNAs that were not recognized by a riboprobe specific to the BMV 3′ UTR. This may be due to the truncated RNAs lacking the 3′ UTR or to the 4R packaging a significant proportion of cellular RNAs. RNAs from heated virions confirmed that the truncated RNAs or the additional RNA4 were co-packaged with RNA3 and RNA4 (Fig. 4D), where 2H15, 2H7 and 4R all exhibited a band or smear larger than the 3 kb RNA3-RNA4 complex.
Fig. 4. Analysis of mutant virions that had 360 fewer positive charges.

A) Schematic of the N-terminal sequences of the four mutants characterized. The basic residues are in bold and underlined. Only the substituted residues were typed out in the mutants. B) Spectroscopic analysis of the number of nucleotides per virion. C) RNAs from the four purified BMV mutants. For each set of mutants, the left image was ethidium bromide stained glyoxal gel and the right was Northern blot image that detect BMV RNAs. The ratios of the full-length RNA4 to RNA3 are shown at the bottom of the Northern blot image. D) RNA complex formed by heating the virions. The RNAs were separated on a TBE gel and stained by ethidium bromide.
Curiously, mutants 4S and 2HA15, which did not alter the number of positive charges within the capsid, packaged a reduced amount of RNA4 relative to RNA3 (Fig. 3C). Since this defect was not observed in 2H7, and the substitutions in the 4S overlapped with the site of insertion in 2HA15, it is likely that this location in the CP N-terminal tail affects RNA4 packaging. In analysis of RNAs from heated virions, the control virions did not exhibit RNA with electrophoretic mobility higher than 3 kb (Fig. 4D). However, the ca. 3 kb band from the 4S and 2HA15 appeared more heterogeneous compared to that of the WT, suggesting the co-packaging of truncated RNAs with RNA3 in some of the particles. This scenario can also account for the observation by spectroscopic analysis that 4S and 2HA15 virions contain the same overall number of nucleotides as do WT virions.
All six mutants and 1H described above were also produced in the three-particle form. The results were highly consistent with the results for the viruses in R3/4 form analyzed in this work. A summary of the results from the three-particle BMV is in sFig. 5C.
We also constructed mutants 3H and 4H, that had, respectively, either three or four repeats of the KATR motif inserted after residue 15 of the CP. In N. benthamiana 3H and 4H produced the expected amount and ratios of the BMV RNAs, suggesting that their RNA replication was unaffected by the mutations (data not show). However, the yield of 3H and 4H particles were at less than 5% of the wild type, thus precluding further analysis of the virions. These results suggest that there is an upper limit of either residues and/or positively-charged residues that can be inserted in the N-terminal tail before affecting virion integrity.
Mutants with 360 fewer positive charges packaged less amounts of RNAs
To examine the relationship between electrostatic charge and RNA encapsidation further, we made mutants 1415AA and 1920AA that had pairs of arginines substituted with alanines (Fig. 4A). The number of positive charges thereby decreased by 360 within each mutant capsid. Substitutions of the pairs of arginine to lysines at the same positions generated mutants 1415KK and 1920KK.
All four mutants virions purified as single bands in CsCl density gradients with comparable yields to that of the WT. In negative-stained electron micrographs, the virions were indistinguishable from the WT. Furthermore, CP exhibited the expected electrophoretic mobilities in SDS-PAGE (sFig. 3A, 3B). Mutants 1415KK and 1920KK both had TMapp of 78°C in the DSF assay, comparable to that of the WT virions (sFig. 3B, 3A). Mutants 1415AA and 1920AA had a predominant denaturation peak with a TMapp of 65.5°C and a minor one with a TMapp of 72°C (sFig. 4A, 4B). Spectroscopic analysis revealed that the 1415AA and 1920AA contained, respectively, 25% and 28% less RNA relative to the WT. Interestingly, although 1415KK was unaffected in its morphology and in its thermodenaturation, it packaged 14% less RNA (Fig. 4B). Mutant 1920KK had RNA levels that are comparable to that of WT BMV,
The RNAs encapsulated in the four mutants were examined by gel electrophoresis and Northern blots (Fig. 4C). Mutant 1920KK had the expected complement of RNA3 and RNA4. Mutant 1415KK, 1415AA, and to a lesser extent, 1920AA, had reduced amounts of RNA4 compared to RNA3. 1415KK also contained a truncated RNA that was slightly smaller than RNA4 and was detectable by the BMV-specific riboprobe. Heat-treated virions revealed that, where present, RNA4 and possibly the truncated BMV RNA in 1415KK, were packaged together with RNA3 (Fig. 4D). These results reinforce our earlier observations that the amount of RNA packaged was could be affected by both the number and the location of positive charges in the BMV CP.
Theoretical calculation of the optimal amounts of RNAs in BMV mutants
We sought to determine whether the variation observed in RNA packaging by BMV mutants could be understood by thermodynamic models that incorporate electrostatics, RNA and N-terminal tail conformational entropy, and excluded volume effects. Specifically, we calculated, as a function of the number of charges in the peptide tails, the free energy of a theoretical virion due to confinement of the RNA and interactions among the RNA, the N-terminal tail, and ions. We employed the method of Scheutjens and Fleer (SF) in which electrostatics are treated with the nonlinear Poisson Boltzmann (PB) equation with the spatial distributions of RNA, N-terminal tails, ions, and water solved numerically with a self consistent field approximation on a lattice.32–35 With the exception of the N-terminal tails, the assembled capsid was treated as a sphere, which is impermeable to N-terminal tails and RNA, but permeable to water and small ions. The N-terminal tails were explicitly represented as flexible polymers, whose segments carry electric charge dictated by the amino acid sequence. The RNA molecule was treated as a linear polyelectrolyte, and we imposed spherical symmetry on the complex. We assumed that the capsid geometry is invariant to packaged RNA length, and hence the free energy due to attractive protein subunit-subunit binding interactions is a constant in our model. However, the free energy due to interactions among the N-terminal tails depends on protein sequence and RNA length in our model.
We calculated encapsidation free energies under three sets of assumptions to determine their effect on the results. In the first case, we considered one charge per RNA nucleotide and we modeled the virion in the presence of 130 mM monovalent salt. In the second case, we considered one charge per RNA nucleotide and we modeled the virion in the presence of a negatively charged macromolecule to which the capsid is impermeable. As noted by Ting et al.,36 negatively charged macromolecules are ubiquitous within the cell, and if they are selectively excluded from the interior of the capsid their presence establishes a Donnan equilibrium and thus a potential gradient across the capsid shell. Specifically, we considered the capsid in the presence of small permeable cations and ions with respective concentrations of 130 mM and 30 mM and impermeable negative charge (on the macromolecules) of 100 mM. In the third case, we considered the same ion concentrations as for case 2, but we assumed that the charge density of the RNA molecule is limited by counterion condensation.37 Under that approximation the bare charge on the RNA molecule (1 charge/0.5 nm in our model) is reduced to a linear density of 1 charge/lB with lB the Bjerrum length. Counterion condensation cannot be investigated explicitly within our model due to the continuum approximation and imposition of spherical symmetry.
The free energy is shown as a function of RNA length for the third case (100 mM impermeable macromolecule and counterion condensation) for the WT capsid and the five mutants, 1415AA, 1920AA, 1H, 2H15 and 4R (Fig. 5A, 5B). 2H7 was not examined since it has the same inserted sequence as 2H15. We find that in all cases the free energy varies nonmonotonically with RNA length, with a minimum that corresponds to the length of RNA around which the assembled capsid is most thermodynamically stable. Under these assumptions the optimal RNA length is approximately 2400-nt for the WT capsid. This value is comparable to but less than the 2900–3000 nt typically packaged by BMV capsids. In addition, the volume fraction of RNA segments (Fig. 5D) is nonmonotonic with distance from the capsid surface, which is qualitatively consistent with the experimental observation.
Fig. 5. Theoretical prediction of the relationship between the capsid charge and RNA length.

A) The free energy is shown as a function of RNA length for the WT and the five mutants in which additional charges were inserted, deleted, or substituted into the N-terminal tails. Results are shown for case 3 described in the text, where there is a concentration cmacro=100 mM of anionic macromolecules excluded from the capsid and the RNA charge is renormalized to 0.7 charges/nucleotide by counterion condensation. B) The same data is plotted to identify the driving force for RNA encapsidation, ΔFEX, which is the free energy difference between the capsid with the RNA molecules enclosed and the empty capsid. C) The predicted optimal length of packaged RNA is shown as a function of capsid charge for all peptide sequences (1420AA, 1920AA, WT, 1HA, 2HA, 1H, and 2H15) for each of the three sets of assumptions considered in the text: cmacro=0, cmacro=100mM, and cmacro=100 mM with charge renormalization due to counterion condensation. D) Theoretical prediction of the RNA density as a function of radial position within the capsid.
The predicted encapsidation free energies are qualitatively similar for cases 1 and 2 described above. The predicted optimal packaged length is shown as a function of total capsid charge for each case and each sequence in Fig. 5C. In all cases the optimal length increases approximately linearly with the amount of capsid charge, with small variations due to changes in capsid N-terminal tails. For case 1 the amount of charge on the packaged RNA corresponds to nearly the amount of positive charge on the capsid. For case 2 the amount of packaged RNA increases by approximately 300 nucleotides for each sequence in comparison to case 1. The magnitude of this increase can be understood by simple arguments described in the supporting information. Finally, for case 3 the ratio of RNA charge to capsid charge is approximately the same as for case 2 after accounting for charge renormalization due to counterion condensation.
The predicted optimal length increases by approximately 20% for the 1H mutant and 40% for the 2H15 mutant. These changes in optimal RNA length with respect to the WT capsid are about two-fold larger than the experimental observations, which are an approximately 13% increase for 1H and a 21% increase for 2H15. The predicted decreases for the 1415AA and 1920AA mutants of −21% are close to the experimental observations, which are −25% and −28%. Furthermore, the minimum free energy RNA length for the control sequence, 2HA15, is identical to that for WT (data not shown), consistent with experimental observations. In contrast, the predicted increase for the 4R sequence is approximately equal to that of 2H15 (42%), while in experiments the 4R sequence packaged only 9% more RNAs. In all cases, the amount of charge on the packaged RNA increases linearly with the amount of positive charge on the capsid N-terminal tails, indicating that electrostatics is a dominant effect in these calculations. These results suggest, however, that additional effects accounting for the sequence of charges in the N-terminal tails would be required for a theoretical model to agree with the full set of experimental results.
BMV packages extra RNAs did not change the capsid structure
Our observations with the mutant virions suggest that the BMV can sterically accommodate additional RNAs. To better analyze the densities of the RNAs within the virion, we selected 2H15 for further characterization. Cryo-electron micrographs of virions frozen in vitreous were used to reconstruct a 3D density map by applying icosahedral symmetry. The map was refined to a resolution of 5 Å with well-defined pentameric and hexameric capsomeres (sFig. 6A). The lateral section of the density showed an outer diameter of approximately 286 Å (Fig. 6A), consistent with averaging diameter of the WT.18 The inner radius of the 2H15 is about 96.4 Å, which is about 6 Å larger than that of the WT. This is probably because larger segment on the N-terminal tails in 2H15 are disordered. The class average of the 2H15 particles exhibited that the center of virion was filled with additional density when compared to the WT, which is likely to be associated with the additional RNA4 (sFig. 6B). Results from small angle X-ray diffraction studies also revealed that 2H15 had a thicker layer of the encapsidated RNAs and capsid (sFig. 7).
Fig. 6. The cryo-EM structure of 2H15 BMV.
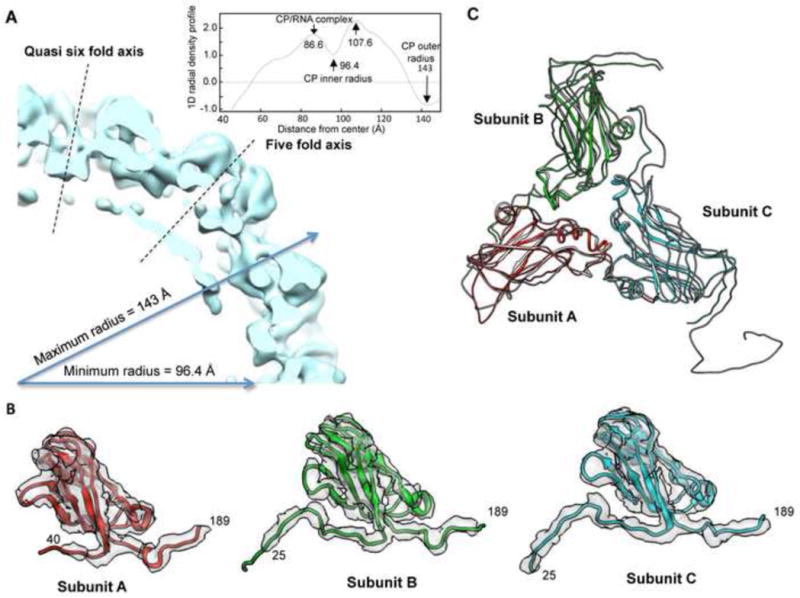
A) A quarter cross-section through the 2H15 capsid. The plane of the cross-section includes the 5-fold and quasi 6-fold axes which cut in the centers of the pentameric and hexameric capsomeres. The insert is the averaged 1D radial density profile of the 2H15. B) Models for subunits. Subunit A was shown in red (residues 40–189), subunit B in green (residues 25–189) and subunit C in blue (residues 25–189), respectively. The inserted 8 residues were not included in the numbering of the residues. The density map is shown in transparent grey and displayed at slightly higher isosurface threshold. C) Backbone representation of the 2H15 trimer superimposed on the corresponding part from the crystal model of the wild type BMV (pdb:1js9), which is presented as a dark gray overlay.
The cryo-EM map fitted well with the 3.3 Å crystal structure of the WT BMV and enabled the tracing of the Ca backbone for A, B and C capsid subunits (Fig. 6B and 6C). With the guide of the crystal structure, we assigned the model of pentameric subunit A to residues 41–189, the model of hexameric subunit B and C to residues 25–189 and 25–189, respectively (the numbers do not include the inserted residues to allow comparison to the X-ray structure of the WT BMV (Fig. 6B).18 The densities of the remainder of the N-terminal tail were not resolved in the models. These observations agree with the assumption that the N-terminal tails are complexed with the RNA. A further superposition of the backbone of the 2H15 trimer on that of the crystal structure of the WT revealed that the globular domains of the capsid subunits were largely unchanged relative to the comparable structures from the crystal structure of BMV (rmsd < 2Å), while the N-terminal and C-terminal tails displayed the most significant variations. We did not find any substantial change in diameter of the center pore of the quasi-3-fold axes at current resolution level. Overall, our cryo-EM model of 2H15 indicated that the inserted residues in the N-terminal tail and the packaging of additional RNA4 did not affect the folding or the arrangement of the capsid subunits. However, in this reconstruction of 2H15 we were unable to visualize the density of RNA.
DISCUSSION
A common motif in positive-strand RNA viruses is that their CPs contains highly positively-charged tails that extend into the interior of the capsid. Presumably, the electrostatic attraction between the RNA and positively-charged tails guides virion assembly, and maintains virus integrity. This work seeks to examine whether electrostatic interaction is sufficient to explain BMV RNA packaging using a combination of mechanical and molecular genetic approaches. The results are also compared to models of RNA encapsidation based on thermodynamic modeling of capsid-RNA interaction. This comparison leads to the conclusion that, while electrostatics can guide RNA packaging, changes in the amount and the species of RNA encapsidated varied for different mutations with the same capsid charges, and that there are sequence-specific effects on RNA recognition related to the location of positively-charged residues in the BMV CP.
One prediction of an electrostatic model is that mechanical resistance to pressure will correlate with the length of the RNA encapsidated. However, the three-particle BMV, a natural mix of the three subpopulations of BMV, exhibits a rather wide range of elastic constants in the nanoindentation experiment, as opposed to a narrow one observed for R3/4 BMV. The heterogeneity in elastic property of the three-particle BMV could not solely come from the different orientation of the particles or the packaging of non-genomic material from host cells, which should also be valid for R3/4 particles as well. Most importantly, the difference in elastic constant due to capsid stabilization by the electrostatic interaction of different RNAs and CP could not account for the observed drastic variation, suggesting that there are forces other than the electrostatics that also contribute substantially to the RNA-CP interaction within capsid.
Molecular genetic analysis of BMV mutants with altered N-terminal sequences further supported that the role for the positively-charged N-terminal tail is more complex than a simple bearer of electrostatic charges. Twelve purified mutant BMV R3/4 virions with increased, decreased or unchanged numbers of capsid charges were analyzed for the amount of encapsidated RNA by spectroscopic analysis. The correlation between the total number of positive changes on the N-terminal tails to the calculated number of RNA nucleotides was plotted in Fig. 7. In general, the RNA length was affected by the capsid charge. For example, all four BMV mutants with increased capsid charges, 1H, 2H7, 2H15 and 4R, packaged more RNA than the WT. However, several features were notable. First, the sum of the RNAs packaged by virions that have the same number of charges could differ by ca. 400-nt (e.g. compare the results for the 4R and 2H7, or the WT and 1415KK). This distribution is underscored by the results from the 1H virions, which formed two distinct particles between which the amount of RNA varied by an estimated 400-nt. Second, we note that the 4R, whose charge is increased by substitution instead of insertion, had less encapsidated RNAs than did 2H7 and 2H15, which had the same number of capsid charges. This may arise because 2H7 and 2H15 have longer extended N-termini with more evenly dispersed positive charges. Third, a non-linear correlation for the twelve mutants was observed. The incorporation of additional RNA in response to increased numbers of positive charges in the capsid was lower than the loss of RNA that resulted when the number of capsid charges was decreased. While this effect might be attributed to excluded volume, its magnitude was not captured in any of the theoretical models that we considered, all of which do account for excluded volume.
Fig. 7. The correlation between the charges in the N-terminal tails and the amount of RNA packaged.

The net charges on N-terminal tails per capsid was calculated by multiplying the number of positive charges within the first 49 residues by 180, the number of CP subunits per T=3 capsid. The RNA length for each mutant was estimated by applying the corresponding percentage change from Fig. 2B, 3B and 4B to 2987-nt, the expected size of RNA3 and RNA4 in WT. The mutants with increase, unchanged, and decreased capsid charges were colored in green, yellow and red, respectively.
We note that there appear to be other constraints on RNA encapsidation. For example, BMV virions with insertions of three or four repeats of the residues inserted in 1H were unable to efficiently form virions although the accumulation of viral RNAs in N. benthamiana leaves were largely unaffected by the mutations. Based on the currently available results, however, we cannot determine whether the defect in virion formation is due to steric issues that arose from the increased length of the capsid tails, or from the increased amount of RNA that is associated with the increased capsid charges.
A position-dependent effect of residues in the N-terminal tail on RNA encapsidation was also suggested by our results. Mutant 1920KK is competent at packaging RNA4 while mutant 1415KK packaged reduced amounts of RNA4 and a truncated BMV RNA. It is possible that a bidentate contact involving arginines at residues 14 and 15 is required for proper interaction between the capsid and RNA4. Other mutants with unchanged capsid charges but sequence alterations close to residue 14 and 15, namely 4S and 2HA15, also reduced RNA4 encapsidation, A similar specific interaction between the N-terminal tail and RNA4 was also observed with mutant 2H15, which have four positively-charged residues inserted after position 15. 2H15 packaged two copies of RNA4 per RNA3 in the virion. Rao and colleagues have also observed that the N-terminal tail of BMV is involved in the selective encapsidation of BMV RNA4.23
The mutations we analyzed had dramatic variations in the packaging of RNA4 in contrast to the minimal effect on the encapsidation of RNA3. These results suggest that the RNAs play a role beyond that of a simple polyelectrolyte in the assembly of virions. The BMV capsid may have evolved mechanisms to differentially recognize the four BMV RNAs. A greater flexibility for the encapsidation for RNA4 is logical for BMV since it could be produced from a copy of RNA3 and is not needed to initiate RNA synthesis.16,17,26 By contrast, RNA2 is preferentially encapsidated by several mutants when the three-particle virions were examined (sFig. 4). We have also demonstrated previously that the RNA2-containing virion is the most resistant to degradation.28 Perhaps it is more important for BMV to protect RNA2 since it encodes the polymerase needed for propagation of the virus.
Although our experimental data argue against the existence of a fixed correlation between the genome size and capsid charge, the charge ratio and optimal RNA length predicted by thermodynamic models incorporating electrostatics still provide valuable guidance. We need to bear in mind that these charge ratios and optimal RNA lengths vary when different models are considered. For instance, Hu and Shklovskii assumed that RNA wrap around peptide tails and found that the free energy is minimized when the RNA contour length is approximately equal to the total contour length of peptide tails, resulting in an optimal charge ratio of 2;11 Belyi and Muthukumar treated the RNA as a polyelectrolyte and the peptide tails as oppositely charged brushes and used the ground state dominance approximation to predict an optimal charge ratio of 1:1.10 Upon renormalizing the charge on RNA and the peptide tails according to counterion condensation theory, the predicted ratio of packaged nucleotides to capsid charges was 1.6:1. Calculations that place the capsid charge entirely at the surface have predicted charge ratios of <1, 1, and 2.38–40 Ting et al. found that the optimal ratio of packaged charge to capsid charge was sensitive to details such as the charge density on the peptides but was less than 1:1.36 They showed however that the Donnan potential due to anionic macromolecules which are selectively excluded from the capsid could increase this ratio to larger than 1:1. In this work we applied a continuum version of the self-consistent field theory with explicit representation of the N-terminal tail as flexible polymers with the sequences of charge considered in our experiments. We found that for all sets of approximations that we considered the optimal amount of packaged charge was relatively insensitive to the location of charges within the peptide sequence and increased linearly with the total positive charge on the capsid. Upon incorporating the Donnan potential and charge renormalization due to counterion condensation our theory predicted an optimal charge ratio of approximately 2.
Based on our experimental data describing the amount of RNA packaged by each mutant, the amount of packaged RNA tends to increase with increasing positive charges on the capsid, and the ratio of packaged nucleotides to positive charge on the capsid ranges from 0.33 to 2.22. These results are in line with the predictions of the various theoretical models listed above. However, the fact that the amounts of RNA packaged vary nonlinearly with capsid charge suggests that the sensitivity of RNA recognition by the peptide sequence is not fully captured by any of the theoretical approaches.
There are several factors that could account for the imperfect agreement between theory and experimental observations. First, some discretion is required when counting the number of capsid charges when comparing the predicted charge ratio to that of real viruses. The theoretical calculation is based on a model in which the peptide arms protrude inwards from capsid and interact with RNA. However, defining the part of the peptide arm which extends inward and hence interacts with the RNA is relatively arbitrary, which directly affects the counting of charges. Second, the disposition of charges across the secondary or tertiary structure of the peptide tails is not well captured in the theoretical models, including ours. This simplification could contribute to the fact that the theoretical calculations predict that the RNA packaging free energy depends strongly on the net charge of the peptide tails, but is relatively insensitive to the location of the charges within this sequence. This consideration may be especially relevant for the 4R mutant, where the mutated residues are more clustered than in other mutants or the WT. Furthermore, the theory does not include sequence-specific peptide structures which may affect the recognition of particular RNA sequences and/or structures (see below). Other approximations in the theory, such as the assumption of spherical symmetry within the virion or the neglect of ion-ion correlations, could also contribute to the discrepancy between theory and experimental results. Finally, it is possible that, in addition to the thermodynamic considerations described here, kinetics or other nonequilibrium effects also control the amount of RNA packaged in the experiments.41,42 The capsid assembly is an out-of-equilibrium process, so that equilibrium is only reached, if ever, once the final product has assembled.
There are several caveats in the experimental data that we want to highlight. First, the genome size derived from spectroscopic measurement only reflects the average length of RNA for a population of virions. Although all the mutants in our study, except 1H, purified as a single band in the CsCl gradient, the RNA content within each mutant is not necessarily homogenous given the gradient resolution, especially when truncated RNAs of various lengths are packaged. This variation is also evident in the profiles of RNAs from the heat-treated virions, where the co-packaged RNAs migrated in a smear. In other occasions, a minor population of virus that is probably too diluted to form a visible band in CsCl gradient may be included in the purified virion. For instance, a small fraction (less than 10%) of the 1415AA and 1920AA virions packaged RNA4 together with RNA3 while the majority packaged RNA3 alone.
Second, it is important to note that, with the exception of 4R, a BMV specific riboprobe complementing to 3′ UTR recognized most of the truncated RNAs encapsulated by the mutants. Although it is unclear how these truncated RNAs are produced, the prevalence of 3′ UTR suggests that this region may contain a sequence or assume a conformation that can be specifically recognized by BMV CP. In vitro RNA encapsidation experiments have revealed that the BMV capsid preferentially encapsidates RNAs containing a tRNA-like structure, which is found in the BMV 3′ UTR.43 It is also possible that the tRNA-like structure, a motif required for BMV replication,44 contributes to the specific recognition by the CP through coordinating RNA replication and encapsidation.45–47
Lastly, the BMV mutants in our study accommodated RNAs that ranged from 2000-nt to 4000-nt in sizes without any obvious change in the capsid sizes or conformations. The cryo-EM reconstruction of the 2H15 virion yielded models comparable to the WT virions. These results indicate that BMV capsid is a highly adaptable cage for RNA. However, the volume within the capsid is finite, as seen by the low yields of the 3H and 4H mutant virions in our studies. A self-assembly study using the CP of CCMV, a close relative of BMV, and RNA of different size found that when RNA exceeded 4500-nt, it was assembled into two or more connecting capsids.48
In summary, comparison of theoretical predictions with our experimental observations virions with varying amounts of positive charge indicates that electrostatics plays an important role in the amount of RNA which is packaged. However, our results also indicate that the specific amino acid sequence of the capsid N-terminal tails contributes to RNA packaging in a manner which cannot be entirely described by models which treat them as simple polyelectrolytes, possibly due to secondary or tertiary structure of the peptide and its corresponding ability to recognize particular structural motifs found within the viral genome.
Materials and Methods
Plasmid constructs and agroinfiltration
An Agrobacterium-mediated gene delivery system was used to express the three BMV genomic RNAs as well as the individual BMV replication proteins.26 cDNA for BMV RNA3 expressing the mutant CP was made by PCR-mediated site-directed mutagenesis with appropriate primers that will be available upon request. The sequences of the cDNAs were confirmed to have no other mutations. N. benthamiana plants used to produce virions were grown in an environmentally regulated growth chamber kept at a constant 25°C, 70 to 75% humidity and a 16/8 h light/dark cycle.
Virus purification
The purification of BMV was modified from the method of Bujarski.49 Briefly, N. benthamiana leaves were homogenized in virus buffer I (250 mM NaOAc, 10 mM MgCl2, pH 4.5), and the supernatants were clarified by 10% chloroform. The supernatant was then layered on a 10% sucrose cushion prepared in virus buffer I, and centrifuged for 3 h at 28,000 rpm using Beckman SW32 rotor to pellet the virus. The pellets were dissolved in virus buffer II (50 mM NaOAc, 10 mM MgCl2, pH 5.2) with a 50% cesium chloride (w/v) and banded by centrifuging for 20 h at 65,000 rpm using a Beckman TLA110 rotor. The band of virions, which appeared opalescent white, was collected and dialyzed with three changes of virus buffer II and stored at −80°C until use.
Nanoindentation by atomic force microscope
To mount the sample, a droplet of 50 μL virus was deposited on a freshly cleaved highly ordered pyrolytic graphite (HOPG) disk, mounted on the peizo holder, and incubated for 10 minutes before probing. A droplet holder was used to minimize evaporation and maintain a stable buffer system.
AFM imaging and mechanical indentation measurements were recorded using a Cypher AFM (Asylum Research) instrument in liquid, at room temperature. Silicon cantilevers (BioLeverMini, Olympus) with a tip radius of ~ 9 nm and spring constant of ~0.1 N/m were used. The cantilever spring constant was calibrated using the thermal-oscillation method.50 AFM images were obtained in the alternative contact mode using low set point amplitudes (< 10 mV). This way, damage to the virus by the tip was minimized. Samples were first imaged at low resolution (500 nm × 500 nm) to identify a single virus. Higher-resolution topographic images (80 nm × 80 nm) were then recorded, followed by force mapping in contact mode. Force maps consisted of arrays of 6 × 6 force-distance curves on 80 nm × 80 nm areas, using a maximum applied force of 700 pN and a loading rate of 1 μm/s. After force mapping, a topographic image was always collected to check the integrity of the particle. When notable morphological differences were observed, the data was rejected.
To estimate the instrumental response, a series of force curves were collected at the same position (middle of the virus particle) from the same particle, until a large deviation of spring constant was observed. The variance in the elastic constants obtained this way characterizes the accuracy of measurements in the absence of heterogeneous broadening. Images and force maps were analyzed using Igor Pro 6.12 software (Wavemetrics).
Spring constant calculation
The compression of the virus by the tip can be represented as two springs connected in series represented by the virus and the cantilever [Michel 2006]. The elastic constant of a virus is related to that of the cantilever by:
Where, Keff = effective spring constant obtained by a least-square fit of the linear region in the force-distance curve. Kc = the cantilever spring constant.
RNA extraction and Northern blot
virion RNAs were extracted from the CsCl gradient purified virions by a 1:1 mixture of phenol/chloroform after treating virions with 2X lysis buffer (200 mM Tris, 2 mM EDTA, 2% SDS, pH 8.5). Northern blots were performed following the separation of glyoxal-treated RNA on a 1.2% agarose-BPTE gel.51 An in vitro-transcribed 3′UTR riboprobe was used to detect the positive-strand BMV RNAs.30 Bands in the blots were quantified using ImageQuant software.
Analysis of encapsidated RNAs
To determine whether distinct RNAs were present in one virion, purified virions was heated to 65°C for 10 min followed by an immediate cooling on ice for 10 min. Virion RNA was extracted as described above and analyzed by electrophoresis on a 0.9% agarose-TBE gel.52 Some samples were also transferred to a Nylon membrane and probed with the riboprobe specific to the BMV 3′UTR described above to determine whether the RNAs contained the nearly identical 3′ UTR of the BMV RNAs
Differential scanning fluorimetry
DSF was performed in a 96-well plate in a Stratagene real-time PCR machine. Each reaction had a total volume of 25 μl containing 4 μg virion, and SYPRO orange (Molecular Probes) at a 2.5X final concentration. The temperature ramp was set at a rate of 1.0°C/min from 25 to 95°C. The fluorescence intensity was measured with Ex470/Em550 and plotted as the first derivative of fluorescence versus temperature. The temperature with the fastest change in fluorescence intensity was extrapolated to be the apparent virus denaturing temperature.
Quantification of RNAs per virion
Spectrophotometry analysis of the virions was performed according to Porterfield et al.29 Briefly, the UV absorption spectra were collected using an Agilent 8453 UV-Visible spectrophotometer after blanking with virus buffer II. The concentration of nucleotide ([RNA]) and CP monomer ([protein]) was calculated using equations
where Acorrected, 260 and Acorrected, 280 are the corrected absorbances of virus at wavelengths 260nm and 280nm after subtracting the light scattering part. L is the length of the light path. The values used for εRNA260, εRNA280, εprotein260, εprotein280 were 8,000 M−1 cm−1, 4,000 M−1 cm−1, 14,400 M−1 cm−1, 24,000 M−1 cm−1, respectively. The concentration of capsid was obtained from that of the CP monomer with the knowledge that each capsid was made up by 180 monomers. The concentration of the RNA divided by that of the capsid was extrapolated to be the length of the RNA per capsid.
Electron microscopy and image processing
For negative staining electron microscopy, samples were applied to glow-discharged carbon coated copper grids and stained with 1% uranyl acetate. The grids were visualized by a magnification of 30,000 on a JEOL JEM 1010 electron microscope at an operating voltage of 80 kV. Images were collected on a 4K × 4K CCD Gatan Camera.
The WT BMV (1 mg/ml), or 2H15 (0.8 mg/ml) was applied onto a R1.2/1.3 Quantifoil grid. The grids were previously washed and glow-discharged. After blotting and being rapidly frozen in liquid ethane using a Vitrobot, the grids were stored in liquid nitrogen. JEOL JEM3200FC electron microscope (JEOL, Tokyo, Japan), operated at 300kV, and 4K × 4K CCD Gatan Camera (Gatan, Pleasanton, CA) were used for imaging and data collection. The particles were selected from the electron micrographs using EMAN’s Boxer routine.53 The particles were translationally and rotationally aligned, classified, and averaged without applying symmetry using EMAN’s Refine2d command.
Modeling
To calculate the thermodynamic driving force for RNA packaging, we employed the method of Scheutjens and Fleer (SF).32,54–57 Our calculation is comparable to the self-consistent field calculations performed in Siber et al.58 but we explicitly model the N-terminal arms. The lattice approach facilitates representation of different sequences. The capsid is modeled as a sphere with an inner radius 11 nm which is impenetrable to RNA and protein segments but permeable to water and small ions. The N-terminal tails are treated as flexible polymers grafted to the inner surface of the capsid. The N-terminal tail is considered to include the first 49 amino acid residues of the WT protein, and any residues inserted into this region for mutant sequences. The WT sequence thus includes 10 positive charges and one negative charge for a net charge of nine. The RNA molecule is represented as a linear polyelectrolyte. For the N-terminal tails and the RNA, the statistical segment length and segment diameters are set equal to the lattice size, l = 0.5nm. This value is consistent with the average excluded volume of amino acids and small ions (including their solvent shells). It underestimates the excluded volume of RNA, but we find that the results are insensitive to varying l. The length l is roughly consistent with the distance between unpaired nucleotides in single-stranded RNA and thus we consider that each RNA segment corresponds to approximately 1.5 nucleotides. We neglect the effects of base pairing and tertiary structure on the RNA architecture; repeating the calculation with the RNA represented as a branched polymer yielded almost identical free energies.
We consider one RNA molecule confined to the capsid interior; the results would change only slightly if we considered the case of several smaller RNA molecules with the same overall contour length. Except for charge, all species have identical interactions which account for excluded volume. For each protein sequence and RNA length considered, the spatial distribution of densities for each species is calculated by minimizing the semi-grand free energy,54,55,59 where the water and small ions are in equilibrium with a bulk solution. As described in the main text, we consider three sets of assumptions. In the first two cases, the RNA charge density is not renormalized. In the first case, the virion is in equilibrium with a bulk solution containing 130 mM 1:1 salt. In the second case, the bulk solution contains 130 mM monovalent cations, 30 mM monovalent anions, and 100 mM negative charges on macromolecules. The capsid is permeable to the monovalent anions and cations but the macromolecule charges are selectively excluded from the capsid interior. In case three, we consider the same bulk solution as for case 2, but we assume that the RNA charge density is renormalized by counterion condensation to a linear charge density of e/lB with lB=0.714 nm the Bjerrum length.60–65 Our implementation follows those described in Refs. 54,55,59 full details are given in the supporting online material.
Reconstructing and modeling cryo-electron microscopic map of 2H15
A total of 11,576 particles were selected from the micrographs for CTF correction and 3-D reconstruction. To generate an initial model for reconstruction, about 3,000 particles were selected by MPSA to build a sub nanometer model of 2H15.66 The map was then refined by EMAN.53 A 5.0 Å resolution of the cryo-reconstruction density map was determined by 0.5 FSC curve provided by EMAN program (sFig. 6A). An atomic model of the BMV capsid derived from the 3.3 Å crystal structure was then fitted into this cryo-TEM map and used as a guide to manually trace the Cα backbone of the 2H15 using program COOT.67
Supplementary Material
RESEARCH HIGHLIGHTS.
The role of electrostatic interaction in viral RNA encapsidation was examined
Viral capsids with extra (+)-charged residues packaged increased amounts of RNA
One mutant virion encapsidated an additional copy of its four RNAs
The RNA encapsidated were compared with those from theoretical calculations
This work provides information on a basic requirement for viral RNA encapsidation
Acknowledgments
We thank the IU and Texas A&M Cereal Killers past and present. We thank A. Murali for help with the EMAN program and the Office of the Vice Provost for Research at Indiana University for supporting the Small Angle X-ray Scattering Measurements. This work was funded by the NIH NIAID grant 1R01AI090280 (CK) and the NIH NIAID grant R01AI080791 (MFH) and NIH P41RR002250 (WC).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References Cited
- 1.Polisky B, McCarthy B. Location of Histones on Simian Virus 40 DNA. Proc Natl Acad Sci USA. 1975;72:2895–2899. doi: 10.1073/pnas.72.8.2895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sung MT, Cao TM, Coleman RT, Budelier KA. Gene and protein sequences of adenovirus protein VII, a hybrid basic chromosomal protein. Proc Natl Acad Sci USA. 1983;80:2902–2906. doi: 10.1073/pnas.80.10.2902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ruigrok RW, Crépin T, Kolakofsky D. Nucleoproteins and nucleocapsids of negative-strand RNA viruses. Curr Opin Microbiol. 2011;14:504–510. doi: 10.1016/j.mib.2011.07.011. [DOI] [PubMed] [Google Scholar]
- 4.Smith TJ, Chase E, Schmidt T, Perry KL. The structure of cucumber mosaic virus and comparison to cowpea chlorotic mottle virus. J Virol. 2000;74:7578–7586. doi: 10.1128/jvi.74.16.7578-7586.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Silva AM, Rossmann MG. The Refinement of Southern Bean Mosaic Virus in Reciprocal Space. Acta Crystallogr, Sect B. 1985;41:147–157. [Google Scholar]
- 6.Choi HK, Lu G, Lee S, Wengler G, Rossmann MG. Structure of Semliki Forest virus core protein. Proteins. 1996;27:345–359. doi: 10.1002/(sici)1097-0134(199703)27:3<345::aid-prot3>3.0.co;2-c. [DOI] [PubMed] [Google Scholar]
- 7.Fisher AJ, Johnson JE. Ordered duplex RNA controls capsid architecture in an icosahedral animal virus. Nature. 1993;361:176–179. doi: 10.1038/361176a0. [DOI] [PubMed] [Google Scholar]
- 8.Tong L, Wengler G, Rossmann MG. Refined structure of Sindbis virus core protein and comparison with other chymotrypsin-like serine proteinase structures. J Mol Biol. 1993;230:228–247. doi: 10.1006/jmbi.1993.1139. [DOI] [PubMed] [Google Scholar]
- 9.Speir JA, Munshi S, Wang G, Baker TS, Johnson JE. Structures of the native and swollen forms of cowpea chlorotic mottle virus determined by X-ray crystallography and cryo-electron microscopy. Structure. 1995;3:63–77. doi: 10.1016/s0969-2126(01)00135-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Belyi VA, Muthukumar M. Electrostatic origin of the genome packing in viruses. Proc Natl Acad Sci USA. 2006;103:17174–17178. doi: 10.1073/pnas.0608311103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hu T, Zhang R, Shkovskii BI. Electrostatic theory of viral self-assembly. Physica A. 2008;387:3059–3064. [Google Scholar]
- 12.Kaplan IB, Zhang L, Palukaitis P. Characterization of Cucumber Mosaic Virus. Virology. 1998;246:221–231. doi: 10.1006/viro.1998.9192. [DOI] [PubMed] [Google Scholar]
- 13.Venter PA, Marshall D, Schneemann A. Dual roles for an arginine-rich motif in specific genome recognition and localization of viral coat protein to RNA replication sites in flock house virus-infected cells. J Virol. 2009;83:2872–2882. doi: 10.1128/JVI.01780-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dong XF, Natarajan P, Tihova M, Johnson JE, Schneemann A. Particle Polymorphism Caused by Deletion of a Peptide Molecular Switch in a Quasiequivalent Icosahedral Virus. J Virol. 1998;72:6024–6033. doi: 10.1128/jvi.72.7.6024-6033.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Marshall D, Schneemann A. Specific Packaging of Nodaviral RNA2 Requires the N-Terminus of the Capsid Protein. Virology. 2001;285:165–175. doi: 10.1006/viro.2001.0951. [DOI] [PubMed] [Google Scholar]
- 16.Rao ALN. Genome packaging by spherical plant RNA viruses. Annu Rev Phytopathol. 2006;44:61–87. doi: 10.1146/annurev.phyto.44.070505.143334. [DOI] [PubMed] [Google Scholar]
- 17.Kao CC, Sivakumaran K. Brome mosaic virus, good for an RNA virologist’s basic needs. Mol Plant Pathol. 2000;1:91–97. doi: 10.1046/j.1364-3703.2000.00017.x. [DOI] [PubMed] [Google Scholar]
- 18.Lucas RW, Larson SB, McPherson A. The crystallographic structure of brome mosaic virus. J Mol Biol. 2002;317:95–108. doi: 10.1006/jmbi.2001.5389. [DOI] [PubMed] [Google Scholar]
- 19.Sacher R, Ahlquist P. Effects of deletions in the N-terminal basic arm of brome mosaic virus coat protein on RNA packaging and systemic infection. J Virol. 1989;63:4545–4552. doi: 10.1128/jvi.63.11.4545-4552.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Larson SB, Lucas RW, McPherson A. Crystallographic structure of the T=1 particle of brome mosaic virus. J Mol Biol. 2004;346:815–831. doi: 10.1016/j.jmb.2004.12.015. [DOI] [PubMed] [Google Scholar]
- 21.Calhoun SL, Speir JA, Rao ALN. In vivo particle polymorphism results from deletion of a N-terminal peptide molecular switch in brome mosaic virus capsid protein. Virology. 2007;364:407–421. doi: 10.1016/j.virol.2007.03.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rao AL, Grantham GL. Molecular studies on bromovirus capsid protein. II Functional analysis of the amino-terminal arginine-rich motif and its role in encapsidation, movement, and pathology. Virology. 1996;226:294–305. doi: 10.1006/viro.1996.0657. [DOI] [PubMed] [Google Scholar]
- 23.Choi YG, Rao AL. Molecular studies on bromovirus capsid protein. VII Selective packaging on BMV RNA4 by specific N-terminal arginine residuals. Virology. 2000;275:207–217. doi: 10.1006/viro.2000.0513. [DOI] [PubMed] [Google Scholar]
- 24.Michel JP, et al. Nanoindentation studies of full and empty viral capsids and the effects of capsid protein mutations on elasticity and strength. Proc Natl Acad Sci USA. 2006;103:6184–6189. doi: 10.1073/pnas.0601744103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Roos WH, et al. Mechanics of bacteriophage maturation. Proc Natl Acad Sci USA. 2012;109:2342–2347. doi: 10.1073/pnas.1109590109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gopinath K, Dragnea B, Kao C. Interaction between Brome mosaic virus proteins and RNAs: effects on RNA replication, protein expression, and RNA stability. J Virol. 2005;79:14222–14234. doi: 10.1128/JVI.79.22.14222-14234.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Argos P. Secondary structure prediction of plant virus coat proteins. Virology. 1981;110:55–62. doi: 10.1016/0042-6822(81)90007-6. [DOI] [PubMed] [Google Scholar]
- 28.Hema M, et al. Effects of amino-acid substitutions in the Brome mosaic virus capsid protein on RNA encapsidation. Mol Plant Microbe Interact. 2010;23:1433–1447. doi: 10.1094/MPMI-05-10-0118. [DOI] [PubMed] [Google Scholar]
- 29.Porterfield JZ, Zlotnick A. A simple and general method for determining the protein and nucleic acid content of viruses by UV absorbance. Virology. 2010;407:281–288. doi: 10.1016/j.virol.2010.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hema M, Gopinath K, Kao C. Repair of the tRNA-like CCA sequence in a multipartite positive-strand RNA virus. J Virol. 2005;79:1417–1427. doi: 10.1128/JVI.79.3.1417-1427.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Krishna NK, Schneemann A. Formation of an RNA heterodimer upon heating of nodavirus particles. J Virol. 1999;73:1699–1703. doi: 10.1128/jvi.73.2.1699-1703.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Scheutjens J, Fleer GJ. Statistical-Theory of the Adsorption of Interacting Chain Molecules. 2 Train, Loop, and Tail Size Distribution. J Phys Chem. 1980;84:178–190. [Google Scholar]
- 33.Scheutjens JMHM, Fleer GJ. Statistical theory of the adsorption of interacting chain molecules. 1 Partition function, segment density distribution, and adsorption isotherms. The Journal of Physical Chemistry. 1979;83:1619–1635. [Google Scholar]
- 34.Bohmer MR, Evers OA, Scheutjens JMHM. Weak polyelectrolytes between two surfaces: adsorption and stabilization. Macromolecules. 1990;23:2288–2301. [Google Scholar]
- 35.Bohmer MR, Koopal LK, Lyklema J. Micellization of ionic surfactants: calculations based on a self-consistent field lattice model. The Journal of Physical Chemistry. 1991;95:9569–9578. [Google Scholar]
- 36.Ting CL, Wu J, Wang ZG. Thermodynamic basis for the genome to capsid charge relationship in viral encapsidation. Proc Natl Acad Sci USA. 2011;108:16986–16991. doi: 10.1073/pnas.1109307108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Manning GS. Limiting Laws and Counterion Condensation in Polyelectrolyte Solutions I. Colligative Properties. J Chem Phys. 1969;51:924. [Google Scholar]
- 38.Siber A, Podgornik R. Nonspecific interactions in spontaneous assembly of empty versus functional single-stranded RNA viruses. Phys Rev E. 2008;78:051915. doi: 10.1103/PhysRevE.78.051915. [DOI] [PubMed] [Google Scholar]
- 39.Angelescu DG, Bruinsma R, Linse P. Monte Carlo simulations of polyelectrolytes inside viral capsids. Phys Rev E Stat Nonlin Soft Matter Phys. 2006;73:041942. doi: 10.1103/PhysRevE.73.041921. [DOI] [PubMed] [Google Scholar]
- 40.van der Schoot P, Bruinsma R. Electrostatics and the assembly of an RNA virus. Phys Rev E Stat Nonlin Soft Matter Phys. 2005;71:061928. doi: 10.1103/PhysRevE.71.061928. [DOI] [PubMed] [Google Scholar]
- 41.Elrad OM, Hagan MF. Encapsulation of a polymer by an icosahedral virus. Phys Biol. 2010;7:045003. doi: 10.1088/1478-3975/7/4/045003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kivenson A, Hagan MF. Mechanisms of Capsid Assembly around a Polymer. Biophys J. 2010;99:619. doi: 10.1016/j.bpj.2010.04.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Choi YG, Dreher TW, Rao ALN. tRNA elements mediate the assembly of an icosahedral RNA virus. Proc Natl Acad Sci USA. 2002;99:655–660. doi: 10.1073/pnas.022618199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sivakumaran K, Kim CH, Tayon R, Jr , Kao CC. RNA sequence and secondary structural determinants in a minimal viral promoter that directs replicase recognition and initiation of genomic plus-strand RNA synthesis. J Mol Biol. 1999;294:667–682. doi: 10.1006/jmbi.1999.3297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Annamalai P, Rao ALN. Replication-independent expression of genome components and capsid protein of brome mosaic virus in planta: a functional role for viral replicase in RNA packaging. Virology. 2005;338:96–111. doi: 10.1016/j.virol.2005.05.013. [DOI] [PubMed] [Google Scholar]
- 46.Annamalai P, Rao ALN. Packaging of brome mosaic virus subgenomic RNA is functionally coupled to replication-dependent transcription and translation of coat protein. J Virol. 2006;80:10096–10108. doi: 10.1128/JVI.01186-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Annamalai P, Rofail F, Demason DA, Rao ALN. Replication-coupled packaging mechanism in positive-strand RNA viruses: synchronized coexpression of functional multigenome RNA components of an animal and a plant virus in Nicotiana benthamiana cells by agroinfiltration. J Virol. 2008;82:1484–1495. doi: 10.1128/JVI.01540-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cadena-Nava RD, et al. Self-assembly of viral capsid protein and RNA molecules of different sizes: requirement for a specific high protein/RNA mass ratio. Journal of Virology. 2011 doi: 10.1128/JVI.06566–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bujarski JJ. Bromovirus isolation and RNA extraction. Methods Mol Biol. 1998;81:183–188. doi: 10.1385/0-89603-385-6:183. [DOI] [PubMed] [Google Scholar]
- 50.Proksch R, Schäffer TE, Cleveland JP, Callahan RC, Viani MB. Finite optical spot size and position corrections in thermal spring constant calibration. Nanotechnology. 2004;15:1344–1350. [Google Scholar]
- 51.Sambrook J, Russell DW. Separation of RNA According to Size: Electrophoresis of Glyoxylated RNA through Agarose Gels. Cold Spring Harb Protoc. 2006;2006:pdb.prot4057. doi: 10.1101/pdb.prot4057. [DOI] [PubMed] [Google Scholar]
- 52.Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor Laboratory Pr; 1989. [Google Scholar]
- 53.Ludtke SJ, Baldwin PR, Chiu W. EMAN: semiautomated software for high-resolution single-particle reconstructions. J Struct Biol. 1999;128:82–97. doi: 10.1006/jsbi.1999.4174. [DOI] [PubMed] [Google Scholar]
- 54.Israels R, Leermakers FAM, Fleer GJ. On the Theory of Grafted Weak Polyacids. Macromolecules. 1994;27:3087–3093. [Google Scholar]
- 55.Bohmer MR, Evers OA, Scheutjens J. Weak Polyelectrolytes between 2 Surfaces - Adsorption and Stabilization. Macromolecules. 1990;23:2288–2301. [Google Scholar]
- 56.van Lent B, Israels R, Scheutjens J, Fleer GJ. Interaction between Hairy Surfaces and the Effect of Free Polymer. J Colloid Interface Sci. 1990;137:380–394. [Google Scholar]
- 57.Bohmer MR, Koopal LK, Lyklema J. Micellization of Ionic Surfactants - Calculations Based on a Self-Consistent Field Lattice Model. J Phys Chem. 1991;95:9569–9578. [Google Scholar]
- 58.Siber A, Podgornik R. Nonspecific interactions in spontaneous assembly of empty versus functional single-stranded RNA viruses. Phys Rev E Stat Nonlin Soft Matter Phys. 2008;78:051915. doi: 10.1103/PhysRevE.78.051915. [DOI] [PubMed] [Google Scholar]
- 59.Hagan MF. A theory for viral capsid assembly around electrostatic cores. J Chem Phys. 2009;130:114902. doi: 10.1063/1.3086041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Manning GS. Limiting Laws and Counterion Condensation in Polyelectrolyte Solutions. 2 Self-Diffusion of Small Ions. J Chem Phys. 1969;51:934–938. [Google Scholar]
- 61.Manning GS. Limiting Laws and Counterion Condensation in Polyelectrolyte Solutions. 3 an Analysis Based on Mayer Ionic Solution Theory. J Chem Phys. 1969;51:3249–3252. [Google Scholar]
- 62.Manning GS. Counterion condensation on charged spheres, cylinders, and planes. J Phys Chem B. 2007;111:8554–8559. doi: 10.1021/jp0670844. [DOI] [PubMed] [Google Scholar]
- 63.Muthukumar M. Double screening in polyelectrolyte solutions: Limiting laws and crossover formulas. J Phys Chem. 1996;105:5183–5199. [Google Scholar]
- 64.Patra CN, Yethiraj A. Density functional theory for the distribution of small ions around polyions. J Phys Chem B. 1999;103:6080–6087. [Google Scholar]
- 65.Kundagrami A, Muthukumar M. Theory of competitive counterion adsorption on flexible polyelectrolytes: divalent salts. J Chem Phys. 2008;128:244901. doi: 10.1063/1.2940199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Liu X, Jiang W, Jakana J, Chiu W. Averaging tens to hundreds of icosahedral particle images to resolve protein secondary structure elements using a Multi-Path Simulated Annealing optimization algorithm. J Struct Biol. 2007;160:11–27. doi: 10.1016/j.jsb.2007.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


