Abstract
B-DNA becomes unstable under superhelical stress and is able to adopt a wide range of alternative conformations including strand-separated DNA and Z-DNA. Localized sequence-dependent structural transitions are important for the regulation of biological processes such as DNA replication and transcription. To directly probe the effect of sequence on structural transitions driven by torque, we have measured the torsional response of a panel of DNA sequences using single molecule assays that employ nanosphere rotational probes to achieve high torque resolution. The responses of Z-forming d(pGpC)n sequences match our predictions based on a theoretical treatment of cooperative transitions in helical polymers. “Bubble” templates containing 50–100 bp mismatch regions show cooperative structural transitions similar to B-DNA, although less torque is required to disrupt strand–strand interactions. Our mechanical measurements, including direct characterization of the torsional rigidity of strand-separated DNA, establish a framework for quantitative predictions of the complex torsional response of arbitrary sequences in their biological context.
Keywords: helix-coil transition, magnetic tweezers, nucleic acid, torque spectroscopy
Structural transitions of the DNA double helix are essential for accessing genomic information. Strand separation is required for the initiation of both transcription and replication, and it is favored by negative superhelical stress (1). The sequence dependence of stress-induced destabilization (2) has been hypothesized to affect the specific localization of replication initiation (3, 4) and the variable response of transcriptional promoters (5). Supercoiling can lead to either activation or repression of transcription, and promoter-specific responses are exploited by organisms that use supercoiling as a global regulator of changing transcriptional programs (6). In addition to strand separation, superhelical tension may also be released by the formation of Z-DNA, a left-handed double-helical structure (7, 8). Propensity for each of these structural transitions is strongly sequence-dependent; Z-DNA formation is favored by GC repeats and more generally by alternation of purines and pyrimidines (9, 10).
Bulk supercoiling and single molecule twisting experiments have been used to characterize the response of DNA to negative superhelical stress (4, 8, 9, 11). In single stretched DNA molecules, B-DNA ceases to display linear torsional elasticity at a critical torque of approximately -10 pNnm, entering an apparent cooperative structural transition to an underwound form (11, 12). This transition has been proposed to represent strand separation (13), but it has been noted that other noncanonical structures such as Z-DNA may also be simultaneously populated in random sequences under negative torques (14, 15). The critical torque may thus reflect a complex underlying response in which (for example) strand separation in AT-rich regions coexists with Z-DNA formation in GC repeats. Saturation of the underwinding transition leads to a net left-handed twist (14–16), which may reflect an average property of multiple contributing states. The corresponding region of DNA phase space has been labeled “L-DNA” (16) solely to recognize this overall handedness. Torque measurements on homogeneous sequences are needed to dissect the distinct contributions of specific structural transitions to the overall response of heterogeneous DNA molecules.
Mechanical perturbations can be used to obtain detailed sequence-dependent structural, thermodynamic, and kinetic properties of nucleic acids (17). Forces measured during unzipping of long double-stranded DNAs show clear sequence-dependent features that can largely be reproduced using calculations based on the primary sequence and assuming thermal equilibrium (18); recent measurements have been used to determine salt-dependent base pair free energies (19). Unfolding of RNA and DNA hairpins has been extensively studied using force spectroscopy, and high-resolution optical trapping measurements of a panel of small DNA hairpins allowed the exploration of the full sequence-dependent folding landscape of a nucleic acid (20). Despite the natural utility of torque as an alternative perturbation to tension for double-helical nucleic acids, no detailed sequence-dependent torque spectroscopy studies have been completed to date.
The rotor bead tracking assay (11) was the first of a number of methods introduced over the past decade (11, 12, 21–24) for measuring torque on single stretched DNA molecules. Torque is measured by observing the angular velocity of a submicron bead attached to the side of the DNA. This assay lacks versatility because of its baroque geometry and inability to impose fixed twist, and it has not been widely applied for torque measurements beyond its initial introduction. We have developed variants of the rotor bead tracking assay that overcome its original limitations, enabling routine torque spectroscopy of specific DNA sequences.
Here, we use our torque-measurement techniques to investigate specific states that can contribute to localized sequence-dependent responses to negative supercoiling. We initially characterize a long random DNA sequence, establishing the sequence-averaged mechanical properties of torque-destabilized DNA. These properties are consistent with a mixed state, which we investigate further by analyzing a panel of different sequences of interest (SOIs) chosen to test individual structural transitions. GC repeats were chosen to investigate the B-Z transition. Mismatch sequences were chosen as low-stability substrates to investigate strand separation and characterize the mechanical properties of strand-separated DNA. The use of homogeneous simple repeat sequences facilitates meaningful comparisons of experiment with theory and allows us to extract thermodynamic and structural parameters for sequence-dependent transitions. All of the transitions we have investigated are well described by a single statistical mechanical model for cooperative structural transitions in polymers.
Results
Feedback Enhanced Rotor Bead Tracking Assay for Torque Measurements.
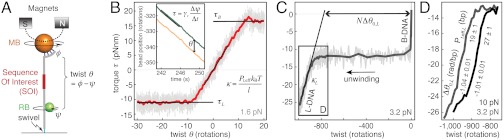
We have investigated sequence-dependent structural transitions using an extension of the rotor bead tracking (RBT) assay (11, 25) that allows torque to be measured as a function of imposed twist (Fig. 1). A single DNA molecule is stretched between a cover slip surface and a magnetic bead, while a second fluorescent “rotor” bead is attached to the side of the molecule (Fig. 1A). The DNA attachment to the cover slip surface is unconstrained and acts as a swivel. The attachment to the magnetic bead is torsionally constrained so that twist can be introduced by rotating the magnets. The rotor will spin to relieve this twist, but a fixed twist condition can be imposed by rotating the magnets to counteract this relaxation, using a “twist clamp” feedback algorithm. The torque τ can be dynamically measured from the angular velocity of the rotor as in previous work (11) using τ = ωγr, where ω is the angular velocity of the rotor bead and γr its rotational drag coefficient (Fig. 1B, Inset). γr is measured in situ for each rotor bead by analyzing Brownian fluctuations (see SI Text). In this work, we report torque-twist relationships obtained by linearly ramping the twist clamp setpoint up and down to sample twist values during unwinding and rewinding.
Fig. 1.
Feedback enhanced rotor bead tracking assay and torsional response of control DNA. (A) Experimental setup for dynamic torque measurements. The DNA molecule is attached to the cover slip surface by a single digoxigenin:α-digoxigenin interaction (light blue), which acts as a swivel. The magnetic bead (MB, orange) is attached via multiple fluorescein:α-fluorescein interactions (dark blue), producing a torsional constraint. The fluorescent rotor bead (RB, green) is attached to the side of the DNA molecule via biotin:neutravidin interactions. A specific “sequence of interest” (SOI, red) is placed between the MB and the RB attachment of the DNA molecule. The angular position ϕ of the MB is controlled by rotating the magnets, and the angle ψ of the RB is measured using videomicroscopy. The twist θ in the DNA is obtained as θ = ϕ - ψ with the equilibrium twist defined as θ = 0. Feedback control of MB angle can be used to impose a twist clamp. (B) Torque-twist diagram for 4.6-kb control DNA tethers. Unless stated otherwise, all such curves are obtained from four independent twist clamp experiments, and unwinding curves are superimposable with rewinding. Twist was ramped at 0.05 turns/s, torque was obtained from rotor angular velocities averaged over a 3-s window, and data were subsequently binned along the twist axis and averaged (red line). The postbuckling torque τB, critical torque for the B-L transition τL, and torsional spring constant  (where l is the length of the DNA and Pt,eff is the effective twist persistence length) can be extracted from the torque-twist plot. DNA twist and rotor angular velocities were obtained from raw traces (insert) showing the cumulative angles of the magnets (orange trace) and RB (green trace) as a function of time. (C) Torque-twist plot showing complete conversion of B-DNA to L-DNA under negative torque for an N base pair long control DNA held under 3.2 pN tension. A faster twist ramp and proportionately larger bin sizes have been used for data in the plateau region. By saturating the transition, the extrapolated change in helicity per base pair Δθ0,L can be obtained. From the slope κL the twist persistence length
(where l is the length of the DNA and Pt,eff is the effective twist persistence length) can be extracted from the torque-twist plot. DNA twist and rotor angular velocities were obtained from raw traces (insert) showing the cumulative angles of the magnets (orange trace) and RB (green trace) as a function of time. (C) Torque-twist plot showing complete conversion of B-DNA to L-DNA under negative torque for an N base pair long control DNA held under 3.2 pN tension. A faster twist ramp and proportionately larger bin sizes have been used for data in the plateau region. By saturating the transition, the extrapolated change in helicity per base pair Δθ0,L can be obtained. From the slope κL the twist persistence length  of L-DNA can be calculated. (D) L-DNA helicity and elasticity vary with tension. Δθ0,L and effective twist persistence length Pt,eff,L of the L state are displayed ± SEM, based on linear fits to the data spanning −15 and -20 pNnm. Averaged traces are each based on three independent measurements.
of L-DNA can be calculated. (D) L-DNA helicity and elasticity vary with tension. Δθ0,L and effective twist persistence length Pt,eff,L of the L state are displayed ± SEM, based on linear fits to the data spanning −15 and -20 pNnm. Averaged traces are each based on three independent measurements.
Structural Transitions and Elasticity of Random DNA.
To test our dynamic torque measurement method and establish baseline torque-twist relationships for a random DNA sequence, we tested a perfectly complementary double-stranded DNA segment, absent of obvious special sequence features. In a plot of torque as a function of twist (Fig. 1B), the nearly linear region around zero torque reflects the twist elasticity of B-DNA, from which we can extract the effective twist persistence length Pt,eff. The constant-torque plateau (τL) at approximately -10 pNnm reflects the structural transition from B-form to L-form (Fig. 1 B and C). After saturating this transition, a second linear region reflects the twist elasticity of L-DNA (Fig. 1 C and D). Under positive torque, a force-dependent torque plateau (τB) reflects the plectonemic buckling transition of B-DNA. We observe a torque “overshoot” at the beginning of this buckling transition (Fig. 1B and Fig. S1 A and D), which was previously inferred from indirect measurements (23, 26).
Our baseline measurements are consistent with established values for the high-force limiting value of the twist persistence length Pt,eff of DNA (approximately 100 nm or approximately 300 bp) (11, 12, 23) and the critical torque of the B-L transition (approximately -10 pNnm) (11, 12) and are summarized in supplementary Table S1. We have further discovered ionic strength effects (Fig. S1 B and C and Table S1), including a decrease in the critical torque for the B-L transition by approximately 15% at high ionic strength (Fig. S1B and Table S1). This latter effect of salt is in the opposite direction from the expected result if the B-L transition consisted solely of strand separation.
Initiation of the B-L transition occurs at low twist densities near σ = -0.02, reflecting local formation of alternative structures under supercoiling conditions that are biologically accessible. To measure the sequence-averaged mechanical properties of structures that form at the critical torque, the DNA molecule can be unwound through the entire torque plateau, so that no B-DNA remains (Fig. 1 C and D). Complete transformation of kilobase-scale DNA segments is unlikely to occur in biology, but knowledge of the overall helicity and rigidity of the resulting “L-DNA” places important constraints on any detailed model for sequence-dependent responses to negative supercoiling.
The length of the B-L plateau shows a difference in helicity of approximately 1.1 rad/bp between B-DNA and L-DNA at the critical torque, agreeing with previous reports (11, 14–16). We fit the linear region of L-DNA elasticity to obtain the torsional rigidity of L-DNA, which is 10–15 times softer than B-DNA (Fig. 1D and Table S1). Extrapolating from this region, we obtain a difference in helicity between B-DNA and L-DNA at zero torque, Δθ0,L = 1 rad/bp. L-DNA thus has the external properties of a left-handed structure, with an average extrapolated unstressed helical pitch of approximately 15 bp. The twist persistence length of L-DNA Pt,eff = 20–30 bp (Fig. 1D) is substantially larger than previous estimates of a few bp for strand-separated DNA (27, 28).
Our measurements of significant helicity and relatively high rigidity for L-DNA, together with a reduction in the coexistence torque at high ionic strength, are difficult to reconcile with a model in which L-DNA consists solely of strand-separated DNA. Our results are consistent with a model in which L-DNA consists of a mixture of strand-separated DNA and other noncanonical DNA structures, such as Z-DNA (11). To directly investigate the energetics of the B-Z transition and the properties of strand-separated DNA, we investigated sequences that are expected to form Z-DNA or strand-separated DNA at low torques.
GC Tracts Form Z-DNA Under Mild Negative Torques.
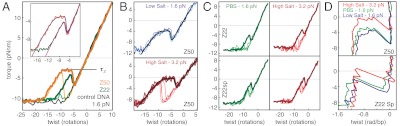
High salt conditions and negative torque induce Z-DNA formation in GC tracts (9, 29, 30). To investigate Z-DNA formation, we used SOIs containing either a 50 bp or a 22 bp GC repeat. Torque-twist plots of Z50 and Z22 collected at 1.6 pN of tension show clear transitions to a second state upon application of negative torque (Fig. 2A). B-Z transitions for Z50 show a negative torque overshoot followed by a plateau. The coexistence plateau occurs at τZ = -3 pNnm in PBS. For the shorter Z22 construct, the plateau is absent and the torque-twist diagram resembles the two-state behavior seen in RNA hairpin unzipping experiments (17). Under moderate or high salt conditions, the unwinding and rewinding curves show hysteresis, implying that B-Z transitions are not in equilibrium on the time scale of the experiment (Fig. 2 A and B). However, the plateau and overshoot motifs are seen in both the unwinding and rewinding curves. In low salt, unwinding and rewinding curves are in close agreement (Fig. 2B). Within the B-Z coexistence plateau region, torque-twist curves are reversible under all conditions. In subsequent analysis of the B-Z transition, we use rewinding curves as approximations for equilibrium torque-twist relationships.
Fig. 2.
Torsional response of GC repeats: Z-DNA formation. (A) Templates containing 50 bp (yellow, Z50) or 22 bp (green, Z22) GC repeat SOIs were assayed in PBS under 1.6 pN. Negative control DNA (black line) under identical conditions serves as a reference. Individual unwinding curves (fine lines) were variable and differed from the consistent behavior of rewinding curves (averaged, bold lines). Fits to a cooperative transition model (see SI Text) are displayed (inset) along with the corresponding experimental data (violet, Z50; red, Z22). (B) Overlay of individual unwinding (dark blue) and rewinding (light blue) experiments for Z50 under low salt conditions (Upper), showing reversibility. Under high salt conditions (Lower) the B-Z coexistence plateau occurs at lower absolute torques, and unwinding (bold dark red) and rewinding (fine light red) traces show hysteretic behavior. (C) The response of 22 bp GC repeats is affected by the inclusion of atomic spacers (Z22sp) and by increases in ionic strength (HS). Comparison of unwinding (fine lines) with averaged rewinding curves (bold lines) shows reduced hysteresis in Z22sp. (D) The torsional response of the DNA handles has been subtracted from averaged rewinding curves in order to show the inferred response of the SOI alone, plotted as torque vs. twist/bp.
Z-DNA formation is cooperative (9, 10, 30), which may be explained by a high nucleation penalty due to the formation of unfavorable conformations at junctions between Z-DNA and B-DNA (10, 30, 31). Once a nucleus of Z-DNA has been formed in a GC tract, it may be extended readily due to the small free energy difference between B-form and Z-form for d(pGpC). We fit the measured torque-twist relationships to a model for cooperative structural transitions in polymers (Fig. 2 A and B and SI Text) to verify our physical understanding and extract structural, mechanical, and thermodynamic parameters for alternative structural forms. In this model, each basepair within the SOI can exist in one of two states with differences in free energy (ΔG0) and twist (Δθ0) as well as a change in compliance associated with the transition, accounting for differing twist persistence lengths. Cooperativity in the model arises because junctions between differing states are assessed a free energy penalty J.
Fits to our model (see SI Text) reproduce the measured overshoot and plateau motifs (Fig. 2 A and B) and explain the torsional response of GC tracts: As negative twists are added to the DNA, negative torque initially accumulates linearly until a nucleus of Z-DNA is formed, upon which torque is partially relieved and the molecule enters a B-Z coexistence torque plateau, followed by a further linear accumulation of torque once the entire GC tract has been converted to Z-DNA (Fig. S2).
Our fit values for ΔG0 and J are close to bulk measurements for the B-Z transition in GC repeats (Table 1). Z22 and Z50 display changes in helicity Δθ0 = -0.9 rad/bp, which is smaller than the expected Δθ0 = -1.12 rad/bp based on gel electrophoresis assays (10) and X-ray crystal structures of Z-DNA (7). A possible explanation for this discrepancy is the contribution of helical distortions at B/Z junctions, which are believed to be formed within the GC-repeat region and have been estimated to occupy approximately 3 bp (30). Junctions may also contribute additional compliance to the Z-state, explaining unexpected posttransition softening for Z50 (Fig. 2 and Table S2).
Table 1.
Structural and thermodynamic parameters of sequence-dependent transitions induced by torque
Fits to model predictions (SI Text) were used to obtain changes in twist and free energy per bp, and the domain wall penalty J that accounts for the cooperativity of the transition. Buffers used were PBS, high salt (HS) and low salt (LS). Z22 and Z22sp show two-state behavior that is insufficient to simultaneously constrain J and ΔG0, so J was fixed to the value observed for Z50.
*Held constant during fitting. Value based on Z50.
To mitigate the effect of B/Z junctions, we created new constructs incorporating 9-atom-long spacers within the phosphate backbone at all 3′ and 5′ ends of the Z22 GC tract (Fig. 2C). The spacer-containing Z22sp constructs show increased changes in helicity as expected (Tab. 1) and also display reduced hysteresis (Fig. 2C). A comparison of Z22 and Z22sp provides an independent test of our measurement of the domain wall penalty J. Unlike Z22, the Z22sp construct lacks B/Z junctions after complete conversion to Z-DNA, leading to a shift in the torque-twist curve as expected (Fig. 2C).
A dramatic change in behavior is observed for B-Z transitions under high salt conditions of 1.8 M NaCl. We measure Δθ0 = -1.11 ± 0.02 rad/bp, indistinguishable from previously measured values of -1.12 rad/bp, and the coexistence plateau occurs at a very small negative torque of only -0.4 pNnm (Fig. 2 B and D and Table 1). The difference in compliance between B- and Z-states is also smaller than seen at lower ionic strength. The stiffness and Δθ0 values are consistent with a model in which high salt mitigates any end effects due to B/Z junctions, in addition to stabilizing Z-DNA.
Positive Torque Induces a Structural Transition in poly(dT) Bubbles.
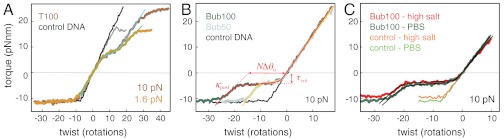
L-DNA has been proposed to be partially or entirely composed of strand-separated DNA, and thermodynamic models of supercoiling-induced strand separation depend on knowledge of the mechanical properties of strand-separated DNA. To investigate these properties, we tested the torsional response of DNA mismatch bubbles. SOIs created by inserting 100 dTs into both strands (T100) show a unique response (Fig. 3A). Near zero torque, the slope of the torque-twist curve is shallower than for negative control DNA, indicating softening of poly(dT) by a factor of > 10 relative to B-form (Table 1 and Table S2). When the molecules are overwound to a critical torque of approximately 8 pNnm, a reversible transition occurs to a second structural state. The changes in stiffness and helicity during this transition are tension-dependent, but in all cases the transition involves an extrapolated Δθ0 > 0 and a decrease in rigidity (Table 1, Fig. 3A and Table S2). This transition between distinct structural states within T100 suggests that there are weak but significant strand–strand interactions in the absence of torque, which are disrupted when torque is applied. Assuming that the high-energy softer state corresponds to strand-separated DNA, the positive values of Δθ0 imply that the unstressed conformation of poly(dT)•poly(dT) is slightly left-handed. We fit the structural transition using the same general model as for B-Z transitions (Fig. 3A), notably yielding a lower value for the boundary penalty J than is generally obtained for denaturation of matched duplexes (2, 27, 32), and lower than we obtained for B-Z transitions (Table 1 and Table S2). The twist persistence length of strand-separated DNA inferred from these measurements is approximately 7 bp at 1.6 pN of tension.
Fig. 3.
Torsional responses of mismatch bubbles. (A) Torque-twist curve for d(pT)100•d(pT)100 SOI (T100) and negative control DNA (gray tones) in PBS, together with fits to the cooperative transition model (light blue and green lines). (B) Response of 100 bp (dark green, Bub100) and 50 bp (light green, Bub50) d(pGpApCpT)n•d(pGpApCpT)n SOIs assayed in PBS under 10 pN of tension. Red and yellow lines show transition model fits. Characteristic curves show a plateau at a critical torque τcrit, a posttransition torsional spring constant κpost, and a change in helicity Δθ0 for the transition. (C) The response of Bub100 is insensitive to increased ionic strength. Torque-twist curves are shown for Bub100 in PBS (green) and in high ionic strength (red line) alongside control DNA (light green and orange, respectively) under 10 pN of tension. Transition model fits are shown in black.
Mismatch Bubbles form B-Like Helical Structure.
Long stretches of mismatched DNA have been used to investigate in vitro DNA helicase loading. Mismatch bubbles of the type d(pGpApCpT)n•d(pGpApCpT)n serve as a synthetic origin of DNA replication and enable helicase loading, whereas canonical double-stranded DNA does not (33). We have measured torque-twist curves for constructs containing 100 bp (Bub100) or 50 bp (Bub50) of mismatched DNA. In contrast to poly(dT) bubbles, torsionally relaxed GACT mismatch bubbles are not detectably softer than B-DNA. At low negative torques, Bub50 and Bub100 exhibit a reversible plateau corresponding to a structural transition (Fig. 3B). The length of the plateau is proportional to the length of the mismatch, with Δθ0 close to 1 rad/bp (Table 1). These measurements suggest a low-stability B-DNA-like structure for the mismatch inserts. The base pair geometry of GACT mismatch bubbles, in which pyrimidine bases oppose purines, may help conserve a Watson–Crick-like structure with a reduced number of hydrogen bonds in comparison with complementary B-DNA. From model fits, the change in free energy per base pair ΔG0 for disrupting GACT mismatches is approximately 0.55 kcal/mol, comparable to previous measurements based on van’t Hoff analysis of single base pair mismatches in oligonucleotides (34). In contrast to B-Z transitions in GC repeats, the critical torque for disruption of Bub100 is not significantly affected by increasing the ionic strength (Fig. 3C).
To confirm the Bub100 signatures and further validate the twist clamp feedback assay, we repeated the measurements using an alternative variant of rotor bead tracking (Fig. S4). In contrast to dynamic torque measurements based on the angular velocity of the rotor, this second approach uses a static experimental setup in which torque is quantified by measuring the SOI against a calibrated transducer DNA segment.
Discussion
Several assays have previously been described for measuring torque in single stretched DNA molecules (11, 12, 21–23). All methods employ at least one microparticle as a handle for applying pN-scale tensions to the DNA using optical or magnetic forces. In most methods, this handle also serves as a rotational probe for determining the torque, employing soft torsional potentials generated by either magnetic fields (12, 21, 22) or polarized optical traps (23, 35). For any torque measurement method, resolution is ultimately limited by the Brownian torque noise (4kBTγrB)1/2, where γr is the rotational drag coefficient of the torque probe and B is the bandwidth of the measurement (22). For a spherical probe, γr scales with the cube of the radius, and so a reduction in probe size can dramatically improve torque resolution. If the rotational probe is also used as a handle for applying tension, its volume cannot be reduced arbitrarily without sacrificing accessible force ranges. Compromises between the conflicting requirements of torque probes and force handles have been a factor in the poor torque resolution of some previous methods, necessitating hours or even days (22) of noise-averaging data collection to determine torque-twist relationships.
Our methods based on the rotor bead tracking assay (11) avoid compromises by segregating the force handle from the torque probe. The dynamic implementation relies on the viscous drag of the rotor bead and allows a direct torque measurement with similar performance to assays employing an angular optical trap and nanofabricated quartz cylinders (23, 24): Using approximately 400 nm rotor beads, we measure torque to within approximately 1 pNnm by integrating over 3 s of data. For the static implementation (Fig. S4), which relies on a calibrated molecular spring, there are no barriers in principle to using smaller rotational probes that could further improve the resolution by orders of magnitude. Neither assay relies on a micron-sized rotational probe (12, 21, 22), allowing torque resolutions sufficient for routine sequence-dependent torque spectroscopy. Unlike angular optical trap measurements (23), the torque measurements reported here do not include simultaneous high-resolution extension measurements, which may be incorporated in future improvements to the assays. In an additional tradeoff with competing methods, our assays based on rotor bead tracking (11) require specialized molecular constructs with three distinct attachment chemistries. These constructs are readily prepared using standard molecular biology techniques, as described in SI Text.
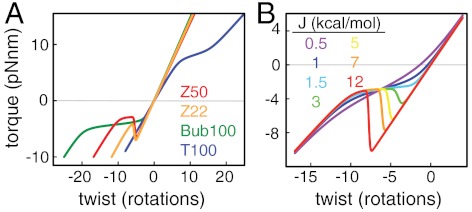
Accurate quantitative models of sequence-dependent structural transitions are needed to explain sophisticated cellular functions such as supercoiling-dependent regulation of gene expression profiles (5, 6). In three divergent examples, we found that the behaviors of short regions of homogenous sequences were well described by a model for cooperative structural transitions in polymers, allowing the extraction of thermodynamic parameters relevant to sequence-dependent torsional responses (Fig. 4). In addition to quantifying base pair stabilities, measurements of torque as a function of imposed twist allow sensitive determination of the cooperativity parameter J that arises from junctions between differing states (Fig. 4B). We have also introduced an experimental strategy that rigorously challenges transition theory by changing the boundary conditions: Atomic spacers can be introduced at the edge of an SOI to create a boundary that thermodynamically approximates a free end but still permits the application of a torsional constraint. Finally, our analysis enables compliance measurements for soft high-energy states such as strand-separated DNA, providing an important (2, 27) but often poorly quantified parameter needed for predicting mechanical transitions. Our theory and methods establish a framework for understanding the torsional response of arbitrary sequences, which ultimately should be predictable from reductionist measurements on simple sequences such as those described here.
Fig. 4.
A model for cooperative structural transitions in homogeneous sequences under torque. (A) Fits are shown for Z50, Z22, Bub100, and T100, demonstrating the range of behaviors explained by the model described in SI Text. (B) Visualization of the effect of cooperativity in our model, shown by a series of calculated torque-twist curves, using all the parameters from Z50 except for the domain wall penalty J, which was varied from 0.5 to 12 kcal/mol.
Z-DNA formation can have specific effects on both the initiation (36) and elongation (37) phases of transcription, and Z-forming sequences may also act as efficient buffers to absorb changes in linking number without changing torque, avoiding perturbations of biological processes. The thermodynamic parameters of Z-DNA formation calculated from our measurements are in good agreement with previous measurements based on gel electrophoresis (10) (Table 1). For B-Z transitions under moderate or high salt conditions, we observe hysteretic effects reflecting kinetic barriers to Z-DNA formation (Fig. 2), consistent with bulk kinetics: at a constant supercoiling density of σ = -0.05, the half time for the B-Z transition of d(GC)16 was measured to be approximately 20 min (38).
Duplexes with extended mismatch regions are intermediates in homologous DNA recombination (39, 40) and other hybridization-driven processes. Hybrid destabilization due to individual basepair mismatches has been extensively characterized (34), but little is known about the structure of long mismatched stretches or the cooperativity of mismatched strand–strand interactions. Extended mismatches have been assumed to function as open bubbles in helicase loading experiments (33). Our measurements of cooperative structural transitions in Bub50 and Bub100 change our physical understanding of these experiments: Rather than an open bubble, the DNA may form a region of weak double helix that requires energy input from protein binding to separate the strands.
Both of the mismatch sequences we investigated show a surprising degree of cooperative structure formation, but the two sequences also form strikingly different structures, shedding new light on the plasticity of double-stranded DNA. GACT bubbles seem to adopt a B-DNA-like structure (Fig. 3B), while the properties of the poly(dT) bubble clearly diverge from B-form (Fig. 3A). Hydrogen bonds within dT•dT base pairs have been observed in single dT•dT mismatches with NMR spectroscopy (41) and predicted based on a X-ray crystal structure of two stemloop DNAs (42). Our results suggest that poly(dT)•poly(dT) forms a soft ladder-like structure with a slight left-handed helicity; interactions between the strands are disrupted under mild positive torques to form a second state that is even softer and presumably corresponds to strand-separated DNA. At 1.6 pN, the twist persistence length of this state is approximately 7 bp (Fig. 3A and Table S2), slightly larger than previous estimates of approximately 2 bp for strand-separated DNA (27, 28).
Models of biological site-specific responses to supercoiling must account for the interplay between multiple available underwound structural forms (43). For long random sequences, the single “B-DNA to L-DNA” plateau at -10 pNnm may obscure a complex underlying response, in which specific sequences preferentially undergo differing structural transitions. Our direct measurements of B-Z transitions underscore the ease with which high-propensity sequences can form Z-DNA in preference to strand separation, and we have measured properties of L-DNA that are difficult to reconcile with a model in which L-DNA consists of strand-separated DNA alone. L-DNA has a torsional persistence length significantly greater than either our estimates or previous estimates for strand-separated DNA, is stabilized rather than destabilized by high ionic strength, and is confirmed to be highly helical. These measurements are all consistent with a previous proposal (11, 16) in which L-DNA consists of a mixture of strand-separated DNA and noncanonical structures such as Z-DNA. In biological contexts, trans-acting factors can bias the equilibrium between available structural transitions, and it should be emphasized that stress-induced destabilization of the duplex (1) is important even when spontaneous strand separation is absent: Linking number deficits may be absorbed by stabilizing a bubble upon protein binding (44) or invasion of a homologous strand to form a D loop (45), without ever forming an open bubble as a stable intermediate in this transaction.
Our measurements of the helicity and torsional rigidity of L-DNA are in good agreement with another study (46) that appeared while our manuscript was under review. This study also concluded that L-DNA has a low bending rigidity and used two different heterogeneous sequences to show that the mechanical properties of L-DNA can differ depending on sequence composition. All of these measurements are consistent with the “mixed state” hypothesis.
Our measurements of mechanics and thermodynamics of short homogeneous sequences have begun to describe the sequence-dependent folding landscape for superhelical double-stranded DNA. Application of our experimental and theoretical methods to a larger panel of DNA sequences will enable explorations of the full conformational range of DNA under torque and allow us to relate molecular properties to localized responses with functional roles in supercoiling-dependent regulation of replication, recombination, and transcription. The sequence-dependent torsional response of paired and mispaired duplexes is also of direct relevance to understanding and designing the mechanical behavior of DNA nanostructures. DNA nanotechnology has already benefited from mechanical models parameterized using previous single molecule measurements (47). The high-resolution torque measurements techniques we have established may be adapted to mechanochemical studies of DNA-associated enzymes and are broadly applicable to mechanical investigations of multistranded biopolymers.
Methods
Detailed methods are described in SI Text. DNA molecules, beads, and flow cells were prepared using similar procedures to previous work (11). The following three imaging buffers were used for rotor bead tracking experiments. PBS: 137 mM NaCl, 2.7 mM KCl, 0.5 mg/mL BSA, 10 mM Na2HPO4, 2 mM KH2PO4, pH 7.4. High salt: 1.8 M NaCl, 0.5 mg/mL BSA, 50 mM Tris, pH 7.4. Low salt: 0.5 mg/mL BSA, 10 mM Tris, pH 7.4. All buffers also contained 5 mM EDTA and an oxygen scavenging system. Rotor bead tracking experiments were performed on an inverted fluorescence microscope equipped with magnetic tweezers. Data analysis, models for fitting and statistics are described in SI Text.
Supplementary Material
Acknowledgments.
We would like to thank J. Berger, J. Marko, A. Spakowitz, A. Vologodskii, A. Basu, P. Lebel, M. Elting, P. Pease, and the Bryant and Berger groups for useful discussions and comments on the manuscript. This work was supported by a Pew Scholars Award to Z.B.; by a Morgridge Family Stanford Graduate Fellowship to L.E.F.; and by an EMBO long-term fellowship, a Stanford University Dean’s Fellowship, and an Swiss National Science Foundation fellowship for advanced researchers to F.C.O.
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1113532109/-/DCSupplemental.
References
- 1.Benham CJ. Torsional stress and local denaturation in supercoiled DNA. Proc Natl Acad Sci USA. 1979;76:3870–3874. doi: 10.1073/pnas.76.8.3870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Benham CJ. Energetics of the strand separation transition in superhelical DNA. J Mol Biol. 1992;225:835–847. doi: 10.1016/0022-2836(92)90404-8. [DOI] [PubMed] [Google Scholar]
- 3.Ak P, Benham CJ. Susceptibility to superhelically driven DNA duplex destabilization: A highly conserved property of yeast replication origins. PLoS Comput Biol. 2005;1:e7. doi: 10.1371/journal.pcbi.0010007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kowalski D, Natale DA, Eddy MJ. Stable DNA unwinding, not “breathing,” accounts for single-strand-specific nuclease hypersensitivity of specific A+T-rich sequences. Proc Natl Acad Sci USA. 1988;85:9464–9468. doi: 10.1073/pnas.85.24.9464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Peter BJ, et al. Genomic transcriptional response to loss of chromosomal supercoiling in Escherichia coli. Genome Biol. 2004;5:R87. doi: 10.1186/gb-2004-5-11-r87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vijayan V, Zuzow R, O’Shea EK. Oscillations in supercoiling drive circadian gene expression in cyanobacteria. Proc Natl Acad Sci USA. 2009;106:22564–22568. doi: 10.1073/pnas.0912673106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang AHJ, et al. Molecular-structure of a left-handed double helical DNA fragment at atomic resolution. Nature. 1979;282:680–686. doi: 10.1038/282680a0. [DOI] [PubMed] [Google Scholar]
- 8.Peck LJ, Nordheim A, Rich A, Wang JC. Flipping of cloned d(pCpG)n. d(pCpG)n DNA sequences from right- to left-handed helical structure by salt, Co(III), or negative supercoiling. Proc Natl Acad Sci USA. 1982;79:4560–4564. doi: 10.1073/pnas.79.15.4560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nordheim A, et al. Negatively supercoiled plasmids contain left-handed Z-DNA segments as detected by specific antibody binding. Cell. 1982;31:309–318. doi: 10.1016/0092-8674(82)90124-6. [DOI] [PubMed] [Google Scholar]
- 10.Peck LJ, Wang JC. Energetics of B-to-Z transition in DNA. Proc Natl Acad Sci USA. 1983;80:6206–6210. doi: 10.1073/pnas.80.20.6206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bryant Z, et al. Structural transitions and elasticity from torque measurements on DNA. Nature. 2003;424:338–341. doi: 10.1038/nature01810. [DOI] [PubMed] [Google Scholar]
- 12.Lipfert J, Kerssemakers JW, Jager T, Dekker NH. Magnetic torque tweezers: Measuring torsional stiffness in DNA and RecA-DNA filaments. Nat Methods. 2010;7:977–980. doi: 10.1038/nmeth.1520. [DOI] [PubMed] [Google Scholar]
- 13.Allemand JF, Bensimon D, Lavery R, Croquette V. Stretched and overwound DNA forms a Pauling-like structure with exposed bases. Proc Natl Acad Sci USA. 1998;95:14152–14157. doi: 10.1073/pnas.95.24.14152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Leger JF, et al. Structural transitions of a twisted and stretched DNA molecule. Phys Rev Lett. 1999;83:1066–1069. [Google Scholar]
- 15.Sarkar A, Leger JF, Chatenay D, Marko JF. Structural transitions in DNA driven by external force and torque. Phys Rev E Stat Nonlin Soft Matter Phys. 2001;63:051903. doi: 10.1103/PhysRevE.63.051903. [DOI] [PubMed] [Google Scholar]
- 16.Bustamante C, Bryant Z, Smith SB. Ten years of tension: Single-molecule DNA mechanics. Nature. 2003;421:423–427. doi: 10.1038/nature01405. [DOI] [PubMed] [Google Scholar]
- 17.Tinoco I, Li PT, Bustamante C. Determination of thermodynamics and kinetics of RNA reactions by force. Q Rev Biophys. 2006;39:325–360. doi: 10.1017/S0033583506004446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bockelmann U, Thomen P, Essevaz-Roulet B, Viasnoff V, Heslot F. Unzipping DNA with optical tweezers: High sequence sensitivity and force flips. Biophys J. 2002;82:1537–1553. doi: 10.1016/S0006-3495(02)75506-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Huguet JM, et al. Single-molecule derivation of salt dependent base-pair free energies in DNA. Proc Natl Acad Sci USA. 2010;107:15431–15436. doi: 10.1073/pnas.1001454107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Woodside MT, et al. Direct measurement of the full, sequence-dependent folding landscape of a nucleic acid. Science. 2006;314:1001–1004. doi: 10.1126/science.1133601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Celedon A, et al. Magnetic tweezers measurement of single molecule torque. Nano Lett. 2009;9(4):1720–1725. doi: 10.1021/nl900631w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mosconi F, Allemand JF, Croquette V. Soft magnetic tweezers: A proof of principle. Rev Sci Instrum. 2011;82:034302. doi: 10.1063/1.3531959. [DOI] [PubMed] [Google Scholar]
- 23.Forth S, et al. Abrupt buckling transition observed during the plectoneme formation of individual DNA molecules. Phys Rev Lett. 2008;100:148301. doi: 10.1103/PhysRevLett.100.148301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Deufel C, Forth S, Simmons CR, Dejgosha S, Wang MD. Nanofabricated quartz cylinders for angular trapping: DNA supercoiling torque detection. Nat Methods. 2007;4:223–225. doi: 10.1038/nmeth1013. [DOI] [PubMed] [Google Scholar]
- 25.Gore J, et al. DNA overwinds when stretched. Nature. 2006;442:836–839. doi: 10.1038/nature04974. [DOI] [PubMed] [Google Scholar]
- 26.Brutzer H, Luzzietti N, Klaue D, Seidel R. Energetics at the DNA supercoiling transition. Biophys J. 2010;98:1267–1276. doi: 10.1016/j.bpj.2009.12.4292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bauer WR, Benham CJ. The free energy, enthalpy and entropy of native and of partially denatured closed circular DNA. J Mol Biol. 1993;234:1184–1196. doi: 10.1006/jmbi.1993.1669. [DOI] [PubMed] [Google Scholar]
- 28.Marko JF. Torque and dynamics of linking number relaxation in stretched supercoiled DNA. Phys Rev E Stat Nonlin Soft Matter Phys. 2007;76:021926. doi: 10.1103/PhysRevE.76.021926. [DOI] [PubMed] [Google Scholar]
- 29.Lee M, Kim SH, Hong SC. Minute negative superhelicity is sufficient to induce the B-Z transition in the presence of low tension. Proc Natl Acad Sci USA. 2010;107:4985–4990. doi: 10.1073/pnas.0911528107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fuertes MA, Cepeda V, Alonso C, Perez JM. Molecular mechanisms for the B-Z transition in the example of poly[d(G-C)center dot d(G-C)] polymers. A critical review. Chem Rev. 2006;106:2045–2064. doi: 10.1021/cr050243f. [DOI] [PubMed] [Google Scholar]
- 31.Ha SC, Lowenhaupt K, Rich A, Kim YG, Kim KK. Crystal structure of a junction between B-DNA and Z-DNA reveals two extruded bases. Nature. 2005;437:1183–1186. doi: 10.1038/nature04088. [DOI] [PubMed] [Google Scholar]
- 32.Amirikyan BR, Vologodskii AV, Lyubchenko Yu L. Determination of DNA cooperativity factor. Nucleic Acids Res. 1981;9:5469–5482. doi: 10.1093/nar/9.20.5469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Smelkova NV, Borowiec JA. Synthetic DNA replication bubbles bound and unwound with twofold symmetry by a simian virus 40 T-antigen double hexamer. J Virol. 1998;72:8676–8681. doi: 10.1128/jvi.72.11.8676-8681.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Aboul-ela F, Koh D, Tinoco I, Martin FH. Base-base mismatches. Thermodynamics of double helix formation for dCA3XA3G + dCT3YT3G (X, Y = A,C,G,T) Nucleic Acids Res. 1985;13:4811–4824. doi: 10.1093/nar/13.13.4811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.La Porta A, Wang MD. Optical torque wrench: Angular trapping, rotation, and torque detection of quartz microparticles. Phys Rev Lett. 2004;92:190801. doi: 10.1103/PhysRevLett.92.190801. [DOI] [PubMed] [Google Scholar]
- 36.Wittig B, Wolfl S, Dorbic T, Vahrson W, Rich A. Transcription of human c-myc in permeabilized nuclei is associated with formation of Z-DNA in three discrete regions of the gene. EMBO J. 1992;11:4653–4663. doi: 10.1002/j.1460-2075.1992.tb05567.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ditlevson JV, et al. Inhibitory effect of a short Z-DNA forming sequence on transcription elongation by T7 RNA polymerase. Nucleic Acids Res. 2008;36:3163–3170. doi: 10.1093/nar/gkn136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Peck LJ, Wang JC, Nordheim A, Rich A. Rate of B to Z structural transition of supercoiled DNA. J Mol Biol. 1986;190:125–127. doi: 10.1016/0022-2836(86)90082-3. [DOI] [PubMed] [Google Scholar]
- 39.Kowalczykowski SC, Dixon DA, Eggleston AK, Lauder SD, Rehrauer WM. Biochemistry of homologous recombination in Escherichia coli. Microbiol Rev. 1994;58:401–465. doi: 10.1128/mr.58.3.401-465.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.San Filippo J, Sung P, Klein H. Mechanism of eukaryotic homologous recombination. Annu Rev Biochem. 2008;77:229–257. doi: 10.1146/annurev.biochem.77.061306.125255. [DOI] [PubMed] [Google Scholar]
- 41.Kouchakdjian M, Li BF, Swann PF, Patel DJ. Pyrimidine. pyrimidine base-pair mismatches in DNA. A nuclear magnetic resonance study of T.T pairing at neutral pH and C.C pairing at acidic pH in dodecanucleotide duplexes. J Mol Biol. 1988;202:139–155. doi: 10.1016/0022-2836(88)90526-8. [DOI] [PubMed] [Google Scholar]
- 42.Chattopadhyaya R, Ikuta S, Grzeskowiak K, Dickerson RE. X-ray structure of a DNA hairpin molecule. Nature. 1988;334:175–179. doi: 10.1038/334175a0. [DOI] [PubMed] [Google Scholar]
- 43.Anshelevich VV, Vologodskii AV, Frank-Kamenetskii MD. A theoretical study of formation of DNA noncanonical structures under negative superhelical stress. J Biomol Struct Dyn. 1988;6:247–259. doi: 10.1080/07391102.1988.10507711. [DOI] [PubMed] [Google Scholar]
- 44.Revyakin A, Ebright RH, Strick TR. Promoter unwinding and promoter clearance by RNA polymerase: Detection by single-molecule DNA nanomanipulation. Proc Natl Acad Sci USA. 2004;101:4776–4780. doi: 10.1073/pnas.0307241101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Strick TR, Croquette V, Bensimon D. Homologous pairing in stretched supercoiled DNA. Proc Natl Acad Sci USA. 1998;95:10579–10583. doi: 10.1073/pnas.95.18.10579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sheinin MY, Forth S, Marko JF, Wang MD. Underwound DNA under tension: Structure, elasticity, and sequence-dependent behaviors. Phys Rev Lett. 2011;107:108102. doi: 10.1103/PhysRevLett.107.108102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Dietz H, Douglas SM, Shih WM. Folding DNA into twisted and curved nanoscale shapes. Science. 2009;325:725–730. doi: 10.1126/science.1174251. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.