Abstract
The development, evaluation, and implementation of new and improved diagnostics have been identified as critical needs by human immunodeficiency virus (HIV) and tuberculosis researchers and clinicians alike. These needs exist in international and domestic settings and in adult and pediatric populations. Experts in tuberculosis and HIV care, researchers, healthcare providers, public health experts, and industry representatives, as well as representatives of pertinent US federal agencies (Centers for Disease Control and Prevention, Food and Drug Administration, National Institutes of Health, United States Agency for International Development) assembled at a workshop proposed by the Diagnostics Working Group of the Federal Tuberculosis Taskforce to review the state of tuberculosis diagnostics development in adult and pediatric populations.
A workshop proposed by the Diagnostics Working Group of the Federal Tuberculosis Taskforce was convened in Silver Spring, Maryland, in June 2011 to review the state of tuberculosis diagnostics development in adult and pediatric populations. The objectives of the workshop were to initiate discussion and facilitate the identification and evaluation of diagnostic tools for tuberculosis and tuberculosis/human immunodeficiency virus (HIV) coinfection. This article, which provides a summary of the key points discussed in the Clinical Research and Development of Tuberculosis Diagnostics track of the workshop, is divided by technologies and platforms currently under development or optimization, including (1) culture-based technologies, (2) molecular-based technologies, and (3) nonmolecular, novel technologies for diagnosis. The objective of the Clinical Research and Development Track was to bring together principal groups and researchers in the field of tuberculosis diagnostics to (1) identify and prioritize critical and important clinical research studies for the evaluation of current and future tuberculosis diagnostics, (2) identify and disseminate information regarding resources available to researchers, and (3) coordinate research efforts to ensure expediency of research critical to this field and to maximize efficient use of available resources. The main topics of discussion included: improving diagnostic tests; moving from silos to synergy; current barriers and challenges with existing platforms; increasing productivity; and collaboration. In the nearly 3 days of presentations and discussions, 2 primary themes emerged: building and maintaining momentum and moving from silos to synergy (see the viewpoint article from Kim et al, this supplement) .
BUILDING MOMENTUM AND MOVING FROM SILOS TO SYNERGY FOR IMPROVED DIAGNOSTIC TESTING
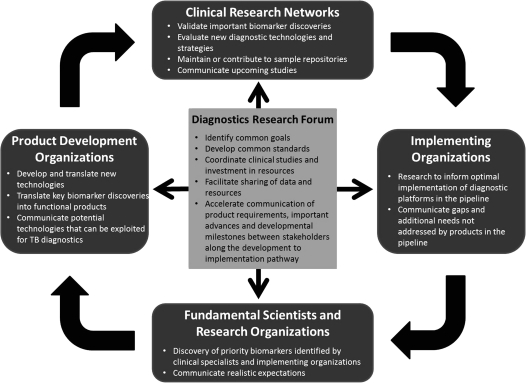
After a long drought in the tuberculosis diagnostics pipeline, the timing is right to take advantage of a powerful range of innovative technologies, along with increased potential for new molecular approaches, making it more critical than ever to work together efficiently and productively. The World Health Organization (WHO) objectives for tuberculosis test development include the need to (1) simplify and improve detection of tuberculosis cases, particularly point-of-care (POC) tests with same-day results; (2) enable more effective monitoring of tuberculosis treatment for latent and active cases; (3) rapidly identify drug resistance to first- and second-line antituberculosis medicines; and (4) reliably identify latent tuberculosis infection and determine the risk of progression to active disease. This workshop highlighted that no single group has enough resources to take on research in tuberculosis diagnostics alone. Adaptation and enhancement of existing resources and coordinated strategies and research agendas will be needed for efficient development of novel diagnostics (Figure 1). Prioritization of the scientific agenda in the context of information and data sharing among the stakeholders should occur at all levels, possibly through the development of a Tuberculosis Diagnostics Research Forum that would prevent redundancy and bring researchers and clinicians closer to synergy in the collective search for improved tuberculosis diagnostics. Table 1 shows selected leading tuberculosis diagnostics, as well as those in advanced stages of development.
Figure 1.
Conceptual framework for building momentum and moving from silos to synergy for improved diagnostic tests.
Table 1.
Tuberculosis Diagnostics: Approved Modalities and Those in Advanced Stages of Development
| Modality | Example | Strength | Weakness | Sensitivity | Affordability | Simplicity | Principal Setting | Speed of Detection |
| Smear microscopy | Ziehl-Neelsen, Auramine | Minimal equipment required; low cost | Poor sensitivity, especially for extrapulmonary tuberculosis, children, and HIV coinfection; operator-dependent; no DST | Low | Inexpensive | Simple | Local labs | Rapid |
| Traditional solid culture on egg-based media | Ogawa, Lowenstein-Jensen | Low cost; simple; robust; can provide DST | Some biosafety concerns; DST usually delayed after detection; poorly standardized; moderate sensitivity | Moderate | Moderate | Simple | Regional labs | Slow |
| Modern culture on synthetic media | BACTEC MGIT, thin-layer agar | Sensitive; provides DST; well validated for extrapulmonary tuberculosis | Biosafety concerns; training and equipment requirements; cross-contamination and inability to detect polyclonal infection (liquid); DST usually delayed after detection; requires international cold chain supplies | High | Moderate/Expensive | Moderate | Regional labs | Intermediate |
| MODS | In-house protocol; Hardy kit | Simultaneous DST; sensitive; rapid for culture | Biosafety concerns; training and equipment requirements; requires international cold chain supplies | High | Moderate/Expensive | Moderate | Regional labs | Intermediate |
| NAAT | Amplicor, Gen-Probe, LAMP, GeneXpert | Simultaneous DST; most tests are simple; biosafety | Genotypic DST has uncertain clinical implications; moderate sensitivity | Moderate | Moderate/Expensive | Moderate/Simple | Local/Regional labs | Rapid |
| Line probe | INNO-LiPARif, MTBDR | Provides rapid DST for patients known to have tuberculosis | Expensive; complex; principally used for DST, not tuberculosis diagnosis; genotypic DST has uncertain clinical implications; requires international cold chain supplies | Low | Expensive | Complex | Regional labs | Rapid |
| LAM | TB ELISA | Nonsputum based | Low sensitivity in HIV-uninfected patients | Low | Undefined | Moderate | Regional labs? | Rapid |
Abbreviations: DST, drug-susceptibility testing; HIV, human immunodeficiency virus; LAM, lipoarabinomannan; LAMP, loop-mediated isothermal amplification; MODS, microscopic-observation drug susceptibility; NAAT, nucleic acid amplification tests.
CULTURE-BASED TECHNOLOGIES
Worldwide, acid-fast bacilli (AFB) sputum smear microscopy is the most widely used method for identifying tuberculosis cases. It is inexpensive and rapid. Overall, however, AFB smear-based diagnosis has several major drawbacks, including low sensitivity and insufficient specificity, particularly in individuals who are HIV-infected; the absence of drug-susceptibility information; and, importantly, significant differences in performance depending on the operator [1–3]. As a consequence, sputum culture remains the recognized gold standard for confirming diagnosis of tuberculosis, despite the delays and complexities involved with culture-based diagnostics. Culture improves the sensitivity and specificity of mycobacteriology for the diagnosis of tuberculosis; it provides live bacteria by which drug susceptibility and genotyping can be assessed as a method for treatment monitoring; it is the most studied; and it is applicable broadly to sputum, urine, pleural fluid, cerebrospinal fluid, and biopsied or excised tissue, among other specimen types. However, the need for biosafety facilities and specially trained staff to perform the necessary procedures, issues of cross-contamination, lack of standardized methodology, delays in diagnosis, and lack of access in high-burden countries remain flaws of conventional culture methods [4–6]. Using this paradigm, globally around one-third of tuberculosis cases go undetected, detection in children remains poor, as many as 20% of tuberculosis cases in HIV infection are culture negative, and only a small proportion of multidrug-resistant (MDR) tuberculosis is recognized at the time of initial tuberculosis diagnosis [4, 7].
Cultures using liquid media are more sensitive and faster in reaching a diagnosis than most traditional techniques, which use egg-based solid media, although culture using thin-layer synthetic agar improves the performance of solid culture media [8, 9]. Efforts to automate and streamline components of culturing Mycobacterium tuberculosis have led to development of several commercial liquid-culture-based technologies. These include the BacT/Alert 3D System (bioMérieux), the Versa TREK system (Trek Diagnostic Systems), the BACTEC MGIT 960 system (Becton Dickinson), and the microscopic-observation drug-susceptibility assay (MODS; Hardy Diagnostics). Confirmation of M. tuberculosis speciation and differentiation from other mycobacteria using these systems is commonly achieved with nonmolecular approaches including the MGIT TBc Identification Test (TBc ID, Becton Dickinson) and Capilia TB (TAUNS), both MTP64-based immunochromatographic assays, and concurrent culture or subculture in selective agents such as p-nitrobenzoic acid [10–12]. The MODS assay is based on the detection of the characteristic morphology of tuberculosis under an inverted light microscope, is accurate and rapid [13], and usually involves the identification of M. tuberculosis and MDR tuberculosis concurrently, which is infrequently included in other liquid-culture techniques [14, 15]. The first-line antituberculosis drugs isoniazid and rifampicin are incorporated into the MODS assay, permitting simultaneous direct testing for MDR tuberculosis. A low-cost, all-reagents-provided MODS test kit has recently been developed for commercial sale, addressing a common limitation of all noncommercial diagnostic tests, that of lack of standardization.
Despite the advances in and automation of liquid-culture-based technologies and the fact that positive culture represents the gold standard for diagnosis, there continue to be significant challenges with these techniques, including cost, logistics of specimen handling and specimen transport, biosafety precautions, and the need for extensive personnel training [6]. These concerns notwithstanding, a recent WHO policy statement recommends implementation of liquid-culture systems as part of a country-specific comprehensive plan for laboratory capacity strengthening [8]. Field and cost-effectiveness studies are required to assess the impact of such practice on tuberculosis control worldwide [16]. The development of an assay, culture based or otherwise, ends with assessments of implementation and outcomes, and innumerable challenges must be considered and addressed on the road to successfully rolling out new diagnostics [6]. In addition to the need for field studies, there remain a number of procedural issues as well as scientific questions related to smear-based diagnosis and culture-based technologies that deserve further study [2]. As an example, it is well recognized that liquid cultures are generally more prone to contamination [17]. Specimen handling, particularly as it relates to decontamination procedures, needs to be further studied and better standardized to optimize M. tuberculosis culture yield, while minimizing contamination and the risk of M. tuberculosis cross-contamination, which causes false-positive cultures. The impact of polyclonal infection on the accuracy of diagnostics and disease outcomes deserves further study. In HIV-associated tuberculosis, polyclonal infection has been reported both as 2 strains in concurrent sputum as well as the isolation of different sputum and blood strains, and may represent a combination of reactivation disease and newly acquired infection [12]. Existing liquid-culture systems do not identify polyclonal infection. Another area of interest relates to the role of resuscitation promoting factor (RPF) dependence within a population of M. tuberculosis cells on culture-based detection [18]. RPFs are a family of secreted proteins produced by M. tuberculosis that stimulate mycobacterial growth. RPF-dependent M. tuberculosis cells in sputum may vary widely between patients and during times of treatment. It has been hypothesized that M. tuberculosis populations detected by RPF supplementation and the identification of lipid-body-rich cells by microscopy may provide a view into the persister population and could open up new possibilities for monitoring treatment response [19].
MOLECULAR-BASED DETECTION OF M. TUBERCULOSIS
Diagnostic molecular technologies have improved since the 1990s, with more and better molecular biology techniques, among them polymerase chain reaction (PCR)–based technologies, fluorescent in situ hybridization, peptide nucleic acids, electrochemical detection of DNA, biochips, nanotechnology, and proteomic technologies [20]. Most of the molecular-based technologies, designed with fast-growing organisms in mind, are now applied to the diagnosis of slow-growing pathogens such as M. tuberculosis. Nucleic acid amplification tests (NAATs) are routine procedures in many settings because they are specific and reliable, with detection of M. tuberculosis in specimens several weeks earlier than culture (results available within 24–48 hours of sample receipt) [21]. NAATs can be developed in-house or are available commercially, based on PCR or other technologies, and are fully or partially automated. Sensitivity, however, in some commercial and in-house assays has been variable [22, 23], especially in testing of smear-negative samples. The recently published Centers for Disease Control and Prevention (CDC) guidelines suggest that NAATs will become standard practice in the United States for patients with suspected tuberculosis and that all clinicians and public health programs should have NAATs available to lessen time to diagnosis [24]. Commercial direct amplification tests include Amplicor (Roche Diagnostic Systems), based on PCR of 16S ribosomal RNA (rRNA), and the Amplified MTD (M. tuberculosis Direct) test (Gen-Probe) based on transcription-mediated amplification of rRNA. Both are approved by the US Food and Drug Administration (FDA) for direct tuberculosis detection in sputum samples, but only Gen-Probe is commercially available in the United States. Loop-mediated isothermal amplification (LAMP) is another new NAAT that can be used in areas with limited resources because expensive and complex instruments are not needed [25]. LAMP-based assays have targeted gyrB [25, 26], rrs [27], and more recently, the repetitive insertion sequence IS6110 for the detection of M. tuberculosis in clinical sputum samples. The IS6110-based LAMP assay may be a test with higher sensitivity than assays that are based on gyrB and rrs and, if confirmed, would be a good candidate for use in developing countries [28].
The recently developed Xpert MTB/RIF test (hereafter referred to as Xpert) on the GeneXpert platform (produced by Cepheid with support and funding from US federal agencies; the National Institutes of Health [NIH], National Institute of Allergy and Infectious Diseases [NIAID]; and the Foundation for Innovative and New Diagnostics [FIND], funded by the Bill & Melinda Gates Foundation) is an automated molecular-beacons-based approach to diagnosing M. tuberculosis and rifampin resistance [29, 30]. Molecular beacons are hybridization probes that, when attached to their target, emit fluorescence. The Xpert test has been shown to have high sensitivity and specificity for detection of M. tuberculosis and associated rifampin resistance in high-incidence settings, and plans are in place to pursue FDA approval for use of the test in the United States. Capable of providing results in less than 2 hours, Xpert may also reliably diagnose extrapulmonary tuberculosis [31]. Due to the automated, rapid, and sensitive nature of the test, Xpert has been endorsed by WHO and is to be rolled out as part of national plans for tuberculosis and MDR tuberculosis care and control [32]. A recent implementation of Xpert in South Africa highlighted the need for clinical pathways and algorithms for the optimal integration of the test into tuberculosis programs. For example, management of HIV-infected persons with suspected tuberculosis who test negative, among other clinical scenarios, warrants the study and institution of such algorithms.
The development of molecular diagnostics into automated systems, such as the Xpert assay, has identified new areas of research. These include assessing how to optimize sensitivity in nonrespiratory specimens for use in extrapulmonary and pediatric tuberculosis; determining whether semiquantitative test outputs can be used for treatment monitoring; determining the optimal settings in which these tests might replace AFB smears, culture, or both; evaluating the use of Xpert as an infection control tool; determining the frequency of false positives for rifampin resistance; and measuring the predictive value of Xpert as a diagnostic in low-incidence settings [33]. In addition, the cost and cost–benefit of Xpert and other potential NAATs need to be studied, with recognition of the fact that actual test costs extend beyond the cost of the cartridges and reagents and include costs for transport, personnel training, maintenance, waste disposal, mechanisms for assuring quality, and secure locations for storage. Similarly, when comparing these expenses to the use of the broadly available and inexpensive AFB smear, the costs of repeat testing as well as the societal costs of delayed and missed diagnoses given the low sensitivity of this method for diagnosis also need to be considered.
MOLECULAR-BASED DETECTION OF DRUG RESISTANCE
Two rapid molecular tests have recently been implemented to screen patients at high risk for drug-resistant tuberculosis. Also known as line probe assays, the rapid tests are the INNO-LiPARif.TB assay (Innogenetics) and the GenoType MTBDR assay (Hain Lifescience). They are available globally but are not yet FDA approved for use in the United States. INNO-LiPARif.TB can detect the presence of M. tuberculosis in addition to mutations associated with rifampin resistance [16]. The assay does not detect mutations associated with isoniazid resistance, but it is believed that the majority of isolates with mutations conferring rifampin resistance also have mutations that confer isoniazid resistance. A recent study, however, has shown that the presence of rifampin resistance alone may not be a reliable marker to diagnose MDR tuberculosis [34].
The GenoType MTBDR is a DNA strip assay developed for the rapid detection of gene mutations associated with rifampin and isoniazid resistance (rpoB and katG) in clinical isolates [35]. Evaluations of GenoType MTBDR assays conducted by several research groups have been published, and generally the reviews have attested to the excellent accuracy for rifampin resistance [36]. Several groups have shown that the specificity of this assay for isoniazid is also excellent, but sensitivity is variable [37–39]. The GenoType MTBDRplus, developed to detect a broader variety of rpoB and inhA gene mutations, appears to have enhanced the assay’s detection of isoniazid resistance [35].
Clinicians are currently using molecular information concerning drug resistance to influence and guide therapeutic decisions. Genotypic methods have the potential to meet the increasing need for fast and accurate assessment of drug-susceptibility testing, but increased research in this area is clearly needed. The CDC offers a molecular detection of drug resistance service to provide rapid results on a wide range of specific mutations and evidence of mixed populations of M. tuberculosis (http://www.cdc.gov/tb/topic/Laboratory/mddr.htm). The platform involves semiautomated conventional PCR and DNA sequencing that can be readily expanded to accommodate additional loci in the sequencing panel as quickly as new mutations associated with resistance are identified.
There are several gaps and challenges in the molecular detection of drug resistance. For instance, not all mutations that account for specific types of drug resistance are known (eg, pyrazinamide, the fluoroquinolones, and ethambutol) [40]. Given the recent renaissance in terms of new drugs under development for drug-resistant tuberculosis, development of tools for the rapid detection of both MDR and extensively drug-resistant tuberculosis is a critical need. Researchers have also recovered heterogeneous populations of bacilli with different resistance mutations from a single patient’s sputum [41]. The implication of genetic heterogeneity may be the simultaneous presence of drug-resistant and drug-susceptible phenotypes, both of which may require targeted treatment. Indeed, there are still challenges in deciphering what the clinical implications of identifying mutations are, how mutations cause drug resistance, and how to incorporate knowledge of mutations into molecular detection methodologies. Drug resistance is probably more complicated than initial paradigms have predicted—a single mutation can confer resistance to multiple drugs or multiple gene mutations could result in resistance to a single antimicrobial [42].
An available resource for tuberculosis researchers is data from whole genome sequencing in almost 50 drug-resistant strains compared with whole genome sequencing of drug-susceptible strains (publicly available at http://www.tbdreamdb.com/). The database is interactive, allowing it to serve as a resource for the development of molecular diagnostics and surveillance tools for tuberculosis. In addition, the database is useful for the structural mapping of mutations to better understand the mechanisms of drug resistance for novel pharmaceutical design. One of the utilities of this database has been a better understanding of the fact that isoniazid-resistant strains often have multiple gene mutations. The results of DNA sequencing of 28 genes associated with drug resistance in 1600 M. tuberculosis strains will also be made public in the near future in usable form. The TubercuList (http://tuberculist.epfl.ch/) is another server constructed around a database of DNA and protein sequences derived from a paradigm M. tuberculosis strain [43]. Information about the genomes of the tubercle bacilli can be retrieved and analyzed using various criteria, for example, keywords, gene names, or locations.
In general, this workshop session concluded that a fuller sequencing of resistant strains was needed, that public databases on mutations causing resistance should be curated on an ongoing basis, and that confirmation of resistance-causing mutations through the creation and phenotyping of point mutants was an important research goal. Drugs identified as high priority for future research and incorporation into rapid diagnostics include pyrazinamide, the fluoroquinolones, and ethambutol. In summary, novel tools are urgently needed for rapid diagnosis of drug-resistant tuberculosis. Knowledge about mutations that lead to antimicrobial resistance as well as an understanding of the relative occurrence of specific mutations can spur the development of better diagnostic tools.
NONMOLECULAR DIAGNOSTIC TECHNOLOGIES
Several nonmolecular approaches to discovery of tuberculosis diagnostic biomarkers are emerging, among them optic methods, automated optics, and nonoptics, including magnetic bead technologies, clustered magnetic nanoparticles, electronic nose assays, lateral flow assays, and -omics. Biomarkers are molecular features—molecules, genes, or characteristics—that can identify and/or monitor a particular physiological process or disease in the host [44]. For example, researchers have screened urine samples, serum, and saliva searching for evaluable markers through any variety of platforms—genomic, proteomic, metabolomic, lipidomic, and glycomic. New technologies are being developed and tested for identification of biomarkers for active tuberculosis, with a particular focus on pathogen-specific markers. For host-based markers, surface-enhanced laser desorption/ionization time-of-flight mass spectrometry, a type of proteomic fingerprint technology, has been used to screen for potential protein biomarkers in serum for the diagnosis of tuberculosis [45]. Matrix-assisted laser desorption/ionization time-of-flight mass spectrometry has also been used to profile the serum proteome and has identified several host biomarkers that differentiate individuals with tuberculosis from controls with some accuracy [46]. FIND and its partners have used a high-throughput cloning and expression method to examine the entire tuberculosis proteome in order to carry out serological profiling against antigen arrays and identify antibody biomarkers that might be targets for POC diagnostic tools [47].
Given the limitations of sputum as a diagnostic specimen, for example, in children or in extrapulmonary disease, the availability of a nonsputum-based, nonculture-based diagnostic in high-burden settings would represent a significant advancement in tuberculosis control (see McNerney et al, this supplement). Lipoarabinomannan (LAM), a 17.5-kD heat-stable glycolipid within the cell wall of M. tuberculosis, is one potential nonculture-based, nonmolecular-based marker of active tuberculosis [48]. To diagnose tuberculosis in HIV-infected patients with advanced immunosuppression, a LAM lateral flow assay (Clearview TBELISA, Inverness Medical Innovations) was developed. It has been tested using urine, sputum, and cerebrospinal fluid and is currently being assessed in clinical trials. A recent study of HIV-coinfected patients in South Africa found that the assay was not sufficiently sensitive to replace culture [49]. A similar assay, the urine LAM-ELISA (Chemogen) was evaluated in tuberculosis patients with and without HIV coinfection. The conclusion was that the assay does not appear to be useful as an independent diagnostic for pulmonary tuberculosis [50]. Whether the assay could serve as a supplemental device in the diagnosis of HIV-associated tuberculosis is still under investigation [51, 52].
An example of optical methods for the diagnosis of tuberculosis is the application of reporter enzyme fluorescence (REF) to whole animal imaging. Recently published studies have demonstrated that the enzyme ß-lactamase, expressed by bacteria but not their eukaryotic hosts, can be used along with near-infrared fluorogenic substrates to detect pulmonary tuberculosis infections in real time in mice [53]. Because the crystal structure of M. tuberculosis ß-lactamase is known, researchers can design specific substrates that are distinctly different from other bacterial ß-lactamases. The primary improvement of REF over other types of systems is that REF does not require recombinant strains that can introduce foreign genes that may unpredictably interfere with bacterial physiology, especially when expressed from plasmids. REF permits sensitive detection of M. tuberculosis whether in vitro or in vivo. Thus, this imaging system has the potential to become a noninvasive diagnostic tool for sputum and other specimens from humans with suspected tuberculosis infections. The AFB smear detects 5000–10 000 bacilli/mL in sputum compared with REF, which can detect a minimum of 100 bacilli under ideal laboratory conditions in just a few hours.
Volatile organic compounds (VOCs) in breath are also being investigated as novel diagnostic biomarkers for active pulmonary tuberculosis [54, 55]. Recently, investigators at Menssana Research analyzed breath VOCs in 226 symptomatic high-risk patients from the United States, the Philippines, and the United Kingdom, using gas chromatography/mass spectroscopy [56]. Breath VOCs contained apparent biomarkers of active pulmonary tuberculosis composed of oxidative stress products (alkanes and alkane derivatives) and volatile metabolites of M. tuberculosis (cyclohexane and benzene derivatives), which identified active pulmonary tuberculosis with 85% accuracy in the symptomatic high-risk subjects evaluated in these field studies. Additional research is needed to fully evaluate the specificity of VOCs for M. tuberculosis in comparison with other disease-causing mycobacteria. Nonetheless, preliminary data are encouraging for this “nose-based” technology for diagnosing active tuberculosis.
Aerosol-based novel diagnostics for tuberculosis are also being explored. Investigators at Livermore Instruments are evaluating a single-particle laser desorption/ionization time-of-flight mass spectrometry tool for diagnosis. This bioaerosol mass spectrometry (BAMS) system was originally designed for environmental and biodefense activities. Pilot preclinical data were presented to suggest that M. tuberculosis particles could be identified in bioaerosol generated by the cough of an infectious tuberculosis patient [57, 58]. At this time, BAMS systems are large and costly. However, with additional engineering, this reagent-free, rapid (<2 minutes per patient) platform could significantly transform current approaches to screening for tuberculosis.
This workshop session discussed a number of paradigm-changing approaches that need additional research. Of importance were research and development efforts to move accurate diagnostics into the primary care setting. Significant additional research is needed before we can shift sites of active tuberculosis diagnosis to communities, pharmacies, and general health clinics. If this is not possible, could ultradecentralization of diagnostics be at least matched to the systems in place for directly observed therapy? Other questions were as follows: Could novel technologies, such as the breath- and aerosol-based rapid diagnostics, provide the opportunity to shift our approach to case detection and screening, from passive to active, from individual to mass? Can simple, affordable, POC diagnostics that are found to have high sensitivity but marginal specificity still be useful as referral tests for additional examination? If so, how might they be implemented? Last, the proportion of research and development in tuberculosis diagnostics that should be directed toward drug-susceptible vs drug-resistant tuberculosis remains unresolved and is highly setting dependent. Clearly, approaches that address these issues together are desirable. In support of this, more and better mechanisms are needed to coordinate the actions of studies and the funding process. The funding mechanisms and agencies that exist should make a greater effort to support and supplement one another in order to implement projects as part of a global architecture in healthcare.
MAXIMIZING AND OPTIMIZING RESEARCH THROUGH COORDINATION AND COLLABORATION
A number of existing resources can assist tuberculosis researchers in their efforts to identify new or optimize existing diagnostic technologies. Biospecimens are available to researchers through the tuberculosis specimen bank of WHO’s Special Programme for Research in Tropical Diseases (http://apps.who.int/tdr/svc/diseases/tuberculosis/specimen-bank). The National Disease Research Interchange (NDRI) is another resource for biospecimens. NDRI, a nonprofit NIH-funded organization, supports biomedical research through the recovery, preservation, and distribution of human biospecimens. NDRI’s network of tissue acquisition centers provides a range of normal and diseased biospecimens that are procured, preserved, and shipped according to investigator-specified criteria. All biospecimens are accompanied by extensive medical and social history data. More recently, a tuberculosis-specific biobanking initiative, the Consortium for TB Biomarkers (CTB2), was launched to facilitate biomarker discovery, in particular in the arena of markers of treatment response. With initial funding from the FDA, CTB2 is being developed by 3 organizations central to tuberculosis clinical drug development: NIAID’s AIDS Clinical Trials Group, CDC’s Tuberculosis Trials Consortium, and the Global Alliance for TB Drug Development. The CTB2 and its affiliated networks have agreed on a core set of samples to be collected, processed, and stored, including use of common data elements obtained from well-characterized patients enrolled in randomized, controlled tuberculosis treatment trials [44].
In order to optimally interpret the contribution of and advancement provided by new tuberculosis diagnostics over existing techniques, the clinical trials and field studies used to evaluate them should be designed, implemented, and reported according to universally accepted standards. Although most tuberculosis diagnostic studies report sensitivity and specificity, reporting is often not standardized and few have a randomized, controlled design. Use of culture as a gold standard for reference is imperfect, and patient outcomes may be missing from evaluations of tuberculosis diagnostics. Heterogeneity of trial design and populations complicate cross-study comparisons or meta-analyses. Standards for the Reporting of Diagnostic accuracy studies (STARD) were designed to improve the accuracy and completeness of diagnostic studies, permit readers to evaluate the potential bias in the study, and assess generalizability [59]. The STARD statement includes a checklist of 25 items and recommends the use of a flow diagram to follow study design and patients. Widespread adoption of such standards, together with harmonized definitions, assessment of clinical impact, and perhaps even standardized quality assurance, would advance the field.
CONCLUSIONS AND RECOMMENDATIONS
Although there is significant work under way to develop new diagnostic assays and devices for tuberculosis, a rapid, accurate, POC, low-cost diagnostic has yet to be realized. Because tuberculosis principally affects people in limited-resource settings, for any new tuberculosis diagnostic to have major public health impact, simplicity and low cost will be as important as analytical accuracy. Tuberculosis diagnostics should also be assessed in terms of clinical impact beyond assessments of microbiological performance. Unfortunately, the current tuberculosis diagnostic research literature largely neglects the impact of diagnostic tests on patient-important diagnoses and outcomes [60].
Through international input and consensus, the Stop TB Partnership of WHO is developing a Global TB Research Roadmap (http://www.stoptb.org/global/research/) that aims to delineate priority research questions that need to be critically addressed for improved tuberculosis control, with the goal of elimination by 2050. The Roadmap recognizes that cross-disciplinary approaches spanning the continuum of research across all disciplines from basic to implementation science are required to achieve this goal. The Roadmap is 1 of the main objectives of the TB Research Movement, a global initiative of the Stop TB Partnership. The TB Research Movement provides leadership and advocacy to mobilize increased resources in support of a coherent and comprehensive global tuberculosis research agenda, as well as a forum for researchers, funders, and implementers of tuberculosis research to coordinate plans and actions that will ensure research needs are addressed, opportunities prioritized, and gaps filled. Within the aegis of this initiative, proactive participation and organization of key groups involved in tuberculosis research are needed to construct and implement strategies to harmonize study design and data definitions and to effectively share information about ongoing and planned studies worldwide. At a minimum, this type of communication and coordination for observational cohort studies and phase 2 and 3 clinical trials are urgently needed. Given the substantial investment from many diverse groups, including US government agencies, coordination of research and clinical evaluations in this field has the potential to accelerate advances efficiently and economically. Rapid development, assessment, and adoption of these diagnostic technologies require defining the critical path and aligning key stakeholders and their respective roles so as to facilitate the progression from development to implementation.
Notes
Acknowledgments.
We thank the meeting organizing committee (Ms Bonnie Plikaytis [CDC], and Drs Gail Jacobs [NIAID], Mamodikoe Makhene [NIAID], and Christine Sizemore [NIAID]), and the sponsors (CDC, NIAID, Office of the Global AIDS Coordinator, and the National Institute of Child Health and Human Development) for organizing and supporting this workshop.
Disclaimers.
The findings and conclusions in this report are those of the author(s) and do not necessarily represent the views of the CDC or the NIH.
Financial support.
This work was supported by the National Heart, Lung, and Blood Institute of the NIH (K23HL092629 to P. N.) and the Wellcome Trust, FIND, and Innovation For Health and Development (to C.A. E.).
Potential conflicts of interest.
M. P. has no commercial associations or other conflicts of interest relevant to the work presented. He was supported by a grant from the Bill and Melinda Gates Foundation (grant number 28766.01). A. D. reports that he and his laboratory receive licensing income for the use of molecular beacons in the GeneXpert MTB/RIF assay. His personal income has been voluntarily and irrevocably capped at $5,000 per year and income to his laboratory has been voluntarily capped at $50,000 per year. All other authors: no reported conflicts.
All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1.Steingart KR, Henry M, Ng V, et al. Fluorescence versus conventional sputum smear microscopy for tuberculosis: a systematic review. Lancet Infect Dis. 2006;6:570–81. doi: 10.1016/S1473-3099(06)70578-3. [DOI] [PubMed] [Google Scholar]
- 2.Steingart KR, Ng V, Henry M, et al. Sputum processing methods to improve the sensitivity of smear microscopy for tuberculosis: a systematic review. Lancet Infect Dis. 2006;6:664–74. doi: 10.1016/S1473-3099(06)70602-8. [DOI] [PubMed] [Google Scholar]
- 3.Steingart KR, Ramsay A, Pai M. Optimizing sputum smear microscopy for the diagnosis of pulmonary tuberculosis. Expert Rev Anti Infect Ther. 2007;5:327–31. doi: 10.1586/14787210.5.3.327. [DOI] [PubMed] [Google Scholar]
- 4.World Health Organization report—global tuberculosis control. Geneva, Switzerland: 2011. [Google Scholar]
- 5.Dorman SE. New diagnostic tests for tuberculosis: bench, bedside, and beyond. Clin Infect Dis. 2010;50(Suppl 3):S173–7. doi: 10.1086/651488. [DOI] [PubMed] [Google Scholar]
- 6.Parsons LM, Somoskovi A, Gutierrez C, et al. Laboratory diagnosis of tuberculosis in resource-poor countries: challenges and opportunities. Clin Microbiol Rev. 2011;24:314–50. doi: 10.1128/CMR.00059-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wilson D, Nachega J, Morroni C, Chaisson R, Maartens G. Diagnosing smear-negative tuberculosis using case definitions and treatment response in HIV-infected adults. Int J Tuberc Lung Dis. 2006;10:31–8. [PubMed] [Google Scholar]
- 8.World Health Organization policy—TB diagnostics and laboratory strengthening. http://www.who.int/tb/laboratory/policy_liquid_medium_for_culture_dst/en/index.html. Accessed 18 October 2011. [Google Scholar]
- 9.Chien HP, Yu MC, Wu MH, Lin TP, Luh KT. Comparison of the BACTEC MGIT 960 with Lowenstein-Jensen medium for recovery of mycobacteria from clinical specimens. Int J Tuberc Lung Dis. 2000;4:866–70. [PubMed] [Google Scholar]
- 10.Wang JY, Lee LN, Lai HC, et al. Performance assessment of the Capilia TB assay and the BD ProbeTec ET system for rapid culture confirmation of Mycobacterium tuberculosis. Diagn Microbiol Infect Dis. 2007;59:395–9. doi: 10.1016/j.diagmicrobio.2007.06.010. [DOI] [PubMed] [Google Scholar]
- 11.Sharma B, Pal N, Malhotra B, Vyas L. Evaluation of a rapid differentiation test for Mycobacterium tuberculosis from other mycobacteria by selective inhibition with p-nitrobenzoic acid using MGIT 960. J Lab Physicians. 2010;2:89–92. doi: 10.4103/0974-2727.72157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brent AJ, Mugo D, Musyimi R, et al. Performance of the MGIT TBc identification test and meta-analysis of MPT64 assays for identification of the Mycobacterium tuberculosis complex in liquid culture. J Clin Microbiol. 2011;49:4343–6. doi: 10.1128/JCM.05995-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Leung E, Minion J, Benedetti A, Pai M, Menzies D. Microcolony culture techniques for tuberculosis diagnosis: a systematic review. Int J Tuberc Lung Dis. 2012;16:16–23. doi: 10.5588/ijtld.10.0065. i--iii. [DOI] [PubMed] [Google Scholar]
- 14.Moore DA, Evans CA, Gilman RH, et al. Microscopic-observation drug-susceptibility assay for the diagnosis of TB. N Engl J Med. 2006;355:1539–50. doi: 10.1056/NEJMoa055524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Moore DA, Mendoza D, Gilman RH, et al. Microscopic observation drug susceptibility assay, a rapid, reliable diagnostic test for multidrug-resistant tuberculosis suitable for use in resource-poor settings. J Clin Microbiol. 2004;42:4432–7. doi: 10.1128/JCM.42.10.4432-4437.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.World Health Organization STP. Pathways to better diagnostics for tuberculosis: a blueprint for the development of TB diagnostics. http://www.stoptb.org/assets/documents/research/2009.pdf. Accessed 18 October 2011. [Google Scholar]
- 17.Anthony RM, Cobelens FG, Gebhard A, et al. Liquid culture for Mycobacterium tuberculosis: proceed, but with caution. Int J Tuberc Lung Dis. 2009;13:1051–3. [PubMed] [Google Scholar]
- 18.Mukamolova GV, Turapov O, Malkin J, Woltmann G, Barer MR. Resuscitation-promoting factors reveal an occult population of tubercle bacilli in sputum. Am J Respir Crit Care Med. 2010;181:174–80. doi: 10.1164/rccm.200905-0661OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Garton NJ, Waddell SJ, Sherratt AL, et al. Cytological and transcript analyses reveal fat and lazy persister-like bacilli in tuberculous sputum. PLoS Med. 2008;5:e75. doi: 10.1371/journal.pmed.0050075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pai M, Minion J, Sohn H, Zwerling A, Perkins MD. Novel and improved technologies for tuberculosis diagnosis: progress and challenges. Clin Chest Med. 2009;30:701–16. doi: 10.1016/j.ccm.2009.08.016. viii. [DOI] [PubMed] [Google Scholar]
- 21.Laraque F, Griggs A, Slopen M, Munsiff SS. Performance of nucleic acid amplification tests for diagnosis of tuberculosis in a large urban setting. Clin Infec Dis. 2009;49:46–54. doi: 10.1086/599037. [DOI] [PubMed] [Google Scholar]
- 22.Flores LL, Pai M, Colford JM, Jr, Riley LW. In-house nucleic acid amplification tests for the detection of Mycobacterium tuberculosis in sputum specimens: meta-analysis and meta-regression. BMC Microbiol. 2005;5:55. doi: 10.1186/1471-2180-5-55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ling DI, Flores LL, Riley LW, Pai M. Commercial nucleic-acid amplification tests for diagnosis of pulmonary tuberculosis in respiratory specimens: meta-analysis and meta-regression. PLoS One. 2008;3:e1536. doi: 10.1371/journal.pone.0001536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Centers for Disease Control and Prevention. Updated guidelines for the use of nucleic acid amplification tests in the diagnosis of tuberculosis. MMWR Morb Mortal Wkly Rep. 2009;58:7–10. [PubMed] [Google Scholar]
- 25.Boehme CC, Nabeta P, Henostroza G, et al. Operational feasibility of using loop-mediated isothermal amplification for diagnosis of pulmonary tuberculosis in microscopy centers of developing countries. J Clin Microbiol. 2007;45:1936–40. doi: 10.1128/JCM.02352-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Iwamoto T, Sonobe T, Hayashi K. Loop-mediated isothermal amplification for direct detection of Mycobacterium tuberculosis complex, M. avium, and M. intracellulare in sputum samples. J Clin Microbiol. 2003;41:2616–22. doi: 10.1128/JCM.41.6.2616-2622.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pandey BD, Poudel A, Yoda T, et al. Development of an in-house loop-mediated isothermal amplification (LAMP) assay for detection of Mycobacterium tuberculosis and evaluation in sputum samples of Nepalese patients. J Med Microbiol. 2008;57:439–43. doi: 10.1099/jmm.0.47499-0. [DOI] [PubMed] [Google Scholar]
- 28.Aryan E, Makvandi M, Farajzadeh A, et al. A novel and more sensitive loop-mediated isothermal amplification assay targeting IS6110 for detection of Mycobacterium tuberculosis complex. Microbiol Res. 2010;165:211–20. doi: 10.1016/j.micres.2009.05.001. [DOI] [PubMed] [Google Scholar]
- 29.Boehme CC, Nabeta P, Hillemann D, et al. Rapid molecular detection of tuberculosis and rifampin resistance. N Engl J Med. 2010;363:1005–15. doi: 10.1056/NEJMoa0907847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Boehme CC, Nicol MP, Nabeta P, et al. Feasibility, diagnostic accuracy, and effectiveness of decentralised use of the Xpert MTB/RIF test for diagnosis of tuberculosis and multidrug resistance: a multicentre implementation study. Lancet. 2011;377:1495–505. doi: 10.1016/S0140-6736(11)60438-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hillemann D, Rusch-Gerdes S, Boehme C, Richter E. Rapid molecular detection of extrapulmonary tuberculosis by the automated GeneXpert MTB/RIF system. J Clin Microbiol. 2011;49:1202–5. doi: 10.1128/JCM.02268-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.World Health Organization PS. Automated real-time nucleic acid amplification technology for rapid and simultaneous detection of tuberculosis and rifampicin resistance: Xpert MTB/RIF system. http://whqlibdoc.who.int/publications/2011/9789241501545_eng.pdf. Accessed 19 October 2011. [PubMed] [Google Scholar]
- 33.Kranzer K. Improving tuberculosis diagnostics and treatment. Lancet. 2011;377:1467–8. doi: 10.1016/S0140-6736(11)60513-8. [DOI] [PubMed] [Google Scholar]
- 34.Smith S, Kurbatova EV, Cavanaugh J, Peter J. Global isoniazid resistance patterns in rifampin-resistant and rifampin-susceptible tuberculosis. Int J Tuberc Lung Dis. 2011 doi: 10.5588/ijtld.11.0445. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hillemann D, Rusch-Gerdes S, Richter E. Evaluation of the GenoType MTBDRplus assay for rifampin and isoniazid susceptibility testing of Mycobacterium tuberculosis strains and clinical specimens. J Clin Microbiol. 2007;45:2635–40. doi: 10.1128/JCM.00521-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bwanga F, Hoffner S, Haile M, Joloba ML. Direct susceptibility testing for multi drug resistant tuberculosis: a meta-analysis. BMC Infect Dis. 2009;9:67. doi: 10.1186/1471-2334-9-67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Makinen J, Marttila HJ, Marjamaki M, Viljanen MK, Soini H. Comparison of two commercially available DNA line probe assays for detection of multidrug-resistant Mycobacterium tuberculosis. J Clin Microbiol. 2006;44:350–2. doi: 10.1128/JCM.44.2.350-352.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Miotto P, Piana F, Penati V, Canducci F, Migliori GB, Cirillo DM. Use of genotype MTBDR assay for molecular detection of rifampin and isoniazid resistance in Mycobacterium tuberculosis clinical strains isolated in Italy. J Clin Microbiol. 2006;44:2485–91. doi: 10.1128/JCM.00083-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wilson ML. Recent advances in the laboratory detection of Mycobacterium tuberculosis complex and drug resistance. Clin Infec Dis. 2011;52:1350–5. doi: 10.1093/cid/cir146. [DOI] [PubMed] [Google Scholar]
- 40.Zhang Y, Yew WW. Mechanisms of drug resistance in Mycobacterium tuberculosis. Int J Tuberc Lung Dis. 2009;13:1320–30. [PubMed] [Google Scholar]
- 41.Post FA, Willcox PA, Mathema B, et al. Genetic polymorphism in Mycobacterium tuberculosis isolates from patients with chronic multidrug-resistant tuberculosis. J Infect Dis. 2004;190:99–106. doi: 10.1086/421501. [DOI] [PubMed] [Google Scholar]
- 42.Campbell PJ, Morlock GP, Sikes RD, et al. Molecular detection of mutations associated with first- and second-line drug resistance compared with conventional drug susceptibility testing of Mycobacterium tuberculosis. Antimicrob Agents Chemother. 2011 doi: 10.1128/AAC.01550-10. 55(5):2032–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lew JM, Kapopoulou A, Jones LM, Cole ST. TubercuList—10 years after. Tuberculosis. 2011;91:1–7. doi: 10.1016/j.tube.2010.09.008. [DOI] [PubMed] [Google Scholar]
- 44.Nahid P, Saukkonen J, MacKenzie WR, et al. CDC/NIH Workshop. Tuberculosis biomarker and surrogate endpoint research roadmap. Am J Respir Crit Care Med. 2011;184:972–9. doi: 10.1164/rccm.201105-0827WS. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Liu JY, Jin L, Zhao MY, et al. New serum biomarkers for detection of tuberculosis using surface-enhanced laser desorption/ionization time-of-flight mass spectrometry. Clin Chem Lab Med. 2011;49:1727–33. doi: 10.1515/CCLM.2011.634. [DOI] [PubMed] [Google Scholar]
- 46.Agranoff D, Fernandez-Reyes D, Papadopoulos MC, et al. Identification of diagnostic markers for tuberculosis by proteomic fingerprinting of serum. Lancet. 2006;368:1012–21. doi: 10.1016/S0140-6736(06)69342-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kunnath-Velayudhan S, Salamon H, Wang HY, et al. Dynamic antibody responses to the Mycobacterium tuberculosis proteome. Proc Natl Acad Sci U S A. 2010;107:14703–8. doi: 10.1073/pnas.1009080107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mutetwa R, Boehme C, Dimairo M, et al. Diagnostic accuracy of commercial urinary lipoarabinomannan detection in African tuberculosis suspects and patients. Int J Tuberc Lung Dis. 2009;13:1253–9. [PMC free article] [PubMed] [Google Scholar]
- 49.Gounder CR, Kufa T, Wada NI, et al. Diagnostic accuracy of a urine lipoarabinomannan enzyme-linked immunosorbent assay for screening ambulatory HIV-infected persons for tuberculosis. J Acquir Immune Defic Syndr. 2011;58:219–23. doi: 10.1097/QAI.0b013e31822b75d4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Reither K, Saathoff E, Jung J, et al. Low sensitivity of a urine LAM-ELISA in the diagnosis of pulmonary tuberculosis. BMC Infect Dis. 2009;9:141. doi: 10.1186/1471-2334-9-141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Minion J, Leung E, Talbot E, Dheda K, Pai M, Menzies D. Diagnosing tuberculosis with urine lipoarabinomannan: systematic review and meta-analysis. Eur Respir J. 2011;38:1398–405. doi: 10.1183/09031936.00025711. [DOI] [PubMed] [Google Scholar]
- 52.Dheda K, Davids V, Lenders L, et al. Clinical utility of a commercial LAM-ELISA assay for TB diagnosis in HIV-infected patients using urine and sputum samples. PLoS One. 2010;5:e9848. doi: 10.1371/journal.pone.0009848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kong Y, Yao H, Ren H, et al. Imaging tuberculosis with endogenous beta-lactamase reporter enzyme fluorescence in live mice. Proc Natl Acad Sci U S A. 2010;107:12239–44. doi: 10.1073/pnas.1000643107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Banday KM, Pasikanti KK, Chan EC, et al. Use of urine volatile organic compounds to discriminate tuberculosis patients from healthy subjects. Anal Chem. 2011;83:5526–34. doi: 10.1021/ac200265g. [DOI] [PubMed] [Google Scholar]
- 55.Phillips M, Cataneo RN, Condos R, et al. Volatile biomarkers of pulmonary tuberculosis in the breath. Tuberculosis (Edinb) 2007;87:44–52. doi: 10.1016/j.tube.2006.03.004. [DOI] [PubMed] [Google Scholar]
- 56.Phillips M, Basa-Dalay V, Bothamley G, et al. Breath biomarkers of active pulmonary tuberculosis. Tuberculosis. 2010;90:145–51. doi: 10.1016/j.tube.2010.01.003. [DOI] [PubMed] [Google Scholar]
- 57.Adams KL, Steele PT, Bogan MJ, et al. Reagentless detection of Mycobacteria tuberculosis H37Ra in respiratory effluents in minutes. Anal Chem. 2008;80:5350–7. doi: 10.1021/ac8002825. [DOI] [PubMed] [Google Scholar]
- 58.Tobias HJ, Schafer MP, Pitesky M, et al. Bioaerosol mass spectrometry for rapid detection of individual airborne Mycobacterium tuberculosis H37Ra particles. Appl Environ Microbiol. 2005;71:6086–95. doi: 10.1128/AEM.71.10.6086-6095.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bossuyt PM, Reitsma JB, Bruns DE, et al. Towards complete and accurate reporting of studies of diagnostic accuracy: the STARD initiative. Ann Intern Med. 2003;138:40–4. doi: 10.7326/0003-4819-138-1-200301070-00010. [DOI] [PubMed] [Google Scholar]
- 60.Pai M, Ramsay A, O’Brien R. Evidence-based tuberculosis diagnosis. PLoS Med. 2008;5:e156. doi: 10.1371/journal.pmed.0050156. [DOI] [PMC free article] [PubMed] [Google Scholar]